Introduction
Repetitive transcranial magnetic stimulation (rTMS),
as a non-invasive stimulation technique delivering a repetitive
pulsed magnetic field, has been widely applied in treating various
neurological diseases, including depression (1), pain (2), epilepsy (3), headache (4), insomnia (5) and Alzheimer's disease (6). Although the relevant mechanisms
remain to be elucidated, rTMS treatment can induce neural
plasticity effects, as evidenced by functional magnetic resonance
imaging (fMRI) (7) and positron
emission tomography (PET) analyses (8). In addition, rTMS has been
demonstrated to influence glucose metabolism (8), long-term potentiation (9), the activity of ion channels
(10), and the expression of
plasticity-associated genes (11).
Neural progenitor cells (NPCs) in the subgranular
zone (SGZ) and subventricular zone (SVZ) of the brain can
self-renew, proliferate, migrate and differentiate (12). Following cerebral ischemia, NPCs
are activated for proliferation and can migrate to the injured
region for neuron repair and regeneration (13). rTMS has been shown to increase NPC
proliferation in the SGZ of healthy rats (14) and in the SVZ of focal cerebral
ischemia rats (15). However, the
underlying mechanism of rTMS remains to be fully elucidated.
MicroRNAs (miRs) are 20-40-bp small non-coding RNAs,
which can inhibit the translation of mRNAs involved in various
physiological and pathological processes (16). Increasing evidence indicates that
miRs modulate the proliferation of NPCs (17,18). Using array analysis, a previous
study identified that miR-106b may promote the proliferation of
NPCs (17). Brett et al
(19) demonstrated that
overexpressing the entire miR106b~25 cluster enhanced the
proliferation of in vitro cultured NPCs. According to the
analysis of targeting gene prediction (www.targetscan.org, and Kyoto Encyclopedia of Genes
and Genomes), p21 of the cyclin-dependent kinase inhibitor (CDKI)
family is negatively regulated by miR-106b, which has been shown to
contribute to cell proliferation through accelerating the G1-to-S
transition (20,21). In addition, the expression of p21
can be regulated by other miRs (22,23) in other types of cells. However,
whether miR-106b can modulate the expression of p21 in NPCs has not
been investigated.
Our previous study (24) indicated that rTMS was able to
directly induce the proliferation of NPCs accompanied with the
upregulation of miR-106b. The present study aimed to further
investigate the effects of rTMS on cultured NPCs transfected with
Lenti-miR-106b or small interfering (si)RNAs to clarify whether
rTMS promotes NPC proliferation by upregulating the expression of
miR-106b and possibly inhibiting the expression of p21.
Materials and methods
Reagents
The primary antibodies and reagents used were as
follows: Dulbecco's modified Eagle's medium/Nutrient Mixture F-12
(DMEM/F12; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), B-27® Supplement (Invitrogen; Thermo Fisher
Scientific, Inc.), basic fibroblast growth factor (b-FGF;
Peprotech, Inc., Rocky Hill, NJ, USA), epidermal growth factor
(EGF; Peprotech, Inc.), TrypLE™ Express Enzyme (Gibco; Thermo
Fisher Scientific, Inc.), poly-L-lysine (Sigma; Merck KGaA,
Darmstadt, Germany), β-actin antibody (cat. no. BM0627; Wuhan
Boster Biological Technology Co., Ltd., Wuhan, China), EdU (Ruibo
Biological Technology Co., Ltd., Guangzhou, China), mouse anti-rat
nestin (cat. no. 556309; BD Biosciences, Franklin Lakes, NJ, USA),
FITC-labeled rabbit anti-mouse IgG (cat. no. 315-005-003; Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA).
Preparation of proliferation medium
For the production of 100 ml of proliferation
medium, 98 ml DMEM/F12 medium, 2 ml B-27® without
vitamin A, 2 µg b-FGF and 2 µg EGF were mixed,
sterilized using a 0.22-µm filter in a laminar flow hood,
and stored in a 4°C refrigerator.
Culture of NPC neurospheres
The NPC neurospheres were cultured as previously
described (25). In brief,
bilateral hippocampal tissues were rapidly dissected from the
brains of 10-15 neonatal Sprague-Dawley rats within 3 days of birth
for each experiment. The neonatal rats (weight, 5-6 g) were
provided by Tongji Medical College Experimental Animal Center of
Huazhong Technology University (Huazhong, China). Rooms were
maintained at 20-24°C (50% relative humidity) and a 12-h light/dark
cycle. The hippocampal tissues were placed into cold Hank's
Buffered Salt Solution (HBSS; Sigma-Aldrich; Merck KGaA), Following
enzyme digestion with TrypLE™ Express (Gibco; Thermo Fisher
Scientific, Inc.) in a 5% CO2 incubator (37°C for 2
min), the tissues were mechanically dissociated using a pipette
several times, and centrifuged (300 x g 5 min, 4°C). The cells were
suspended in the proliferation medium, as described above, and were
seeded (104-5 cells/ml, passage one) in
dishes for culture with DMEM/F12 in a 5% CO2 incubator
at 37°C. The neurospheres were subcultured every 5 days. The second
generation of NPCs was prepared for rTMS. All experimental
procedures were approved by the ethics committee of the Wuhan
Sports University (Wuhan, China).
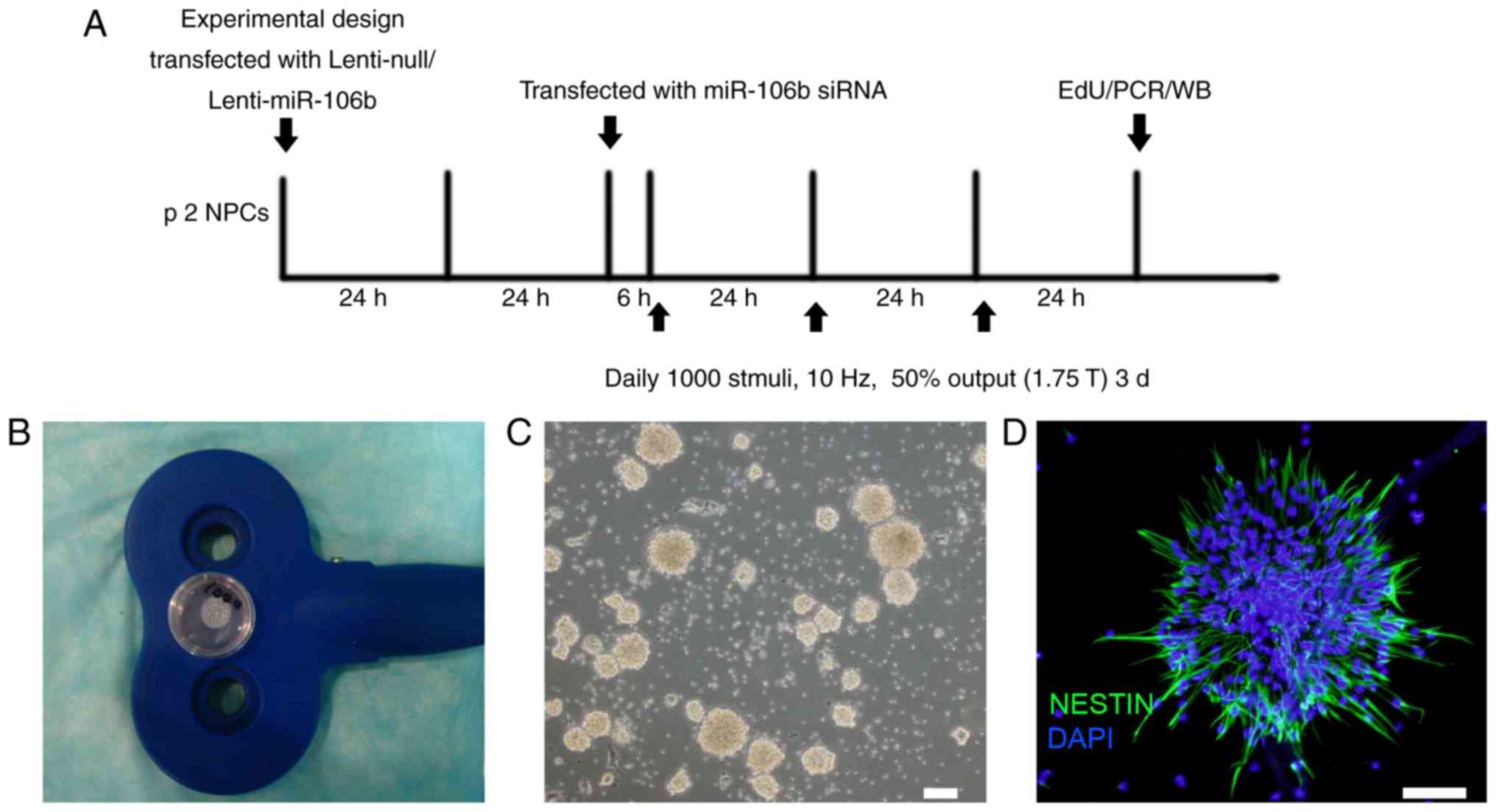
Experimental design
The experimental design is outlined in Fig. 1A. The NPCs were used for rTMS and
miR overexpression/downregulation experiments. For the
over-expression of miR-106b, the NPCs were transfected with
lentivirus (Lenti)-null, or Lenti-miR-106b for 48 h prior to rTMS.
For the downregulation of miR-106b, the NPCs were transfected with
miR-106b siRNA using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 6 h prior to rTMS. Following 3 days of
stimulation, the NPCs were used for EdU staining or miR/protein
analyses. In the sham group, the NPCs were treated with rTMS
without stimuli output. An empty lentivirus or Lipofectamine 2000
without siRNA was used for the respective negative control groups
(Lenti-null + sham: LN; negative control + sham: NC). The groups
were named as follows: Lenti-miR-106b + sham: L106b; Lenti-miR-106b
+ rTMS: L106bS; anti-miR-106b + sham: A106b; anti-miR-106b + rTMS:
A106bS.
Transfection of the NPCs with
Lenti-miR-106b or miR-106b siRNA
The pLVX-ZsGreen-Puro-rno-miR-106b vector (Wuhan
Biofavor Co., Ltd., Wuhan, China) was transfected into 293T cells
(Wuhan Biofavor Co., Ltd.) to generate high-titer lentivirus
(biological titer, 1.0x108 TU/ml) containing miR-106b.
The NPCs were infected with the lentivirus based on the equation
that MOI=30. The cells were re-suspended in 2 ml of complete
medium, and incubated with 1.5x107 TU lentivirus at 37°C
with 5% CO2 for 48 h. Subsequently, the medium
containing the NPCs was replaced with fresh medium to obtain 80%
confluence. The siRNAs for miR-106b-5p (5'-UAA AGU GCU GAC AGU GCA
GAU-3') were synthesized by GenePharma Co., Ltd. (Shanghai, China).
The NPCs were re-suspended at 105 cells/ml in Opti-MEM
medium (Invitrogen; Thermo Fisher Scientific, Inc.), and
transferred into flasks to culture for 2 h. According to the
manufacturer's protocol, the miR-106b siRNAs were transiently
transfected into NPCs using siRNA-Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) and cultured for 48 h at 37°C with
5% CO2. The NPCs were then treated with rTMS.
rTMS
The NPCs with or without miR modification were
treated by sham or rTMS using a CCy-I type transcranial magnetic
stimulation instrument (Wuhan Yiruide Medical Equipment Co., Ltd.,
Wuhan, China) according to a previous study (24). In brief, the culture dish was
placed in the cross-center of an ‘8’-shaped magnetic coil which had
a stimulus distance of rTMS of <1 cm between the cells and the
coil (Fig. 1B). The rTMS was
performed daily at 1,000 stimuli for 3 days at 10 Hz, with 1.75 T
output. The neurospheres were examined under a light microscope
(Fig. 1C).
Immunofluroscence and EdU staining
Following 3 days of rTMS, the cells were stained
with nestin, which is a common marker of NPCs. The resuspended
neurospheres were seeded into the 24-well glass slides coated with
polylysine, and fixed with -20°C methanol for 20 min. Subsequently,
for the immunostaining of nestin, each coverslip was incubated with
20 µl mouse anti-rat nestin antibody (1:100) at 4°C
overnight. The cells were then incubated with secondary
FITC-labeled rabbit anti-mouse IgG (1:400) for 2 h at room
temperature, protected from the light. DAPI was added for nuclear
staining for 15 min at room temperature.
EdU staining was used to determine the proliferative
NPCs. The re-suspended NPCs in each 24-well contained 500 µl
solution which was diluted with the culture medium at a ratio of
1,000:1 (reagent A) and cultured for 2 h. The medium was the
discarded and 500 µl of pre-cooling pure methanol was added
for fixation at room temperature for 20 min. The slides were then
stained with 1X Apollo® staining reaction solution and
1X Hoechst 33342 reaction solution for 30 min respectively at room
temperature (Fig. 1D).
Immunofluorescence images were observed using the
Olympus Bx51 fluorescence microscope. A total of five
randomly-selected fields were counted in a blinded-manner using
image processing software (ImageJ, v.1.6.0; National Institutes of
Health, Bethesda, MD, USA) for quantification.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
According to the manufacturer's protocol, the total
RNA of the cells was isolated using TRIzol reagent (Thermo Fisher
Scientific, Inc.) and RNA concentration was measured using a
spectrophotometer. The reverse transcription of RNA was performed
using a TaqMan MicroRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) at 70°C for 5 min, 42°C
for 60 min, and 95°C for 5 min. To quantify the expression of
miR-106b, a 20-µl reaction system included 100 µM/l
rno-miR-106b forward and rno-miR-106b reverse primer, 10 µl
SYBR Green/Flourescein qPCR Master mix (2X; Takara Bio, Inc., Otsu,
Japan) and 4 µl cDNA (10X). The conditions were as follows:
A cycle of 50°C for 2 min, a 95°C for 10 min, followed by 40 cycles
of 95°C for 30 sec and 60°C for 30 sec. The 2-ΔΔCq
method was used to analyze the relative change in the expression of
miR-106b (26). The primer
sequences were as follows: U6, forward 5'-CGC TTC GGC AGC ACA TAT
AC-3 and reverse 5'-AAA TAT GGA ACG CTT CAC GA-3'; rno-miR-106b,
forward 5'-TGC GCT AAA GTG CTG ACA GTG-3' and reverse 5'-CTC AAG
TGT CGT GGA GTC GGC AA-3'.
Western blot analysis
The lysates of NPCs were extracted using a RIPA
buffer (Beyotime Institute of Biotechnology, Shanghai, China) and
were centrifuged at 12,000 x g for 10 min at 4°C. Then 400
µl the supernatant mixed with 100 µl Laemmli buffer
and was heated at 100°C for 10 min. The protein concentration was
determined by using the Protein Assay kit for bicinchoninic acid
(Thermo Fisher Scientific, Inc.). Electrophoresis was performed
with 50 µg of total protein. Protein was resolved on a 15%
SDS PAGE and transferred on to polyvinylidene difluoride membranes.
Membrane transfer of the p21 protein was achieved under 200 mA for
1 h. The membrane was then immersed in 5% tris-buffered saline and
tween (TBST) and incubated at room temperature for 2 h. The primary
antibody rabbit anti-rat p21 (1:500; cat. no. sc-397; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) was incubated overnight at
4°C for 16 h. The membrane was then fully washed with TBST, and the
goat anti-rat IgG secondary antibody (1:50,000; cat. no. BA1054;
Wuhan Boster Biological Technology Co., Ltd.) conjugated to HRP was
used for incubation of the membrane at room temperature for 2 h.
The Gene Genius Bio-Imaging system gel imager was used to capture
images, and BandScan version 5.0 software (Glyko Inc., Novato, CA,
USA) was used to analyze the optical density signal strips.
Statistical analysis
The experimental data are expressed as the mean ±
standard deviation. All experiments were repeated at least 3 times.
Differences between groups were analyzed by one-way analysis of
variance followed by the LSD test. Differences between two groups
were analyzed using Student's t-test. SPSS 17.0 statistical
software (version 17.0; SPSS, Inc., Chicago, IL, USA) was used for
statistical analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
Characterization of NPCs cultured from
the hippocampus
The hippocampal tissues were separated from the
newborn rats. Following the first passage, the cells started to
form neurospheres, which had grown to almost 100 µm on the
fifth day. The neurospheres at passage 2 exhibited a smooth shiny
surface under light microscopy (Fig.
1C) and positively expressed the NPC-specific marker nestin
(Fig. 1D).
rTMS promotes the proliferation of NPCs
in vitro
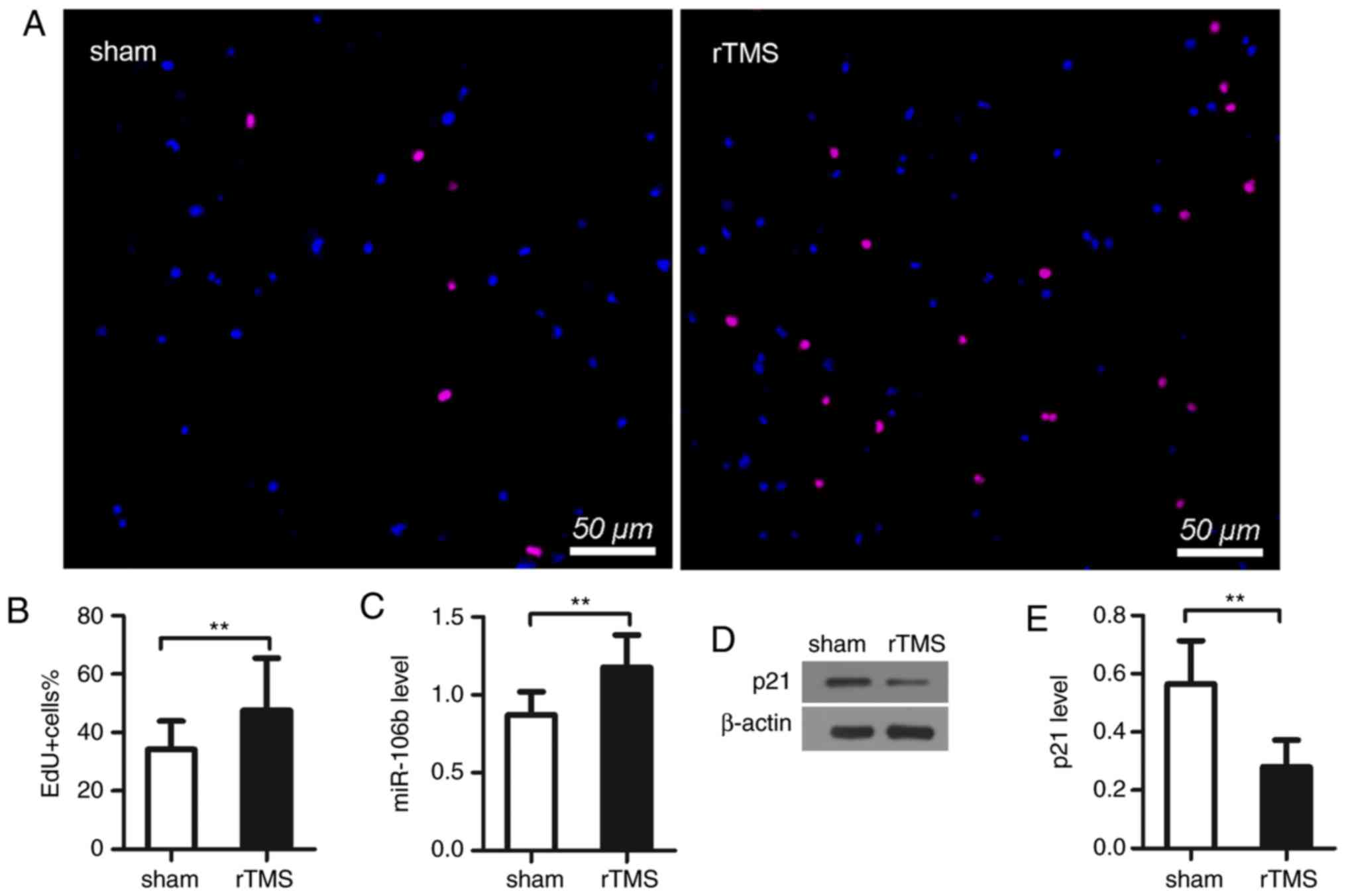
EdU staining was used to analyze NPC proliferation.
The results showed that there was a higher proportion of
EdU-positive cells in the rTMS group than in the sham group cells
(sham, vs. rTMS, 38.1±9.5%, vs. 51.7±25.5%, P<0.01; Fig. 2A and B). These results indicated
that rTMS promoted the proliferation of NPCs.
rTMS increases miR-106b and decreases p21
levels in NPCs in vitro
The results showed that the treatment of rTMS
significantly upregulated the expression of miR-106b (sham, vs.
rTMS, 0.87±0.15, vs. 1.18±0.21, P<0.01; Fig. 2C). As shown in the results of the
western blot analysis, rTMS markedly decreased the level of p21
(sham, vs. rTMS, 0.57±0.15, vs. 0.28±0.09, P<0.05; Fig. 2D and E).
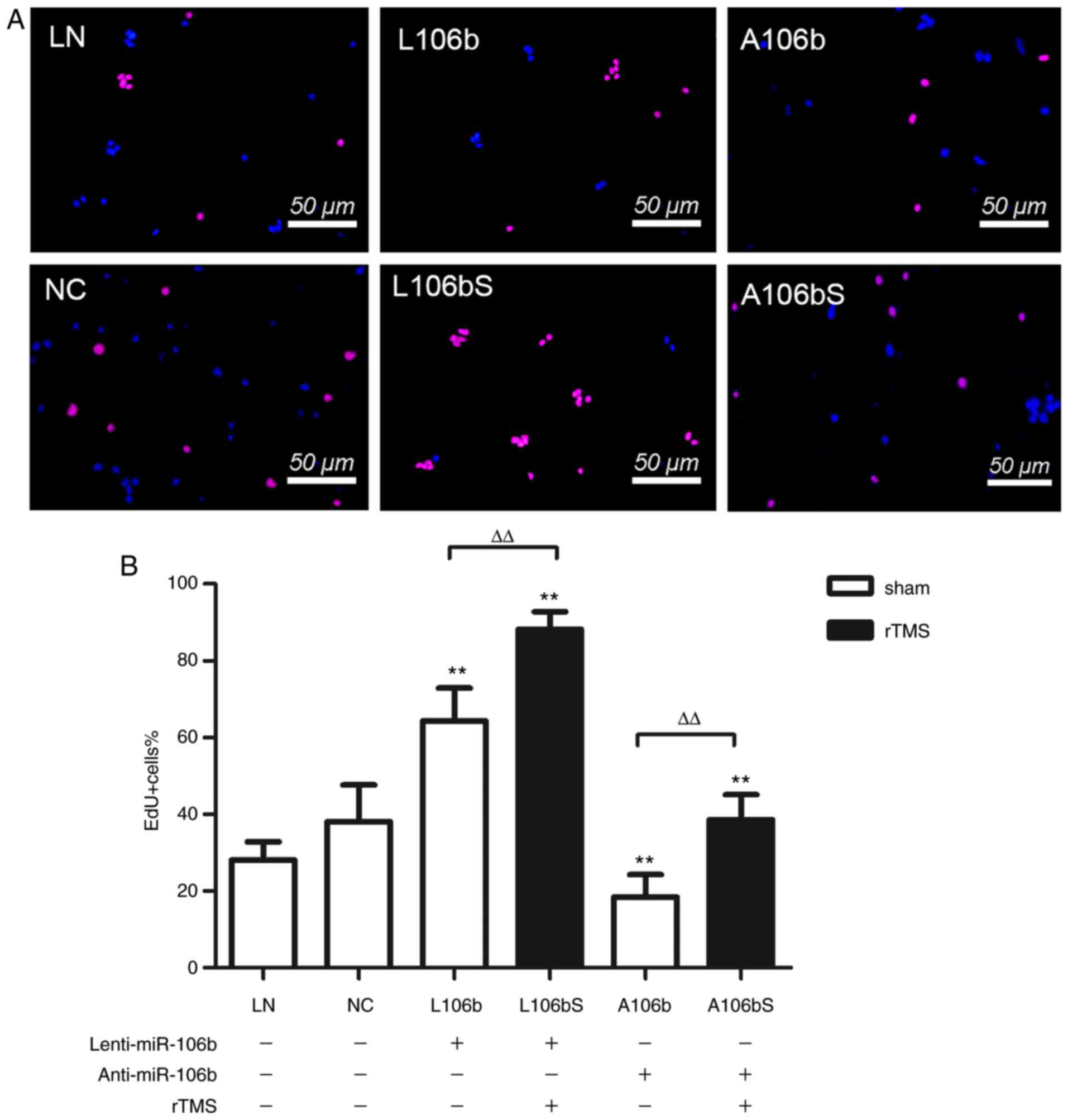
Overexpressing miR-106b further enhances
the proliferation of NPCs induced by rTMS
In order to illustrate whether miR-106b is involved
in the effects induced by rTMS on NPCs, the expression of miR-106b
in NPCs was modulated. As shown in Fig. 3A and B, the overexpression of
miR-106b increased the number of EdU-positive cells compared with
the number in the cells transfected with Lenti-null, the
transfection control (L106b, vs. LN, 64.3±8.6%, vs. 28.1±4.7%,
P<0.01). However, the knockdown of miR-106b reduced the
proliferation of NPCs (A106b, vs. NC, 18.4±5.9%, vs. 38.1±9.5%,
P<0.01). rTMS further increased the proliferation of NPCs in the
miR-106b overexpression group (L106bS, vs. L106b, 88.2±4.6%, vs.
64.3±8.6%, P<0.01), which was eliminated by miR-106b siRNA
(A106bS, vs. A106b, 38.6±6.5%, vs. 18.4±5.9%, P<0.01). Together,
these data indicate that miR-106 modulated the rTMS-induced
proliferation of NPCs.
rTMS upregulates the expression of
miR-106b
Subsequently, the present study examined the
expression of miR-106b in each group, and found that rTMS increased
miR-106b in cells of the overexpression group (L106b, vs. L106bS,
2.09±0.1, vs. 2.43±0.11, P<0.01; Fig. 4A) and knockdown group (A106b, vs.
A106bS, 0.30±0.02, vs. 0.48±0.02, P<0.01; Fig. 4B).
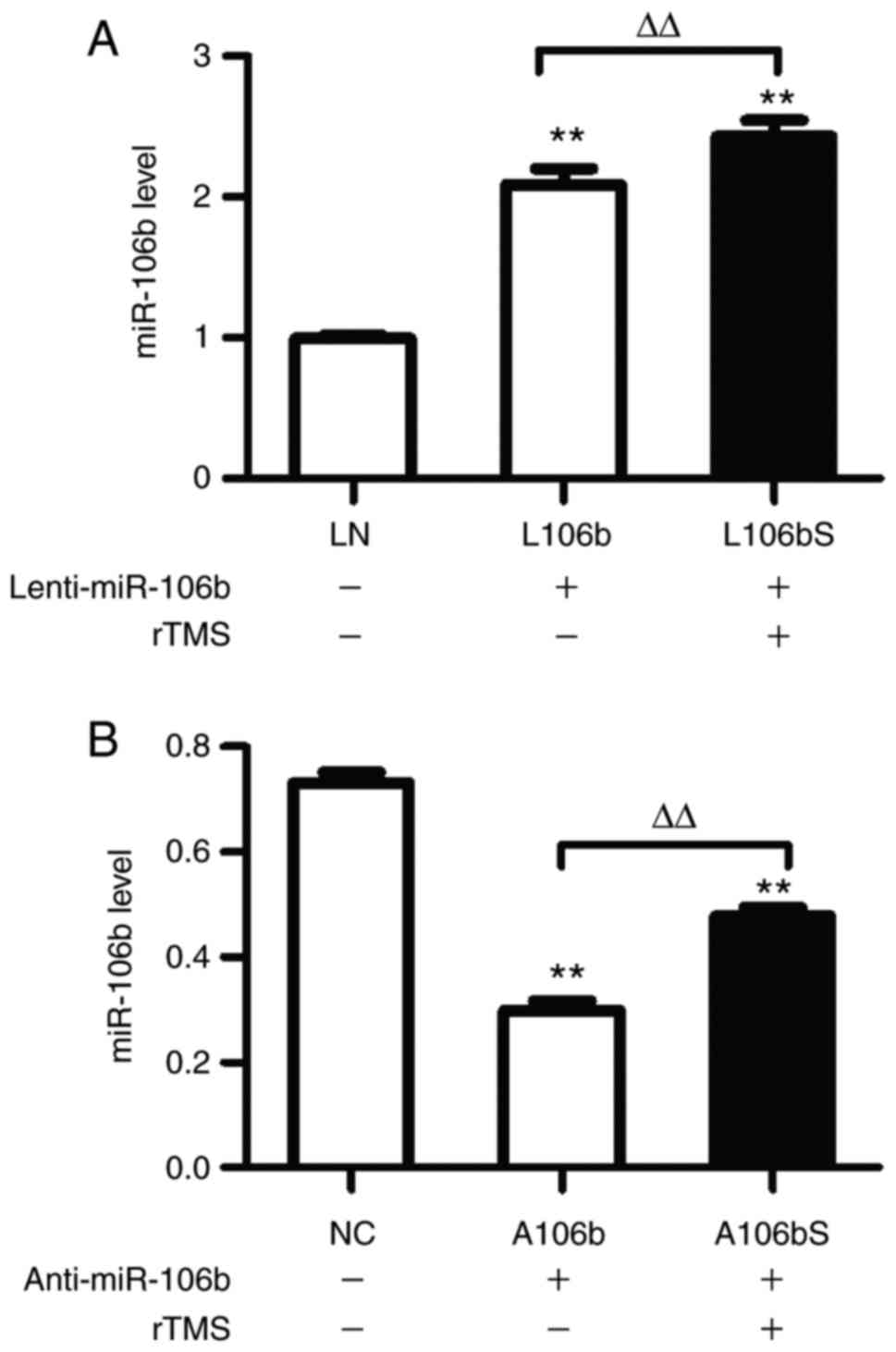 | Figure 4rTMS increases the expression of
miR-106b in neural progenitor cells in miR-106b overexpression and
inhibition. (A) Relative expression levels of miR-106b following
the overexpression of miR-106b were assessed by RT-qPCR analysis.
(B) Expression levels of miR-106b following inhibition of miR-106b
were assessed by RT-qPCR analysis. Relative expression levels are
expressed as the mean ± standard deviation. **P<0.01,
vs. sham group, ΔΔP<0.01. miR, microRNA; rTMS,
repetitive transcranial magnetic stimulation; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; NC, negative
control + sham; L106b, Lenti-miR-106b + sham; L106bS,
Lenti-miR-106b + rTMS; A106b, anti-miR-106b + sham; A106bS,
anti-miR-106b + rTMS. |
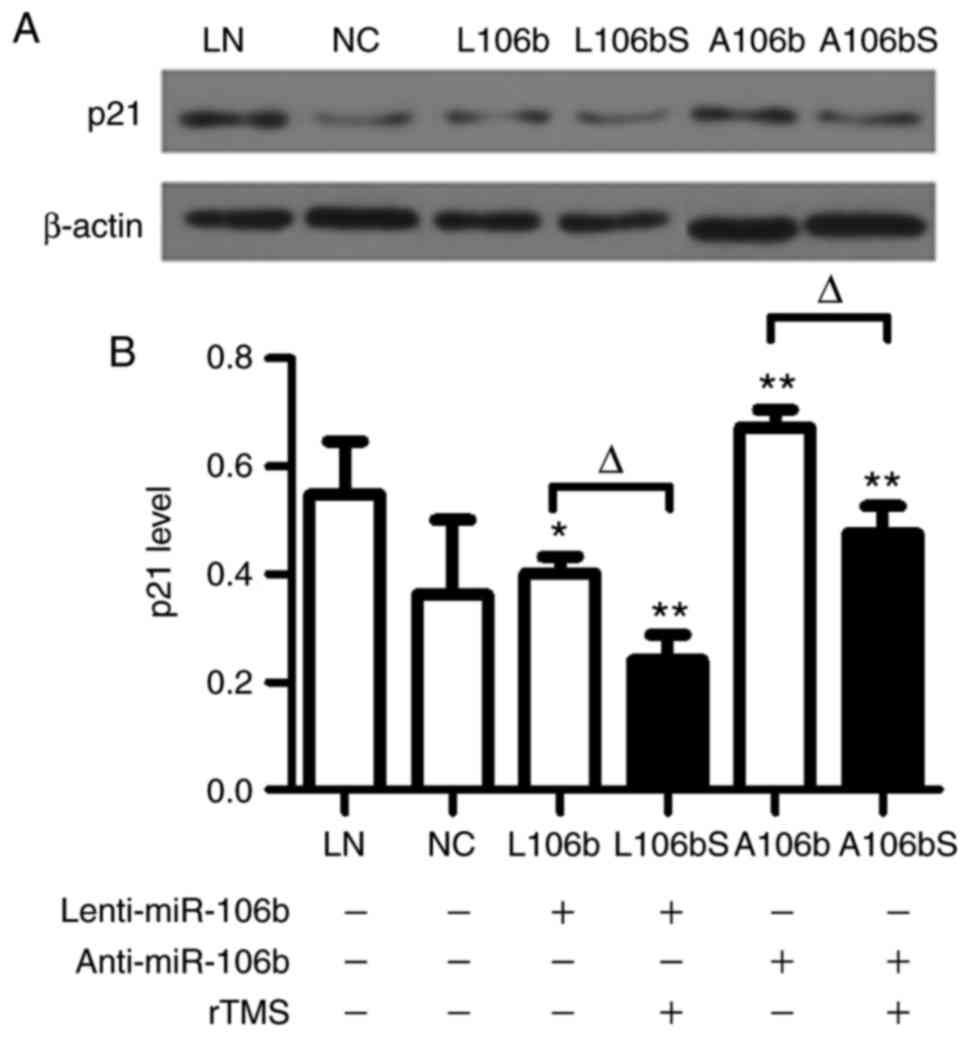
rTMS attenuates the protein expression of
p21 in NPCs
Following lentiviral infection and knockdown of
miR-106b in NPCs, the protein expression of p21 was assessed by
western blot analysis (Fig. 5A and
B). The data showed that the level of p21 was significantly
decreased by rTMS in the overexpression group (L106b, vs. L106bS,
0.40±0.03, vs. 0.24±0.05, P<0.05) and knockdown group (A106b,
vs. A106bS, 0.67±0.03, vs. 0.48±0.05, P<0.05).
The results showed that miR-106b, which promoted the
proliferation of cells via p21, was upregulated by rTMS. These
results suggested that rTMS promotes the proliferation of NPCs via
miR-106b and possibly by inhibiting the expression of p21.
Discussion
It has been shown that rTMS can induce plasticity in
the brain (7,8) and can influence the gene expression
profile of NPCs and cultured neural cells (27-29). As a clinical treatment, evidence
from fMRI (7) and PET (8) analyses has demonstrated that rTMS
alters prefrontal-hippocampal network dynamics in healthy
volunteers and increases glucose metabolism in rats. It has also
been found to modulate miRs in vitro (24). In the present study, it was
observed that rTMS induced EdU-positive NPCs and upregulated the
expression of miR-106b. Subsequently, miR-106b was either stably
overexpressed or its siRNAs were transfected into NPCs, and it was
confirmed that rTMS promoted the proliferation of NPCs through
miR-106b and possibly by inhibiting the expression of kinase
inhibitor p21. The data are presented in Fig. 2.
The protocols of rTMS are generally controversial in
treatment of the nervous system (30). Stimulation frequency is the most
important factor in terms of rTMS parameters. Low frequency rTMS is
considered to have an inhibitory effect on the brain (26), whereas high frequency rTMS has
excitatory effects (31). In
animal experiments, a high frequency (>5 Hz) has been reported
to promote neural plasticity and improve behavior in rats with
depression (32,33) and in rats with focal cerebral
ischemia (15), associated with
plasticity genes, including brain-derived neurotrophic factor
(BDNF) (33-35). In cell experiments, compared with
low frequency (1 Hz) rTMS, high frequency (10 Hz) rTMS induced
neuroprotective and anti-apoptotic effects in a cell model of
hippocampal injury (36,37). In addition, high frequency (10 Hz)
rTMS has been shown to induce neural plasticity in hippocampal
slice cultures (31). In clinical
experiments, high frequency rTMS is generally used for neuropathic
pain (38,39), cognition and motor recovery in
patients with Parkinson's disease and Alzheimer's disease (6), and leads to superior improvements
over low frequency rTMS. The stimulation intensity is another
important parameter; it decreases within the coil distance of 3.5
cm, and 60% of its intensity is maintained at a distance of 1 cm
(40). Although transcranial
magnetic stimulations should not be uniform on the suspended cell
cultures in a dish due to the difference in distance, the
electromagnetic field has been shown to be effective in inducing
NPC proliferation (29). The
results showed that the proliferation of NPCs was promoted by rTMS
daily (1,000 stimuli) for 3 days at 10 Hz, with 1.75 T output.
The expression of miR-106b is high in the adult rat
brain and influences thousands of target genes. One of these,
minichromosome maintenance complex component 7, which is decreased
in the brain of rats with Down's syndrome, suggests that miR-106b
is closely associated with nerve generation (41). In addition, miR-106b influences
the insulin/insulin-like growth factor-1-Forkhead box O pathway
(19), which can promote NPC
proliferation (42). Our previous
study found that protein kinase inhibitor p21 as the target gene of
miR-106b was another proliferative factor through regulating
cyclins (24).
The molecular mechanism of p21 regulating the
proliferation of NPCs remains to be fully elucidated. Cell cycle is
regulated by cyclins, cyclin-dependent kinase (CDK) and CDKI
(43). p21 as one of the CDKIs,
is the direct target gene of miR-106b (20). It combines with CDK2, CDK4/6,
cyclinA, cyclinD and cyclinE to arrest the cell cycle (44). miR-106b-med-ited p21 silencing can
affect the cell cycle and promote the cells to exit the G1 stage
and enter the S stage (44,45). In addition, p21 can be combined
with enhancer SRY-box binding protein-2 (Sox2) regulatory region 2
(46), which is a Sox2 marker in
NPCs (47). Low p21 increasing
the expression of Sox2 can induce the proliferation of NPCs.
Tailless (Txl) is an orphan nuclear receptor specifically expressed
in NPCs and P21, as target gene of Txl, is crucial for the
homeostasis of NPCs (48,49). In addition, Yoon et al
(50) claimed that a therapeutic
effect of rTMS on subacute cerebral ischemia rat was associated
with an anti-apoptotic effect. Liu et al (51) demonstrated that miR-106b modulated
the anti-apoptotic effect through inhibiting p21. Decreasing
apoptosis upregulates neuronal turnover, which is beneficial for
neural plasticity (52). p21 is a
protector preventing premature loss of the NSC population (53); when there is a lack of p21, cells
have a higher proliferative activity. In the present study, it was
found that rTMS decreased the expression of p21, which was
consistent with the EdU-positive cells. These data are supported by
an in vivo study (16),
which showed that 14 days of chronic rTMS increased the number of
BrdU-positive cells in the dentate gyrus of rats. The present study
did not characterize cell differentiation of the cultured NPCs in
the proliferation medium, which requires examination in future
investigations.
There is an equilibrium system in place to balance
the generation, proliferation or differentiation of cells in the
brain, and the pool of stem cells can be depleted due to a weak
proliferation rate (54-56). The results of the present study
suggested that rTMS assisted in maintaining the equilibrium system
by the appropriate continuous growth rate of NPCs in the brain. It
is reported that, in the adult hippocampus, ~700 new neurons
(annual turnover rate 1.75%) are exchanged every day, with a mild
decline during aging (57).
Treatment including regular physical activity has been suggested to
resist aging due to promoting the proliferation of NPCs associated
with increasing BDNF (58,59).
Taken together, neurogenesis induced by rTMS may be another method
to alleviate aging, which has application prospects in future
healthcare and medical treatment.
According to the data, rTMS increases miR-106b and
decreases p21 levels in NPCs in vitro, which is determined
by overexpressing and downregulating miR-106b expression. The
present study showed that rTMS-miR-106b was the main pathway
influencing the action of NPCs. In conclusion, high frequency (10
Hz) rTMS promoted NPC proliferation via upregulating miR-106b, and
possibly by inhibiting the expression of p21.
Acknowledgements
Not applicable.
Funding
The study was supported by the National Natural
Science Foundation of Young Scholars of China (grant no. 81700280),
the Program of Natural Science Foundation of Hubei Province, China
(grant no. 2017CFB361), the Outstanding Young Scientific and
Technological Innovation Team in the Colleges and Universities of
Hubei Province, China (grant no. T201523), the Scientific Research
Project supported by Wuhan Sports University (grant no. 2016XH24),
and the program of China Scholarship Council (grant no.
201708420245).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL and GL performed experiments; HL, GL, JW and CM
wrote the manuscript; all authors contributed to manuscript
preparation, discussed the results, analyzed data and commented on
the manuscript; HL, YC and YY developed the concepts and designed
the study. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by the
ethics committee of the Wuhan Sports University (Wuhan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lewis G: Transcranial magnetic stimulation
for depression. Lancet. 391:1639–1640. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hosomi K, Shimokawa T, Ikoma K, Nakamura
Y, Sugiyama K, Ugawa Y, Uozumi T, Yamamoto T and Saitoh Y: Daily
repetitive transcranial magnetic stimulation of primary motor
cortex for neuropathic pain: A randomized, multicenter,
double-blind, crossover, sham-controlled trial. Pain.
154:1065–1072. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gersner R, Oberman L, Sanchez MJ,
Chiriboga N, Kaye HL, Pascual-Leone A, Libenson M, Roth Y, Zangen
A, Rotenberg A, et al: H-coil repetitive transcranial magnetic
stimulation for treatment of temporal lobe epilepsy: A case report.
Epilepsy Behav Case Rep. 5:52–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kalita J, Laskar S, Bhoi SK and Misra UK:
Efficacy of single versus three sessions of high rate repetitive
transcranial magnetic stimulation in chronic migraine and
tension-type headache. J Neurol. 263:2238–2246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang CG, Zhang T, Yue FG, Yi ML and Gao
D: Efficacy of repetitive transcranial magnetic stimulation in the
treatment of patients with chronic primary insomnia. Cell Biochem
Biophys. 67:169–173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bentwich J, Dobronevsky E, Aichenbaum S,
Shorer R, Peretz R, Khaigrekht M, Marton RG and Rabey JM:
Beneficial effect of repetitive transcranial magnetic stimulation
combined with cognitive training for the treatment of Alzheimer's
disease: A proof of concept study. J Neural Transm (Vienna).
118:463–471. 2011. View Article : Google Scholar
|
|
7
|
Bilek E, Schäfer A, Ochs E, Esslinger C,
Zangl M, Plichta MM, Braun U, Kirsch P, Schulze TG, Rietschel M, et
al: Application of high-frequency repetitive transcranial magnetic
stimulation to the DLPFC alters human prefrontal-hippocampal
functional interaction. J Neurosci. 33:7050–7056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee SA, Oh BM, Kim SJ and Paik NJ: The
molecular evidence of neural plasticity induced by cerebellar
repetitive transcranial magnetic stimulation in the rat brain: A
preliminary report. Neurosci Lett. 575:47–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Touge T, Gerschlager W, Brown P and
Rothwell JC: Are the after-effects of low-frequency rTMS on motor
cortex excitability due to changes in the efficacy of cortical
synapses? Clin Neurophysiol. 112:2138–2145. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Yang H, Tang X, Bai W, Wang G and
Tian X: Repetitive transcranial magnetic stimulation regulates
L-type Ca(2+) channel activity inhibited by early sevoflurane
exposure. Brain Res. 1646:207–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang N, Xing M, Wang Y, Tao H and Cheng
Y: Repetitive transcranial magnetic stimulation enhances spatial
learning and synaptic plasticity via the VEGF and BDNF-NMDAR
pathways in a rat model of vascular dementia. Neuroscience.
311:284–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ming GL and Song H: Adult neurogenesis in
the mammalian central nervous system. Annu Rev Neurosci.
28:223–250. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Müller-Dahlhaus F and Ziemann U:
Metaplasticity in human cortex. Neuroscientist. 21:185–202. 2015.
View Article : Google Scholar
|
|
14
|
Ueyama E, Ukai S, Ogawa A, Yamamoto M,
Kawaguchi S, Ishii R and Shinosaki K: Chronic repetitive
transcranial magnetic stimulation increases hippocampal
neurogenesis in rats. Psychiatry Clin Neurosci. 65:77–81. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo F, Han X, Zhang J, Zhao X, Lou J, Chen
H and Huang X: Repetitive transcranial magnetic stimulation
promotes neural stem cell proliferation via the regulation of
miR-25 in a rat model of focal cerebral ischemia. PLoS One.
9:e1092672014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng Y, Yi R and Cullen BR: Recognition
and cleavage of primary microRNA precursors by the nuclear
processing enzyme Drosha. EMBO J. 24:138–148. 2005. View Article : Google Scholar :
|
|
17
|
Chen H, Qian K, Tang ZP, Xing B, Chen H,
Liu N, Huang X and Zhang S: Bioinformatics and microarray analysis
of microRNA expression profiles of murine embryonic stem cells,
neural stem cells induced from ESCs and isolated from E8.5 mouse
neural tube. Neurol Res. 32:603–613. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anokye-Danso F, Snitow M and Morrisey EE:
How microRNAs facilitate reprogramming to pluripotency. J Cell Sci.
125:4179–4187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brett JO, Renault VM, Rafalski VA, Webb AE
and Brunet A: The microRNA cluster miR-106b~25 regulates adult
neural stem/progenitor cell proliferation and neuronal
differentiation. Aging (Albany NY). 3:108–124. 2011. View Article : Google Scholar
|
|
20
|
Ivanovska I, Ball AS, Diaz RL, Magnus JF,
Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson
AL, et al: MicroRNAs in the miR-106b family regulate p21/CDKN1A and
promote cell cycle progression. Mol Cell Biol. 28:2167–2174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karimian A, Ahmadi Y and Yousefi B:
Multiple functions of p21 in cell cycle, apoptosis and
transcriptional regulation after DNA damage. DNA Repair (Amst).
42:63–71. 2016. View Article : Google Scholar
|
|
22
|
Wang H, Zhu LJ, Yang YC, Wang ZX and Wang
R: MiR-224 promotes the chemoresistance of human lung
adenocarcinoma cells to cisplatin via regulating G1/S
transition and apoptosis by targeting p21(WAF1/CIP1). Br J Cancer.
111:339–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Semaan A, Qazi AM, Seward S, Chamala S,
Bryant CS, Kumar S, Morris R, Steffes CP, Bouwman DL, Munkarah AR,
et al: MicroRNA-101 inhibits growth of epithelial ovarian cancer by
relieving chromatin-mediated transcriptional repression of
p21(waf1/cip1). Pharm Res. 28:3079–3090.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu H, Han XH, Chen H, Zheng CX, Yang Y
and Huang XL: Repetitive magnetic stimulation promotes neural stem
cells proliferation by upregulating MiR-106b in vitro. J Huazhong
Univ Sci Technolog Med Sci. 35:766–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reynolds BA and Weiss S: Generation of
neurons and astrocytes from isolated cells of the adult mammalian
central nervous system. Science. 255:1707–1710. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Casula EP, Tarantino V, Basso D, Arcara G,
Marino G, Toffolo GM, Rothwell JC and Bisiacchi PS: Low-frequency
rTMS inhibitory effects in the primary motor cortex: Insights from
TMS-evoked potentials. Neuroimage. 98:225–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cash RFH, Dar A, Hui J, De Ruiter L,
Baarbé J, Fettes P, Peters S, Fitzgerald PB, Downar J and Chen R:
Influence of inter-train interval on the plastic effects of rTMS.
Brain Stimul. 10:630–636. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stock M, Kirchner B, Waibler D, Cowley DE,
Pfaffl MW and Kuehn R: Effect of magnetic stimulation on the gene
expression profile of in vitro cultured neural cells. Neurosci
Lett. 526:122–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui M, Ge H, Zhao H, Zou Y, Chen Y and
Feng H: Electromagnetic fields for the regulation of neural stem
cells. Stem Cells Int. 2017:98984392017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barker AT, Freeston IL, Jalinous R and
Jarratt JA: Magnetic stimulation of the human brain and peripheral
nervous system: An introduction and the results of an initial
clinical evaluation. Neurosurgery. 20:100–109. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vlachos A, Müller-Dahlhaus F, Rosskopp J,
Lenz M, Ziemann U and Deller T: Repetitive magnetic stimulation
induces functional and structural plasticity of excitatory
postsynapses in mouse organotypic hippocampal slice cultures? J
Neurosci. 21:17514–17523. 2012. View Article : Google Scholar
|
|
32
|
Sun P, Wang F, Wang L, Zhang Y, Yamamoto
R, Sugai T, Zhang Q, Wang Z and Kato N: Increase in cortical
pyramidal cell excitability accompanies depression-like behavior in
mice: A transcranial magnetic stimulation study. J Neurosci.
31:16464–16472. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Samuels BA and Hen R: Neurogenesis and
affective disorders. Eur J Neurosci. 33:1152–1159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bakker N, Shahab S, Giacobbe P, Blumberger
DM, Daskalakis ZJ, Kennedy SH and Downar J: rTMS of the dorsomedial
prefrontal cortex for major depression: Safety, tolerability,
effectiveness, and outcome predictors for 10 Hz versus intermittent
theta-burst stimulation. Brain Stimul. 8:208–215. 2015. View Article : Google Scholar
|
|
35
|
Uhm KE, Kim YH, Yoon KJ, Hwang JM and
Chang WH: BDNF genotype influence the efficacy of rTMS in stroke
patients. Neurosci Lett. 594:117–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Post A, Müller MB, Engelmann M and Keck
ME: Repetitive transcranial magnetic stimulation in rats: Evidence
for a neuroprotective effect in vitro and in vivo. Eur J Neurosci.
11:3247–3254. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Aydin-Abidin S, Trippe J, Funke K, Eysel
UT and Benali A: High- and low-frequency repetitive transcranial
magnetic stimulation differentially activates c-Fos and zif268
protein expression in the rat brain. Exp Brain Res. 188:249–261.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lefaucheur JP, Drouot X, Ménard-Lefaucheur
I, Keravel Y and Nguyen JP: Motor cortex rTMS restores defective
intracortical inhibition in chronic neuropathic pain. Neurology.
67:1568–1574. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cruccu G, Aziz TZ, Garcia-Larrea L,
Hansson P, Jensen TS, Lefaucheur JP, Simpson BA and Taylor RS: A
meta-analysis neurostimulation therapy for neuropathic pain. Eur J
Neurol. 14:952–970. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grehl S, Martina D, Goyenvalle C, Deng ZD,
Rodger J and Sherrard RM: In vitro magnetic stimulation: A simple
stimulation device to deliver defined low intensity electromagnetic
fields. Front Neural Circuits. 10:852016. View Article : Google Scholar :
|
|
41
|
Hewitt CA, Ling KH, Merson TD, Simpson KM,
Ritchie ME, King SL, Pritchard MA, Smyth GK, Thomas T, Scott HS and
Voss AK: Gene network disruptions and neurogenesis defects in the
adult Ts1Cje mouse model of Down syndrome. PLoS One. 5:e115612010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kouroupi G, Lavdas AA, Gaitanou M,
Thomaidou D, Stylianopoulou F and Matsas R: Lentivirus-mediated
expression of insulin-like growth factor-I promotes neural
stem/precursor cell proliferation and enhances their potential to
generate neurons. J Neurochem. 115:460–474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sherr CJ and Roberts JM: Inhibitors of
mammalian G1 cyclin-dependent kinases. Genes Dev. 9:1149–1163.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Denicourt C and Dowdy SF: Cip/Kip
proteins: More than just CDKs inhibitors. Genes Dev. 18:851–855.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ilyin GP, Glaise D, Gilot D, Baffet G and
Guguen-Guillouzo C: Regulation and role of p21 and p27
cyclin-dependent kinase inhibitors during hepatocyte
differentiation and growth. Am J Physiol Gastrointest Liver
Physiol. 285:G115–G127. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Marqués-Torrejón MÁ, Porlan E, Banito A,
Gómez-Ibarlucea E, Lopez-Contreras AJ, Fernández-Capetillo O, Vidal
A, Gil J, Torres J and Fariñas I: Cyclin-dependent kinase inhibitor
p21 controls adult neural stem cell expansion by regulating Sox2
gene expression. Cell Stem Cell. 12:88–100. 2013. View Article : Google Scholar :
|
|
47
|
Arnold K, Sarkar A, Yram MA, Polo JM,
Bronson R, Sengupta S, Seandel M, Geijsen N and Hochedlinger K:
Sox2(+) adult stem and progenitor cells are important for tissue
regeneration and survival of mice. Cell Stem Cell. 9:317–329. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y, Liu HK and Schütz G: Role of the
nuclear receptor Tailless in adult neural stem cells. Mech Dev.
130:388–390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu HK, Belz T, Bock D, Takacs A, Wu H,
Lichter P, Chai M and Schütz G: The nuclear receptor tailless is
required for neurogenesis in the adult subventricular zone. Genes
Dev. 22:2473–2478. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yoon KJ, Lee YT and Han TR: Mechanism of
functional recovery after repetitive transcranial magnetic
stimulation (rTMS) in the subacute cerebral ischemic rat model:
Neural plasticity or anti-apoptosis? Exp Brain Res. 214:549–556.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu Z, Yang D, Xie P, Ren G, Sun G, Zeng X
and Sun X: MiR-106b and MiR-15b modulate apoptosis and angiogenesis
in myocardial infarction. Cell Physiol Biochem. 29:851–862. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chambers RA, Potenza MN, Hoffman RE and
Miranker W: Simulated apoptosis/neurogenesis regulates learning and
memory capabilities of adaptive neural networks.
Neuropsychopharmacology. 29:747–758. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Leslie KF: p21: Protector of progenitor
pools. Science Signal. 6:ec2732013. View Article : Google Scholar
|
|
54
|
Kippin TE, Martens DJ and van der Kooy D:
p21 loss compromises the relative quiescence of forebrain stem cell
proliferation leading to exhaustion of their proliferation
capacity. Genes Dev. 19:756–767. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Orford KW and Scadden DT: Deconstructing
stem cell self-renewal: Genetic insights into cell-cycle
regulation. Nat Rev Genet. 9:115–128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Paik JH, Ding Z, Narurkar R, Ramkissoon S,
Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, et al:
FoxOs cooperatively regulate diverse pathways governing neural stem
cell homeostasis. Cell Stem Cell. 5:540–553. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Spalding KL, Bergmann O, Alkass K, Bernard
S, Salehpour M, Huttner HB, Boström E, Westerlund I, Vial C,
Buchholz BA, et al: Dynamics of hippocampal neurogenesis in adult
humans. Cell. 153:1219–1227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kronenberg G, Reuter K, Steiner B, Brandt
MD, Jessberger S, Yamaguchi M and Kempermann G: Subpopulations of
proliferating cells of the adult hippocampus respond differently to
physiologic neurogenic stimuli. J Comp Neurol. 467:455–463. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nam SM, Kim JW, Yoo DY, Yim HS, Kim DW,
Choi JH, Kim W, Jung HY, Won MH, Hwang IK, et al: Physical exercise
ameliorates the reduction of neural stem cell, cell proliferation
and neuroblast differentiation in senescent mice induced by
D-galactose. BMC Neurosci. 15:1162014. View Article : Google Scholar : PubMed/NCBI
|