Introduction
Esophageal cancer (EC) is one of the most common
malignant tumors worldwide. It consists of two histological types,
adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC),
each with distinct etiological and pathological characteristics.
ESCC is the major histological type of EC in China (1). Despite advances in the treatment of
ESCC, including surgery, chemotherapy, radiation or a combination
of these options, the prognosis of patients with ESCC remains poor,
with an overall 5-year survival rate of 20% following surgery
(2). To improve the overall
outcome for patients with ESCC, using molecular targets to treat
cancer has brought promising results and attracted increasing
attention (3-5). The molecular mechanisms may lead to
the identification of new biomarkers and novel therapeutic
targets.
MicroRNAs (miRNAs) are ~22-nucleotide,
single-stranded, small non-coding RNAs that modulate gene
expression by binding to the 3' untranslated region (3'-UTR) of
their target mRNAs to repress protein translation or promote mRNA
degradation (6). It serves key
roles in various physiological processes, including cell
proliferation, differentiation, apoptosis, invasion, metastasis and
maturation (7). miRNA
(miR)-106b-3p has been reported to be deregulated in some types of
cancer, including increased in laryngeal carcinoma (8), bladder cancer (9), hepatocellular carcinoma (10) and renal cell carcinoma (11). However, the role of miR-106b-3p in
ESCC is unclear.
Epithelial-mesenchymal transition (EMT) is a complex
biological process. Specifically, following EMT, cells have
increased expression of mesenchymal molecules, including Snail,
Slug and Vimentin, and decreased expression of the epithelial
adhesion marker E-cadherin, leading to enhanced motility and
metastasis (12). The activation
of Wnt/β-catenin is a key step in the process of tumorigenesis
(13). Increasing evidence has
demonstrated that aberrant activation of the Wnt/β-catenin
signaling pathway promotes tumor progression in various types of
human cancer (14-17). Zinc and ring finger 3 (ZNRF3) is
part of the E3 ubiquitin ligase family, which negatively regulates
Wnt/β-catenin signaling (18).
ZNRF3 inhibits Wnt signaling by promoting the turnover of Frizzled
and Low-density lipoprotein receptor-related protein 6 (18). Accumulating data have strongly
demonstrated that ZNRF3 is involved in the process of tumorigenesis
(19-21). β-catenin is a dual function
protein that regulates coordinated cell-cell adhesion and gene
transcription. β-catenin has two sub-cellular locations, either
bound to E-cadherin on the cell membrane in adherens junctions in
intercalated disc structures, or free in the cytoplasm. β-catenin
regulates gene expression via the Wnt/β-catenin signaling pathway,
inducing transition from an epithelial phenotype to a mesenchymal
one.
Our previous studies have found that miR-106b-3p is
upregulated in ESCC tissues. Furthermore, miR-106b-3p has been
reported to be deregulated in certain types of cancer, including in
laryngeal carcinoma, bladder cancer, hepatocellular carcinoma and
renal cell carcinoma (8-11). However, the role of miR-106b-3p in
ESCC is unclear. Therefore, the aim of the current study was to
explore the potential mechanism of miR-106b-3p in cell
proliferation and EMT of ESCC via ZNRF3 and the Wnt/β-catenin
signaling pathway, and to determine its utility in ESCC diagnosis
and therapy.
Patients and methods
Patient samples
A total of 50 paired ESCC tumor tissues and matched
normal adjacent tissues were collected at the First Affiliated
Hospital of Soochow University (Suzhou, China) between July 2016
and August 2017 (mean age 57±12 years; 21 female patients and 29
male patients). All the samples were histologically confirmed to be
ESCCs esophageal hyperplasia, dysplasia or non-malignant tissues by
the pathologist (22). All
samples were immediately stored at -80°C. This study was approved
by the Ethics Committee of Nanjing Medical University and each
patient provided written informed consent.
Cell lines and cell culture
The three human ESCC cell lines (KYSE150, ECA-109
and EC9706) and normal epithelial cell line (HET-1A) were purchased
from the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). All cell lines were maintained in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified chamber with 5%
CO2.
Cell transfection
Cells were transfected with miR-106b-3p mimics,
miR-106b-3p inhibitors and NC using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at a final
concentration of 50 nM according to the manufacturer's
instructions. The sequences were as follows: miR-106b-3p mimics,
5'-TAA AGT GCT GAC AGT GCA GAT-3'; miR-106b-3p inhibitors, 5'-AUC
UGC ACU GUC AGC ACU UUA-3'; NC, 5'-CAG UAC UUU UGU GUA GUA CAA-3'.
Subsequently, cells were cultured with fresh medium containing 10%
FBS for 48 h. Transfection efficiencies were validated by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Total RNA extraction and RT-qPCR
Isolation of total RNA from samples was conducted
using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. cDNA was synthesized using the
SYBR® PrimeScript™ RT-PCR kit (Takara Biotechnology Co.,
Ltd., Dalian, China) for 30 min at 37°C according to the
manufacturer's instructions. ZNRF3 and miR-106b-3p expression was
determined by RT-qPCR using SYBR Premix ExTaq (Takara Biotechnology
Co., Ltd., Dalian, China) reagents. The primers used were as
follows: Forward, 5'-CAT CGT CAA CAA GCA GAA AGT G-3' and reverse,
5'-GGA GAC CAC GAC GAA-GAA AG-3' for ZNRF3; forward, 5'-TTT TCG CCC
TTA GCG TGA AGA-3' and reverse, 5'-GAG GCA GTC GAA TTT GCG T-3' for
β-actin. The thermocycling conditions for qPCR were as follows:
95°C for 5 min followed by 40 cycles of 95°C for 15 sec and 60°C
for 60 sec. All of the samples were normalized with small nuclear
RNA U6, the relative quantification and statistical analysis was
performed using the 2-ΔΔCq method (23). The experiments were repeated at
least three times.
Western blot analysis
Total protein was extracted using
radioimmunoprecipitation assay lysis buffer supplemented with
protease inhibitors (Gibco; Thermo Fisher Scientific, Inc.). The
protein concentration was measured using the bicinchoninic acid
method. Protein (50 µg/well) was separated on 10%
SDS-polyacrylamide gels and then blotted onto polyvinylidene
difluoride membranes (EMD Millipore, Bedford, MA, USA).
Subsequently, membranes were blocked with 5% skimmed milk at room
temperature for 1 h, and then incubated with primary antibodies at
4°C overnight. The specific primary antibodies used were as
follows: ZNRF3 (cat. no. ab122353; 1:1,000 dilution), p21 (cat. no.
ab109520; 1:1,000 dilution), p27 (cat. no. ab32034; 1:1,000
dilution), cyclin D1 (cat. no. ab16663; 1:1,000 dilution), Slug
(cat. no. Ab27568; 1:1,000 dilution), Snail (cat. no. Ab53519;
1:1,000 dilution), N-cadherin (cat. no. Ab76057; 1:1,000 dilution),
E-cadherin (cat. no. Ab1416; 1:1,000 dilution), Vimentin (cat. no.
Ab8978; 1:1,000 dilution) and GAPDH (cat. no. ab9485; 1:1,000
dilution; all Abcam, Cambridge, MA, USA). Then, the membranes were
washed with PBS three times and incubated with horseradish
peroxidase-conjugated secondary antibody (cat. no. ab6721; 1:2,000;
Abcam) at room temperature for 1 h. GAPDH used as the internal
reference protein. Signals were detected using an enhanced
chemiluminescence kit (GE Healthcare, Chicago, IL, USA).
Cell proliferation and colony formation
assays
Cell proliferation ability was detected using an MTT
assay. Briefly, 1×106 cells (KYSE150 and ECA-109) were
seeded into 96-well plates and cultured. MTT solution (10
µl) was added to each well, followed by incubation for 4 h.
The culture medium discarded and 150 µl dimethyl sulfoxide
(DMSO; Gibco; Thermo Fisher Scientific, Inc.) was added. Then the
mixture was gently shaken for 15 min and the cells were evaluated
at 24, 48, 72 and 96 h, after seeding. The absorbance was measured
at 490 nm using a microplate reader. For the colony formation
assay, 500 cells per well were seeded into 6-well plates. After
incubation for 3 weeks, cells were fixed with 4% paraformaldehyde
at room temperature for 20 min. Finally, the cells were stained
with 1% crystal violet for 4 h at room temperature and counted.
Flow cytometry
The effects of miR-106b-3p on cell cycle were
explored by flow cytometry in ESCC. After transfection for 48 h,
1×106 cells were seeded into 6-well plates and washed
twice with PBS. Subsequently, cells were digested with 0.25%
trypsin and fixed with 70% ethanol at -4°C overnight. Finally, 500
ml PI/RNase was added to cells and incubated in the dark for 45 min
at 37°C. The multiplication cycle was detected using flow cytometry
(FACSCalibur; BD Biosciences, San Jose, CA, USA).
Cell adhesion assay
Approximately 1×105 cells were harvested
and resuspended in culture medium and then transferred to 96-well
plates were pre-coated with collagen I, collagen IV and fibronectin
(Sigma-Aldrich; Merck KGaA). Following incubation for 2 h at room
temperature, the non-adherent cells were removed by washed twice
with PBS. The adhesive cells were fixed in 4% paraformaldehyde and
then stained with 0.5% crystal violet. The cells were examined
under a light microscopy.
Wound healing assays
Cells which were transfected with miR-106b-3p
mimics, miR-106b-3p mimics NC, miR-106b-3p inhibitors or
miR-106b-3p inhibitors NC were cultured to 80-90% confluence in
24-well plates in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS. Linear scratch wounds
were created on the confluent cell monolayers with a pipette tip
after transfection for 24 h. Images were captured at 0, 24 and 48 h
under an inverted microscope (magnification, ×100).
Transwell assay
Cell migration and invasion assays were performed
using Transwell chambers. Cells were seeded in 24-well plates and
harvested. For invasion or migration assays, the upper chamber was
first coated with or without Matrigel, respectively, then
1×105 KYSE150 or ECA-109 cells were seeded in the upper
chamber containing fresh serum-free culture media and supplemented
medium was added to the lower chamber containing 20% FBS. The cells
on the upper surface were gently removed after culturing for 24 h.
Migrant cells attached to the lower surface were fixed with 4%
paraformaldehyde for 30 min at room temperature and stained with
0.1% crystal violet for another 30 min at room temperature. Cells
were counted under a microscope in four randomly selected fields
(magnification, ×200). All experiments were conducted three
times.
Immunofluorescence
Cells (1×105 cells/well) were seeded on
6-well plates and incubated for 48 h. Subsequently, KYSE150 and
ECA-109 cells were fixed with 4% paraformaldehyde for 30 min at
room temperature and incubated with the primary antibody (ZNRF3;
cat. no. ab122353; 1:1,000 dilution; Abcam) at room temperature for
2 h after blocking cell with 3% bovine serum albumin (Gibco; Thermo
Fisher Scientific, Inc.). Fluorescein isothiocyanate-conjugated
secondary antibody (cat. no. ab205718; 1:2,000; Abcam) was
incubated with cells for 1 h at room temperature. The nuclei were
counted and stained with DAPI for 5 min at room temperature. The
slides were examined under a fluorescence microscopy.
Dual-luciferase reporter assays
A search for putative targets of miR-183 was
performed with TargetScan Human7.2 (www.targetscan.org/vert_72/) and miRanda (www.microrna.org/) software. The targeting gene
(ZNRF3) of miR-106b-3p was predicted using microRNA.org (http://www.microrna.org) and verified in a
dual-luciferase reporter gene assay. The wild-type 3'-UTR of human
ZNRF3 mRNA was amplified from human genomic DNA containing
miR-106b-3p binding site inserted into the
SpeI/HindIII sites of pMIR-REPORT™ (Promega
Corporation, Madison, WI, USA) luciferase reporter plasmid to
generate pMIR-ZNRF3-WT plasmid which were designed and synthesized
by Invitrogen (Thermo Fisher Scientific, Inc.) The complementary
sequence for miR-106b-3p seed sequence in ZNRF3 3'-UTR was mutated
and named as pMIR-ZNRF3-MUT plasmid. The 10 nM pMIR-ZNRF3-MUT or
pMIR-ZNRF3-WT and PRL-TK-Renilla were co-transfected in
KYSE150 and ECA-109 cells with 10 nM miR-106b-3p mimic and negative
control using Lipofectamine® 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). After 48 h, cells were lysed and
assayed for luciferase activity using a dual luciferase reporter
assay (Promega Corporation). Normalized luciferase activity was
reported as luciferase activity/Renilla luciferase
activity.
Statistical analysis
All data are presented as the mean ± standard
deviation. SPSS 21.0 statistical software (IBM Corp., Armonk, NY,
USA) was used to explore the statistical analysis. Comparisons
between two groups were conducted using two-tailed Student's t-test
and multiple group comparisons were conducted via one-way analysis
of variance with Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-106b-3p is upregulated in ESCC
tissues and cell lines
The expression of miR-106b-3p in 50 paired ESCC
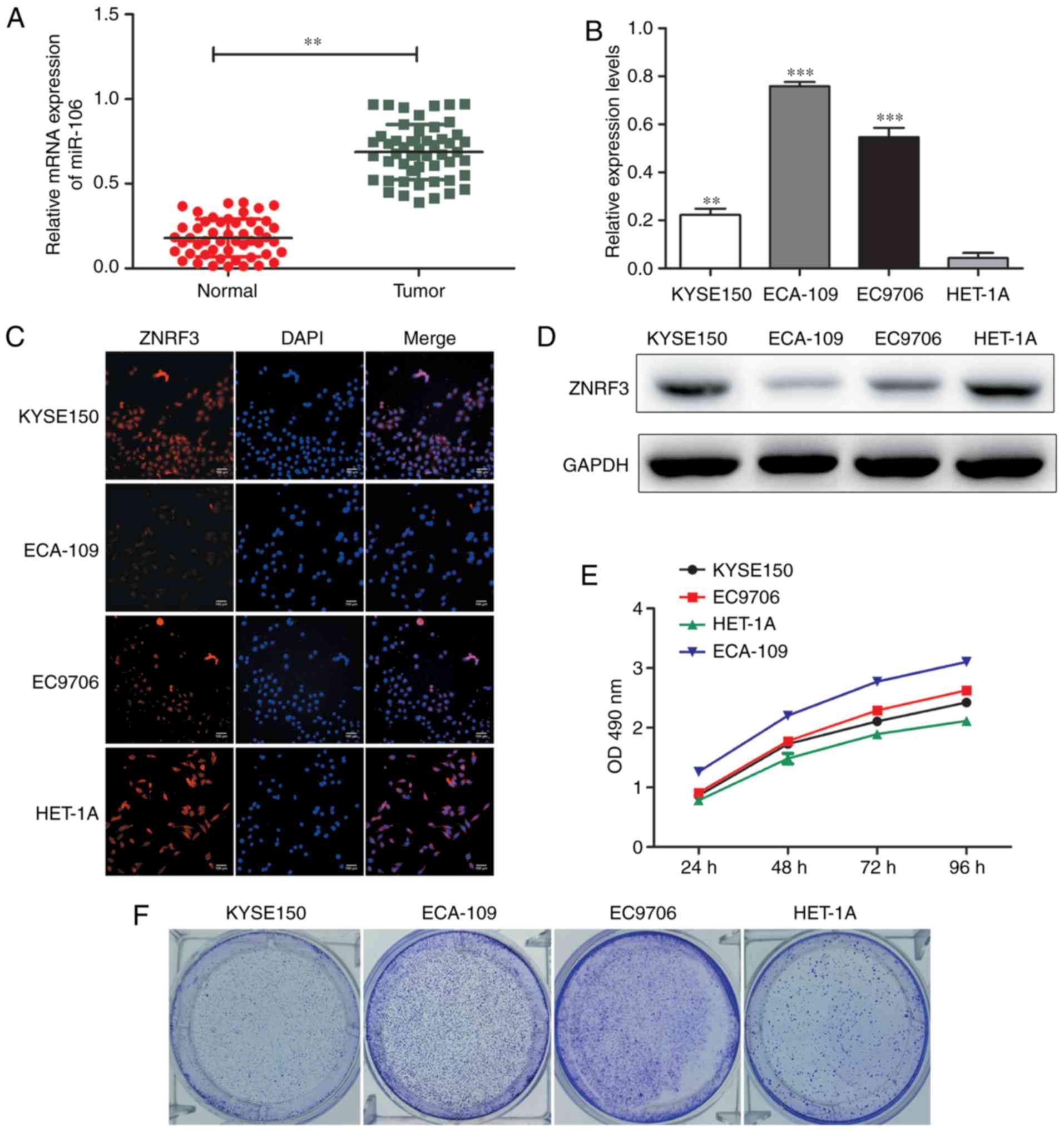
tissues and non-tumor tissues was detected by RT-qPCR (Fig. 1A). We found tThat the expression
levels of miR-106b-3p were significantly up-regulated in ESCC
tissues compared to with non-tumor tissues. Furthermore, the
expression of miR-106b-3p in ESCC cell lines (KYSE150, ECA-109,
EC9706) was significantly increased compared with the normal
epithelial cell line HET-1A (Fig.
1B). ZNRF3 expression was determined by western blot analysis
and immunofluorescence (Fig. 1C and
D). The proliferation abilities of cell lines were performed by
MTT and colony formation assays (Fig.
1E and F). These results suggested that miR-106b-3p may
function as a regulator in the progression of ESCC.
miR-106b-3p promotes cell
proliferation
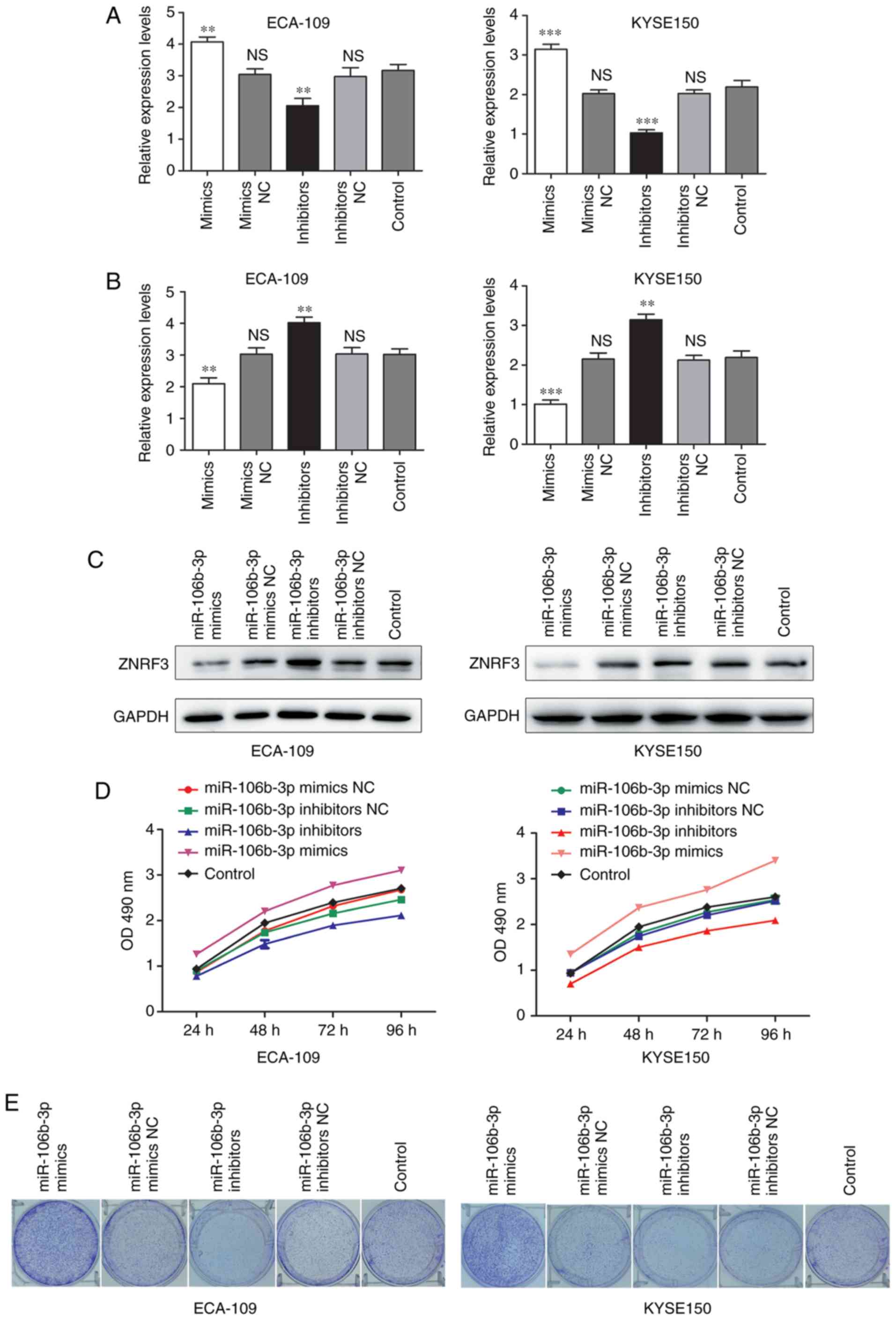
To characterize the function of miR-106b-3p on cell
proliferation in ESCC, miR-106b-3p mimics, inhibitors and
corresponding negative controls were synthesized and transfected
into KYSE150 and ECA-109 cells. The expression of miR-106b-3p was
determined by RT-qPCR (Fig. 2A)
and ZNRF3 expression was detected by RT-qPCR and western blot
(Fig. 2B and C). MTT assay was
used to examine the proliferation of KYSE150 and ECA-109 cells
(Fig. 2D); the data demonstrated
that the proliferation rate of cells was markedly increased by the
transfection of miR-106b-3p mimics compared with the negative
control, while that of cells in the miR-106b-3p inhibitors group
was decreased. Colony formation assays further confirmed the
proliferative function of miR-106b-3p in ESCC cells (Fig. 2E). These results indicated that
miR-106b-3p silencing could suppress the proliferation of ESCC
cells.
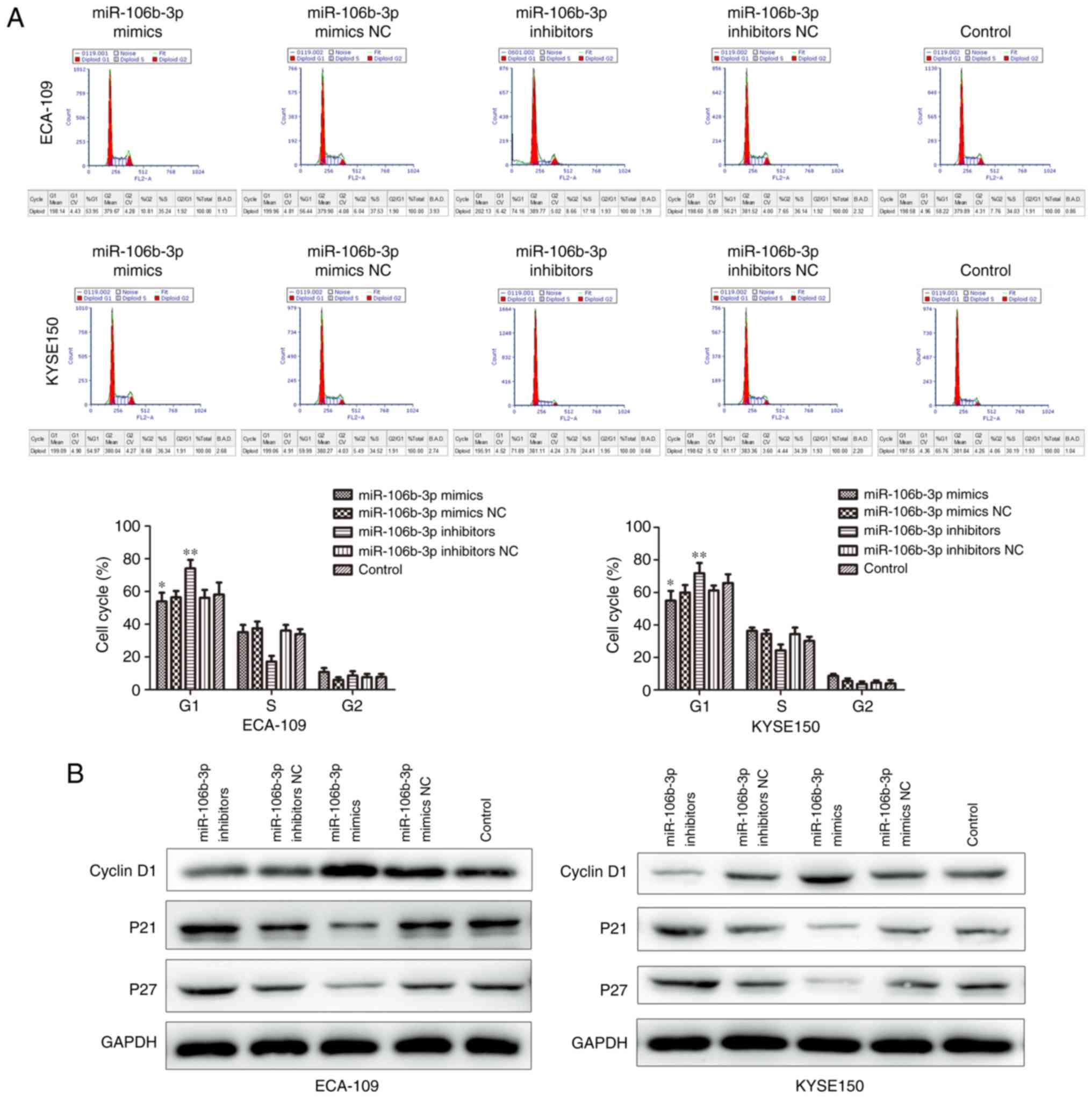
Flow cytometry was used to analyze cell cycle
distribution in KYSE150 and ECA-109 cell lines following mimic and
inhibitor transfection. Downregulation of miR-106b-3p induced G1
cell cycle arrest, which was demonstrated the by reduced percentage
of S and G2/M and the increasing percentage of G1 (Fig. 3A). Additionally, p21 and p27 were
increased by miR-106b-3p inhibitor, and cyclin D1 was decreased by
miR-106b-3p inhibitor (Fig. 3B).
Collectively, these data demonstrated that miR-106b-3p had a
growth-stimulative function in ESCC.
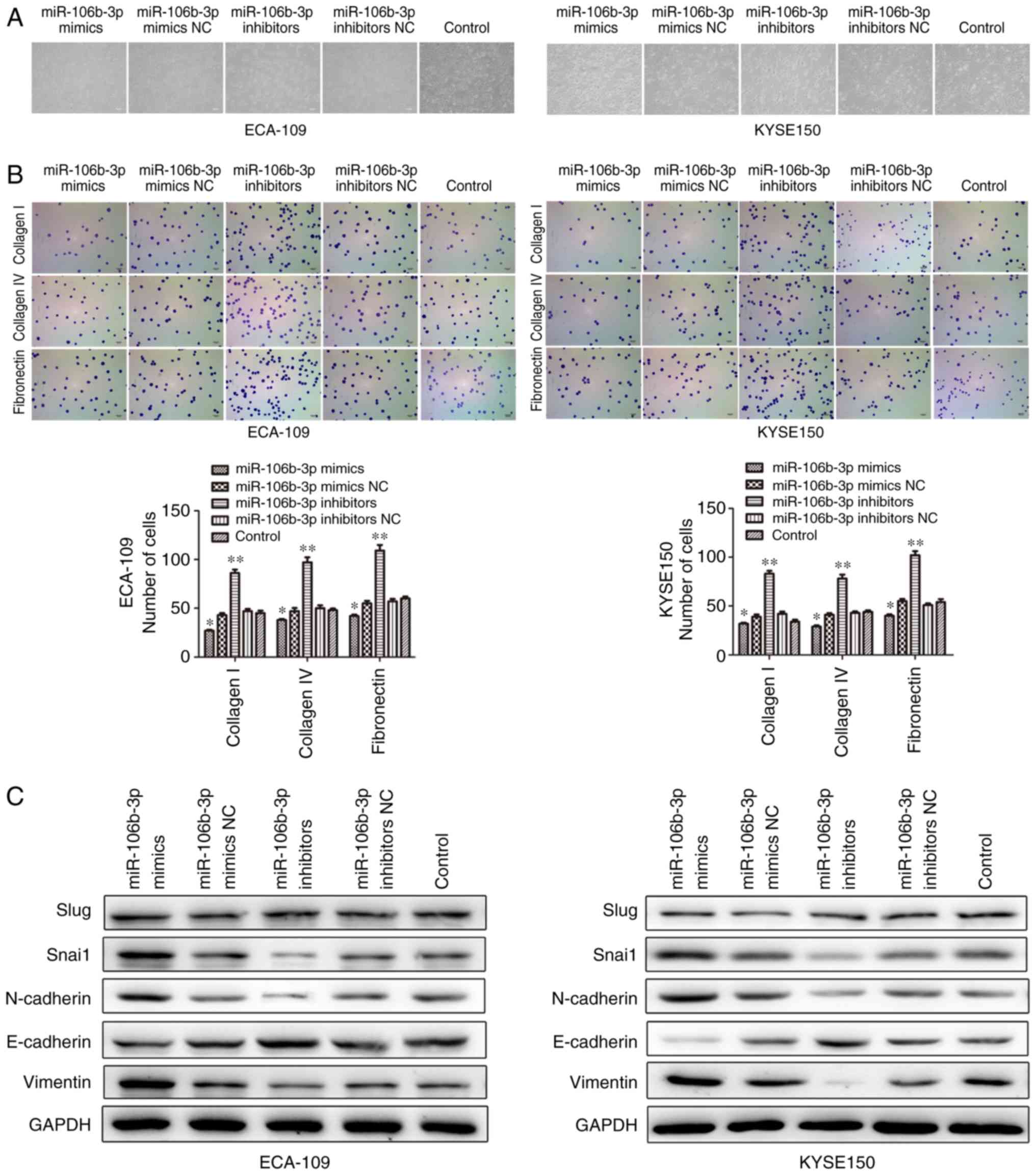
Downregulation of miR-106b-3p suppresses
the adhesion and EMT of ESCC cells in vitro
To functionally investigate the biological role of
miR-106b-3p in ESCC, gain-of-function experiments were performed.
Considering the implication of miR-106b-3p in cell motility and
adhesion, an adhesion assay was conducted. Cell morphology was
observed using a microscope (Fig.
4A) and adhesion ability was detected by adhesion assay. The
results showed that upregulation of miR-106b-3p exhibited a
significant reduction in attachment to collagen I, collagen IV and
fibronectin, which are essential components of the extracellular
matrix (Fig. 4B).
EMT can change cell invasion and migration ability.
To identify whether miR-106b-3p can affect EMT, changes in the
expressions of common EMT markers in miR-106b-3p-expressing cells
were determined by western blot analysis (Fig. 4C). Cells transfected with
miR-106b-3p mimics presented a mesenchymal-like phenotype, and the
mesenchymal marker expressions (Vimentin and N-cadherin) were
enhanced. By contrast, decreased protein expression of Vimentin and
N-cadherin was observed in cells transfected with miR-106b-3p
inhibitors compared with the control group. In addition,
miR-106b-3p inhibitors markedly increased the expression of the
epithelial marker E-cadherin (Fig.
4C). However, miR-106b-3p inhibitor did not affect Slug
expression levels and had suppressive effect on Snail levels in
KYSE150 and ECA-109 cell lines. Collectively, the results indicated
that miR-106b-3p promotes the invasive characteristics of ESCC
cells.
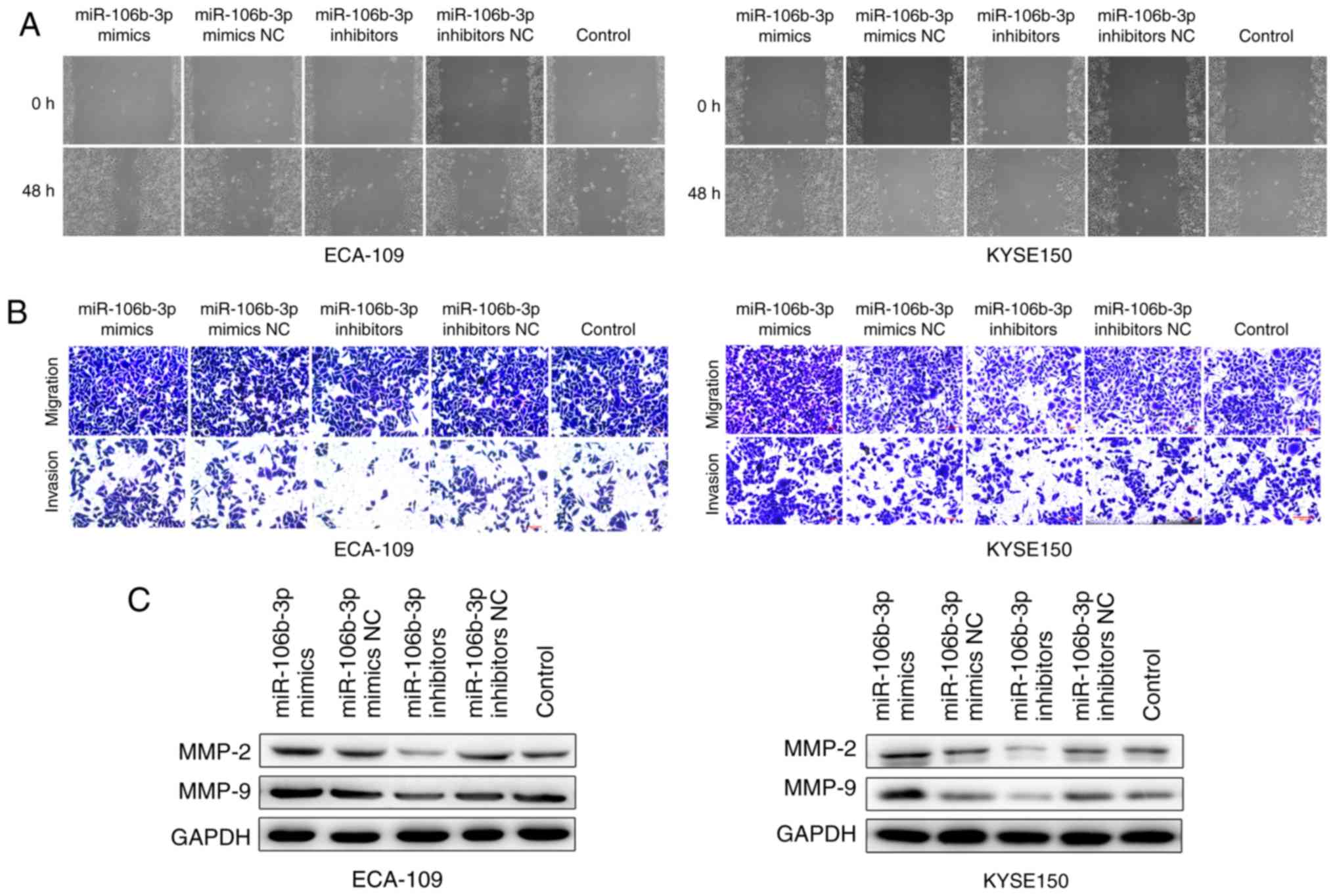
miR-106b-3p induces the migration and
invasion of ESCC
Wound healing and Transwell assays were performed to
further investigate the effects of miR-106b-3p on migration and
invasion of ESCC cells. The wound healing assay demonstrated that
miR-106b-3p expression was positively associated with the rates of
wound healing, and silencing of miR-106b-3p expression with slower
healing (Fig. 5A). In addition,
in vitro migration and invasion assays demonstrated that
miR-106b-3p inhibitor significantly attenuated the migration and
invasion abilities of KYSE150 cells (Fig. 5B). Similar results were obtained
using the ECA-109 cell line. These results demonstrated that
miR-106b-3p promoted ESCC cell migration and invasion. To
investigate cell invasive capacity, proteins associated with
invasion were studied in KYSE150 and ECA-109 cells. MMP-2 and MMP-9
expression was increased in cells transfected with miR-106b-3p
mimics, as detected by western blot analysis (Fig. 5C). The results indicated that
miR-106b-3p may promote the migration and invasion of ESCC
cells.
ZNRF3 is a direct target of
miR-106b-3p
Based on miR target analysis using the websites
TargetScan and miRanda, ZNRF3 identified as a potential target gene
of miR-106b-3p (Fig. 6A). The
predicted binding of miR-106b-3p to the ZNRF3 3'UTR was validated
in a luciferase reporter assay. The reporter plasmids were
co-transfected with miR-106b-3p mimics or NC in KYSE150 and ECA-109
cells. The luciferase activity assay revealed that miR-106b-3p
significantly decreased the luciferase activity of the wild type
3'UTR of ZNRF3, without effect on the mutant (Fig. 6B). Futhermore, immunofluorescence
analysis was used to observe the expression of ZNRF3 regulated by
miR-106b-3p in KYSE150 and ECA-109 cells. The results demonstrated
that overexpression of miR-106b-3p reduced the expression of ZNRF3
in ESCC cells (Fig. 6C). These
results suggested that ZNRF3 was a direct target gene of
miR-106b-3p.
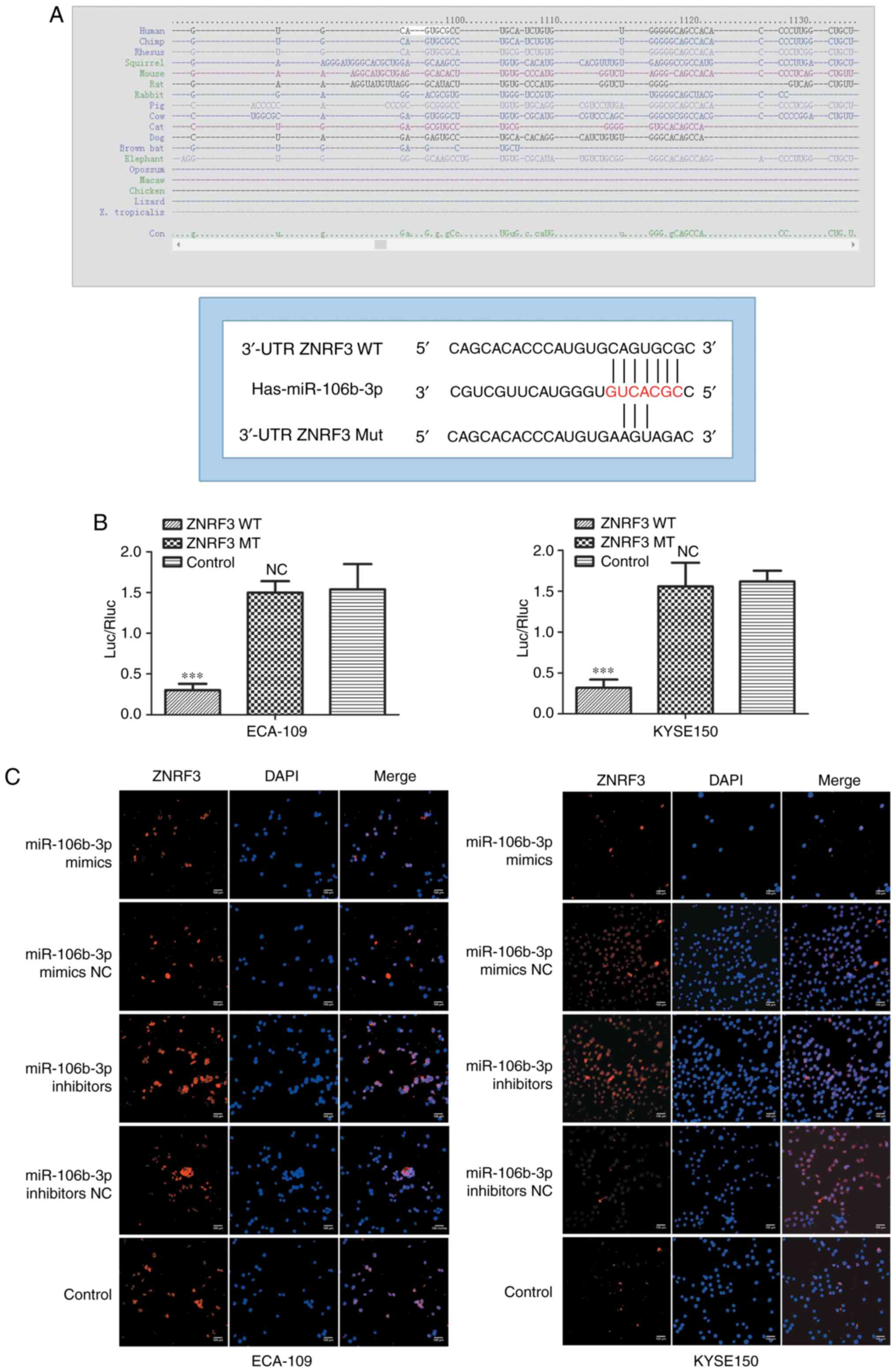 | Figure 6ZNRF3 is a direct target of
miR-106b-3p. (A) Computer prediction of the 3'-UTR of ZNRF3 mRNA
contained a target site for miR-106b-3p. (B) The pMIR-ZNRF3-MUT or
pMIR-ZNRF3-WT and PRL-TK-Renilla were co-transfected in
KYSE150 and ECA-109 cells with miR-106b-3p mimic and negative
control. Luciferase activity assay revealed that miR-106b-3p mimics
suppressed WT ZNRF3 3'-UTR luciferase activity, while it had no
effect on MUT ZNRF3 3'-UTR luciferase activity compared to control
in KYSE150 and ECA-109 cells. (C) Effects of miR-106b-3p
dysregulation on endogenous ZNRF3 expression, which was analyzed by
immunofluorescence. Data are from three independent experiments and
are presented as the mean ± standard deviation.
***P<0.001 vs. control. UTR, untranslated region;
ZNRF3, zinc and ring finger 3; WT, wild type; miR, microRNA; MUT,
mutant; Luc/Rluc, luciferase/Renilla luciferase; NS, no statistical
difference vs. control; NC, negative control. |
miR-106b-3p promoted EMT in ESCC cells by
Wnt/β-catenin signaling pathway
To further elucidate the underlying molecular
mechanisms of miR-106b-3p on promoting cell EMT, western blot and
RT-qPCR assays were performed to explore mRNA and protein levels of
factors associated with the Wnt/β-catenin signaling pathway. As
shown in Fig. 7, the result of
RT-qPCR assay demonstrated that the mRNA expressions of PI3K, AKT,
Wnt and β-catenin were upregulated in the miR-106b-3p mimics group,
while decreased in miR-106b-3p inhibitors group compared with
control group. The result of western blot assay showed that p-PI3K,
p-AKT, Wnt and β-catenin were all upregulated in the miR-106b-3p
mimics group compared with the levels in the control group
(Fig. 8). Inversely, silencing of
miR-106b-3p increased p-GSK3 levels. In summary, the results
demonstrated that miR-106b-3p activated Wnt/β-catenin signaling via
targeting of ZNRF3 and subsequently promoted EMT.
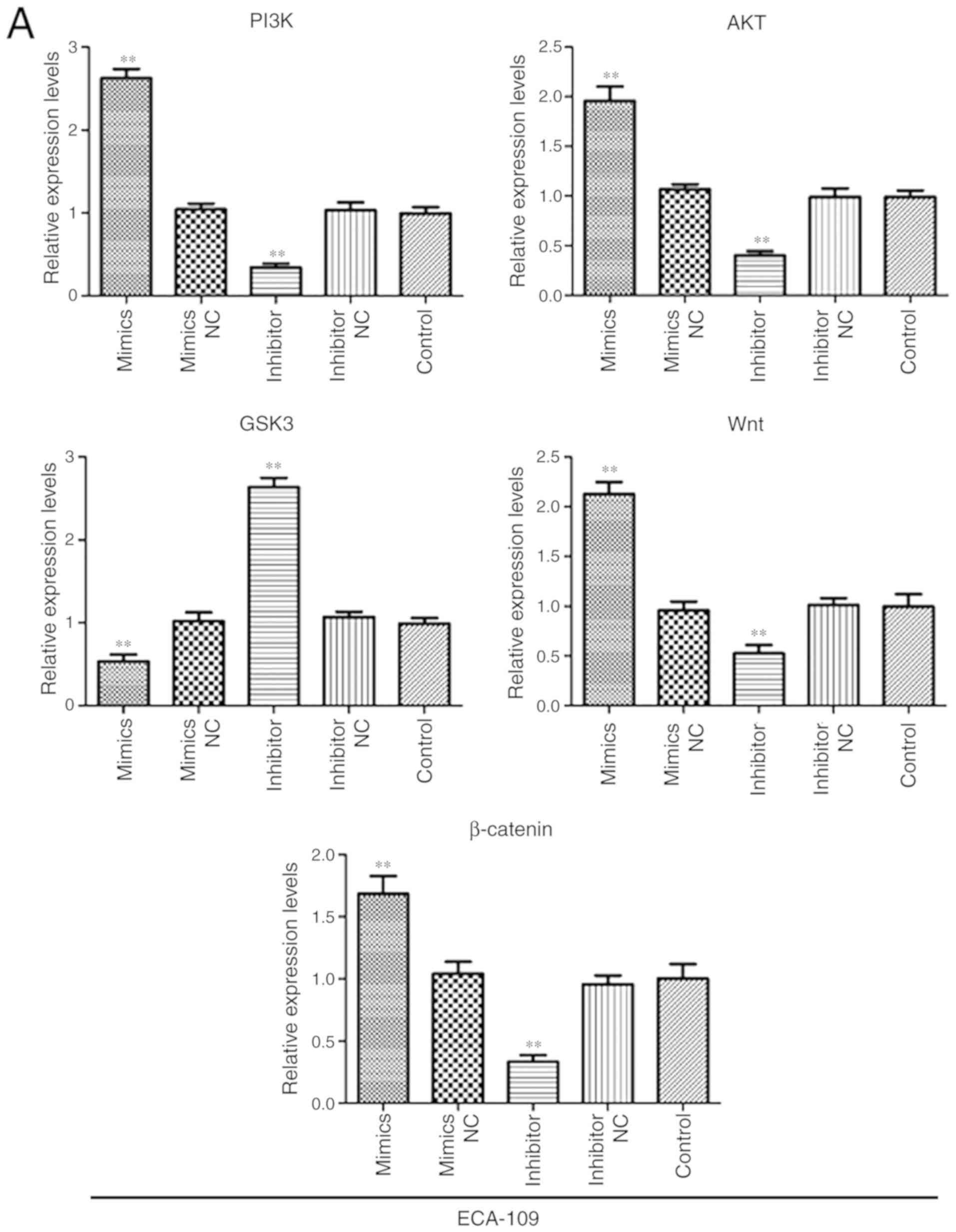 | Figure 7miR-106b-3p activates the
Wnt/β-catenin signaling pathway in ESCC. The gene expressions of,
PI3K, AKT, Wnt, GSK3 and β-catenin in (A) ECA-109 and (B) KYSE150
cells transfected with miR-106b-3p mimics, miR-106b-3p mimics NC,
miR-106b-3p inhibitors, miR-106b-3p inhibitors NC or control using
RT-PCR analysis. The data are presented as the mean ± standard
deviation of three independent experiments. *P<0.05
and **P<0.01 vs. control. miR, microRNA; NC, negative
control; p-, phospho-; PI3K, phosphoinositide 3-kinase; AKT,
protein kinase B; GSK3, glycogen synthase kinase-3. |
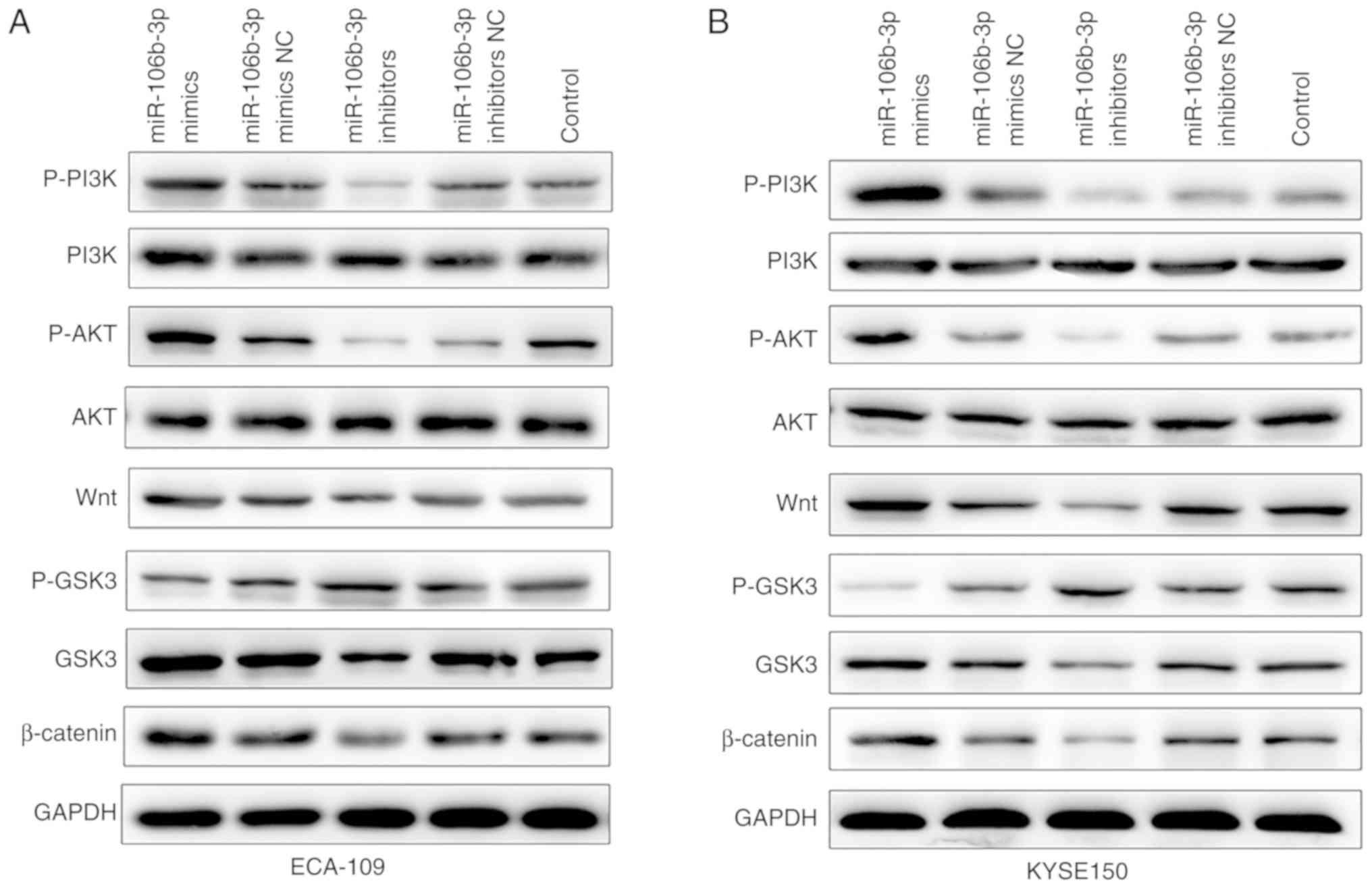 | Figure 8Protein levels of p-PI3K, PI3K,
p-AKT, AKT, Wnt, p-GSK3, GSK3 and β-catenin in (A) ECA-109 and (B)
KYSE150 cells transfected with miR-106b-3p mimics, miR-106b-3p
mimics NC, miR-106b-3p inhibitors, miR-106b-3p inhibitors NC or
control using western blot analysis. GAPDH was used as the internal
control. miR, microRNA; NC, negative control; p-, phospho-; PI3K,
phosphoinositide 3-kinase; AKT, protein kinase B; GSK3, glycogen
synthase kinase-3. |
Discussion
Despite the advancements in treatment options,
improvements in the survival of patients with ESCC have been
limited due to lack of early detection. Recent evidence has
indicated that alterations of specific miRNAs may be promising
translational biomarker candidates in cancer. Furthermore, the
identification of tumor-associated miRNAs and their direct target
genes is critical for understanding the pathogenesis and biological
significance of miRNAs in carcinogenesis and progression of ESCC,
which may provide a novel prognostic and therapeutic targets for
patients with ESCC.
The data of the current study demonstrated that the
levels of miR-106b-3p were significantly upregulated in ESCC
tissues and cells compared with adjacent non-tumor tissues and
normal human epithelial cells, respectively. Overexpression of
miR-106b-3p significantly promoted the cell proliferation and
invasion of ESCC cells. Conversely, miR-106b-3p inhibited the
proliferation and invasion of ESCC cells in other cancer types
(24,25). The data indicated that decreased
expression of miR-106b-3p may have a critical role in the
progression of ESCC.
In order to explore the mechanisms of migration and
invasion by miR-106b-3p of ESCC cells, the potential target genes
of miR-106b-3p were predicted. In a previous study, DAB2 clathrin
adaptor protein was confirmed as a target of miR-106b of cervical
cancer (26,27). In addition, miR-106b was reported
to be expressed at a lower level in osteosarcoma cells and target
the high mobility group AT-hook 2 gene to inhibit the cell
progression (28). Furthermore,
Zhang et al (29) reported
that miR-106b was upregulated in colorectal cancer and promoted
cell invasion and migration by targeting DLC1 Rho GTPase activating
protein. In the current study, ZNRF3 was identified as a direct
target of miR-106b-3p in a luciferase reporter assay. This
observation was validated by the increased ZNRF3 mRNA and protein
expressions in ESCC cells transfected with miR-106b-3p inhibitor.
ZNRF3 is part of the E3 ubiquitin ligase family, which negatively
regulates Wnt signaling. ZNRF3 was associates with the Wnt receptor
complex and inhibits Wnt signaling by promoting the turnover of
Frizzled and LDL receptor related protein 6 receptors. Accumulating
data have strongly suggested that ZNRF3 is involved in cancer
development and exhibits low expression in various human cancer
types (19-21).
The role of EMT in tumor metastasis has received
increasing attention and is a critical process providing
epithelial-derived tumor cells with the increased migration and
invasive abilities, contributing to tumor metastasis (30-32). EMT is a critical process by which
epithelial cells lose their epithelial morphology and acquire a
mesenchymal phenotype, characterized by the decreased expression of
epithelial proteins, including E-cadherin and tight junction
protein ZO-1, and the increased expression of mesenchymal proteins,
including Vimentin and fibronectin (22,30,31). Snail and Slug are members of zinc
finger family and have a central transcriptional role in the
regulation of EMT by directly binding to specific E-boxes on the
E-cadherin promoter (33,34). Furthermore, miR-106b-3p promoted
EMT by downregulating E-cadherin expression and inducing the
expression levels of Snail, N-cadherin and Vimentin in ESCC
cells.
Dysregulation of the Wnt/β-catenin pathway has been
observed in various cancer typess and to regulate cancer metastasis
(35,36). Furthermore, ZNRF3, which was was
identified as a direct target of miR-106b-3p in the current study,
is a negative regulator of Wnt/β-catenin signaling. The
Wnt/β-catenin signaling pathway has an important role in the
maintenance of stem cell properties and tumorigenesis (37,38). Additionally, the nuclear
accumulation of β-catenin is a key step in the activated Wnt
signaling pathway (39). The data
of the current study revealed that overexpression of miR-106b-3p
markedly upregulated the expression levels of Wnt, GSK3 and
β-catenin, and reduced the p-GSK3 level. Thus, these results
suggested for the first time that miR-106b-3p has a role in EMT via
regulation of the Wnt/β-catenin signaling pathway in ESCC.
In conclusion, the findings of the present study
indicated that miR-106b-3p was upregulated, while ZNRF3 was
downregulated in ESCC. It was demonstrated that ZNRF3 was a direct
target of miR-106b-3p and ectopic expression of miR-106b-3p
promoted cell proliferation, migration and invasion capacity of
ESCC cells by targeting ZNRF3. Further research provided evidence
that miR-106b-3p promoted EMT and activated the Wnt/β-catenin
signaling pathway. Taken together, these results suggested that
miR-106b-3p has an important role in invasion and EMT of ESCC
cells.
To the best of our knowledge, this is the first
study to investigate miR-106b-3p in ESCC. These findings expand the
understanding of the molecular mechanisms underlying miR-106b-3p
function and proliferation, invasion and EMT of ESCC, and also
suggest a potential prognostic marker and therapeutic target in
human ESCC.
Funding
This project was funded by the National Nature Fund
(grant no. 81773356).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
GQ and CX conceived and designed the study. CD and
YH performed the experiments. GQ and JS wrote the manuscript and
contributed to the analysis or interpretation of data for the work.
All authors have read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy and integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Nanjing Medical University and every patient provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The present study was supported by Dr Zuo Bin,
Master of Shen Yu, Dr Jian-feng Yang, Dr Wong Zhen, Master of Gao
Fengqing (Hematological Disease Engineering Center of Ministry of
Education of Soochow University, Suzhou, China); Professor Li
Yunman (Department of Physiology, China Pharmaceutical University,
Nanjing, China); Dr Tian Tian (Basic Neurology Teaching and
Research Department of Nanjing Medical University, Nanjing, China);
Dr Zhang Lizhu, Wang Zhen-dong (Nanjing Bairui Biotechnology
Company, Nanjing, China).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mccann J: Esophageal cancers: Changing
character, increasing incidence. J Natl Cancer Inst. 91:497–498.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Y, Da F, Xi J, Ji Z, Liu T, Ma Y,
Zhao Y, Dong L, Wang Q and Shen X: Expression and clinical
significance of UCH37 in human esophageal squamous cell carcinoma.
Dig Dis Sci. 57:2310–2317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kausar T, Ahsan A, Hasan MR, Lin L, Beer
DG and Ralhan R: Sperm protein 17 is a novel marker for predicting
cisplatin response in esophageal squamous cancer cell lines. Int J
Cancer. 126:1494–1503. 2010.
|
|
5
|
Qin YR, Tang H, Xie F, Liu H, Zhu Y, Ai J,
Chen L, Li Y, Kwong DL, Fu L and Guan XY: Characterization of
tumor-suppressive function of SOX6 in human esophageal squamous
cell carcinoma. Clin Cancer Res. 17:46–55. 2011. View Article : Google Scholar
|
|
6
|
Zhang JG, Shi Y, Hong DF, Song M, Huang D,
Wang CY and Zhao G: MiR-148b suppresses cell proliferation and
invasion in hepatocellular carcinoma by targeting WNT1/β-catenin
pathway. Sci Rep. 5:80872015. View Article : Google Scholar
|
|
7
|
Wang Y and Lee CG: MicroRNA and
cancer-focus on apoptosis. J Cell Mol Med. 13:12–23. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cai K, Wang Y and Bao X: MiR-106b promotes
cell proliferation via targeting RB in laryngeal carcinoma. J Exp
Clin Cancer Res. 30:732011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ratert N, Meyer HA, Jung M, Lioudmer P,
Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert
S and Jung K: miRNA profiling identifies candidate mirnas for
bladder cancer diagnosis and clinical outcome. J Mol Diagn.
15:695–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li BK, Huang PZ, Qiu JL, Liao YD, Hong J
and Yuan YF: Upregulation of microRNA-106b is associated with poor
prognosis in hepatocellular carcinoma. Diagn Pathol. 9:2262014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Slaby O, Jancovicova J, Lakomy R, Svoboda
M, Poprach A, Fabian P, Kren L, Michalek J and Vyzula R: Expression
of miRNA-106b in conventional renal cell carcinoma is a potential
marker for prediction of early metastasis after nephrectomy. J Exp
Clin Cancer Res. 29:902010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
J Clin Invest. 139:871–890. 2009.
|
|
13
|
Dieudonné FX, Marion A, Marie PJ and
Modrowski D: Targeted inhibition of T-cell factor activity promotes
syndecan-2 expression and sensitization to doxorubicin in
osteosarcoma cells and bone tumors in mice. J Bone Miner Res.
27:2118–2129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Q, Cao HZ and Zheng PS: LGR5 promotes
the proliferation and tumor formation of cervical cancer cells
through the Wnt/β-catenin signaling pathway. Oncotarget.
5:9092–9105. 2014.PubMed/NCBI
|
|
16
|
Hua HW, Jiang F, Huang Q, Liao Z and Ding
G: MicroRNA-153 promotes Wnt/β-catenin activation in hepatocellular
carcinoma through suppression of WWOX. Oncotarget. 6:3840–3847.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou DS, Wang HB, Zhou ZG, Zhang YJ, Zhong
Q, Xu L, Huang YH, Yeung SC, Chen MS and Zeng MS: TACC3 promotes
stemness and is a potential therapeutic target in hepatocellular
carcinoma. Oncotarget. 6:24163–24177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zebisch M, Xu Y, Krastev C, MacDonald BT,
Chen M, Gilbert RJ, He X and Jones EY: Structural and molecular
basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by
the Wnt agonist R-spondin. Nat Commun. 4:27872013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Y, Lan J, Wang W, Shi Q, Lan Y, Cheng
Z and Guan H: ZNRF3 acts as a tumour suppressor by the Wnt
signalling pathway in human gastric adenocarcinoma. J Mol Histol.
44:555–563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qin H, Cai A, Xi H, Yuan J and Chen L:
ZnRF3 induces apoptosis of gastric cancer cells by antagonizing Wnt
and Hedgehog signaling. Panminerva Med. 57:167–175. 2015.PubMed/NCBI
|
|
21
|
Deng X, Wu B, Xiao K, Kang J, Xie J, Zhang
X and Fan Y: MiR-146b-5p promotes metastasis and induces
epithelial-mesenchymal transition in thyroid cancer by targeting
ZNRF3. Cell Physiol Biochem. 35:71–82. 2015. View Article : Google Scholar
|
|
22
|
Zhang JP, Feng LL, Zhan HF, et al: Stromal
cell-derived factor 1 alpha induce epithelial mesenchymal
transition of Hela cells. Eur J Gynaecol Oncol. 6:933–937.
2017.
|
|
23
|
Wang Z, Wang Y, Ren H, Jin Y and Guo Y:
ZNRF3 inhibits the invasion and tumorigenesis in nasopharyngeal
carcinoma cells by inactivating the Wnt/β-catenin pathway. Oncol
Res. 25:571–577. 2017. View Article : Google Scholar
|
|
24
|
Yau WL, Lam CS, Ng L, Chow AK, Chan ST,
Chan JY, Wo JY, Ng KT, Man K, Poon RT and Pang RW: Over-expression
of miR-106b promotes cell migration and metastasis in
hepatocellular carcinoma by activating epithelial-mesenchymal
transition process. PLoS One. 8:e578822013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang TS, Yang XH, Chen X, Wang XD, Hua J,
Zhou DL, Zhou B and Song ZS: MicroRNA-106b in cancer-associated
fibroblasts from gastric cancer promotes cell migration and
invasion by targeting PTEN. FEBS Lett. 588:2162–2169. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun C, Yao X, Jiang Q and Sun X: miR-106b
targets DAB2 to promote hepatocellular carcinoma cell proliferation
and metastasis. Oncol Lett. 16:3063–3069. 2018.PubMed/NCBI
|
|
27
|
Piao J, You K, Guo Y, Zhang Y, Li Z and
Geng L: Substrate stiffness affects epithelial-mesenchymal
transition of cervical cancer cells through miR-106b and its target
protein DAB2. Int J Oncol. 50:2033–2042. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He QY, Wang GC, Zhang H, Tong DK, Ding C,
Liu K, Ji F, Zhu X and Yang S: miR-106a-5p suppresses the
proliferation, migration, and invasion of osteosarcoma cells by
targeting HMGA2. DNA Cell Biol. 35:506–520. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang GJ, Li JS, Zhou H, Xiao HX, Li Y and
Zhou T: MicroRNA-106b promotes colorectal cancer cell migration and
invasion by directly targeting DLC1. J Exp Clin Cancer Res.
34:732015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kalluri R and Neilson EG:
Epithelial-mesenchymal transition and its implications for
fibrosis. J Clin Invest. 112:1776–1784. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hugo H, Ackland ML, Blick T, Lawrence MG,
Clements JA, Williams ED and Thompson EW: Epithelial-mesenchymal
and mesenchymal-epithelial transitions in carcinoma progression. J
Cell Physiol. 213:374–383. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moreno-Bueno G, Cubillo E, Sarrió D,
Peinado H, Rodríguez-Pinilla SM, Villa S, Bolós V, Jordá M, Fabra
A, Portillo F, et al: Genetic profiling of epithelial cells
expressing E-cadherin repressors reveals a distinct role for Snail,
Slug, and E47 factors in epithelial-mesenchymal transition. Cancer
Res. 66:9543–9556. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Castro Alves C, Rosivatz E, Schott C,
Hollweck R, Becker I, Sarbia M, Carneiro F and Becker KF: Slug is
overexpressed in gastric carcinomas and may act synergistically
with SIP1 and Snail in the down-regulation of E-cadherin. J Pathol.
211:507–515. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View
Article : Google Scholar
|
|
36
|
Choi YS, Shim YM, Kim SH, Son DS, Lee HS,
Kim GY, Han J and Kim J: Prognostic significance of E-cadherin and
beta-catenin in resected stage I non-small cell lung cancer. Eur J
Cardiothorac Surg. 24:441–449. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Leung CO, Mak WN, Kai AK, Chan KS, Lee TK,
Ng IO and Lo RC: Sox9 confers stemness properties in hepatocellular
carcinoma through Frizzled-7 mediated Wnt/β-catenin signaling.
Oncotarget. 7:29371–29386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yuan X, Sun X, Shi X, Wang H, Wu G, Jiang
C, Yu D, Zhang W, Xue B and Ding Y: USP39 promotes colorectal
cancer growth and metastasis through the Wnt/β-catenin pathway.
Oncol Rep. 37:2398–2404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mao Y, Xu J, Li Z, Zhang N, Yin H and Liu
Z: The role of nuclear β-catenin accumulation in the Twist2-induced
ovarian cancer EMT. PLoS One. 8:e782002013. View Article : Google Scholar
|