Introduction
Hepatitis B virus (HBV) is a small (3.2-kb),
partially double-stranded, relaxed circular, enveloped DNA virus,
which specifically infects the hepatocytes of quadrumana (1,2).
Worldwide, ~3.5 billion individuals are affected, among which
>0.4 billion have chronic HBV infection (CHB); HBV is one of the
most common pathogens in humans, which has a major global health
impact (3–6). Persistent infection with HBV results
in serious liver disease, including acute hepatitis, cirrhosis and
hepatocellular carcinoma (7). It
is estimated that 2% of patients with CHB are be likely to develop
cirrhosis each year, and 15–25% of patients with CHB are likely to
succumb to cirrhosis or hepatocellular carcinoma (8,9).
Licensed drugs for the treatment of CHB are nucleos(t)ide analogues
(NAs) and interferon α (IFN-α) (10). However, the emergence of
side-effects poses challenges to the long-term administration of
IFN-α. At the same time, the generation of drug-resistant strains
of HBV with amino acid replacement in the YMDD motif of reverse
transcriptase has been a severe problem in patients with lamivudine
therapy, which may result in virological relapse and biochemical
flare (11–13). Therefore, it is necessary to
explore novel and more effective therapies for HBV.
Transcription factors comprise a deoxyribonucleic
acid binding domain (DBD), and an effector domain (ED) or
transcriptional regulation domain, which may provide a nuclear
localization signal (NLS). The DBD is required to bind to the
target sequence, including the enhancer (Enh) or promoter element,
which is an upward or downward effect field regulating the target
gene, and the NLS delivers a transcription factor into nuclei, as
eukaryotic transcription occurs within the nucleus (14). The characteristics of eukaryotic
transcription factors are the basis of artificial transcription
factor (ATF) technology (15).
The design of a specific recognition DBD is the most difficult and
important part in the establishment of an ATF. Zinc finger proteins
(ZFP) are the most common type of DNA-binding proteins in molecular
biology. Engineered ZFPs bind to an extensive range of DNA
sequences and the finger subunits may also be connected to bind to
long, asymmetric DNA sequences (16–20). The novel Cys2His2 ZFPs are the
most promising candidates for the DBD due to their high specificity
and affinity (21,22). Their functions are diverse and
include DNA recognition, RNA packaging, transcriptional activation,
lipid binding, cell apoptosis and protein folding (23). Approximately 30 amino acids with a
simple, ββα-fold stabilized by hydrophobic interactions and
chelation of a single zinc ion constitute the one-fold ZF domain. A
3-bp DNA fragment is constantly and distinctively recognized by
each ZF domain, and six-ZF proteins were reported to recognize
unique 18-bp fragments due to the canonical TGEKP interfinger
linker between the ZF units (24–26). While engineered ZFPs have been
used in human immunodeficiency virus research (27,28), the potential of engineered ZFPs to
inhibit HBV has remained to be investigated.
In the present study, based on advanced software
development technologies and ZFP design tools on an online
platform, the HBV EnhI-specific ATF was designed, an 18-bp sequence
was selected as the ATF target sequence and corresponding ZF amino
acid sequences were gained; the best ZFPs were fused to an NLS and
an ED to generate the ATF. A series of ATF, ZFP and
Kruppel-associated box (KRAB) eukaryotic expression vectors were
constructed using genetic engineering methods. The fidelity was
confirmed by restriction enzyme digestion and sequence analysis,
and ATF, ZFP and KRAB eukaryotic expression vectors were
transformed or injected into HepG2.2.15 cells and HBV transgenic
mice. At the predetermined times, serum samples, culture
supernatants, hepatic tissues and cells were gathered for
serological and virological detection. The results of the present
study demonstrated that only ATF significantly inhibited HBV
transcription and replication at the viral RNA, protein and viral
progeny level, without any obvious toxic effect in vitro and
in vivo.
Materials and methods
Design and construction of the ATF
expression vector of ZFP
Based on HBV DNA EnhI (1,070–1,234 bp) sequences as
the template, an online platform
(scripps.edu/mb/barbas/zfdesign/zfdesignhome.php) of a ZFP design
tool was used (29) and the
'Search DNA Sequence for Contiguous or Separated Target Sites' and
'Design a Zinc Finger Protein' tools were explored for selecting
the optimal target sequence and the corresponding ZF amino acid
sequences. This meant that the specific target sites of the HBV DNA
EnhI (1,070–1,234 bp) region were optimized, and were recognized to
predict the ZF amino acid sequence. The N-terminal ZF domain was
fused to KRAB repression domains from the human ZFP 10 gene
(30). To facilitate the
localization of ATF in the cell nuclei and detect the expression of
ATF, an NLS from simian virus 40 large T-antigen (31) and the epitope Flag tag were
separately added to the ATF's N-terminal and C terminus. The amino
acid sequence of ATF was then reverse-transcribed into a nucleotide
sequence and optimized by using Primer premier 5.0 software
(Premier Biosoft, Palo Alto, CA, USA). Finally, the ATF nucleotide
sequences were synthesized and cloned into the EcoRI and
BamHI restriction sites of pcDNA3.1(+) expression vector
(Sangon Biotech Co., Ltd., Shanghai, China), Fidelity was confirmed
by restriction enzyme digestion and sequence analysis.
pcDNA3.1(+)-ATF (nls-ZFP-KRAB-Flag) was transformed into competent
DH-5a Escherichia coli cells (Laboratory of Molecular
Biology on Infectious Diseases, Ministry of Education, Chongqing
Medical University, Chongqing, China) and was purified with a
TIANpure Midi Plasmid kit (Tiangen Biotech Co., Ltd., Beijing,
China). Using pcDNA3.1(+)-ATF (nls-ZFP-KRAB-Flag) vector as a
template, the individual primers were designed, and
pcDNA3.1(+)-nls-ZFP-Flag and pcDNA3.1(+)-nls-KRAB-Flag were
constructed according to the above methods.
Cell culture and transfection
The HepG2 and HepG2.2.15 cells were provided by the
Laboratory of Molecular Biology on Infectious Diseases, Ministry of
Education, Chongqing Medical University. The HepG2 cell line, which
is known to be a hepatoblastoma cell line, and HepG2.2.15 cells
[clonal cells derived from HepG2 (32), which was transfected with a
plasmid containing HBV DNA that secretes HB surface antigen (HBsAg)
particles, nucleocapsids and virions] were cultured in Dulbecco's
modified Eagle's medium (HyClone; GE Healthcare, Little Chalfont,
UK) supplemented with 10% fetal bovine serum (HyClone; GE
Healthcare), 100 U/ml penicillin, 100 µg/ml streptomycin
(Beyotime Institute of Biotechnology, Haimen, China) and 380
µg/ml G418 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
at 37°C in a humidified atmosphere containing 5% CO2.
Transient transfections of HepG2.2.15 were performed by using the
X-treme GENE HP DNA Transfection Reagent (Roche Diagnostics, Basel,
Switzerland) according to the manufacturer's instructions with a
final plasmid concentration of 0.01 µg/µl. The
HepG2.2.15 cells (1.5×105/well) were seeded into
six-well plates and cultured overnight. On the second day, 100 ng
(10 µl of a 10 ng/µl solution) pcDNA 3.1(+),
pcDNA-nls-KRAB-flag, pcDNA-ATF or pcDNA-nls-ZFP-flag were added to
the culture medium. At 24, 48 or 72 h post-transfection, the
supernatants and cells were collected for analysis.
Animals
A total of 24 male C57BL/6-HBV-1.3 genome-eq
transgenic mice (age, 6–8 weeks; weight, 18–24 g) were provided by
the Laboratory of Molecular Biology on Infectious Diseases,
Ministry of Education, Chongqing Medical University. The Chongqing
Medical University Medical Research Ethics Committee approved all
animal experiments. All animals were provided sterile water and rat
chow ad libitum, and were kept under a 12-h light/dark cycle
at constant temperature and humidity (temperature, 18–22°C;
humidity, 50–60%). Mice (n=6/group) were injected with 8 µg
pcDNA 3.1(+), pcDNA-nls-KRAB-flag, pcDNA-ATF or pcDNA-nls-ZFP-flag
dissolved in 2 ml PBS via the tail vein within 5–8 sec. Serum
samples were collected via the tail vein after intraperitoneal
injection of 2% pentobarbital sodium (50 mg/kg; cat. no. P3761;
Sigma-Aldrich; Merck KGaA) on days 7, 14, 21 and 28. The
post-anesthetic mice were sacrificed on day 28, and the serum and
liver tissues were collected.
Extraction of HBV replicative
intermediate-DNA (RI-DNA)
Intracellular RI-DNA was extracted at 48 h
post-transfection as previously described (33). In brief, HepG2.2.15 cells were
harvested with trypsin washed twice with ice-cold PBS (pH 7.4).
Cells were then lysed in 200 µl Nonidet P-40 cell lysis
liquid with incubation at 37°C for 15 min, and then centrifuged at
13,000 × g for 5 min. The supernatants were then incubated with 500
µl 35% polyethylene glycol 8000 (1.5 M) in an ice bath for
50 min, and samples were centrifuged again as described above. For
virus precipitation, the sample (supernatants, serum or hepatic
tissue) was incubated with 380 µl proteinase K digestion
liquid, 20 µl proteinase K (Tiangen Biotech Co., Ltd.) in
sterilized ultrapure water (50 µl) overnight at 45°C in a
water bath. HBV DNA was extracted with isovolumetric phenol
chloroform twice, and the supernatants were carefully collected. An
equal volume of isopropyl alcohol was added and the sample was
vortexed. The mixture was centrifuged for 30 min at 15,000 × g and
4°C. After the precipitate was briefly washed with 75% ice-cold
ethanol twice, it was resuspended in 10 µl sterilized
ultrapure water.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was extracted from HepG2.2.15 cells using
the RNAiso Plus kit (Takara Bio Inc., Otsu, Japan) at 72 h
post-transfection following the manufacturer's instructions. A
total of 1 µg RNA was reverse-transcribed to complementary
(c)DNA using the PrimeScript™ RT reagent kit (Takara Bio Inc.). For
analysis of HBV RNA levels, 2 µl of each cDNA were
quantified by real-time PCR with SYBR-Green (Takara Bio Inc.) in a
LightCycler CFX96 (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The PCR condition consisted of three steps as follows: Step
1, pre-degeneration at 95°C for 2 min; step 2, 34 cycles of
denaturation at 94°C for 20 sec, annealing at 60°C for 20 sec, and
extension at 72°C for 20 sec; and step 3, 4°C forever. The
following primers were used: HBV, forward 5′-ATACTGCACTCAGGCAAGC-3′
and reverse 5′-TGCCTCGTCGTCTAACAAC-3′; and β-actin, forward
5′-GGGACCTGACTGACTACCTC-3′ and reverse 5′-TCATACTCCTGCTTGCTGAT-3′.
β-actin mRNA was used as an endogenous control, and the relative
expression levels of HBV mRNA were determined using the
2−∆∆Cq method (34–36).
Detection of HBsAg and HBe antigen
(HBeAg)
At 24, 48 and 72 h post-transfection, the culture
medium was collected and centrifuged at 5,000 × g to remove
cellular debris, followed by storage at −20°C for analysis. At 7,
14, 21 and 28 days post-injection, blood was collected via the tail
vein and then centrifuged at 1,300 × g to collect the serum, which
was stored at −20°C for analysis. The concentrations of HBsAg and
HBeAg were detected with quantitative ELISA kits (Human HBsAg ELISA
kit, E-EL-H1567c; Human HBeAg ELISA kit, CR-018; Elabscience
Biotechnology Co., Ltd., Wuhan, China) following the manufacturer's
instructions.
Quantitative analysis of HBV DNA
At 24, 48 and 72 h post-transfection, HBV DNA was
extracted from the culture medium using a viral DNA extraction kit
(Sangon Biotech Co., Ltd). HBV RI-DNA and HBV DNA from the culture
medium of HepG2.2.15 cells was used as the template for real-time
PCR and quantified using an HBV diagnostic kit (Da-An, Guangzhou,
China) according to manufacturer's instructions.
Cell viability assay
HepG2.2.15 cells were seeded into 96-well plates at
2×104/well and cultured. At 48 h post-transfection, 20
µl Cell Counting Kit-8 (CCK-8) reagent was added to each
well, followed by incubation for 4 h at 37°C with 5%
CO2. The amount of viable sell was determined by
measurement of the absorbance at 450 nm.
Western blot analysis
At 48 h after transfection, the cells were lysed
with 1% radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology) and the protein concentration was
determined using a BCA Assay kit (Beyotime Institute of
Biotechnology). Proteins were separated by 8% SDS-PAGE and then
transferred onto a polyvinylidene difluoride membrane (Beyotime
Institute of Biotechnology). The membranes were incubated with
polyclonal rabbit anti-HBxAg (cat. no. ab39716; 1:800; Abcam,
Cambridge, MA, USA) and rabbit anti-HB core (c)Ag (cat. no. B0586;
1:1,000; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA)
overnight for 4°C. Horseradish peroxidase-labeled goat anti-rabbit
(cat. no. CW0240; 1:5,000; Beijing ComWin Biotech Co., Ltd.,
Beijing, China) was used as a secondary antibody and incubation was
performed for 1 h for 37°C. The signals were detected by the
Enhanced Chemiluminescence Detection system (Pierce; Thermo Fisher
Scientific, Inc.) and β-actin (anti-β actin antibody; cat. no.
ab8226; 1:1,000; Abcam, Cambridge, MA, USA) was used to normalize
the data (37).
Confocal microscopy
HepG2.2.15 cells transfected with ATF eukaryotic
expression vector for 48 h (37°C) were fixed with 4%
paraformaldehyde for 30 min (room temperature), washed three times
with PBS, and were permeabilized with 0.5% Triton X-100 for 5 min
(room temperature). After washing with PBS for three times, the
cells were incubated in 5% goat serum (1:20 in PBS dilution; cat.
no. 16210064; Thermo Fisher Scientific, Inc.) for 60 min. This was
followed by an incubation with anti-Flag polyclonal antibody (cat.
no. YM3001; 1:1,000; ImmunoWay Biotechnology Company, Suzhou,
China) at 4°C for 14 h, followed by rinsing 3 times with PBS and
incubation with fluorescein isothiocyanate-labeled goat anti-rat
secondary antibody (cat. no. sc-2010; 1:100; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at 37°C for 1 h. Finally, the
cells were stained with propidium iodide (Beyotime Institute of
Biotechnology) for 1 min. The expression of ATF protein was
visualized by using a Leica TCS SP2 laser scanning confocal
microscope (Leica Microsystems, Wetzlar, Germany).
Hepatic immunohistochemistry (IHC)
The location and expression of hepatitis B core
antigen (HBcAg) in the hepatic tissues of mice at 28 days post
inoculation was detected by IHC staining (38). The 4% paraformaldehyde-fixed (at
room temperature for 24 h) paraffin-embedded tissue sections
(4.5-µm thickness) were stained by IHC staining. Sections
were submerged in citrate buffer (pH 6.0) at 95°C for 15 min and
then cooled at room temperature. HBcAg was determined in the
hepatic sections by IHC staining with rabbit anti-HBcAg (cat. no.
B0586; 1:150; Dako; Agilent Technologies, Inc.). Incubation was
performed at 37°C for 2 h. Horseradish peroxidase-labeled goat
anti-rabbit (cat. no. CW0240; 1:500; Beijing ComWin Biotech Co.,
Ltd.) was used as a secondary antibody and incubation was performed
for 30 min for 37°C. The IHC images (original magnification, ×400)
were captured using a Leica light microscope (cat. no. DM3000;
Leica Microsystems GmbH, Wetzlar, Germany).
Statistical analysis
Values are expressed as the mean ± standard error of
the mean. Statistical analysis was performed using the one-way
analysis of variance followed by Dunnett's post hoc test, which was
used to assess the differences in numerical variables between the
experimental and control groups, including HBV DNA, HBV mRNA, HBsAg
and HBeAg. All analyses were calculated using SPSS v. 19.0.1 for
Windows (IBM Corp., Armonk, NY, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Target sites of ATF on HBV EnhI
Previous studies have demonstrated that the activity
of EnhI was controlled by the complex interaction of
hepatocyte-specific and immanent transcription factors, including
the tumor suppressor protein p53, the activator protein-1 complex,
retinoid X receptor, hepatocyte nuclear factor 3/4, CCAAT Enh
binding proteins and regulatory factor X-1 (39–43). Transcription of the pre-genomic
(pg)RNA was regulated through the EnhI region and the core
promoter. Besides the pgRNA, EnhI region regulates transcription of
the core and X genes, which has a predominant role in modulating
the expression of the temporal and global HBV gene (44,45). Based on the important functions of
EnhI, the present study demonstrated that inhibition to the
transcriptional activity of HBV reduced the replication of HBV DNA.
The 18-bp sequence 5′-CCCCCACTGGCTGGGGCT-3′ was selected as the ZF
target site and the corresponding amino acid sequence, which
targets the HBV EnhI region and integrates with effector domains to
confer transcriptional repression was successfully determined
(Fig. 1). The resulting ATF was
cloned into the mammalian expression plasmid pcDNA3.1(+).
Homologous sequence alignment of the human genome using the Basic
Local Alignment Search Tool (BLAST) of the National Center for
Biotechnology Information (https://blast.ncbi.nlm.nih.gov/Blast.cgi) allowed for
design of a ZFP with high specificity, and which did not combine
with any other potential target sequence. The BLAST search enabled
the comparison of the query sequence with a database of sequences,
and enabled the identification of library sequences that resemble
the query sequence above a certain threshold. This was used to
align the homology to the human genome nucleotide sequence.
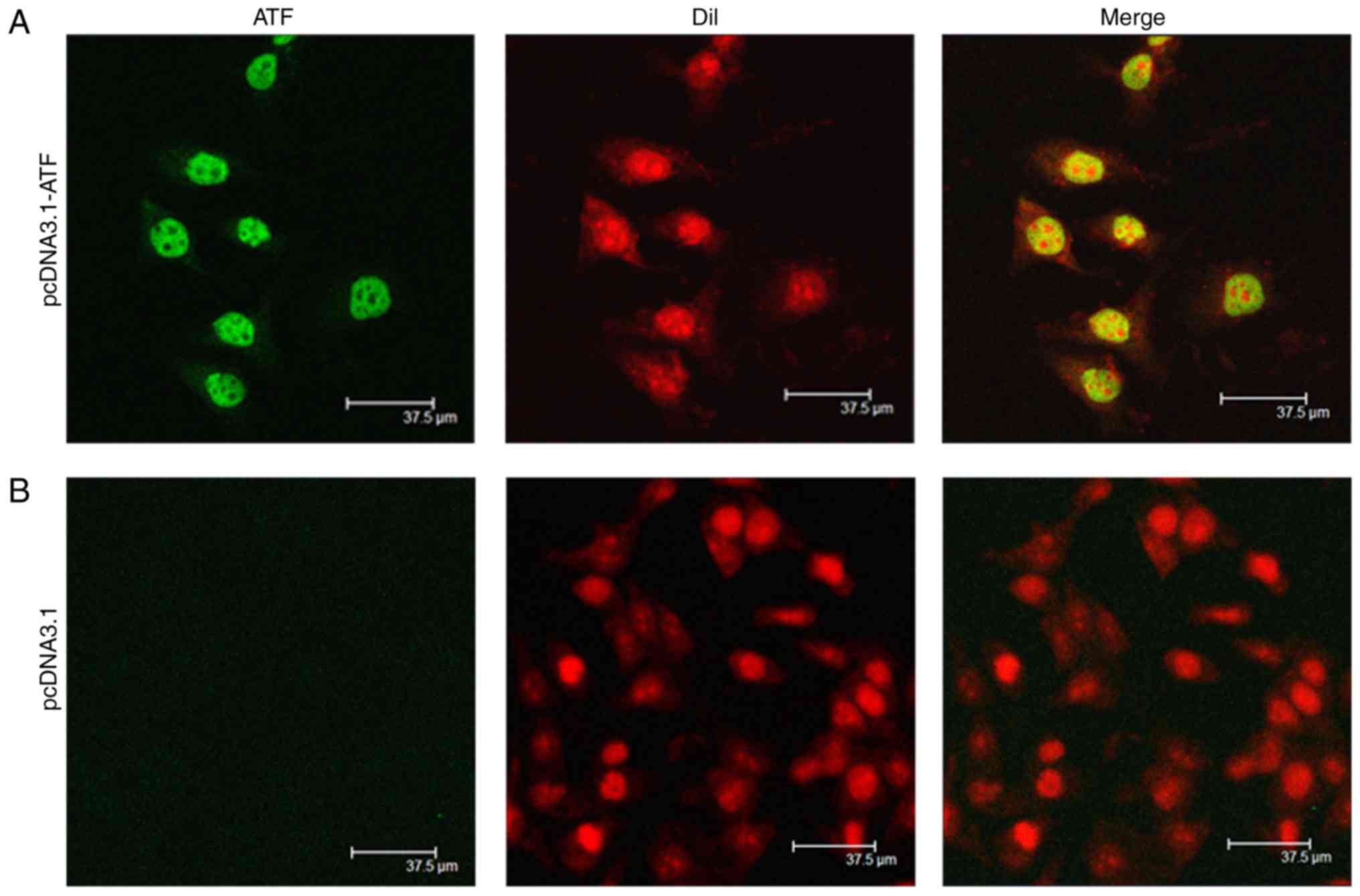
Expression and localization of ATF
To determine the expression of the designed ATF
eukaryotic expression vector after transfection, the ATF protein
was visualized by scanning confocal microscopy. The results
indicated that the designed expression vector normally expressed
ATF, which was mainly located in the nuclei, whereas the control
cells, which were transfected with empty vector pcDNA3.1(+), did
not exhibit any ATF expression (Fig.
2).
Cell viability
To estimate the possibility that the lower protein
expression in HepG2.2.15 cells may be attributed to a cytotoxic
effect caused by ATF, HepG2.2.15 cells were subjected to a CCK-8
cell viability assay at 48 h post-transfection. The viability of
HepG2.2.15 cells after transfection with ATF eukaryotic expression
vector exhibited no difference compared with that of cells
transfected with the empty vector pcDNA3.1(+) (Fig. 3). This result indicated that ATF
had no cytotoxic effect on the HepG2.2.15 cells.
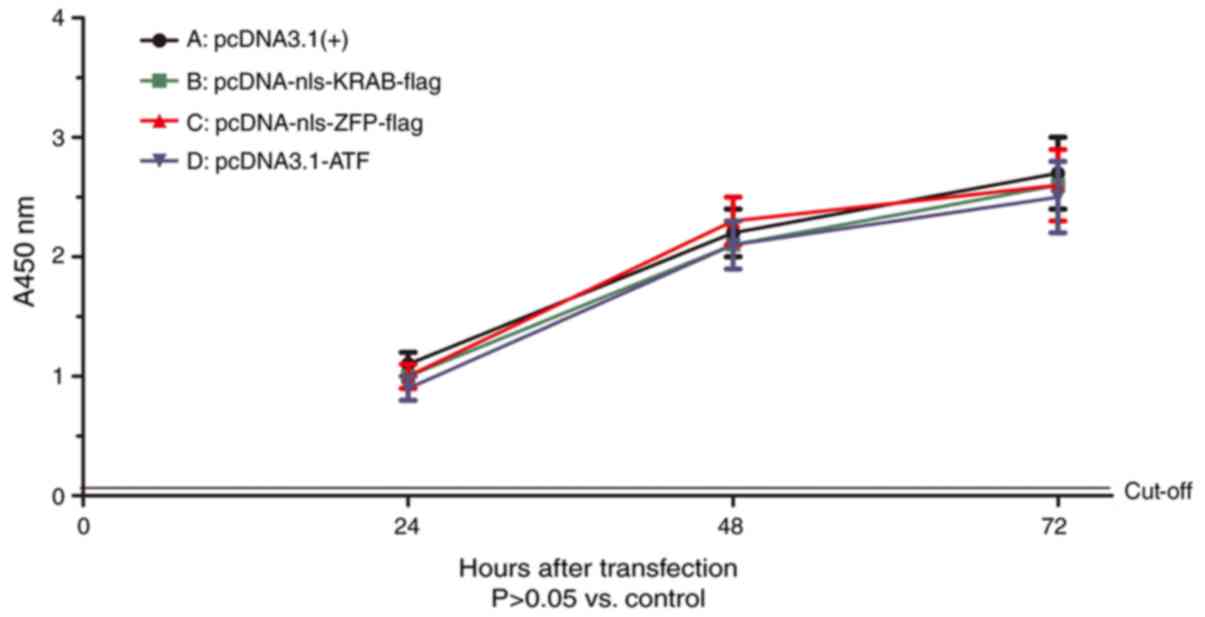 | Figure 3Effect of ATF on the viability of
HepG2.2.15 cells. Cells were seeded into 96-well plates at
2×104/well and cultured. After transfection with (A)
pcDNA3.1(+), (B) pcDNA-nls-KRAB-flag, (C) pcDNA-nls-ZFP-flag or (D)
pcDNA-ATF for 24, 48 or 72 h, the cell viability was determined by
a Cell Counting Kit-8 assay. In terms of the viability of
HepG2.2.15 cells, no statistically significant difference was
present between the groups (P>0.05). The control group (A) value
was set as 1. ATF, artificial transcription factor; ZFP, zinc
finger protein; KRAB, Kruppel-associated box; nls, nuclear
localization signal; A, absorbance. |
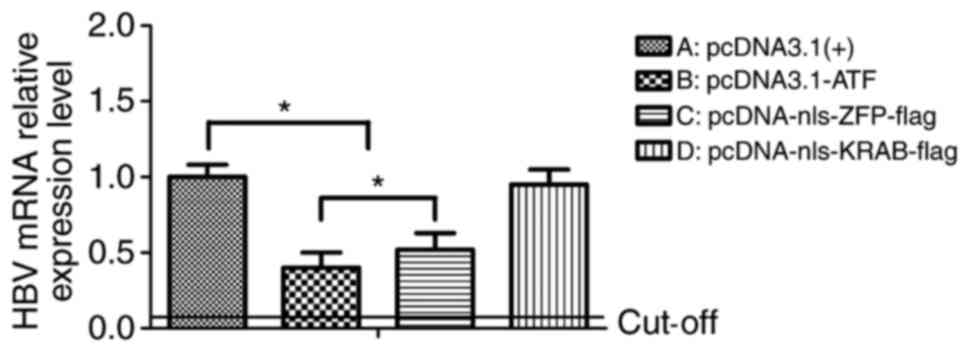
ATF reduces the HBV mRNA in vitro
To investigate the effects of the ATF on HBV mRNA
in vitro, HepG2.2.15 cells were transfected with
pcDNA3.1-ATF (nls-ZFP-KRAB-flag), pcDNA3.1(+)-nls-ZFP-flag,
pcDNA3.1(+)-nls-KRAB-flag and pcDNA3.1(+) as the control. After 72
h, the HBV mRNA was collected and analyzed by RT-qPCR. In cells
transfected with pcDNA3.1-ATF and pcDNA3.1(+)-nls-ZFP-flag, the HBV
mRNA levels were respectively reduced by 63 and 49% compared with
those in the empty vector control (P=0.03), and there was a
significant difference between the expression levels in these two
experimental groups (P=0.04). However, the
pcDNA3.1(+)-nls-KRAB-flag-transfected cells only displayed a slight
decrease in viral RNA production compared with that in the controls
(P=0.24; Fig. 4).
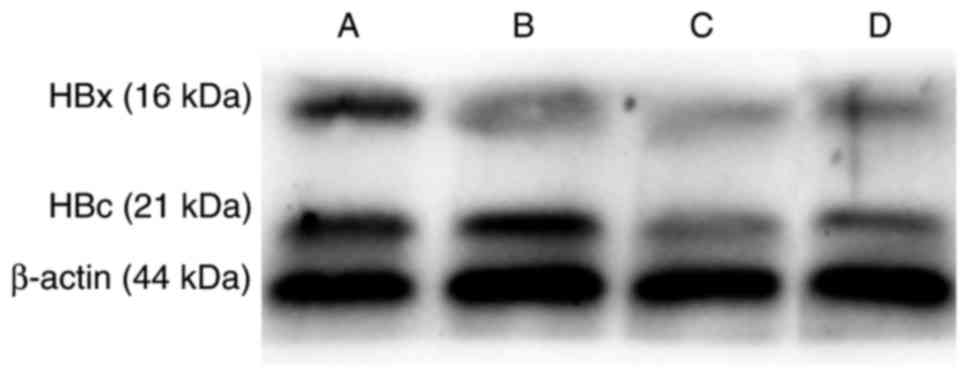
ATF inhibits HBV protein expression in
vitro
To further assess the effect of ATF on viral
transcription in vitro, western blot analysis was performed
to determine the effect of ATF on viral core and x protein
expression (Fig. 5). The results
indicated that pcDNA3.1-ATF and pcDNA3.1(+)-nls-ZFP-flag inhibited
the expression of HBV core and x protein compared with that in the
control group, while the in vitro inhibitory effect of
pcDNA3.1-ATF was significantly stronger than that of
pcDNA3.1(+)-nls-ZFP-flag.
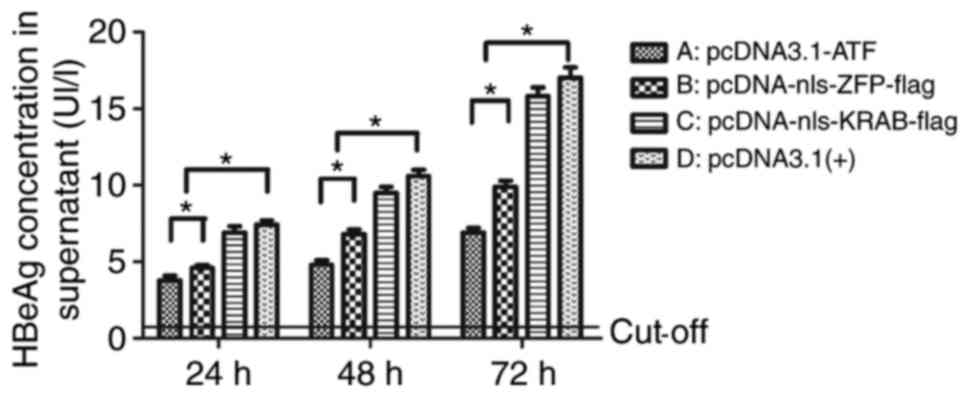
ATF reduces the secretion of HBeAg, but
not HBsAg in vitro
At 24, 48 and 72 h after transfection, the secretion
of HBsAg in HepG2.2.15 cells was not different from that in the
empty vector group (data not shown), while the secretion of HBeAg
was time-dependently reduced in the pcDNA3.1-ATF group when
compared with that in the empty vector group by 52.05, 54.19 and
60.37%, respectively (P<0.01). Furthermore, the inhibition in
the pcDNA3.1(+)-nls-ZFP-flag group was significantly decreased when
compared with that in the empty vector group (P=0.02), and there
was a significant difference between the pcDNA3.1-ATF and
pcDNA3.1(+)-nls-ZFP-flag groups; P=0.03), while no significant
difference was present between the pcDNA3.1(+)-nls-KRAB-Flag and
the empty vector group (P=0.08). This result suggested that
pcDNA3.1-ATF inhibited the expression of HBeAg; while it had no
effect on HBsAg. The inhibition was significantly decreased when
the pcDNA3.1-ATF vector was voided of its effector domain KRAB
(Fig. 6).
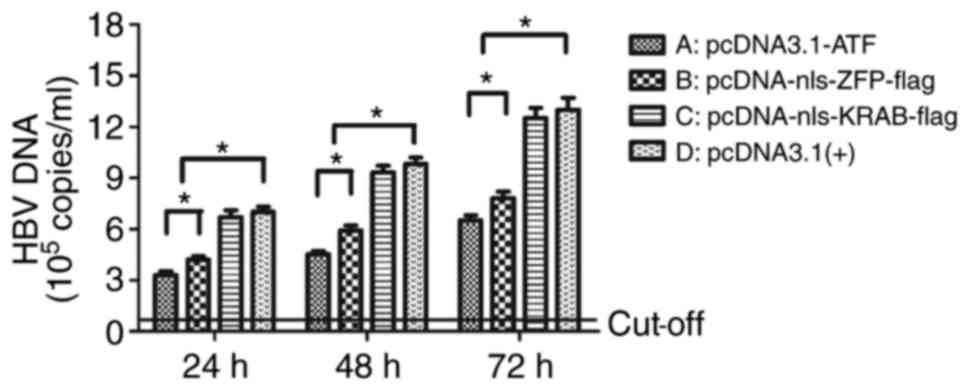
ATF reduces HBV replicative intermediates
in vitro
To determine whether vector-mediated ATF expression
had an impact on HBV replication, the amount of HBV replicative
intermediates was measured at 24, 48 and 72 h post-transfection by
RT-qPCR. Similar to its effect on HBV transcription and protein
expression, pcDNA3.1-ATF significantly inhibited HBV replication
compared with pcDNA3.1(+)-nls-KRAB-Flag and empty vector (P=0.02),
with the inhibitory rate (68.0%) being the highest at 72 h
post-transfection (Fig. 7), which
demonstrated a time-dependency of the inhibition of HBV
replication. The inhibition was slightly decreased, although not
significantly (P=0.27) when the ATF was voided of its effector
domain KRAB, whereas pcDNA-nls-KRAB-flag exerted no inhibitory
effect on HBV replication at 24, 48 and 72 h post-transfection,
compared with the empty vector group (P=0.27).
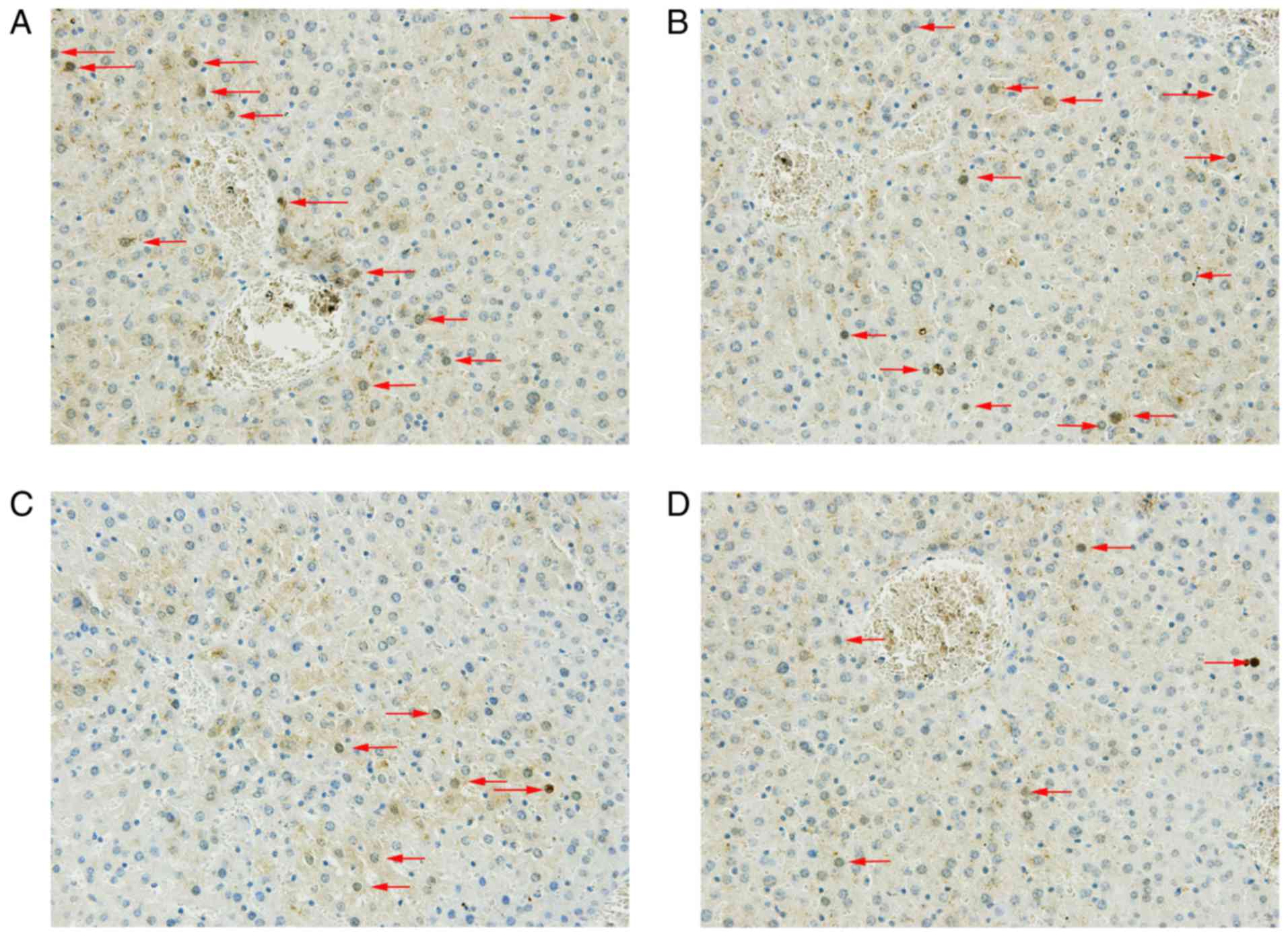
ATF inhibits HBV protein expression in
vivo
To further elucidate the effect of ATF on viral
transcription in vivo, an immunohistochemical assay was
performed to determine the effect of ATF on the expression of core
protein. The results indicated that pcDNA3.1-ATF and
pcDNA3.1(+)-nls-ZFP-flag inhibited the expression of core protein
compared with that in the empty vector group. However,
pcDNA3.1(+)-nls-KRAB-Flag did not inhibit HBV protein expression
in vivo (Fig. 8).
ATF inhibits the secretion of HBeAg but
not HBsAg in vivo
At 1, 3, 7, 14, 21 and 28 days after injection, the
secretion of HBsAg in sera was not different from that in the empty
vector group (data not shown), while the secretion of HBeAg was
time-dependently reduced in the pcDNA3.1-ATF and
pcDNA3.1(+)-nls-ZFP-flag groups when compared with that in the
empty vector group (P=0.02). This result suggested that ATF and ZFP
could inhibit the expression of HBeAg, but had no effect on HBsAg.
The inhibition was significantly decreased when ATF lost its
effector domain KRAB (Fig. 9), as
the secretion of HBeAg was significantly lower in the pcDNA3.1-ATF
group compared with that observed in the pcDNA-nls-KRAB-flag group
(P= 0.03).
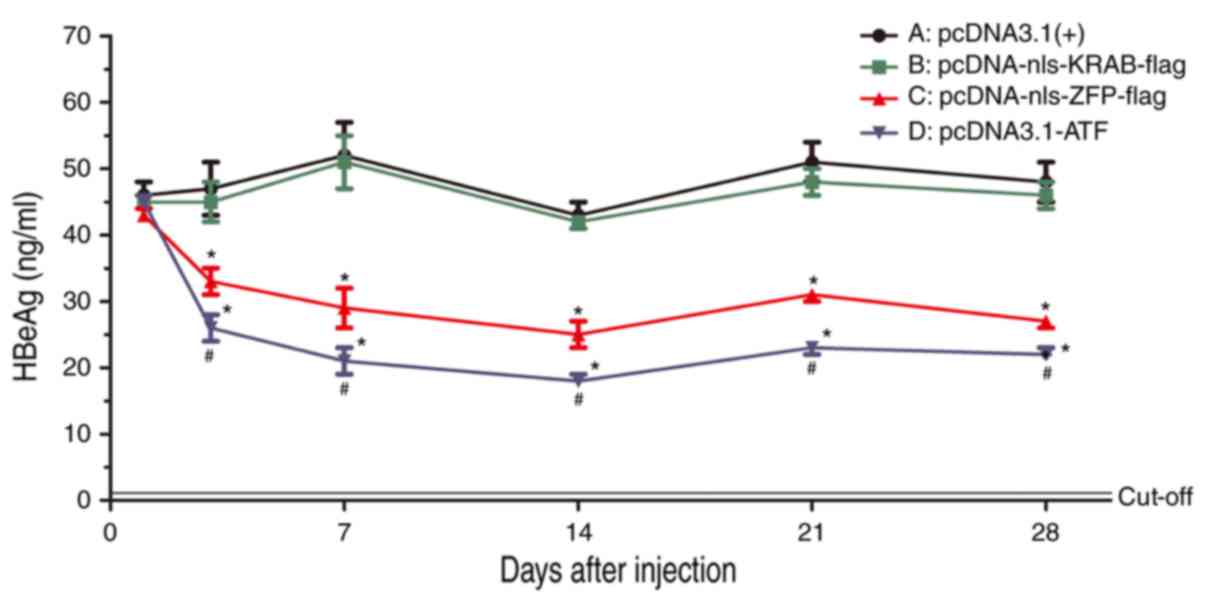 | Figure 9Serum levels of HBeAg were detected
using a radioimmunoassay at 1, 3, 7, 14, 21 and 28 days after
injection of (A) pcDNA3.1(+), (B) pcDNA-nls-KRAB-flag, (C)
pcDNA-nls-ZFP-flag or (D) pcDNA-ATF. *P<0.05 vs. the
control group; #P<0.05 vs. pcDNA-nls-KRAB-flag; ATF,
artificial transcription factor; ZFP, zinc finger protein; KRAB,
Kruppel-associated box; nls, nuclear localization signal; HBeAg,
hepatitis B e antigen. |
ATF reduces HBV replicative intermediates
in vivo
The amount of HBV replicative intermediates was
measured at 1, 3, 7, 14, 21 and 28 days after injection by using
RT-qPCR (Fig. 10). Similar to
its effect on HBV transcription and protein expression, ATF
significantly inhibited HBV replication as compared with that in
the pcDNA3.1(+)-nls-KRAB-Flag and empty vector groups (P=0.03). The
inhibitory rate was highest at 7 days post-injection, and a
time-dependency in the inhibition of HBV replication was observed.
Of note, the pcDNA-nls-KRAB-flag had no inhibitory effect on HBV
replication compared with that in the empty vector group
(P=0.21).
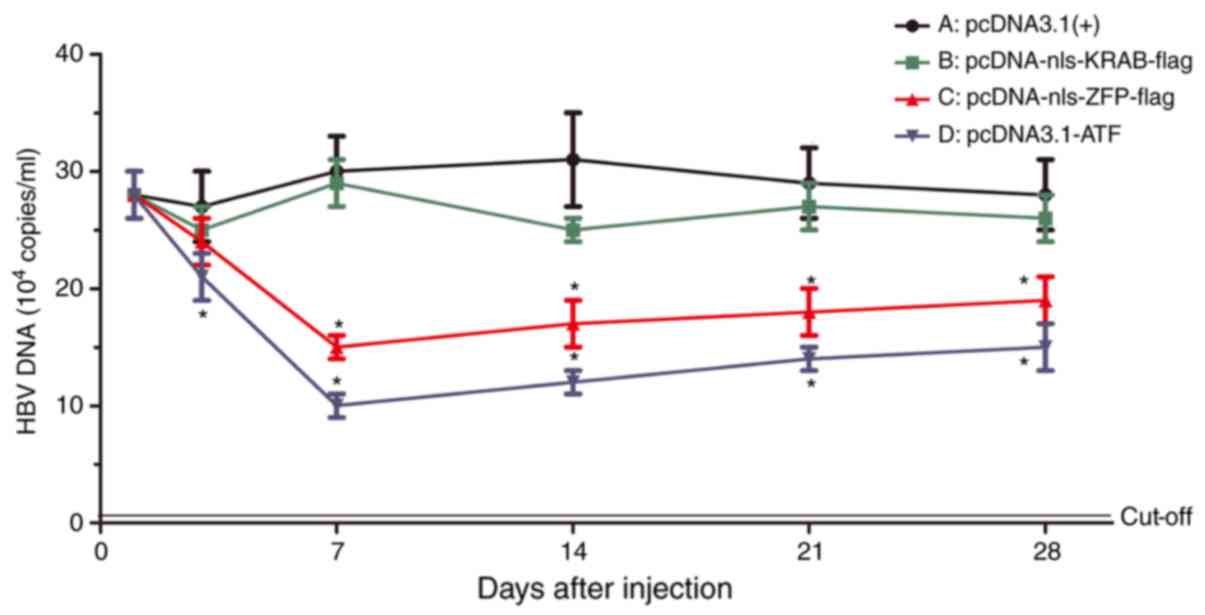 | Figure 10Expression of HBV DNA was detected
in vivo by polymerase chain reaction analysis. HBV
replicative intermediates were measured at 1, 3, 7, 14, 21 and 28
days after injection of (A) pcDNA3.1(+), (B) pcDNA-nls-KRAB-flag,
(C) pcDNA-nls-ZFP-flag or (D) pcDNA-ATF. *P<0.05 vs.
the control group. ATF, artificial transcription factor; ZFP, zinc
finger protein; KRAB, Kruppel-associated box; nls, nuclear
localization signal; HBV, hepatitis B virus. |
Discussion
HBV is a small, coated virus containing a 3,200-bp
partially relaxed circular double-stranded DNA genome (4). Once the hepatocyte is infected, the
core nucleocapsid of HBV is released into the cytoplasm and the
genomic DNA of HBV is transferred to the nucleus of the hepatocyte,
where the partially relaxed circular double-stranded DNA is
transformed into a closed covalently circular DNA (cccDNA)
(46,47). The HBV cccDNA acts as a stencil
for the transcription of all the viral RNAs, which include the
subgenomic RNAs and pgRNA, which is pivotal for maintaining the
replication and infection ability of progeny virus in the host
(48). Transcription is activated
by four promoters: The X, S1, S2 and preC/pg promoters; in
addition, EnhI and EnhII have a decisive role in the regulation of
viral gene transcription (2).
Studies have demonstrated that transcription of pgRNA is regulated
by the core promoter and the EnhI region. In addition, the
transcription of the x and core genes is controlled by the pgRNA
and EnhI region (49,50), which have a predominant role in
modulating the expression of the temporal and global HBV gene
(51). Based on the important
functions of EnhI, inhibition of the transcription activity of HBV
may be a novel strategy to reduce the replication of the progeny of
HBV DNA.
The replication of HBV DNA is precisely controlled
by the expression of the HBV gene. The S1, S2, X and preC/pg
promoters are essential for the transcription of HBV sequences. In
addition, EnhI and EnhII have an important role in the adjustment
of HBV gene expression (2,52).
In the HBV genome, there are partial overlaps between the EnhI
region and X promoter, which may influence the activity of the
promoter. Furthermore, the activity of the core promoter is
regulated by the EnhI, and thereby, EnhI contributes to the high
level of HBV replication (51).
It is likely that inhibition of the transcriptional activity of
EnhI may reduce the replication of HBV DNA.
Along with the rapid development of bioinformatics,
a vast amount of biological information is available in various
databases. Thus, by simulating the structure of natural
transcription factors, artificial proteins that do not exist in
nature, which may be generated to regulate specific genes, provides
a modern tactical concept for the development of novel gene
therapies (53,54). In fact, the conserved regions in
the HBV genome, which included the HBV cccDNA, were specifically
targeted and cleaved by the CRISPR/Cas9 system (55,56). The expression of endogenous target
genes was modulated by the ATFs, which may be used as a potential
powerful molecular tool in living organisms and cells. Numerous
DNA-binding protein molecules have been designed as the DNA-binding
motifs for the ATFs. Among them, the DNA-binding domain consists of
ATFs, which include the Cys2His2-type ZFPs that have been
extensively researched. The targeting sites are specifically
recognized by the ZF-based ATFs in chromosomes, which not only
effectively up- or downregulates the expression of their target
genes by fusing functional domains, but may also be utilized for
antiviral therapies (14,57,58).
In the present study, a genetic engineering method
was applied to construct the pcDNA3.1-ATF eukaryotic expression
vector, which efficiently expressed ATF in vitro and had no
cytotoxic effects, and which was demonstrated to bind to the HBV
Enh and inhibit the replication and expression of HBV DNA in
vivo and in vitro. The DNA binding specificity of the
ATF was unique, unmatched and unparalleled. The HBV EnhI-specific
ATF was designed, constructed and then transformed or injected into
HepG2.2.15 cells and HBV transgenic mice, respectively. The results
demonstrated that the HBV EnhI-specific ATF significantly inhibited
HBV transcription and replication of viral RNA, protein and viral
progeny without any obvious toxic effect in vitro and in
vivo. It is possible that the HBV EnhI-specific ATF is an
important part of advanced combination therapies for eliminating
HBV DNA in infected patients. An efficient treatment of chronic HBV
infection may become feasible by using this bioengineering
technology.
Acknowledgments
This study was supported by the National Science
Foundation of China (grant no. 81471946) and the National Science
Foundation of Chongqing (grant no. cstc2016jcyjA0269).
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Barker LF, Maynard JE, Purcell RH,
Hoofnagle JH, Berquist KR and London WT: Viral hepatitis, type B,
in experimental animals. Am J Med Sci. 270:189–195. 1975.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moolla N, Kew M and Arbuthnot P:
Regulatory elements of hepatitis B virus transcription. J Viral
Hepat. 9:323–331. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Beasley RP, Hwang LY, Lin CC and Chien CS:
Hepatocellular carcinoma and hepatitis B virus. A prospective study
of 22, 707 men in Taiwan. Lancet. 2:1129–1133. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ganem D and Varmus HE: The molecular
biology of the hepatitis B viruses. Annu Rev Biochem. 56:651–693.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ganem D and Prince AM: Hepatitis B virus
infection natural history and clinical consequences. N Engl J Med.
350:1118–1129. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lupberger J and Hildt E: Hepatitis B
virus-induced oncogenesis. World J Gastroenterol. 13:74–81. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chang JJ and Lewin SR: Immunopathogenesis
of hepatitis B virus infection. Immunol Cell Biol. 85:16–23. 2007.
View Article : Google Scholar
|
|
8
|
Pramoolsinsap C: Acute hepatitis A and
acquired immunity to hepatitis A virus in hepatitis B virus (HBV)
carriers and in HBV- or hepatitis C virus-related chronic liver
diseases in Thailand. J Viral Hepat. 7(Suppl 1): S11–S12. 2000.
View Article : Google Scholar
|
|
9
|
Liaw YF, Tsai SL, Sheen IS, Chao M, Yeh
CT, Hsieh SY and Chu CM: Clinical and virologival course of chronic
hepatitis B virus infection with hepatitis C and D virus markers.
Am J Gastroenterol. 93:354–359. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen LP, Zhao J, Du Y, Han YF, Su T, Zhang
HW and Cao GW: Antiviral treatment to prevent chronic hepatitis B
or C-related hepatocellular carcinoma. World J Virol. 12:174–183.
2012. View Article : Google Scholar
|
|
11
|
Dusheiko G: Treatment of HBeAg positive
chronic hepatitis B: Interferon or nucleoside analogues. Liver Int.
33(Suppl 1): S137–S150. 2013. View Article : Google Scholar
|
|
12
|
Craxi A, Antonucci G and Cammà C:
Treatment options in HBV. J Hepatol. 44(Suppl 1): S77–S83. 2006.
View Article : Google Scholar
|
|
13
|
Fung J, Lai CL, Seto WK and Yuen MF:
Nucleoside/nucleotide analogues in the treatment of chronic
hepatitis B. J Antimicrob Chemother. 66:2715–2725. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sera T: Zinc-finger-based artificial
transcription factors and their applications. Adv Drug Deliv Rev.
61:513–526. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Papworth M, Kolasinska P and Minczuk M:
Designer zinc-finger proteins and their applications. Gene.
366:27–38. 2006. View Article : Google Scholar
|
|
16
|
Greisman HA and Pabo CO: A general
strategy for selecting high-affinity zinc finger proteins for
diverse DNA target sites. Science. 275:657–661. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dreier B, Beerli RR, Segal DJ, Flippin JD
and Barbas CF III: Development of zinc finger domains for
recognition of the 5′-ANN-3′ family of DNA sequences and their use
in the construction of artificial transcription factors. J Biol
Chem. 276:29466–29478. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Isalan M, Klug A and Choo Y: A rapid,
generally applicable method to engineer zinc fingers illustrated by
targeting the HIV-1 promoter. Nat Biotechnol. 19:656–660. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Q, Segal DJ, Ghiara JB and Barbas CF
III: Design of polydactyl zinc-finger proteins for unique
addressing within complex genomes. Proc Natl Acad Sci USA.
94:5525–5530. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim JS and Pabo CO: Getting a handhold on
DNA: Design of poly-zinc finger proteins with femtomolar
dissociation constants. Proc Natl Acad Sci USA. 95:2812–2817. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Desjarlais JR and Berg JM: Toward rules
relating zinc finger protein sequences and DNA-binding site
preferences. Proc Natl Acad Sci USA. 89:7345–7349. 1992. View Article : Google Scholar
|
|
22
|
Rebar EJ and Pabo CO: Zinc finger phage:
Affinity selection of fingers with new DNA-binding specificities.
Science. 263:671–673. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Laity JH, Lee BM and Wright PE: Zinc
finger proteins: New insights into structural and functional
diversity. Curr Opin Struct Biol. 11:39–46. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Blancafort P, Magnenat L and Barbas CF
III: Scanning the human genome with combinatorial transcription
factor libraries. Nat Biotechnol. 21:269–274. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jamieson AC, Miller JC and Pabo CO: Drug
discovery with engineered zinc-finger proteins. Nat Rev Drug
Discov. 2:361–368. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lund CV, Blancafort P, Popkov M and Barbas
CF III: Promoter-targeted phage display selections with
preassembled synthetic zinc finger libraries for endogenous gene
regulation. J Mol Biol. 340:599–613. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu H, Yang WP and Barbas CF III: Building
zinc fingers by selection: Toward a therapeutic application. Proc
Natl Acad Sci USA. 92:344–348. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Papworth M, Moore M, Isalan M, Minczuk M,
Choo Y and Klug A: Inhibition of herpes simplex virus 1 gene
expression by designer zinc-finger transcription factors. Proc Natl
Acad Sci USA. 100:1621–1626. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
ZF Tools. http://www.scripps.edu/mb/barbas/zfdesign/zfdesign-home.php.
|
|
30
|
Margolin JF, Friedman JR, Meyer WK,
Vissing H, Thiesen HJ and Rauscher FJ III: Krüppel-associated boxes
are potent transcriptional repression domains. Proc Natl Acad Sci
USA. 91:4509–4513. 1994. View Article : Google Scholar
|
|
31
|
Sadowski I, Ma J, Triezenberg S and
Ptashne M: GAL4-VP16 is an unusually potent transcriptional
activator. Nature. 335:563–564. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Walters KA, Joyce MA, Addison WR, Fischer
KP and Tyrrell DL: Superinfection exclusion in duck heaptitis B
virus infection is mediated by the large surface antigen. J Virol.
78:7925–7937. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
35
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Larionov A, Krause A and Miller W: A
standard curve based method for relative real time PCR data
processing. BMC Bioinformatics. 6:622005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shimizu S, Nomura F, Tomonaga T, Sunaga M,
Noda M, Ebara M and Saisho H: Expression of poly(ADP-ribose)
polymerase in human hepatocellular carcinoma and analysis of biopsy
specimens obtained under sonographic guidance. Oncol Rep.
12:821–825. 2004.PubMed/NCBI
|
|
38
|
Shintani M, Urano M, Takakuwa Y, Kuroda M
and Kamoshida S: Immunohistochemical characterization of pyrimidine
synthetic enzymes, thymidine kinase-1 and thymidylate synthase, in
various types of cancer. Oncol Rep. 23:1345–1350. 2010.PubMed/NCBI
|
|
39
|
Dikstein R, Faktor O and Shaul Y:
Hierarchic and cooperative binding of the rat liver nuclear protein
C/EBP at the hepatitis B virus enhancer. Mol Cell Biol.
10:4427–4430. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Faktor O, Budlovsky S, Ben-Levy R and
Shaul Y: A single element within the hepatitis B virus enhancer
binds multiple proteins and responds to multiple stimuli. J Virol.
64:1861–1863. 1990.PubMed/NCBI
|
|
41
|
Siegrist CA, Durand B, Emery P, David E,
Hearing P, Mach B and Reith W: RFX1 is identical to enhancer factor
C and functions as a transactivator of the hepatitis B virus
enhancer. Mol Cell Biol. 13:6375–6384. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ori A, Zauberman A, Doitsh G, Paran N,
Oren M and Shaul Y: p53 binds and represses the HBV enhancer: An
adjacent enhancer element can reverse the transcription effect of
p53. EMBO J. 17:544–553. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chandra V, Huang P, Potluri N, Wu D, Kim Y
and Rastinejad F: Multidomain integration in the structure of the
HNF-4α nuclear receptor complex. Nature. 495:394–398. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Quasdorff M and Protzer U: Control of
hepatitis B virus at the level of transcription. J Viral Hepat.
17:527–536. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu B, Wen X, Huang C and Wei Y:
Unraveling the complexity of hepatitis B virus: From molecular
understanding to therapeutic strategy in 50 years. Int J Biochem
Cell Biol. 45:1987–1996. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Levrero M, Pollicino T, Petersen J,
Belloni L, Raimondo G and Dandri M: Control of cccDNA function in
hepatitis B virus infection. J Hepatol. 51:581–592. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Beck J and Nassal M: Hepatitis B virus
replication. World J Gastroenterol. 13:48–64. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Christine N, Wei Y and Buendia MA:
Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol.
52:594–604. 2010. View Article : Google Scholar
|
|
49
|
Bock CT, Malek NP, Tillmann HL, Manns MP
and Trautwein C: The enhancer I core region contributes to the
replication level of hepatitis B virus in vivo and in vitro. J
Virol. 74:2193–2202. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Antonucci TK and Rutter WJ: Hepatitis B
virus (HBV) promoters are regulated by the HBV enhancer in a
tissue-specific manner. J Virol. 63:579–583. 1989.PubMed/NCBI
|
|
51
|
Doitsh G and Shaul Y: Enhancer I
predominance in hepatitis B virus gene expression. Mol Cell Biol.
24:1799–1808. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Soussan P, Garreau F, Zylberberg H, Ferray
C, Brechot C and Kremsdorf D: In vivo expression of a new hepatitis
B virus protein encoded by a spliced RNA. J Clin Invest. 105:55–60.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pavletich NP and Pabo CO: Crystal
structure of a five-finger GLI-DNA complex: New perspectives on
zinc fingers. Science. 261:1701–1707. 1993. View Article : Google Scholar
|
|
54
|
Urnov FD and Rebar EJ: Designed
transcription factors as tools the therapeatics and functional
genomics. Biochem Pharmacol. 64:919–923. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ramanan V, Shlomai A, Cox DB, Schwartz RE,
Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN, et
al: CRISPR/Cas9 cleavage of viral DNA efficiently suppresses
hepatitis B virus. Sci Rep. 5:108332015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Karimova M, Beschorner N, Dammermann W,
Chemnitz J, Indenbirken D, Bockmann JH, Grundhoff A, Lüth S,
Buchholz F, Schulze zur Wiesch J and Hauber J: CRISPR/Cas9
nickase-mediated disruption of hepatitis B virus open reading frame
S and X. Sci Rep. 5:137342015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mino T, Hatono T, Matsumoto N, Mori T,
Mineta Y, Aoyama Y and Sera T: Inhibition of DNA replication of
human papilloma-virus by artificial zinc finger proteins. J Virol.
80:5405–5412. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zimmerman KA, Fischer KP, Joyce MA and
Tyrrell DL: Zinc finger proteins designed to specifically target
duck hepatitis B virus covalently closed circular DNA inhibit viral
transcription in tissue culture. J Virol. 82:8013–8021. 2008.
View Article : Google Scholar : PubMed/NCBI
|