1. Introduction
The epithelial cell adhesion molecule (EpCAM, also
known as cluster of differentiation 326 or Tacstd1), is a 40 KD
trans-membrane glycoprotein, consisting of 314 amino acids, firstly
identified in colon cancer in 1979. Its molecular structure
includes: Extracellular domain (EpEX), single transmembrane domain
and intracellular domain (EpICD) (1). EpCAM is a homophilic
Ca2+-independent cell-cell adhesion molecule. In
addition to a role in intercellular adhesion, in vitro and
in vivo studies have revealed that EpCAM plays important
roles in cell signaling, proliferation, differentiation, formation
and maintenance of organ morphology (2).
EpCAM is expressed in many kinds of epithelial
tissues (3) and is also a cell
surface marker on various stem and progenitor cells (4,5).
Mutations in human EpCAM have been identified to be associated with
congenital tufting enteropathy (CTE) (6). To investigate the in vivo
functions of EpCAM, several mutant animal models have been
generated in recent years, including mice, zebrafish and
Xenopus. At least 4 types of global EpCAM knockout mice
suggest that the phenotype is similar to the symptoms of human CTE
(7–9), however one model leads to embryonic
lethality due to placental defect (10). In addition, a conditional knockout
mouse model exhibits impaired motility of the skin Langerhans cells
(11). Zebrafish (12) and Xenopus (13,14) models demonstrate the functions of
EpCAM in morphogenic movements during gastrulation. Hepatic
development has also been affected in EpCAM mutant zebrafish
(15). These studies have
confirmed the important functions of EpCAM in physiological
processes and development.
In addition to important roles in developmental
processes, EpCAM is also highly expressed in epithelial tumor
tissues, and promotes the proliferation of tumors (16,17). Its functions in oncogenesis and
cancer cells have been studied by many groups, and therapeutic
approaches targeting EpCAM are also currently being developed.
Here, the authors summarize the functions of EpCAM
in physiology, development and diseases, and review current
progresses in identifying the underlying molecular mechanisms.
2. Expression pattern and subcellular
localization of EpCAM
EpCAM can be detected in many tissues from very
early embryos to adult animals and human beings, and it is also
highly expressed in numerous types of tumor tissue.
Expression of EpCAM during embryo
development
EpCAM mRNA can be detected in the fertilized zygotes
of zebrafish (12), indicating it
is present maternally. In 1981, EpCAM was defined as one of the
human trophoblast cell-surface antigens (18). In the developing mouse embryo,
EpCAM is also detected in the inner cell mass of the blastocyst
(7), the epiblast (7,19),
and the gonads at E12.5 (7). At
early gastrulation stages including E6.5 and E7.5, EpCAM is
expressed in the ectoderm and endoderm, however may be used as a
marker of the endoderm at and after E8.25 (19). Nagao et al (10) revealed that the expression of
EpCAM is most prominent in the facial primordia, gut, branchial
arches, and otocyst at E9.5. EpCAM protein may be detected in the
allantois, the labyrinthine layer, and the spongiotrophoblasts from
E8.5 and E9.5 placentas of mice, however the highest expression
levels are detected in the labyrinth. In addition, the expression
of EpCAM is readily detected in various epithelial components of a
variety of organs including hair follicles, the nasal plexus,
lungs, kidneys, and pancreas examined at E14.5 (10). EpCAM mRNA is detected in the
developing gut of mice from E9.5 to E15.5, as well as throughout
the intestine, from the duodenum to the colon, at E18.5 (7). EpCAM protein is detected in the
villi and intervillus domains, and is localized to cell-cell
junctions of the intestinal epithelium at E18.5 and at P0. EpCAM is
highly expressed in the epithelia of stomach, lungs, pancreas, and
kidneys at E18.5 (7). During the
embryogenesis of zebrafish, maternally provided EpCAM mRNA is
uniformly distributed in all cells of cleavage and early blastula
stage embryos. Following the onset of epiboly, EpCAM mRNA is
restricted to the enveloping layer (EVL) and basal epidermis,
indicating the zygotic expression of EpCAM is restricted to the
epithelial structures (12).
EpCAM has also been demonstrated to be expressed in migrating
neuromast primordia, otic vesicles and olfactory placodes (12).
Expression pattern of EpCAM in adult
organs and tumor tissues
EpCAM is expressed in many adult organs, just as
Balzar et al (3) and
Schnell et al (2) have
summarized in detail. Similar to what they have mentioned in rats,
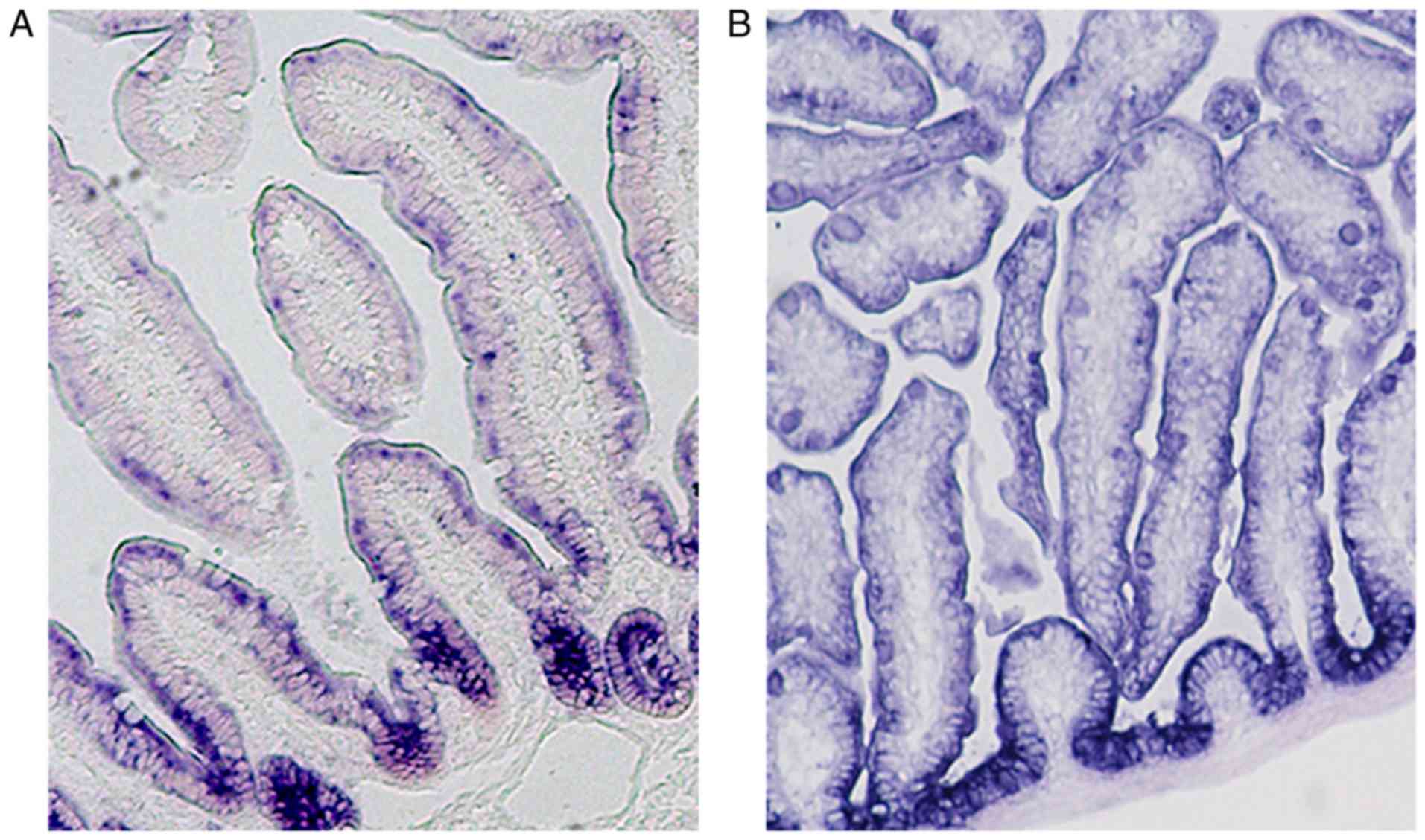
we also observed a gradient of EpCAM from crypts to villi in small
intestines of adult mice (Fig. 1;
unpublished data). It confirmed that the expression level of EpCAM
was high in the proliferative cells and low in the differential
cells. Balzar et al reported that in the skin of adult
humans, the expression level of EpCAM is low in hair follicles and
sweat glands (3). Recently, it
has been reported that EpCAM is expressed in the skin Langerhans
cells (11). In addition,
according to reverse transcription-polymerase chain reaction
analysis, EpCAM mRNA is also present in the skin of adult zebrafish
(12). Recently, Poon et
al (20) studied the
expression pattern of EpCAM in rat uterus, and it was revealed that
prior to implantation, EpCAM mediates intercellular adhesion in the
uterine epithelium, but during implantation, when the uterine
luminal epithelial cells lose the majority of their intercellular
and cell-matrix adhesions, the expression of EpCAM is decreased,
although still present for the maintenance of mucosal
integrity.
EpCAM was originally identified as a
tumor-associated antigen on the basis of its high expression level
in the tumors of epithelial origin (16). EpCAM-positive cells have been
suggested to serve as cancer stem cells for various human cancers,
including colorectal and hepatocellular carcinoma (21,22). Numerous antibody-based therapeutic
approaches targeting EpCAM are currently being developed (23). Further information regarding EpCAM
expression in tumors can be found in Balzar et al (3) and Schnell et al (2).
Subcellular distribution of EpCAM
It has been verified that EpCAM is enriched at the
basolateral membrane of mouse and human intestinal epithelium.
Immunohistochemistry (IHC) staining of the intestinal epithelium of
mice with both EpCAM antibody and either E-cadherin antibody or
ZO-1 antibody, markers of adherens junctions (AJs) and tight
junctions (TJs), respectively, revealed that EpCAM is localized to
TJs, AJs and the lateral membranes of epithelial cells lining the
mouse intestines (7). In normal
human intestinal epithelium, anti-EpCAM antibody stains the region
of TJs and the lateral membrane; the EpCAM signal can even be
detected on the brush border by immunostaining and immunoelectron
microscopy (24). Using
domain-specific antibodies, it was demonstrated that EpICD-specific
staining is speckled in the cytoplasm, perinucleus and nuclei of
FaDu hypopharynx and HCT-8 colon carcinoma cells (25). EpICD may also be detected in the
nuclei of carcinoma samples, but does not appear to localize to the
nuclei in samples from normal colon biopsies and is detected as
distinct speckles in the cell cytoplasm of normal tissues (25).
3. Functions of EpCAM in physiological
processes
EpCAM and cell junctions
The name EpCAM-epithelial cell adhesion
molecule-describes cell-cell adhesion protein. Like most cell
adhesion molecules, the primary function of EpCAM appears to be
cell-cell interaction (26). This
has been supported by the studies with L929 fibroblasts, which are
normally not involved in cellular adhesion. The L929 fibroblasts
form multicellular aggregates of cells when expressing EpCAM,
suggesting that EpCAM is involved in homotypic cell-cell
interactions (27). Although a
previous study revealed that cells expressing EpCAM were only
loosely interconnected (28),
further reports have revealed that EpCAM is essential for cell
junctions: It interacts with several important cell adhesion
molecules (CAMs) and regulates adhesive structures between cells
and cell-matrix, including TJs, AJs, desmosomes, and
hemi-desmosomes.
EpCAM regulates CAMs
EpCAM and classical cadherins. The E-cadherin gene
is extensively studied as a classical cadherin for the whole
cadherin superfamily. The early stage results have revealed that
E-cadherin exerts opposing effects to EpCAM. E-cadherin acts as a
tumor suppressor protein (29–31). Mutational disruption of the
E-cadherin gene has been observed in invasive lobular breast cancer
and gastric carcinoma (31–33). E-cadherin germline mutations are
detectable in families with a high frequency of early-onset gastric
cancer (32–34). In contrast, EpCAM is highly
expressed in a variety of carcinomas and currently considered to be
an important carcinoma marker. In cultured murine fibroblast L
cells, the expression of EpCAM suppresses the E-cadherin-mediated
cell aggregation, because EpCAM disrupts the association of
E-cadherin with the cytoskeleton (35,36). It has also been reported that
EpCAM affects the cadherin-mediated junctions in HBL-100 cells
(35). From these results, it was
hypothesized that EpCAM may act as an antagonist of E-cadherin.
However, this conclusion has recently been
challenged by EpCAM genetic animal models. In contrast to cell
culture studies, Slanchev et al (12) revealed that in the EpCAM mutant
enveloping layer (EVL) of zebrafish, the expression level of
E-cadherin is reduced; EpCAM and E-cadherin tightly interact for an
enhanced effect to promote EVL integrity as well as deep cell
epiboly. Each of them is indispensable for EVL integrity, whereas
combined loss of both EpCAM and E-cadherin leads to severe
layer-autonomous EVL disassembly during early gastrulation stages
(12). Several reports have
revealed that EpCAM contributes to the formation of functional TJs
in the mouse and human intestinal epithelium (7,9,37).
Notably, when E-cadherin is deleted from the mouse intestine, the
barrier function of the intestines is compromised (38), moreover, loss of E-cadherin in
mouse epidermis may lead to improper localization of the key tight
junctional proteins, resulting in permeable TJs and altered
epidermal resistance (39). From
these reports, it was concluded that EpCAM and E-cadherin enhance
each other's physiological functions in vivo.
The conclusions from the reduced expression of EpCAM
in vivo and in vitro are completely different. The
expression of E-cadherin in the CTE children's biopsy specimens is
normal (40). The expression
level and expression pattern of E-cadherin and β-catenin are normal
in the EpCAM mutant intestines of mouse embryos (7), but their expression levels are
disrupted and the intracellular accumulation rapidly increases
after birth (8). As these
alterations occur following birth, they may be secondary effects of
EpCAM mutation. In contrast, in vitro studies suggest that
the reduction of EpCAM expression does not change the expression or
localization of E-cadherin and β-catenin in T84 cells (41).
Various EpCAM knockout mice exhibit abnormal
placental development and die in utero by E12.5, and the expression
of E-cadherin and P-cadherin in E9.5 placentas of these mutant mice
are not affected (10). EpCAM is
co-expressed with E-cadherin, but the expression of EpCAM is
inversely correlated with P-cadherin in mouse placentas, developing
guts, lungs and hair follicles (10). However, Wu et al (41) reported that EpCAM and E-cadherin
did not precisely co-localize in T84 cell monolayers detected by
confocal microscopy, and co-immunoprecipitation studies did not
indicate that EpCAM and E-cadherin were tightly associated in T84
cells.
There are two noteworthy exceptions. Global
depletion of EpCAM by injection of antisense Morpholino
oligonucleotides (EpCAM MO) in Xenopus embryos results in a
marked decrease in C-cadherin protein levels, but does not affect
mRNA levels (the same was observed for the associated tested
molecules E-cadherin, α- and β-catenin), indicating that the
regulation occurs at a posttranscriptional level (14). Cadherin downregulation results
from protein kinase C (PKC) overactivation in EpCAM mutant
Xenopus embryos. In EpCAM-depleted Caco-2 cells, a human
colon cancer cell line, PKC activation increases and the E-cadherin
protein is decreased (14) or
mislocalized (42).
EpCAM and claudins
Claudin-7 is the first member of the family of
claudins which was reported in a study investigating the
association between EpCAM and claudins in both non-transformed
tissues and metastasizing tumor cell lines (24). Claudin-7 was first detected in a
CD44v6-tetraspanis-EpCAM complex, and later it was confirmed that
claudin-7 associates directly with EpCAM (24,43). Claudin-7 has been observed in TJs
and basolateral membranes; the co-localization of EpCAM and
claudin-7 has also been observed on both TJs and basolateral
membranes (7,24). Mutant experiments identified that
the AxxxG motif in the transmembrane domain of EpCAM is required
for association with claudin-7, and claudin-7 is unable to
associate with EpCAM if A279 and G282 are changed to I279 and I282
(44).
Claudin-7 is one of the primary claudin proteins
expressed in the intestine (45).
In EpCAM mutant embryos, claudin-7 protein is downregulated to
undetectable levels in all regions of the intestine examined, but
claudin-7 mRNA is still normal in the mutant intestine (7). EpCAM and claudin-7 have been
demonstrated to colocalize at the basolateral surface of the
intestine, pancreas, stomach, lungs and kidneys in wild type (WT)
mice, detected by fluorescent immunohistochemistry. Claudin-7
protein is also downregulated to undetectable levels in the
pancreas, lungs and stomach of EpCAM knockout mice, but there are
still some weak claudin-7 protein signals in the EpCAM mutant
kidneys (7). Deletion of the exon
4 in EpCAM leads to decreased expression and mislocalization of
EpCAM in the intestinal epithelium rather than along the plasma
membrane, meanwhile, the expression of claudin-7 protein is also
decreased, and the co-localization with EpCAM ∆4 protein is lost,
although claudin-7 mRNA levels remain unaltered (9). Mueller et al (9) also observed a similar situation of
claudin-7 expression in CTE patients. Furthermore, in WT or ∆4
FLAG-tagged EpCAM transfected HEK293 cells, endogenous claudin-7
combines with WT EpCAM, however not with EpCAM ∆4. Knockdown of
EpCAM in T84 and Caco-2 cells using short hairpin RNA leads to
decreased claudin-7 proteins in these cells (41).
As mentioned above, the transmembrane domain of
EpCAM is directly associated with claudin-7. Claudin-7 protein is
decreased when EpCAM is lost. Claudin-1 and claudin-7 are closely
related (~50% homologous at the amino acid level) (46), and their sub-localization is
similar. Wu et al (41)
revealed that the expression of claudin-1 protein is also decreased
following knockdown of EpCAM in T84 and Caco-2 cells, however
claudin-1 mRNA remains normal. Notably, the expression of claudin-1
is also reduced in claudin-7 knockout mice (47). It has been confirmed that
claudin-7 is associated with claudin-1 and facilitates the
incorporation of claudin-1 into EpCAM-containing complexes.
The fact that decreased expression levels of
claudin-7 and claudin-1 are associated with the reduction of EpCAM
expression indicates that the alterations are post-transcriptional.
Inhibition of proteosomes by lactacystin or MG132 are unable to
increase the expression of claudin-7 and claudin-1 proteins in
EpCAM knockout samples or EpCAM knockdown cells (7,41).
Wu et al (41) indicated
that claudin-7, claudin-1, and EpCAM turnover occurs via lysosomes,
and EpCAM may protect claudin-7 and claudin-1 from lysosomal
degradation in Caco-2 cells, detected by immunofluorescence
microscopy and chloroquine treatment experiments (41).
Claudin-7 was not detected in the intestine, lungs,
stomach, and pancreas of EpCAM mutant mice, however is still
detected in the kidney epithelium, albeit at reduced levels
(7). The kidney epithelium may
express an EpCAM-like protein that associates with claudin-7.
EpCAM-2/Tacstd-2 is a candidate for such an EpCAM-like molecule,
given that it is not only structurally related to EpCAM, but also
interacts with claudin-1 and claudin-7 in human tissues and
cultured cells (48).
In addition to claudin-7, the levels of claudins 2,
3, and 15 are also reduced in the intestine of EpCAM mutant mice
(7); claudins 2, 3, 7, and 15
co-precipitate with each other from mouse intestinal lysates.
However, claudin molecules associate with each other in a
heterotypic as well as homotypic manner in cultured cell lines
(49–51). EpCAM may indirectly interact with
these claudins and recruit them to the cell-cell junctions, in
particular the sub-localization of claudin-2, 3, and 15 is only at
the TJs (7). As the distribution
and expression levels of claudins 2, 3, 4, and 8 in the kidneys are
not affected in claudin-7 knockout mice (52), the precise manner of the
interaction between EpCAM and various claudins remains to be
determined. AJs have been found to be necessary for the
establishment of TJs (53);
certain TJ proteins are mislocalized and functional TJs are not
formed in the absence of α-catenin or cadherin (39,54,55). The structure of AJs appears normal
in EpCAM mutant animals, but as mentioned above, their function in
maintaining TJs may be affected, which may be the reason for
decreased levels of other claudins.
In the mouse intestines, claudin-7, claudin-1 and α2
-inte-grin form a complex (47),
therefore it may be hypothesized that this also occurs in other
organs. EpCAM directly binds claudin-7, which in turn facilitates
the incorporation of claudin-1 into the EpCAM-containing complexes.
In many tumor cells, the complex changes to EpCAM-claudin-7-CD4
4v6-tetraspanis (43), which may
be important during tumori-genesis.
EpCAM regulates adhesive structures
It has previously been demonstrated that exogenous
EpCAM mediates Ca2+-independent homophilic cell-cell
adhesion in cultured EpCAM-negative cells. Here, the authors
discuss its in vivo cell-cell adhesion functions from the
reports of genetic animal models and CTE patients.
Apical junctional complex (AJC)
In vertebrate epithelial cells, the apical TJ and
the more basally localized AJ form the AJC (56–58). EpCAM mutant zebrafish display a
persistent basal extension of AJCs in EVL cells under ultrathin
electronic microscopy (12).
Ultrathin electronic microscopy results also indicate that TJs of
the small intestinal epithelia in EpCAM knockout mice are extended,
but freeze-fracture electronic microscopy results suggest that TJs
in the EpCAM mutant mouse intestinal epithelia are scattered and
dispersed (7). It may therefore
be hypothesized that the AJCs in EpCAM mutant EVL cells of
zebrafish are also scattered and dispersed. If the TJs scatter,
their functions will be affected.
The morphology of AJs in EpCAM mutant intestines is
normal, and this may be due to the fact that E-cadherin and
β-catenin are still maintained (7). The mislocalization of TJs indicates
that the AJs' function of supporting TJs may be affected; which may
be because EpCAM and claudin-7 proteins are lost in AJs. The
authors speculate that EpCAM and claudin-7, together with
E-cadherin, are essential to keep the balance between AJs and
cortical tension (Fig. 2A). The
loss of EpCAM and claudin-7 would result in loss of balance between
AJs and cortical tension, affecting the function of AJs leading to
the mislocalization of TJs (Fig.
2B). As mentioned above, in the EpCAM mutant enveloping layer
(EVL) of zebrafish, the expression level of E-cadherin is
decreased, and the EpCAM mutant zebrafish display a persistent
basal extension of AJCs in EVL cells. In summary, these reports
suggest that EpCAM regulates the formation, maintenance and
functions of AJCs.
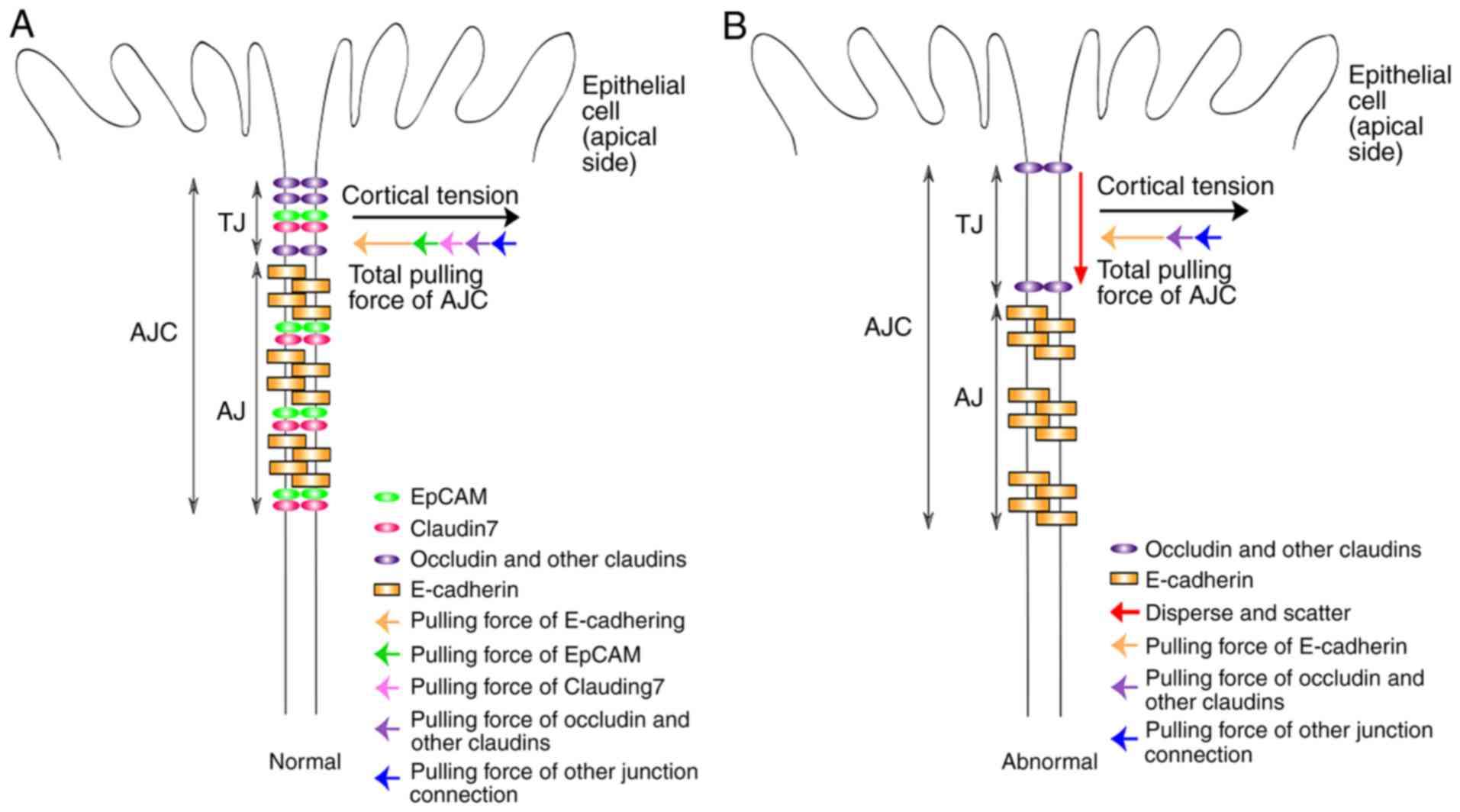 | Figure 2Schematic representation of the
function of EpCAM in cell-cell adhesive structures. (A) In the
normal intestinal epithelium, EpCAM and claudin-7 complex, together
with E-cadherin, is essential to keep the balance between AJs and
cortical tension. The strength of the total pulling force from
E-Cadherin, EpCAM, claudin-7, other claudins, occludin and other
junction connections is equal with the cortical tension between two
epithelial cells; but the direction of the total pulling force is
opposite to that of cortical tension. With this balance, AJs
support TJs to keep the normal structure and functions of TJs. (B)
In EpCAM knockout intestinal epithelium, the EpCAM and claudin-7
complex is completely lost; as is the balance between AJs and
cortical tension. Therefore, the support of AJs to TJs becomes
weak. TJs will therefore disperse and scatter with the EpCAM
mutation and functions of TJs are also affected. EpCAM, epithelial
cell adhesion molecule; AJs, adherens junctions; TJs, tight
junctions. |
Desmosomes
Electron microscopic examination has revealed that
the length and number of desmosomes between the enterocytes of CTE
patients is increased, and some of them exhibit a distorted
structure (9,40). Mueller et al (9) also suggested that the intestines of
EpCAM∆4/∆4 mice have sporadic irregularity, with
crowding and lengthening desmosomes, as determined using an
electron microscope (EM) (9).
Using an immunohistochemical assay, Patey et al (40) found that the expression of
desmoglein is restricted to the upper part of the intercellular
membrane of the epithelial cells lining villi and crypts in the
normal small bowel, whereas the expression of desmoglein is
expanded with staining all over the lateral membrane of the
epithelial cells lining villi and crypts in the intestines of CTE
patients. However, the expression levels of the other components of
desmosomes, plakoglobin and desmoplakin, were normal. EpCAM
knockdown does not lead to marked alteration of the expression and
distribution of desmoglein 2 in T84 cells (41). The association between EpCAM and
desmosomes in the enterocytes remains unclear. The authors
speculate that if the balance between AJs and cortical tension is
broken, the components of desmosomes would move to the basal parts
of the lateral membrane.
Cell-matrix adhesion
The abnormalities in the composition of the basement
membrane of the intestines from CTE patients have been identified,
including the enhanced deposition of heparan sulfate proteoglycan
(HSPG) and type IV collagen, and extremely faint laminin in the
crypt region (59,60). Distribution of α2β1 integrin
adhesion molecules along the crypt-villous axis of CTE patients is
abnormal; in the normal small bowel, the α2 subunit is expressed on
the epithelial cells lining crypts and is absent on the epithelial
cells lining villi, whereas in epithelial dysplasia, the
α2-integrin is expressed on the basolateral membranes of the
epithelial cells lining both the villi and the crypts (40,59,60).
The cell-matrix adhesion situation in EpCAM mutant
animal models remains to be elucidated. As claudin-7 protein levels
are decreased to undetectable levels in EpCAM mutant mice, it has
been hypothesized that claudin-7 knockout mice may be useful for
future investigations. Deletion of claudin-7 alters the normal
distribution pattern of α2-integrin; α2-integrin in WT intestines
is clearly visualized at the basal membrane, whereas in
Claudin7−/− intestines, α2-integrin either forms
clusters or moves toward the apical lateral. Deletion of claudin-7
also significantly increases the expression levels of matric
metallopeptidase (MMP)-3 and MMP-7, which may result in the
degradation of extracellular matrix components (47).
The primary reason for abnormalities of cell-matrix
adhesion in CTE patients or claudin-7 mutant mice remains to be
determined. Basement membrane molecules are involved in the
epithelial mesenchymal cell interactions, which are instrumental in
the development and differentiation of the intestine (61–66). The changes in the cell-matrix
adhesion in EpCAM mutant humans and animals contribute to the
behavioral and morphological alterations of the intestine.
Cell transport
The intestinal TJs of EpCAM mutant mice are
affected. TJs form a barrier that separates the apical from the
basolateral membrane of the epithelium, allowing the selective
passage of ions and solutes (7,58).
Therefore, EpCAM is a very important molecule that regulates
movement of materials across the intestinal epithelium and other
epithelial tissues.
Injection of sulfo-NHS-biotin, a probe that
physically labels cell membrane proteins, into the intestinal lumen
of E18.5 WT and EpCAM mutant embryos revealed that this probe
penetrates the mutant intestinal epithelium easily, and the flux of
lucifer yellow probe from apical to basal as well as from the basal
to apical is increased in the intestine of EpCAM mutant mice at
P3.5 (7). EpCAM∆4/∆4
mice show intestinal permeability defects, with the FITC-dextran
crossing the mutant intestinal barrier into the bloodstream more
quickly compared with WT mice (9). Together, these reports suggest that
the barrier function of the intestine is impaired in EpCAM mutant
mice.
As mentioned above, the results of electron
microscopy showed that TJs mislocalize in EpCAM mutant intestines,
and the combined downregulation of claudins-2, 3, 7, and 15 may be
observed in the intestinal epithelium of EpCAM mutant mice
(7). Claudin-2 and claudin-15 are
responsible for paracellular permeability of Na+
(67). Lei et al (7) found that in EpCAM mutant mice,
NaCl-dilution potential is lowered, and Na+-selective
paracellular permeability is reduced while Cl−-selective
permeability remains normal (7),
like in the claudin-15 mutant mice (67). Combined downregulation of claudins
may result in diarrhea in EpCAM mutant mice and CTE patients.
Knockdown of EpCAM may also lead to significant reduction of
Trans-epithelial Electrical Resistance in T84 and Caco-2 cell lines
(41).
Cell exosomes
Extracellular vesicles (EVs) detach from the cell
membrane or bilayer membrane secreted by cells, and are mainly
composed of apoptotic bodies, ectosomes, extranuclear particles,
cell microbubbles and exosomes (68). Exosomes are released to the
outside of the cell by extracellular secretions from the
intracellular multivesicular bodies after fusion with the cell
membrane, with a diameter of ~40 to 100 nm. EVs carry a variety of
proteins, lipids, DNA, mRNA and miRNA, which are involved in
intercellular communication, cell migration, angiogenesis and
immune regulation (69,70). Increased levels of EVs may act as
diagnostic markers of diabetes, cardiovascular disease, AIDS,
chronic inflammatory diseases and cancers, therefore the accurate
characterization and research of EVs is particularly important
(71).
It has been demonstrated that EpCAM is essential for
the gastrointestinal localization of some EVs secreted from the
intestinal epithelia cells (IECs) (72). Under physiological conditions,
IECs produce EVs with transforming growth factor (TGF)-β1-dependent
immunosuppressive activity (72).
By inducing regulatory T cells and immunosuppressive dendritic
cells, the transfer of EVs into inflammatory bowel disease (IBD)
mice induced by dextran sulfate sodium could improve the symptoms
of IBD. Conversely, reduced production of endogenous EVs may
promote the development of IBD (73). During the development of IBD, IECs
produce EVs with elevated TGF-β1 levels in an extracellular signal
regulated kinase-dependent manner. These EVs tend to localize in
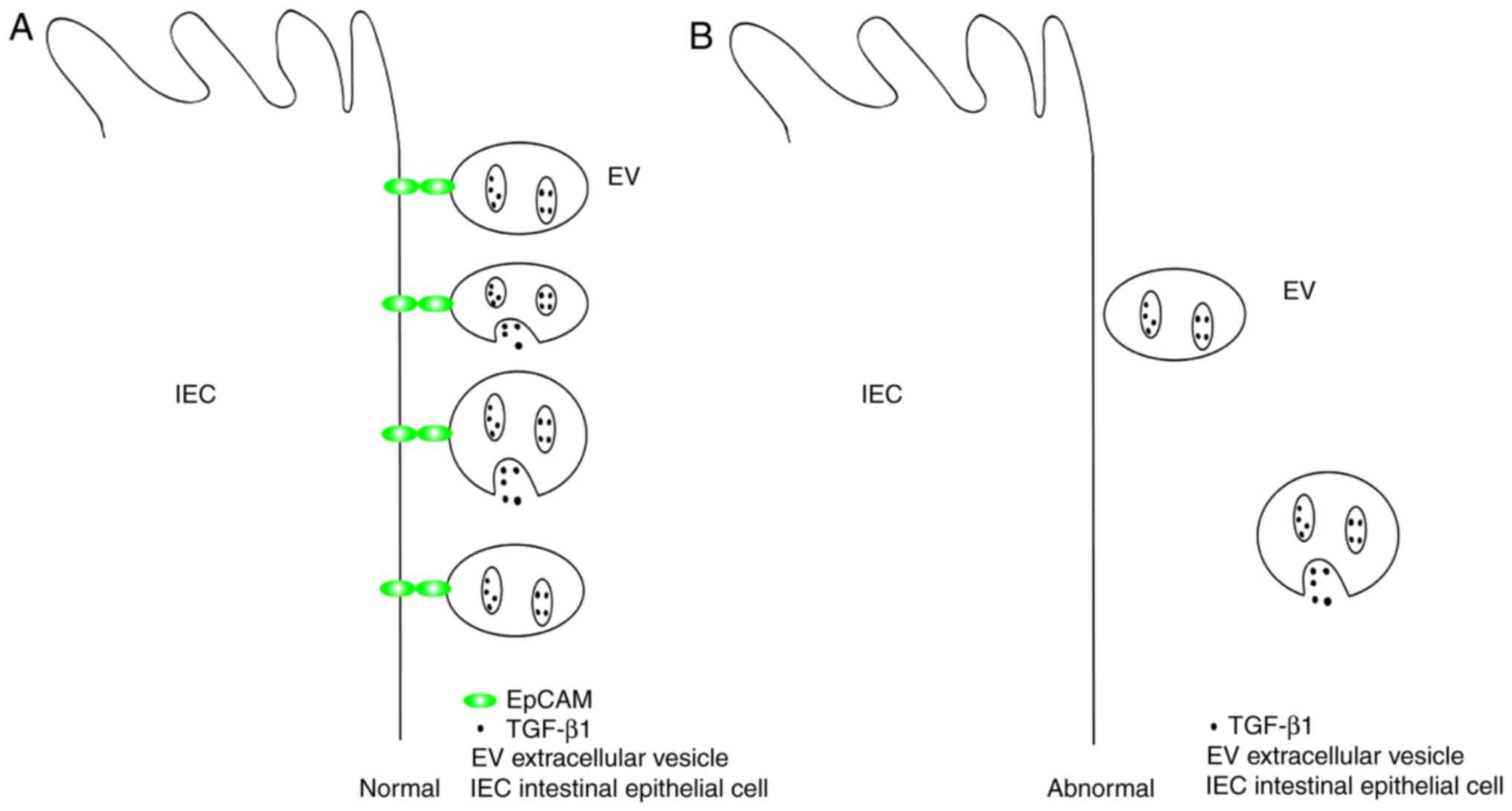
the gut through associating with EpCAMs of IECs (Fig. 3). Knockdown of EpCAM in
vivo increases the severity of IBD in mice, and as the
expression level of EpCAM decreases, the protective effect of EVs
on murine IBD is attenuated (74). Therefore, the function of EpCAM in
the localization of EVs is very important for numerous
physiological or pathological processes.
Cell polarity
The AJCs play crucial roles in the formation of the
polarity and the maintenance of tissue architecture (58). Since EpCAM contributes to the
formation of functional AJCs, it may be important for the polarity
of epithelial cells.
Salomon et al (42) reported that brush border
components of the enterocytes, such as villin and ezrin, partially
disappear, but relocate at the lateral membranes in CTE patients.
They further revealed that apical polarity proteins, such as crumbs
3, cell polarity complex component, protein kinase C and par-3
family cell polarity regulator, relocate laterally at the
tricellular contacts of EpCAM silenced Caco-2 cells, but the basal
polarity is normal. Wu et al (41) found that EpCAM knockdown does not
change the expression or distribution of CD26 (on apical membrane
surfaces), Na/K-ATPase (on basolateral surfaces), or the subapical
localization of myosin IIA, and they concluded that effects of
EpCAM on TJs are selective, and do not cause gross abnormalities in
the cell polarity. Na/K-ATPase proteins are also normal in the
enterocytes of CTE patients (42). It has been suggested that more
cellular polarity markers should be investigated in EpCAM mutant
animal models.
Signaling
From the available data, EpCAM emerged as a crucial
signaling molecule, controlling four independent pathways (Fig. 4). One pathway regulates cell
proliferation via nuclear activities in tumor cells, which is
involved in β-catenin-dependent transcription (25). Here, the authors discuss three
signaling pathways in the developmental processes.
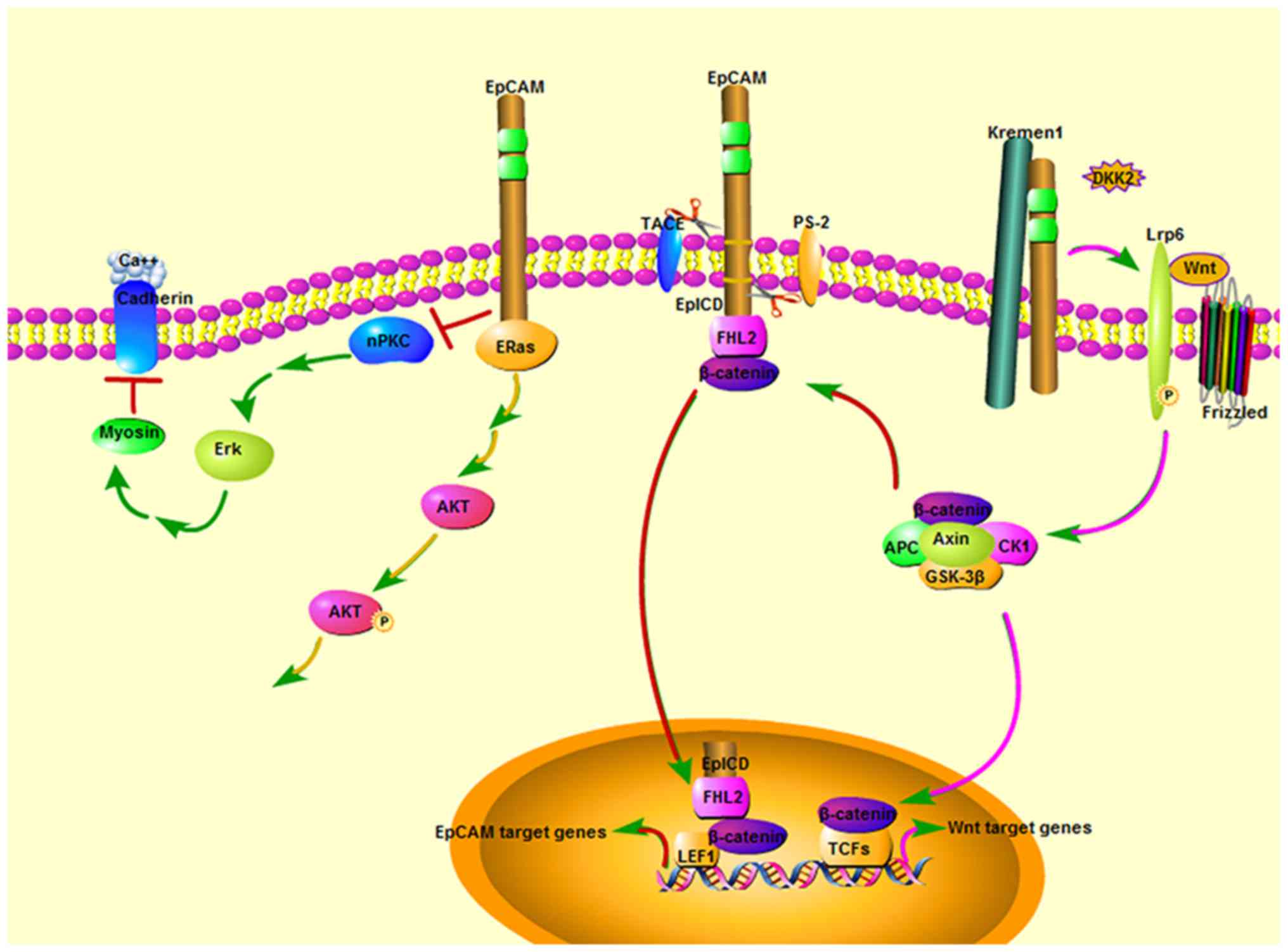 | Figure 4Overview of the role of EpCAM in cell
signaling. From the available data, EpCAM has emerged as a crucial
signaling molecule, controlling four independent pathways. i) The
nPKC-dependent pathway: The EpCAM cytoplasmic tail inhibits the
nPKC activity and ERK pathway to protect cadherin-mediated
adhesion. ii) Wnt signal pathway: EpCAM extracellular domain
directly binds to Kremen1 and disrupts the Kremen1-Dkk2
interaction, which prevents Kremen1-Dkk2-mediated removal of Lrp6
from the cell surface. EpCAM derepresses Lrp6 and cooperates with
Wnt ligand to activate the Wnt signaling through stabilizing
membrane Lrp6 and allowing Lrp6 to cluster into active
signalosomes. iii) ERas/AKT pathway: EpCAM fosters the activating
phosphory-lation of AKT at serine473 by interacting with
hyperactive Ras GTPase ERas, which induces the activation of AKT.
iv) In tumor cells or ES cells, EpICD forms a transcription
activator complex with FHL2, β-Catenin and LEF-1 to increase
transcription of EpCAM target genes, such as c-Myc and Cyclins A/E.
Dkk2, Dickkopf2; Lrp6, Lipoprotein-receptor-related protein 6;
EpCAM, epithelial cell adhesion molecule; PKC, protein kinase C;
ERK, extracellular signal regulated kinase; ERas, embryonic Ras;
AKT, RAC-α serine/threonine-protein kinase; EpICD, intracellular
domain of EpCAM; FHL, four and a half LIM domains; LEF-1, lymphoid
enhancer binding factor 1. |
The nPKC-dependent pathway
Loss- and gain-of function of EpCAM in
Xenopus embryos revealed that EpCAM acts as a potent
inhibitor of novel protein kinase C (nPKC) during development
(13,14). PKC inhibition is caused by a short
segment of the EpCAM cytoplasmic tail. This motif resembles the
pseudosubstrate inhibitory domains of PKCs and bound nPKCs with
high affinity. The effects of loss of EpCAM on the amphibian embryo
tissues have been observed, including sequentially strong
overstimulation of PKC activity and the Erk pathway, exacerbated
myosin contractility, loss of cadherin-mediated adhesion, tissue
dissociation, and, ultimately, cell death (14). Elevated EpCAM levels in either the
ectoderm or the mesoderm result in tissue mixing, and this property
relies on a novel signal function through downregulation of nPKC
activity (13). This activity of
EpCAM does not require its extracellular domain, but its
cytoplasmic motif is essential. A bioinformatics search revealed
the existence of similar motifs in other plasma membrane proteins,
most of which were cell-cell adhesion molecules. Thus, direct
inhibition of PKC by EpCAM represents a general mode of regulation
of signal transduction by cell-surface proteins (14).
Wnt signal pathway
EpCAM has been identified as an endoderm-specific
Wnt activator in zebrafish; EpCAM mutants exhibit defective liver
development similar to prt/wnt2bb mutants (15). It has been revealed that EpCAM
that is directly bound to Kremen1 disrupts the Kremen1-Dickkopf2
(Dkk2) interaction, which prevents Kremen1-Dkk2-mediated removal of
lipoprotein-receptor-related protein 6 (Lrp6) from the cell
surface. EpCAM activates Lrp6 and cooperates with Wnt2bb ligand to
activate the Wnt signaling through stabilizing membrane Lrp6 and
allowing Lrp6 to cluster into active signalosomes. Thus, EpCAM
autonomously promotes and cooperatively activates Wnt2bb signaling
in the endodermal cells of zebrafish embryos (15).
It has also been revealed that in EpCAM mutant
embryos, injection of EpCAM or EpCAM extracellular domain (ECD)
mRNA rescues the diminished expressions levels of fgf3 and dkk1b,
the downstream factors of Wnt signaling in the lateral line
primordia. Therefore, it has been speculated that similar
mechanisms may also be present in the development of other cellular
systems such as lateral line primordia in which Wnt and EpCAM are
active (75,76).
Embryonic Ras (Eras)/RAC-α
serine/threonine-protein kinase (AKT) pathway
It has previously been reported that EpCAM regulates
murine ES cell differentiation via ERas/AKT pathway. ERas has been
identified as a novel EpCAM interactor through
co-immunoprecipitation, and these two proteins have a similar
expression pattern in murine ES cells and embryos. The expression
of EpCAM fosters the activating phosphorylation of AKT at serine473
by interacting with hyperactive Ras GTPase ERas, which induces the
activation of AKT. It has been demonstrated that full-length EpCAM
is essential for this signaling pathway, but the detailed mechanism
is still unclear. EpCAM may orchestrate the differentiation of ES
cells through this signaling pathway (77).
Proliferation
Reports regarding the function of EpCAM on the
regulation of proliferation are contradictory. It has been reported
that enhanced expression of EpCAM is associated with active
proliferation of cancerous or normal tissues (78). EpCAM has also been found to
increase the proliferation of tumor cell lines (79,80). Maetzel et al (25) revealed that knockdown of EpCAM in
FaDu cells results in decreased proliferation, and they concluded
that the proliferative action of EpCAM was attributed to the
ability of EpICD to form a transcription activator complex with
four and a half LIM domains 2, β-Catenin and lymphoid enhancer
binding factor 1 (LEF1) (Fig. 4).
However, this mechanism could not be verified by EpCAM mutant
animal models. Lei et al (7) found that the number of Ki67-positive
cells in the intervillus domains of EpCAM mutant mice at E18.5 was
similar to that in WT animals throughout the intestine. Mueller
et al (9) assessed
enterocyte proliferation using BrdU at a 4 h time point following
intra-peritoneal injection, and found that EpCAM∆4/∆4
mice demonstrate a significantly higher proliferation index
compared with WT mice. Furthermore, knockdown of EpCAM in T84 and
Caco-2 cells increases cell proliferation (41). Overexpression of EpCAM in invasive
colon cancer cell lines may reduce cell proliferation. These
reports indicate that EpCAM may inhibit the proliferation of
progenitor cells of the intestinal epithelium, but the molecular
mechanism remains to be fully elucidated.
Motility
EpCAM is a cell adhesion molecule, however it has
been revealed that it may function to enhance the mobility of cells
and tissues. In 2004, Osta et al (80) reported that EpCAM has a
stimulatory effect on in vitro cell migration. Moreover, it
was found that in both Zebrafish and Xenopus embryos, EpCAM
is required to enable cells to rearrange during epiboly, a
morphogenetic process through which the ectoderm thins and expands
during gastrulation (12,13). Furthermore, in conditional
knockout mice, loss of EpCAM impairs the migration of skin
Langerhans cells (11). The fact
that the migration of cells and tissues may be promoted by a cell
adhesion molecule is quite counterintuitive. EpCAM may act to
increase the expression of cadherins (12–14), which further adds to the puzzle.
Maghzal et al (13)
reported that EpCAM positively regulates both cell movements and
cadherin levels in Xenopus embryos, but they demonstrated
that the regulation of cell motility occurs independently of the
stabilization of cadherins. Motility and normal epiboly may be
rescued in EpCAM-depleted tissues of Xenopus embryos by
expression of an EpCAM construct lacking the whole extracellular
domain (13). EpICD may control
the activity of nPKCs, and thus moderate actomyosin-based
contractility (14).
However, a contrasting result was observed in the
intestinal epithelium of mice. To assess the migration of
enterocytes in EpCAM mutant mice, BrdU positive cells were assessed
at 24 h, and the result revealed that EpCAM∆4/∆4 mice
have significantly higher migration rates of BrdU positive
enterocytes compared with WT mice (9).
4. Functions of EpCAM in development and
stem cells
EpCAM is highly expressed in many embryonic tissues
of vertebrates and in numerous types of progenitor cells and stem
cells. Research has been focused on its functions in developmental
processes and stem cells.
Development
Previously it has been demonstrated that EpCAM is
required to form cells of all germ layers using EpCAM mutant murine
ES cells (77); but EpCAM
knockouts in mice and EpCAM null mutants in Zebrafish give
relatively weak embryonic phenotypes (7–9,12,75). It was speculated that the EpCAM-2
gene which presents in both species functions redundantly (7,14).
It has been confirmed that EpCAM is essential for intestinal
epithelium homeostasis in mice and humans (7–9),
and is also required for the integrity of skin in adult zebrafish
(12).
One EpCAM mutant could cause mouse embryonic
lethality through placental defects, and it was demonstrated that
EpCAM is required for the differentiation or survival of parietal
trophoblast giant cells, normal development of the placental
labyrinth and establishment of a competent maternal-fetal
circulation (10). Lei et
al (7) also demonstrated that
the frequency of homozygous mutant mice is slightly smaller than
expected (22.43% for EpCAM−/− and 13.76% for
EpCAMβgeo/βgeo), suggesting that the mutant alleles are
embryonic lethal in a small proportion of mutant homozygotes.
Maghzal et al (14)
reported that global depletion of EpCAM results in Xenopus
embryonic lethality with strong penetrance (>90%), and EpCAM is
crucially required for tissue integrity. EpCAM therefore has
important functions at different developmental stages of vertebrate
embryos.
EpCAM is highly expressed in human hepatic stem
cells and progenitor cells, however is absent in mature hepatocytes
(78,81,82). Knockout of EpCAM in zebrafish
induces defective early hepatic development; but hepatocytes keep
proliferating in EpCAM mutants, and the liver becomes relatively
normal at 8 days postfertilization (15). Overexpression of EpCAM in murine
ES cells may increase the transcription of hepatocytic markers Afp
and Fn1 (77). Therefore, it is
suggested that liver development in EpCAM knockout mice at early
stages should be investigated.
Stem cells
In 1993, it was demonstrated that EpCAM is highly
expressed in undifferentiated mouse P19 embryonal carcinoma cells,
however is downregulated in differentiated P19 cells (83). It has been confirmed that EpCAM is
implicated in the maintenance of pluripotency in embryonic stem
cells (ESCs) (77,84,85) as well as in somatic stem cells
such as hepatic stem cells (4,81,86,87). The functions of EpCAM in stem
cells have been explored for many years. Regulated intramembrane
proteolysis (RIP) and nuclear translocation of the EpICD of EpCAM
was reported in murine and human ESCs (85,88). It has been demonstrated that in
human and porcine ESCs, EpICD supports pluripotency through
activation of the transcription of reprogramming factors, such as
SRY-Box 2, Oct3/4 and Nanog (85,89,90). The molecular functions of EpCAM in
somatic stem cells remain unknown.
It was revealed that the EpCAM and claudin-7
complex has important functions in human and murine somatic cell
reprogramming processes (89).
EpEX/EpCAM, combined with Oct3/4 or Kruppel like factor 4, is
sufficient to generate human induced pluripotent stem cells (iPSCs)
(91). The EpCAM complex may
activate the transcription of Oct4 and suppress the p53-p21 pathway
to enhance the pluripotency reprogramming (89). EpEX/EpCAM may additionally
activate signal transducer and activator of transcription 3
(STAT3), and the activated STAT3 may lead to the
nuclear-translocation of hypoxia inducible factor 1a to promote
somatic cell reprogramming (91).
5. Functions of EpCAM in diseases
EpCAM and CTE
CTE was firstly reported in 1994 (55), which is a disease characterized by
a classical congenital disorder of the intestinal mucosa, including
subtotal villous atrophy with crypt hyperplasia and focal crowding
of surface enterocytes, resembling tufts. With an incidence
estimated at 1 per 50,000–100,000 in Western Europe, there is a
high rate of mortality in young children with CTE (92). Non-syndromic CTE has been
associated with the mutations of EPCAM, mainly loss-of-function
mutations, in 73% of patients. In 21% of patients with CTE, a
syndromic form is associated with the missense mutations of serine
peptidase inhibitor, kunitz type 2 (SPINT2), an inhibitor of HGF
activator serine protease (also known as HAI-2) (92). It has been demonstrated that
SPINT2 inhibits the activity of cell surface serine protease
matriptase, and mutations of SPINT2 lead to efficient cleavage of
EpCAM by matriptase (41).
Microvillus atrophy has been demonstrated to be present in the
biopsy samples of patients with CTE have mutations in EPCAM or
SPINT2, which may explain, at least in part, the intestinal failure
(93).
CTE causes lethal diarrhea in newborns, and it may
be caused by impaired barrier function of TJs in the intestinal
epithelium of patients. It has been confirmed that molecular
probes, such as sulfo-NHS-biotin, lucifer yellow and FITC-dextran,
cross the EpCAM mutant intestinal epithelium more easily compared
with in control animal models. This leads to a continuous water
leak in the intestines of CTE patients and EpCAM mutant mice.
Claudin-2, 3, 7 and 15 are reduced in the
intestinal epithelium of EpCAM mutant mice (7). Some of these claudins are required
for the nutrient absorption of intestines. Both claudin-2 and
claudin-15 function in the paracellular permeability to
Na+ in the small intestinal epithelium (94–98). This permeability permits
Na+ to access the lumen to support the
Na+-dependent absorption of nutrients, including
glucose, amino acids, vitamin C and others (99–101). The changes in the expression
patterns of claudin-2 and claudin-15 in the small intestine occur
age-dependently. The claudin-2 level is higher than that of
claudin-15 in the infant small intestine, but in adults, the
expression level of claudin-15 is higher than that of claudin-2
(67,102). Knockout of claudin-15 could lead
to Na+ deficiency and glucose malabsorption in the small
intestines of adult mice (67).
Knockout of both claudins 2 and 15 would cause defects in
paracellular Na+ flow and nutrient transport in the
small intestines and finally lead to mortality from malnutrition,
and the mice only survive 25 days after birth (103). From these reports, it may be
suggested that the nutrient absorption in the small intestines of
CTE patients may be affected.
The mechanisms for the tuft-like structure
formations in CTE are not completely clear. The apical polarity of
the enterocytes from CTE is affected, which may result in tuft
formation. The proliferation and mobility of EpCAM mutant
intestinal epithelial cells is also altered, which may be an
additional cause of tuft formation. There is no curative treatment
for CTE yet, therefore elucidation of the mechanisms underlying
tuft formation is very important for the treatment of CTE.
EpCAM and tumors
Originally identified as a novel tumor-specific
cell surface antigen following immunization of mice with cancer
cells in 1970s (1), EpCAM has
been known to be highly expressed in a variety of epithelial
carcinomas (104). EpCAM is also
highly expressed in acute myeloid leukemia (AML), and
EpCAM-positive cells may be leukemia stem cells which could promote
leukemic progression (105). It
has been reported that EpCAM is involved in tumorigenesis,
metastasis, and cancer stem cells (25,106,107).
Previous research has focused on the function of
EpCAM in cancers. Maetzel et al (25) firstly demonstrated that a
proteolytic fragment of EpCAM containing EpICD forms a complex with
β-catenin and LEF-1, which translocates to the nucleus and
activates the transcription of genes associated with cell
proliferation, such as c-Myc and cyclins A and E, thereby promoting
oncogenesis (25) (Fig. 4). This mechanism depends on the
cellular interaction to provide initial signals of regulating
intramembrane proteolysis (108). A complex of EpCAM, claudin-7,
CO-029, and CD44v6 is frequently formed in colorectal cancer, and
this complex, but not the individual molecules, may promote the
progression of colorectal cancer and increase the apoptosis
resistance of cancer cells (43).
EpCAM may promote the chemotherapeutic resistance of myeloid
leukemia via the Wnt5b singling pathway (105). Previously, Wang et al
(107) demonstrated that EpCAM
regulates epithelial-mesenchymal transition (EMT), stemness and
metastasis of nasopharyngeal carcinoma cells through the
phosphatase and tensin homolog (PTEN)/AKT/mechanistic target of
rapamycin kinase (mTOR) pathway. They revealed that overexpression
of EpCAM reduces the expression level of PTEN, and then the
phosphorylation levels of AKT, mTOR, p70S6K and 4EBP1 are increased
with the reduction of PTEN. It may be hypothesized that EpCAM has
similar functions in other types of cancers. The elucidation of
these molecular mechanisms is an important guide in the effective
development of therapies for tumors.
EpCAM has become a therapeutic target in many types
of cancers. The emergence of chemotherapeutic drug resistance in
cancer stem cells is a key factor hindering the effective treatment
of many cancers. A novel system by conjugating cancer stem cells
targeting EpCAM aptamer with a chemotherapeutic drug can eliminate
chemotherapeutic drug resistance. Incubation of the chemotherapy
drug doxorubicin with colorectal cancer cells could lead to the
long-term retention and enrichment of adriamycin in the nucleus,
thus weakening the therapeutic effect (109). Doxorubicin targeted EpCAM
aptamer therapy in tumor-bearing nude mice may significantly
inhibit tumor growth, prolong survival time and lead to a prolonged
dose-dependent tumor latency. In general, conjugation of cancer
stem cells targeting EpCAM aptamer with a chemotherapeutic drug
could transform conventional chemotherapeutic drugs into tumor
killers to overcome the drug resistance in tumors (110). Zheng et al (105) also found that the EpCAM antibody
sensitizes the chemoresistant myeloid leukemia to innate immune
cells. It has been reported that exosomes in the circulatory system
derived from tumors, called extracellular vesicles or vesicles, may
easily be isolated using anti-EpCAM antibodies in combination with
magnetic beads. The exosomes are isolated by various downstream
assays such as sandwich immunization assay and RT-qPCR. The
isolated exosomes could be used to study the distribution of
tumor-specific extracellular body surface proteins and associated
miRNAs (111). Dendritic cells
play a crucial role in the host immune response and antigen
presentation. EpCAM peptide-primed dendritic cell vaccinations
exhibit significant anti-tumor immunity in hepatocellular carcinoma
cells (107).
However, it has been revealed that EpCAM could be a
tumor suppressive protein in certain types of cancers. Hwang et
al (112) suggests that
decreased expression of EpCAM is an early event in oral
carcinogenesis. Gosens et al (113) observed that reduced EpCAM
expression at the invasive margin of rectal carcinomas, and
reduction of EpCAM expression may increase the risk of local
recurrence. A recent report revealed that loss of EpCAM increases
the malignancy of endometrial carcinoma (EC), and high EpCAM
expression favors the survival of patients with EC (114). The molecular mechanisms of the
tumor suppressive function of EpCAM in these cancers are not yet
clear.
EpCAM and inflammatory bowel disease
Inflammatory bowel disease (IBD) refers to chronic
inflammatory disorders in the gastrointestinal tract (115). Crohn's disease and ulcerative
colitis are two clinical forms of IBD (116), and the precise aetiology is
unclear. It has been reported that intestinal barrier function of
patients with IBD is compromised (117,118). The reduction of expression of
several cell adhesion molecules has been found in IBD patients
(119–123). As mentioned above, knockdown of
EpCAM in large intestines of mice increase the severity of murine
IBD induced by dextran sulfate sodium salt (124). However, EpCAM is present at
normal levels and location in the tissue specimens from patients
with Crohn's disease and ulcerative colitis (9). There is no clear evidence that EpCAM
mutation could cause IBD.
The claudin-7 protein is reduced to undetectable
levels in the intestines of EpCAM mutant mice and CTE patients. The
inflammatory and immune responses are clearly observed in
Claudin7−/− intestines, including an increased number of
leukocytes and macrophages, increased mRNA level of interleukin
(IL)-1β, IL-8 receptor β, IL-6, AP-1 and tumor necrosis factor
(TNF)-α, increased protein expression levels of nuclear factor κB
(NF-κB), c-Jun, c-Fos, and cyclooxygenase 2 (COX-2), and elevated
phosphorylations of NF-κB and c-jun (47,125). Claudin-7 mutant mouse models
therefore exhibit IBD-like symptoms.
In fact, an inflammatory infiltration may be
detected at day 4 in the intestines of EpCAM∆4/∆4 mice
(9). However, absent or mild
inflammation may be observed in the intestinal biopsies from CTE
patients (126,127). In JAM-A-deficient mice,
TGF-β-producing CD4+ T cells promote IgA secretion to
protect intestinal inflammation (128), confirming that impaired
intestinal barriers induce adaptive immune compensation. The
intestinal barrier function is affected in EpCAM mutant mice, but
it is normal in claudin-7 knockout mice (47,125). The impaired intestinal barrier
of EpCAM mutant mice or CTE patients may induce adaptive immune
compensation to protect the intestine from inflammation. EpCAM may
also protect intestines from inflammation via binding EVs with
TGF-β1 (124).
CTE patients may be more sensitive to IBD induced
factors, and the EpCAM expression reduction would increase the
severity of human IBD.
EpCAM and cholestatic liver diseases
Cholestatic liver injury is a very big clinical
problem, but underlying specific pathological processes are
unknown. Recently, Song et al (129) reported that overexpression of
long non-coding RNA H19 promotes cholestatic liver fibrosis through
zinc finger E-box binding homeobox 1 (ZEB1)/EpCAM pathway. H19RNA
interacts with the transcriptional suppressor ZEB1 and inhibits its
binding to the EpCAM promoter, thus promoting the expression of
EpCAM. The increased EpCAM may result in development of cholestatic
liver fibrosis. It was additionally revealed that H19 and EpCAM are
both highly expressed in liver specimens from primary sclerosing
cholangitis and primary biliary cholangitis patients. As EpCAM is a
downstream gene of H19, the overexpression of EpCAM may promote
cholestatic liver injury. The exact molecular mechanisms of EpCAM
in promoting cholestatic liver injury are still unclear, but EpCAM
is a marker of biliary hyperplasia (130) and may drive cholangiocyte
proliferation to promote the development of cholestatic liver
fibrosis.
6. Conclusions and perspectives
EpCAM has been recognized to exhibit broad spectrum
functions in multiple physiological and pathological processes. It
is essential for the homeostasis of epithelial tissues through
regulating cell-cell junctions, signaling pathways, cellular
proliferation and mobility. It is a very important molecule that
maintains the pluripotency of ES cells and promotes the iPSCs
process. Mutations or aberrant expression levels of EpCAM are
associated with numerous diseases including many types of
cancers.
EpCAM is a cell adhesion molecule that plays
important roles in the formation and functions of adhesive
structures and polarity. However, EpCAM is highly expressed in
tumor tissues, and the tumor tissues usually lose organized
adhesive structures and cell polarity. These two things seem
contradictory. The clear understanding of different functions of
EpCAM in normal and cancer tissues will play an important role in
exploring the potential therapeutic strategies for cancers.
Tissue-specific mutant and overexpression animal models would be
very useful to explore these molecular mechanisms.
From mutant animal models, it was concluded that
EpCAM has important roles in both the intestinal epithelium and the
placental labyrinth. These two tissues both function on the
absorption of nutrients. Knockout EpCAM in mice impairs the
intestinal barrier function and causes ion transport dysfunction in
the intestinal epithelium (7,9,37).
The nutrient absorption ability of the placental labyrinth may also
be affected, which may be a cause of embryonic lethality. The EpCAM
expression level is higher in the crypts of the small intestine
than in the villi (2,3), and it has been demonstrated that the
EpCAM expression level is also higher in the multi-potent labyrinth
trophoblast progenitor cells than other parts of the placental
labyrinth in mice (131). EpCAM
may have similar functions in regulating the development of these
two tissues.
The different mechanisms underlying the roles of
EpCAM in cancer tissues and the trophectoderm have yet to be
elucidated. EpCAM is highly expressed in both types of tissue.
Trophoblasts can invade and branch out within uterine epithelium
(132), but the invasive ability
of trophoblasts is very limited. EpCAM is associated with the
metastasis of cancer cells (106), and the invasive ability of
cancer cells is uncontrollable. Elucidation of these different
mechanisms would be helpful in exploring the therapeutic ways to
control the metastasis of cancers.
EpCAM is expressed in many types of epithelia in
adult animals and humans, and may have important functions in these
tissues. Therefore, conditional knockout animal models are required
to study these functions. EpCAM is under the control of
transcriptional factor grainyhead like transcription factor 2
(Grhl2), and in the otic epithelium of Grhl2 mutant zebrafish,
EpCAM protein is markedly reduced (133). Grhl2 mutation is associated with
hearing loss in humans, and the hearing and balance system is
severely disrupted in EpCAM mutant zebrafish (133). EpCAM may have functions in otic
epithelium of humans and animals. It has been reported that CTE
patients also suffer from various other condition, including
chronic arthritis (2,134,135). These reports confirmed that
EpCAM has various functions in multiple organs of adults. The
molecular mechanisms of these functions remain unclear. Elucidation
of the molecular functions of EpCAM will be useful for its role as
a therapeutic target in the future.
Acknowledgments
The authors would like to thank Professor Xianglu
Rong, Professor Weijian Bei, and Professor Dewei Ye from Guangdong
Metabolic Disease Research Center of Integrated Chinese and Western
Medicine, Guangdong Pharmaceutical University for their fruitful
discussions.
Funding
This work was supported by grants from the National
Natural Science Foundation of China (grant no. 31671520), the
International Cooperation Project of the Science and Technology
Department of Guangdong province (grant no. 2015A050502050) and the
Construction Project of International Cooperation Base of the
Science and Technology Department of Guangdong province (grant no.
2016B050501003).
Availability of data and materials
Not applicable.
Authors' contributions
ZL and JG conceived the review and analyzed the
relevant literature. LH collected literature and wrote the first
draft of the manuscript. YY collected literature and critically
revised the manuscript. FY and SL collected literature. ZZ created
the figures.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Herlyn M, Steplewski Z, Herlyn D and
Koprowski H: Colorectal carcinoma-specific antigen: Detection by
means of monoclonal antibodies. Proc Natl Acad Sci USA.
76:1438–1442. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schnell U, Cirulli V and Giepmans BN:
EpCAM: Structure and function in health and disease. Biochim
Biophys Acta. 1828:1989–2001. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balzar M, Winter MJ, De Boer CJ and
Litvinov SV: The biology of the 17-1A antigen (Ep-C AM). J Mol Med
(Berl). 77:699–712. 1999. View Article : Google Scholar
|
|
4
|
Schmelzer E, Zhang L, Bruce A, Wauthier E,
Ludlow J, Yao HL, Moss N, Melhem A, McClelland R, Turner W, et al:
Human hepatic stem cells from fetal and postnatal donors. J Exp
Med. 204:1973–1987. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kamimoto K, Kaneko K, Kok CY, Okada H,
Miyajima A and Itoh T: Heterogeneity and stochastic growth
regulation of biliary epithelial cells dictate dynamic epithelial
tissue remodeling. Elife. 5:e150342016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sivagnanam M, Mueller JL, Lee H, Chen Z,
Nelson SF, Turner D, Zlotkin SH, Pencharz PB, Ngan BY, Libiger O,
et al: Identification of EpCAM as the gene for congenital tufting
enteropathy. Gastroenterology. 135:429–437. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lei Z, Maeda T, Tamura A, Nakamura T,
Yamazaki Y, Shiratori H, Yashiro K, Tsukita S and Hamada H: EpCAM
contributes to formation of functional tight junction in the
intestinal epithelium by recruiting claudin proteins. Dev Biol.
371:136–145. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guerra E, Lattanzio R, La Sorda R, Dini F,
Tiboni GM, Piantelli M and Alberti S: mTrop1/Epcam knockout mice
develop congenital tufting enteropathy through dysregulation of
intestinal E-cadherin/β-catenin. PLos One. 7:e493022012. View Article : Google Scholar
|
|
9
|
Mueller JL, McGeough MD, Peña CA and
Sivagnanam M: Functional consequences of EpCam mutation in mice and
men. Am J Physiol Gastrointest Liver Physiol. 306:G278–G288. 2014.
View Article : Google Scholar :
|
|
10
|
Nagao K, Zhu J, Heneghan MB, Hanson JC,
Morasso MI, Tessarollo L, Mackem S and Udey MC: Abnormal placental
development and early embryonic lethality in EpCAM-null mice. PLos
One. 4:e85432009. View Article : Google Scholar :
|
|
11
|
Gaiser MR, Lämmermann T, Feng X, Igyarto
BZ, Kaplan DH, Tessarollo L, Germain RN and Udey MC:
Cancer-associated epithelial cell adhesion molecule (EpCAM; CD326)
enables epidermal Langerhans cell motility and migration in vivo.
Proc Natl Acad Sci USA. 109:E889–E897. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Slanchev K, Carney TJ, Stemmler MP,
Koschorz B, Amsterdam A, Schwarz H and Hammerschmidt M: The
epithelial cell adhesion molecule EpCAM is required for epithelial
morphogenesis and integrity during zebrafish epiboly and skin
development. PLos Genet. 5:e10005632009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maghzal N, Vogt E, Reintsch W, Fraser JS
and Fagotto F: The tumor-associated EpCAM regulates morphogenetic
movements through intracellular signaling. J Cell Biol.
191:645–659. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maghzal N, Kayali HA, Rohani N, Kajava AV
and Fagotto F: EpCAM controls actomyosin contractility and cell
adhesion by direct inhibition of PKC. Dev Cell. 27:263–277. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu H, Ma J, Yang Y, Shi W and Luo L: EpCAM
is an endoderm-specific Wnt derepressor that licenses hepatic
development. Dev Cell. 24:543–553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trzpis M, McLaughlin PM, De Leij LM and
Harmsen MC: Epithelial cell adhesion molecule: More than a
carcinoma marker and adhesion molecule. Am J Pathol. 171:386–395.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cirulli V, Crisa L, Beattie GM, Mally MI,
Lopez AD, Fannon A, Ptasznik A, Inverardi L, Ricordi C, Deerinck T,
et al: KSA antigen Ep-CAM mediates cell-cell adhesion of pancreatic
epithelial cells: Morphoregulatory roles in pancreatic islet
development. J Cell Biol. 140:1519–1534. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lipinski M, Parks DR, Rouse RV and
Herzenberg LA: Human trophoblast cell-surface antigens defined by
monoclonal antibodies. Proc Natl Acad Sci USA. 78:5147–5150. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sherwood RI, Jitianu C, Cleaver O,
Shaywitz DA, Lamenzo JO, Chen AE, Golub TR and Melton DA:
Prospective isolation and global gene expression analysis of
definitive and visceral endoderm. Dev Biol. 304:541–555. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Poon CE, Madawala RJ, Day ML and Murphy
CR: EpCAM is decreased but is still present in uterine epithelial
cells during early pregnancy in the rat: Potential mechanism for
maintenance of mucosal integrity during implantation. Cell Tissue
Res. 359:655–664. 2015. View Article : Google Scholar
|
|
21
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamashita T, Ji J, Budhu A, Forgues M,
Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, et al:
EpCAM-positive hepatocellular carcinoma cells are tumor-initiating
cells with stem/progenitor cell features. Gastroenterology.
136:1012–1024. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baeuerle PA and Gires O: EpCAM (CD326)
finding its role in cancer. Br J Cancer. 96:417–423. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ladwein M, Pape UF, Schmidt DS, Schnölzer
M, Fiedler S, Langbein L, Franke WW, Moldenhauer G and Zöller M:
The cell-cell adhesion molecule EpCAM interacts directly with the
tight junction protein claudin-7. Exp Cell Res. 309:345–357. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maetzel D, Denzel S, Mack B, Canis M, Went
P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M and Gires O:
Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell
Biol. 11:162–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dollé L, Theise ND, Schmelzer E, Boulter
L, Gires O and van Grunsven LA: EpCAM and the biology of hepatic
stem/progenitor cells. Am J Physiol Gastrointest Liver Physiol.
308:G233–G250. 2015. View Article : Google Scholar :
|
|
27
|
Litvinov SV, Velders MP, Bakker HA,
Fleuren GJ and Warnaar SO: Ep-CAM: A human epithelial antigen is a
homophilic cell-cell adhesion molecule. J Cell Biol. 125:437–446.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang C, Liu LW, Sun WJ, Qin SH, Qin LZ
and Wang X: Expressions of E-cadherin, p120ctn, β-catenin and NF-κB
in ulcerative colitis. J Huazhong Univ Sci Technolog Med Sci.
35:368–373. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Frixen UH, Behrens J, Sachs M, Eberle G,
Voss B, Warda A, Löchner D and Birchmeier W: E-cadherin-mediated
cell-cell adhesion prevents invasiveness of human carcinoma cells.
J Cell Biol. 113:173–185. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berx G, Nollet F and van Roy F:
Dysregulation of the E-cadherin/catenin complex by irreversible
mutations in human carcinomas. Cell Adhes Commun. 6:171–184. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Handschuh G, Candidus S, Luber B, Reich U,
Schott C, Oswald S, Becke H, Hutzler P, Birchmeier W, Höfler H and
Becker KF: Tumour-associated E-cadherin mutations alter cellular
morphology, decrease cellular adhesion and increase cellular
motility. Oncogene. 18:4301–4312. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guilford P, Hopkins J, Harraway J, McLeod
M, McLeod N, Harawira P, Taite H, Scoular R, Miller A and Reeve AE:
E-cadherin germline mutations in familial gastric cancer. Nature.
392:402–405. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gayther SA, Gorringe KL, Ramus SJ,
Huntsman D, Roviello F, Grehan N, Machado JC, Pinto E, Seruca R,
Halling K, et al: Identification of germ-line E-cadherin mutations
in gastric cancer families of European origin. Cancer Res.
58:4086–4089. 1998.PubMed/NCBI
|
|
34
|
Corso G, Marrelli D and Roviello F:
Familial gastric cancer and germline mutations of E-cadherin. Ann
Ital Chir. 83:177–182. 2012.PubMed/NCBI
|
|
35
|
Litvinov SV, Balzar M, Winter MJ, Bakker
HA, Briaire-De Bruijn IH, Prins F, Fleuren GJ and Warnaar SO:
Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell
interactions mediated by classic cadherins. J Cell Biol.
139:1337–1348. 1997. View Article : Google Scholar
|
|
36
|
Winter MJ, Nagelkerken B, Mertens AE,
Rees-Bakker HA, Briaire-De Bruijn IH and Litvinov SV: Expression of
Ep-CAM shifts the state of cadherin-mediated adhesions from strong
to weak. Exp Cell Res. 285:50–58. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kozan PA, McGeough MD, Peña CA, Mueller
JL, Barrett KE, Marchelletta RR and Sivagnanam M: Mutation of EpCAM
leads to intestinal barrier and ion transport dysfunction. J Mol
Med (Berl). 93:535–545. 2015. View Article : Google Scholar
|
|
38
|
Bondow BJ, Faber ML, Wojta KJ, Walker EM
and Battle MA: E-cadherin is required for intestinal morphogenesis
in the mouse. Dev Biol. 371:1–12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tunggal JA, Helfrich I, Schmitz A, Schwarz
H, Günzel D, Fromm M, Kemler R, Krieg T and Niessen CM: E-cadherin
is essential for in vivo epidermal barrier function by regulating
tight junctions. EMBO J. 24:1146–1156. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Patey N, Scoazec JY, Cuenod-Jabri B,
Canioni D, Kedinger M, Goulet O and Brousse N: Distribution of cell
adhesion molecules in infants with intestinal epithelial dysplasia
(tufting enteropathy). Gastroenterology. 113:833–843. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu CJ, Mannan P, Lu M and Udey MC:
Epithelial cell adhesion molecule (EpCAM) regulates claudin
dynamics and tight junctions. J Biol Chem. 288:12253–12268. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Salomon J, Gaston C, Magescas J,
Duvauchelle B, Canioni D, Sengmanivong L, Mayeux A, Michaux G,
Campeotto F, Lemale J, et al: Contractile forces at tricellular
contacts modulate epithelial organization and monolayer integrity.
Nat Commun. 8:139982017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kuhn S, Koch M, Nübel T, Ladwein M,
Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L,
Franke WW, et al: A complex of EpCAM, claudin-7, CD44 variant
isoforms, and tetraspanins promotes colorectal cancer progression.
Mol Cancer Res. 5:553–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nübel T, Preobraschenski J, Tuncay H,
Weiss T, Kuhn S, Ladwein M, Langbein L and Zöller M: Claudin-7
regulates EpCAM-mediated functions in tumor progression. Mol Cancer
Res. 7:285–299. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fujita H, Chiba H, Yokozaki H, Sakai N,
Sugimoto K, Wada T, Kojima T, Yamashita T and Sawada N:
Differential expression and subcellular localization of claudin-7,
-8, -12, -13, and -15 along the mouse intestine. J Histochem
Cytochem. 54:933–944. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hewitt KJ, Agarwal R and Morin PJ: The
claudin gene family: Expression in normal and neoplastic tissues.
BMC Cancer. 6:1862006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ding L, Lu Z, Foreman O, Tatum R, Lu Q,
Renegar R, Cao J and Chen YH: Inflammation and disruption of the
mucosal architecture in claudin-7-deficient mice. Gastroenterology.
142:305–315. 2012. View Article : Google Scholar :
|
|
48
|
Nakatsukasa M, Kawasaki S, Yamasaki K,
Fukuoka H, Matsuda A, Tsujikawa M, Tanioka H, Nagata-Takaoka M,
Hamuro J and Kinoshita S: Tumor-associated calcium signal
transducer 2 is required for the proper subcellular localization of
claudin 1 and 7: Implications in the pathogenesis of gelatinous
drop-like corneal dystrophy. Am J Pathol. 177:1344–1355. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Daugherty BL, Ward C, Smith T,
Ritzenthaler JD and Koval M: Regulation of heterotypic claudin
compatibility. J Biol Chem. 282:30005–30013. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Furuse M, Sasaki H and Tsukita S: Manner
of interaction of heterogeneous claudin species within and between
tight junction strands. J Cell Biol. 147:891–903. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Piontek J, Winkler L, Wolburg H, Müller
SL, Zuleger N, Piehl C, Wiesner B, Krause G and Blasig IE:
Formation of tight junction: Determinants of homophilic interaction
between classic claudins. FASEB J. 22:146–158. 2008. View Article : Google Scholar
|
|
52
|
Tatum R, Zhang Y, Salleng K, Lu Z, Lin JJ,
Lu Q, Jeansonne BG, Ding L and Chen YH: Renal salt wasting and
chronic dehydration in claudin-7-deficient mice. Am J Physiol Renal
Physiol. 298:F24–F34. 2010. View Article : Google Scholar :
|
|
53
|
Gladden AB, Hebert AM, Schneeberger EE and
McClatchey AI: The NF2 tumor suppressor, Merlin, regulates
epidermal development through the establishment of a junctional
polarity complex. Dev Cell. 19:727–739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tinkle CL, Pasolli HA, Stokes N and Fuchs
E: New insights into cadherin function in epidermal sheet formation
and maintenance of tissue integrity. Proc Natl Acad Sci USA.
105:15405–15410. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vasioukhin V, Bauer C, Degenstein L, Wise
B and Fuchs E: Hyperproliferation and defects in epithelial
polarity upon conditional ablation of alpha-catenin in skin. Cell.
104:605–617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shin K, Fogg VC and Margolis B: Tight
junctions and cell polarity. Annu Rev Cell Dev Biol. 22:207–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tepass U: Claudin complexities at the
apical junctional complex. Nat Cell Biol. 5:595–597. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Van Campenhout CA, Eitelhuber A, Gloeckner
CJ, Giallonardo P, Gegg M, Oller H, Grant SG, Krappmann D, Ueffing
M and Lickert H: Dlg3 trafficking and apical tight junction
formation is regulated by nedd4 and nedd4-2 e3 ubiquitin ligases.
Dev Cell. 21:479–491. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Goulet O, Kedinger M, Brousse N, Cuenod B,
Colomb V, Patey N, De Potter S, Mougenot JF, Canioni D,
Cerf-Bensussan N, et al: Intractable diarrhea of infancy with
epithelial and basement membrane abnormalities. J Pediatr.
127:212–219. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Goulet O, Salomon J, Ruemmele F, De Serres
NP and Brousse N: Intestinal epithelial dysplasia (tufting
enteropathy). Orphanet J Rare Dis. 2:202007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Beaulieu JF: Differential expression of
the VLA family of integrins along the crypt-villus axis in the
human small intestine. J Cell Sci. 102:427–436. 1992.PubMed/NCBI
|
|
62
|
Simon-Assmann P, Bouziges F, Vigny M and
Kedinger M: Origin and deposition of basement membrane heparan
sulfate proteoglycan in the developing intestine. J Cell Biol.
109:1837–1848. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Simon-Assmann P, Duclos B, Orian-Rousseau
V, Arnold C, Mathelin C, Engvall E and Kedinger M: Differential
expression of laminin isoforms and alpha 6-beta 4 integrin subunits
in the developing human and mouse intestine. Dev Dyn. 201:71–85.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Simon-Assmann P and Kedinger M:
Heterotypic cellular cooperation in gut morphogenesis and
differentiation. Semin Cell Biol. 4:221–230. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Simo P, Simon-Assmann P, Bouziges F,
Leberquier C, Kedinger M, Ekblom P and Sorokin L: Changes in the
expression of laminin during intestinal development. Development.
112:477–487. 1991.PubMed/NCBI
|
|
66
|
Simo P, Bouziges F, Lissitzky JC, Sorokin
L, Kedinger M and Simon-Assmann P: Dual and asynchronous deposition
of laminin chains at the epithelial-mesenchymal interface in the
gut. Gastroenterology. 102:1835–1845. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tamura A, Hayashi H, Imasato M, Yamazaki
Y, Hagiwara A, Wada M, Noda T, Watanabe M, Suzuki Y and Tsukita S:
Loss of claudin-15, but not claudin-2, causes Na+
deficiency and glucose malabsorption in mouse small intestine.
Gastroenterology. 140:913–923. 2011. View Article : Google Scholar
|
|
68
|
Yáñez-Mó M, Siljander PR, Andreu Z, Zavec
AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J,
et al: Biological properties of extracellular vesicles and their
physiological functions. J Extracell Vesicles. 4:270662015.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Colombo M, Raposo G and Théry C:
Biogenesis, secretion, and intercellular interactions of exosomes
and other extracellular vesicles. Annu Rev Cell Dev Biol.
30:255–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Iwai S, Kurosu S, Sasaki H, Kato K and
Maekawa T: Trapping and proliferation of target cells on C60
fullerene nano fibres. Heliyon. 3:e003862017. View Article : Google Scholar :
|
|
72
|
Jiang L, Shen Y, Guo D, Yang D, Liu J, Fei
X, Yang Y, Zhang B, Lin Z, Yang F, et al: Corrigendum:
EpCAM-dependent extracellular vesicles from intestinal epithelial
cells maintain intestinal tract immune balance. Nat Commun.
8:160062017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cai Z, Zhang W, Yang F, Yu L, Yu Z, Pan J,
Wang L, Cao X and Wang J: Immunosuppressive exosomes from TGF-β1
gene-modified dendritic cells attenuate Th17-mediated inflammatory
autoimmune disease by inducing regulatory T cells. Cell Res.
22:607–610. 2012. View Article : Google Scholar
|
|
74
|
Yu L, Yang F, Jiang L, Chen Y, Wang K, Xu
F, Wei Y, Cao X, Wang J and Cai Z: Exosomes with
membrane-associated TGF-β1 from gene-modified dendritic cells
inhibit murine EAE independently of MHC restriction. Eur J Immunol.
43:2461–2472. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Villablanca EJ, Renucci A, Sapède D, Lec
V, Soubiran F, Sandoval PC, Dambly-Chaudière C, Ghysen A and
Allende ML: Control of cell migration in the zebrafish lateral
line: Implication of the gene 'tumour-associated calcium signal
transducer,' tacstd. Dev Dyn. 235:1578–1588. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Aman A and Piotrowski T: Wnt/beta-catenin
and Fgf signaling control collective cell migration by restricting
chemokine receptor expression. Dev Cell. 15:749–761. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Sarrach S, Huang Y, Niedermeyer S,
Hachmeister M, Fischer L, Gille S, Pan M, Mack B, Kranz G, Libl D,
et al: Spatiotemporal patterning of EpCAM is important for murine
embryonic endo- and mesodermal differentiation. Sci Rep.
8:18012018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
De Boer CJ, Van Krieken JH, Janssen-Van
Rhijn CM and Litvinov SV: Expression of Ep-CAM in normal,
regenerating, metaplastic, and neoplastic liver. J Pathol.
188:201–206. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Münz M, Kieu C, Mack B, Schmitt B, Zeidler
R and Gires O: The carcinoma-associated antigen EpCAM upregulates
c-myc and induces cell proliferation. Oncogene. 23:5748–5758. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Osta WA, Chen Y, Mikhitarian K, Mitas M,
Salem M, Hannun YA, Cole DJ and Gillanders WE: EpCAM is over
expressed in breast cancer and is a potential target for breast
cancer gene therapy. Cancer Res. 64:5818–5824. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Schmelzer E, Wauthier E and Reid LM: The
phenotypes of pluripotent human hepatic progenitors. Stem Cells.
24:1852–1858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang L, Theise N, Chua M and Reid LM: The
stem cell niche of human livers: Symmetry between development and
regeneration. Hepatology. 48:1598–1607. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Shimazaki T, Okazawa H, Fujii H, Ikeda M,
Tamai K, McKay RD, Muramatsu M and Hamada H: Hybrid cell extinction
and re-expression of Oct-3 function correlates with differentiation
potential. EMBO J. 12:4489–4498. 1993.PubMed/NCBI
|
|
84
|
González B, Denzel S, Mack B, Conrad M and
Gires O: EpCAM is involved in maintenance of the murine embryonic
stem cell phenotype. Stem Cells. 27:1782–1791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Lu TY, Lu RM, Liao MY, Yu J, Chung CH, Kao
CF and Wu HC: Epithelial cell adhesion molecule regulation is
associated with the maintenance of the undifferentiated phenotype
of human embryonic stem cells. J Biol Chem. 285:8719–8732. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Okabe M, Tsukahara Y, Tanaka M, Suzuki K,
Saito S, Kamiya Y, Tsujimura T, Nakamura K and Miyajima A:
Potential hepatic stem cells reside in EpCAM+ cells of
normal and injured mouse liver. Development. 136:1951–1960. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Tanaka M, Okabe M, Suzuki K, Kamiya Y,
Tsukahara Y, Saito S and Miyajima A: Mouse hepatoblasts at distinct
developmental stages are characterized by expression of EpCAM and
DLK1: Drastic change of EpCAM expression during liver development.
Mech Dev. 126:665–676. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hachmeister M, Bobowski KD, Hogl S,
Dislich B, Fukumori A, Eggert C, Mack B, Kremling H, Sarrach S,
Coscia F, et al: Regulated intramembrane proteolysis and
degradation of murine epithelial cell adhesion molecule mEpCAM.
PLos One. 8:e718362013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Huang HP, Chen PH, Yu CY, Chuang CY, Stone
L, Hsiao WC, Li CL, Tsai SC, Chen KY, Chen HF, et al: Epithelial
cell adhesion molecule (EpCAM) complex proteins promote
transcription factor-mediated pluripotency reprogramming. J Biol
Chem. 286:33520–33532. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yu T, Ma Y and Wang H: EpCAM intracellular
domain promotes porcine cell reprogramming by upregulation of
pluripotent gene expression via beta-catenin signaling. Sci Rep.
7:463152017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Kuan II, Liang KH, Wang YP, Kuo TW, Meir
YJ, Wu SC, Yang SC, Lu J and Wu HC: EpEX/EpCAM and Oct4 or Klf4
alone are sufficient to generate induced pluripotent stem cells
through STAT3 and HIF2α. Sci Rep. 7:418522017. View Article : Google Scholar
|
|
92
|
Salomon J, Goulet O, Canioni D, Brousse N,
Lemale J, Tounian P, Coulomb A, Marinier E, Hugot JP, Ruemmele F,
et al: Genetic characterization of congenital tufting enteropathy:
Epcam associated phenotype and involvement of SPINT2 in the
syndromic form. Hum Genet. 133:299–310. 2014. View Article : Google Scholar
|
|
93
|
Slae MA, Saginur M, Persad R, Yap J,
Lacson A, Salomon J, Canioni D and Huynh HQ: Syndromic congenital
diarrhea because of the SPINT2 mutation showing enterocyte tufting
and unique electron microscopy findings. Clin Dysmorphol.
22:118–120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Furuse M, Furuse K, Sasaki H and Tsukita
S: Conversion of zonulae occludentes from tight to leaky strand
type by introducing claudin-2 into Madin-Darby canine kidney I
cells. J Cell Biol. 153:263–272. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Van Itallie CM, Fanning AS and Anderson
JM: Reversal of charge selectivity in cation or anion-selective
epithelial lines by expression of different claudins. Am J Physiol
Renal Physiol. 285:F1078–F1084. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hou J, Gomes AS, Paul DL and Goodenough
DA: Study of claudin function by RNA interference. J Biol Chem.
281:36117–36123. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Amasheh S, Meiri N, Gitter AH, Schöneberg
T, Mankertz J, Schulzke JD and Fromm M: Claudin-2 expression
induces cation-selective channels in tight junctions of epithelial
cells. J Cell Sci. 115:4969–4976. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yu AS, Cheng MH, Angelow S, Günzel D,
Kanzawa SA, Schneeberger EE, Fromm M and Coalson RD: Molecular
basis for cation selectivity in claudin-2-based paracellular pores:
Identification of an electrostatic interaction site. J Gen Physiol.
133:111–127. 2009. View Article : Google Scholar :
|
|
99
|
Schultz SG and Curran PF: Coupled
transport of sodium and organic solutes. Physiol Rev. 50:637–718.
1970. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Kapus A and Szászi K: Coupling between
apical and paracellular transport processes. Biochem Cell Biol.
84:870–880. 2006. View Article : Google Scholar
|
|
101
|
Tsukaguchi H, Tokui T, Mackenzie B, Berger
UV, Chen XZ, Wang Y, Brubaker RF and Hediger MA: A family of
mammalian Na+-dependent L-ascorbic acid transporters.
Nature. 399:70–75. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
102
|
Holmes JL, Van Itallie CM, Rasmussen JE
and Anderson JM: Claudin profiling in the mouse during postnatal
intestinal development and along the gastrointestinal tract reveals
complex expression patterns. Gene Expr Patterns. 6:581–588. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wada M, Tamura A, Takahashi N and Tsukita
S: Loss of claudins 2 and 15 from mice causes defects in
paracellular Na+ flow and nutrient transport in gut and
leads to death from malnutrition. Gastroenterology. 144:369–380.
2013. View Article : Google Scholar
|
|
104
|
Zhou N, Wang H, Liu H, Xue H, Lin F, Meng
X, Liang A, Zhao Z, Liu Y and Qian H: MTA1-upregulated EpCAM is
associated with metastatic behaviors and poor prognosis in lung
cancer. J Exp Clin Cancer Res. 34:1572015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zheng X, Fan X, Fu B, Zheng M, Zhang A,
Zhong K, Yan J, Sun R, Tian Z and Wei H: EpCAM inhibition
sensitizes chemo-resistant leukemia to immune surveillance. Cancer
Res. 77:482–493. 2017. View Article : Google Scholar
|
|
106
|
Patriarca C, Macchi RM, Marschner AK and
Mellstedt H: Epithelial cell adhesion molecule expression (CD326)
in cancer: A short review. Cancer Treat Rev. 38:68–75. 2012.
View Article : Google Scholar
|
|
107
|
Wang MH, Sun R, Zhou XM, Zhang MY, Lu JB,
Yang Y, Zeng LS, Yang XZ, Shi L, Xiao RW, et al: Epithelial cell
adhesion molecule overexpression regulates epithelial-mesenchymal
transition, stemness and metastasis of nasopharyngeal carcinoma
cells via the PTEN/AKT/mTOR pathway. Cell Death Dis. 9:22018.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Denzel S, Maetzel D, Mack B, Eggert C,
Bärr G and Gires O: Initial activation of EpCAM cleavage via
cell-to-cell contact. BMC Cancer. 9:4022009. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Xiang D, Shigdar S, Bean AG, Bruce M, Yang
W, Mathesh M, Wang T, Yin W, Tran PH, Al Shamaileh H, et al:
Transforming doxorubicin into a cancer stem cell killer via EpCAM
aptamer-mediated delivery. Theranostics. 7:4071–4086. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Hoe SLL, Tan LP, Abdul Aziz N, Liew K,
Teow SY, Abdul Razak FR, Chin YM, Mohamed Shahrehan NA, Chu TL,
Mohd Kornain NK, et al: CD24, CD44 and EpCAM enrich for
tumour-initiating cells in a newly established patient-derived
xenograft of nasopharyngeal carcinoma. Sci Rep. 7:123722017.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Choi YJ, Park SJ, Park YS, Park HS, Yang
KM and Heo K: EpCAM peptide-primed dendritic cell vaccination
confers significant anti-tumor immunity in hepatocellular carcinoma
cells. PLos One. 13:e01906382018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Hwang EY, Yu CH, Cheng SJ, Chang JY, Chen
HM and Chiang CP: Decreased expression of Ep-CAM protein is
significantly associated with the progression and prognosis of oral
squamous cell carcinomas in Taiwan. J Oral Pathol Med. 38:87–93.
2009. View Article : Google Scholar
|
|
113
|
Gosens MJ, Van Kempen LC, Van de Velde CJ,
Van Krieken JH and Nagtegaal ID: Loss of membranous Ep-CAM in
budding colorectal carcinoma cells. Mod Pathol. 20:221–232. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wen KC, Sung PL, Chou YT, Pan CM, Wang PH,
Lee OK and Wu CW: The role of EpCAM in tumor progression and the
clinical prognosis of endometrial carcinoma. Gynecol Oncol.
148:383–392. 2018. View Article : Google Scholar
|
|
115
|
Maloy KJ and Powrie F: Intestinal
homeostasis and its breakdown in inflammatory bowel disease.
Nature. 474:298–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Kaser A, Zeissig S and Blumberg RS:
Inflammatory bowel disease. Annu Rev Immunol. 28:573–621. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Mankertz J and Schulzke JD: Altered
permeability in inflammatory bowel disease: Pathophysiology and
clinical implications. Curr Opin Gastroenterol. 23:379–383. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Welcker K, Martin A, Kölle P, Siebeck M
and Gross M: Increased intestinal permeability in patients with
inflammatory bowel disease. Eur J Med Res. 9:456–460.
2004.PubMed/NCBI
|
|
119
|
Doğan A, Wang ZD and Spencer J: E-cadherin
expression in intestinal epithelium. J Clin Pathol. 48:143–146.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Gassler N, Rohr C, Schneider A, Kartenbeck
J, Bach A, Obermüller N, Otto HF and Autschbach F: Inflammatory
bowel disease is associated with changes of enterocytic junctions.
Am J Physiol Gastrointest Liver Physiol. 281:G216–G228. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Jankowski JA, Bedford FK, Boulton RA,
Cruickshank N, Hall C, Elder J, Allan R, Forbes A, Kim YS, Wright
NA and Sanders DS: Alterations in classical cadherins associated
with progression in ulcerative and Crohn's colitis. Lab Invest.
78:1155–1167. 1998.PubMed/NCBI
|
|
122
|
Karayiannakis AJ, Syrigos KN, Efstathiou
J, Valizadeh A, Noda M, Playford RJ, Kmiot W and Pignatelli M:
Expression of catenins and E-cadherin during epithelial restitution
in inflammatory bowel disease. J Pathol. 185:413–418. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Kucharzik T, Walsh SV, Chen J, Parkos CA
and Nusrat A: Neutrophil transmigration in inflammatory bowel
disease is associated with differential expression of epithelial
intercellular junction proteins. Am J Pathol. 159:2001–2009. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Jiang L, Shen Y, Guo D, Yang D, Liu J, Fei
X, Yang Y, Zhang B, Lin Z, Yang F, et al: EpCAM-dependent
extracellular vesicles from intestinal epithelial cells maintain
intestinal tract immune balance. Nat Commun. 7:130452016.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Tanaka H, Takechi M, Kiyonari H, Shioi G,
Tamura A and Tsukita S: Intestinal deletion of Claudin-7 enhances
paracellular organic solute flux and initiates colonic inflammation
in mice. Gut. 64:1529–1538. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sherman PM, Mitchell DJ and Cutz E:
Neonatal enteropathies: Defining the causes of protracted diarrhea
of infancy. J Pediatr Gastroenterol Nutr. 38:16–26. 2004.
View Article : Google Scholar
|
|
127
|
Ranganathan S, Schmitt LA and Sindhi R:
Tufting enteropathy revisited: The utility of MOC31 (EpCAM)
immunohisto-chemistry in diagnosis. Am J Surg Pathol. 38:265–272.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Khounlotham M, Kim W, Peatman E, Nava P,
Medina-Contreras O, Addis C, Koch S, Fournier B, Nusrat A, Denning
TL and Parkos CA: Compromised intestinal epithelial barrier induces
adaptive immune compensation that protects from colitis. Immunity.
37:563–573. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Song Y, Liu C, Liu X, Trottier J, Beaudoin
M, Zhang L, Pope C, Peng G, Barbier O, Zhong X, et al: H19 promotes
cholestatic liver fibrosis by preventing ZEB1-mediated inhibition
of epithelial cell adhesion molecule. Hepatology. 66:1183–1196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zen Y, Vara R, Portmann B and Hadzic N:
Childhood hepatocellular carcinoma: A clinicopathological study of
12 cases with special reference to EpCAM. Histopathology.
64:671–682. 2014. View Article : Google Scholar
|
|
131
|
Ueno M, Lee LK, Chhabra A, Kim YJ,
Sasidharan R, Van Handel B, Wang Y, Kamata M, Kamran P, Sereti KI,
et al: c-Met-dependent multipotent labyrinth trophoblast
progenitors establish placental exchange interface. Dev Cell.
27:373–386. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Nakaya Y and Sheng G: EMT in developmental
morphogenesis. Cancer Lett. 341:9–15. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Han Y, Mu Y, Li X, Xu P, Tong J, Liu Z, Ma
T, Zeng G, Yang S, Du J and Meng A: Grhl2 deficiency impairs otic
development and hearing ability in a zebrafish model of the
progressive dominant hearing loss DFNA28. Hum Mol Genet.
20:3213–3226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Al-Mayouf SM, Alswaied N, Alkuraya FS,
Almehaidib A and Faqih M: Tufting enteropathy and chronic
arthritis: A newly recognized association with a novel EpCAM gene
mutation. J Pediatr Gastroenterol Nutr. 49:642–644. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Bird LM, Sivagnanam M, Taylor S and
Newbury RO: A new syndrome of tufting enteropathy and choanal
atresia, with ophthalmologic, hematologic and hair abnormalities.
Clin Dysmorphol. 16:211–221. 2007. View Article : Google Scholar : PubMed/NCBI
|