Introduction
Myocardial infarction (MI), also known as a heart
attack, is one of the leading causes of mortality globally
(1). Death of cardiomyocytes,
angiogenesis, fibrosis, hypertrophy and contractile dysfunction
during MI result in impaired cardiac function (2-5).
In particular, cardiac fibrosis following MI serves a key function
in regulating cardiac function (6). In addition, it has been identified
as a key factor in the incidence of heart failure, as excessive
fibrosis results in ventricular dilation, infarct expansion and
hypertension (7). For this
reason, inhibition of cardiac fibrosis progression in infarcted
hearts may preserve cardiac function and prevent heart failure.
However, at present, there is no efficient therapy available
(8).
Exosomes, membrane-bound vesicles of 30-100 nm, are
secreted by various cell types and are present in the majority of
body fluids (9,10). An exosome is a 'nanosphere' with a
bilayered membrane, containing various types of lipids and
proteins. For example, the majority of exosomes contain membrane
transport and fusion proteins, heat-shock proteins, multivesicular
body biogenesis proteins [including programmed cell death 6
interacting protein (Alix)], tetraspanin [including cluster of
differentiation (CD)9, CD63 and CD81] and endosomal/lysosomal
proteins [including lysosomal associated membrane protein 2
(LAMP2)]. In addition, exosomes are comprised of different types of
lipids, including cholesterol, sphingolipids, phosphoglycerides,
ceramides and saturated fatty acid chains. These exosome
compositions function as cargo for cell-to-cell communication
(11,12). Lately, exosomes have emerged as a
promising drug delivery system with lower toxicity compared with
conventional delivery systems and high therapeutic efficacy
(13). However, two of the major
challenges in developing exosome-based drug delivery methods are
the scale up procedure and utilization without side effects
(14). As human peripheral blood
is widely used for transfusion and is easily obtained, the use of
human peripheral blood-derived exosomes as a microRNA (miRNA/miR)
delivery system, has substantial potential as a therapeutic
tool.
Previous studies have demonstrated that certain
miRNAs function as essential modulators in a number of
physiological and pathological MI processes, including fibrosis
(15-17). Among various miRNAs, miR-21 serves
a crucial role in the development of cardiac fibrosis in response
to MI through numerous target genes, including SMAD family member 7
(Smad7), sprout RTK signaling antagonist 1 and phosphatase and
tensin homolog (PTEN) (18-20). Thus, the function of miR-21 in
regulating gene expression during fibrosis makes miR-21 an ideal
candidate for therapeutic applications.
The present study aimed to investigate whether
exosomes isolated from human peripheral blood may function as
efficient vehicles for miRNA delivery. To achieve this aim, miR-21,
a well-known therapeutic target for cardiac diseases, was
transfected into human peripheral blood-derived exosomes and
whether the exosomes may deliver the miRNA in in vitro and
in vivo models was evaluated. Furthermore, whether exosomes
loaded with an miR-21 inhibitor may modulate the fibrotic
remodeling in an MI mouse model was investigated. Therefore, the
present study may provide a potential, effective approach for the
treatment of cardiac disease.
Patients and methods
Exosome isolation and
characterization
Human peripheral blood was obtained from three
patients without serious or progressive cardiac disease at Yonsei
University Health System (Seoul, Korea) from August 2015 to July
2016; the clinical profile of the patients is presented in Table I. Written informed consent was
obtained from all patients and the study protocol was ethically
approved by the local ethics committee (Institutional Review Board
of Severance Hospital (Seoul, Korea) of the Yonsei University
Health System; approval no. YUMC 4-2011-0872). Exosomes were
isolated from human peripheral blood using the Exoquick exosome
precipitation kit (System Biosciences, Palo Alto, CA, USA)
according to the manufacturer's protocol. Briefly, human peripheral
blood was centrifuged at 3,000 × g for 15 min at 4°C to remove
cells and cellular debris. Exoquick Exosome Precipitation Solution
(63 µl) was added to 250 µl supernatant, transferred
to a sterile tube, and incubated for 12 h at 4°C. Following
incubation, the mixtures were centrifuged at 1,500 × g for 30 min
at 4°C and the supernatant was removed by aspiration. Exosome
pellets were resuspended in 200 µl Dulbecco's
phosphate-buffered saline (PBS), aliquoted to avoid freeze and thaw
cycles, and stored at −80°C.
 | Table IClinical profile of patients from
whom exosomes were isolated. |
Table I
Clinical profile of patients from
whom exosomes were isolated.
| Case | Sex/age | Diagnosis | Medication |
|---|
| 1 | Female/66 | Atrial flutter | Amiodarone |
| 2 | Female/77 | Atrial flutter,
hypertension | Verapamil |
| 3 | Male/67 | Paroxysmal
supraventricular tachycardia, hypertension | Perindopril |
For transmission electron microscopy (TEM), exosomes
were prepared in PBS. The samples were adsorbed to a
Formvar-carbon-coated electron microscope grid (Leica Microsystems,
Inc., Buffalo Grove, IL, USA) for ~1 min and the grid surface was
then touched for 15 sec with 2% uranyl acetate. Exosome size and
morphology were observed using a JEM-1011 electron microscope (JEOL
Ltd., Tokyo, Japan).
Nanopaticle tracking analysis (NTA) was performed to
measure the size distribution and concentration of exosomes using a
NanoSight LM10 instrument (Malvern Instruments, Ltd., Malvern, UK).
The samples were captured for 60 sec at room temperature with a
flow rate of 50 ml/min, and the data was obtained using NTA v.3.2
software (Malvern Panalytical, Ltd., Malvern, UK), which allows
videos of particles moving under Brownian motion to be captured and
automatically analyzed to generate high-resolution size
distribution and concentration data.
Isolated exosomes were labeled using the PKH26 Red
Fluorescent Cell Linker kit (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) according to the manufacturer's protocol. Next, unlabeled
or PKH26 red fluorescent dye-labeled exosomes were added to H9C2
and HL-1 cell lines obtained from the American Type Culture
Collection (ATCC, Manassas, VA, USA) and incubated for 24 h at 37°C
in 5% CO2. The cardiac cells were washed twice with PBS,
fixed with 4% paraformaldehyde for 30 min at room temperature and
stained with DAPI for 5 min at room temperature. The uptake of
exosomes by cardiac cell lines was observed using a confocal
microscope (Carl Zeiss, Jena, Germany).
miRNA transfection into exosomes
The negative control miRNA (cat. no. SMC-2003),
miR-21 mimic (5′-UAGCUUAUCAGACUGAUGUUGA-3′; cat. no. SMM-002) and
miR-21 inhibitor (5′-AUCGAAUAGUCUGACUACAACU-3′; cat. no. SMI-002)
were obtained from Bioneer Corporation (Daejoen, Korea). The miRNAs
were transfected into human peripheral blood-derived exosomes using
the Exo-Fect™ Exosome Transfection kit (System Biosciences)
according to the manufacturer's protocol. Next, the exosomes were
washed twice using Amicon ultra centrifugation tubes (EMD
Millipore, Billerica, MA, USA) with cold PBS. The transfected
exosomes were directly used for further experiments.
Cell culture and hypoxia
H9C2 cardiac muscle cells, purchased from the
American Type Culture Collection (ATCC), were grown in Dulbecco's
modified Eagle's medium (WelGENE, Inc., Daegu, Korea) supplemented
with 10% fetal bovine serum (FBS; Young In Frontier Co., Ltd.,
Seoul, Korea) and 1% penicillin-streptomycin (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). HL-1 cardiac muscle cells were
obtained from ATCC. Cells were maintained in Complete Claycomb
Medium (SAFC Biosciences, Lenexa, KS, USA) supplemented with 10%
FBS, 1% penicillin-streptomycin, 100 µM norepinephrine
(Sigma-Aldrich; Merck KGaA) and 4 mM L-glutamine (Gibco; Thermo
Fisher Scientific, Inc.) in plates coated with 12.5 µg/ml
fibronectin and 0.02% gelatin (both Sigma-Aldrich; Merck KGaA). To
induce hypoxia, cells were placed in Forma™ Series II 3131
Water-Jacketed CO2 incubators (Thermo Fisher Scientific,
Inc.) with 94% N2, 5% CO2 and 1%
O2 at 37°C for 24 h. The corresponding normoxic control
cells were maintained at 37°C in a humidified incubator with 5%
CO2 and 21% O2. Following treatment with
transfected exosomes into H9C2 and HL-1 cells for 24 h at 37°C, the
cells were exposed to hypoxia for a further 24 h.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from cultured cells and mouse heart
tissues was extracted using the miRNeasy Mini kit (Qiagen GmbH,
Hilden, Germany) according to the manufacturer's protocols.
Complementary DNA was synthesized using the miRNA 1st-Strand cDNA
Synthesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA)
and the High Capacity cDNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. RT-qPCR was performed on an AriaMx
Realtime PCR System using the Brilliant III Ultra-Fast
SYBR®-Green QPCR Master Mix (both Agilent
Technologies, Inc.). The reaction conditions were as follows: 95°C
for 3 min followed by 40 cycles at 95°C for 3 sec and 55°C for 20
sec. The PCR primers were synthesized by Cosmo Genetech Co., Ltd.
(Daejeon, Korea) and are listed in Table II. The relative expression levels
of the miRNAs and the miR-21 target genes were calculated using the
2−ΔΔCq method (21)
and normalized against U6 and GAPDH, respectively.
 | Table IIPrimers used for reverse
transcription-quantitative polymerase chain reaction. |
Table II
Primers used for reverse
transcription-quantitative polymerase chain reaction.
| Gene name | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) |
|---|
| mmu-miR-21 |
GGGGTAGCTTATCAGACTGATG | |
| mmu-U6 |
GCTTCGGCAGCACATATACTAAAAT | |
| mmu-Smad7 |
GTGGCATACTGGGAGGAGAA |
GATGGAGAAACCAGGGAACA |
| mmu-PTEN |
GAGGGATAAAACACCATG |
AGGGGTAGGATGTGAACCAGTA |
| mmu-MMP2 |
CACATACAGGATCATTGGTTACAC |
ACAGGAAGGGGAACTTTGAGTA |
| mmu-Collagen I |
CATGTTCAGCTTTGTGGACCT |
GCAGCTGACTTCAGGGATGT |
| mmu- Collagen
III |
TCCCCTGGAATCTGTGAATC |
TGAGTCGAATTGGGGAGAAT |
| mmu-GAPDH |
GGGTTCCTATAAATACGGACTGC CC |
ATTTTGTCTACGGGACGA |
| rno-miR-21 C |
GCCGTAGCTTATCAGACTG | |
| rno-U6 |
CTCGCTTCGGCAGCACA | |
| rno-Smad7 |
TTTTGAGGTGTGGTGGG |
GAGGCAGTAAGACAGGGATGA |
| rno-PTEN |
AGAACTTATCAAACCCTT |
GTCCTTACTTCCCCAT |
| rno-MMP2 |
AGAAGGCTGTGTTCTTCGCA |
AAAGGCAGCGTCTACTTGCT |
| rno-GAPDH |
GGCACAGTCAAGGCTGAGAATG |
ATGGTGGTGAAGACGCCAGTA |
Western blot analysis
Total protein from cultured cells and mouse heart
tissues was extracted with radioimmunoprecipitation assay Lysis and
Extraction Buffer containing Protease and Phosphatase Inhibitor
Cocktail (both Thermo Fisher Scientific, Inc.). The protein
concentration of cell and tissue lysates was determined using a
Pierce™ 660 nm Protein Assay reagent (Thermo Fisher Scientific,
Inc.). Equal amounts of cell (20 µg) and tissue lysates (60
µg) were separated on a 10% Gradi-Gel™ II gradient gel
(Elpis Biotech, Inc., Daejeon, Korea) and transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were subsequently blocked for 1 h at room
temperature in tris-buffered saline (TBS)-0.1% Tween 20 (TBS-T)
containing 5% bovine serum albumin and incubated overnight at 4°C
with primary antibodies against the following proteins: Alix (cat.
no. sc-53540; 1:1,000), LAMP2 (cat. no. sc-18822; 1:1,000; both
Santa Cruz Biotechnology Inc., Dallas, TX, USA), CD9 (cat. no.
EXOAB-CD9A-1; 1:1,000), CD63 (cat. no. EXOAB-CD63A-1; 1:1,000; both
System Biosciences), CD81 (cat. no. sc-166029; 1:1,000), Smad7
(cat. no. sc-365846; 1:1,000), PTEN (cat. no. sc-7974; 1:1,000; all
Santa Cruz Biotechnology Inc.), matrix metalloproteinase 2 (MMP2;
cat. no. ab37150; 1:1,000; Abcam, Cambridge, UK) and GAPDH (cat.
no. sc-166574; 1:1,000; Santa Cruz Biotechnology Inc.). GAPDH was
used as the loading control. Following three washes with TBS-T, the
membranes were incubated with horseradish peroxidase-conjugated
goat anti-mouse IgG-HRP (cat. no. sc-2005; 1:5,000) and goat
anti-rabbit IgG-HRP (cat. no. sc-2004; 1:5,000; both Santa Cruz
Biotechnology Inc.) for 1 h at room temperature. Blotted proteins
were visualized using an enhanced chemiluminescence kit (Santa Cruz
Biotechnology Inc.).
Animal experiments
A total of 20 adult male C57BL/6 mice (8 weeks old;
20-25 g) were purchased from the Orient Bio Inc. (Seongnam, Korea).
All mice were maintained under standard conditions (temperature,
20±0.5°C; humidity, 60±5%; light/dark cycle, 12 h), and allowed
free access to food and water. The mice were anesthetized by
intraperitoneal injections of ketamine (100 mg/kg) and xylazine
(Rompun®, 10 mg/kg) and maintained with a positive
pressure ventilation using a ventilator (Harvard Apparatus Co.,
Millis, MA, USA). MI was then induced by ligation of the left
anterior descending coronary artery with a 6-0 silk suture
(Ethicon, Inc., Cincinnati, OH, USA). Prior to injecting into the
MI mice, negative control miRNA, miR-21 mimic or miR-21 inhibitor
at a final concentration of 5 µmol/ml were loaded into a 200
µl exosome sample containing 1 µg/µl exosomal
proteins using the Exoquick exosome precipitation kit (System
Biosciences) according to the manufacturer's protocol. Next,
exosomes containing negative control miRNA, miR-21 mimic or
inhibitor were administered by three intra-myocardial injections
(total 100 µl) into the infarct border zone, as previously
described (22,23). The mice were divided into the
following five groups (four mice/group): i) non-MI, ii) MI, iii) MI
+ negative control miRNA-loaded exosome injection, iv) MI + miR-21
mimic-loaded exosome injection and v) MI + miR-21 inhibitor-loaded
exosome injection. Cardiac tissues were obtained from the left
ventricular (LV) infarcted zone, infarct border zone and remote
zone in the MI mice and from the LV apex in the non-MI mice. All
procedures were performed in accordance with the ethical approval
of the Institutional Animal Care and Use Committee of Yonsei
University College of Medicine (approval reference no. 2011-0136)
and the Guide for the Care and Use of Laboratory Animals published
by the National Institutes of Health (24).
Echocardiography was performed with a Vevo 2100
system (VisualSonics, Inc., Toronto, ON, Canada). LV ejection
fraction (LVEF) and fractional shortening (FS) were calculated
according to the following formulas: LVEF (%) = [left ventricular
end-diastolic volume (LVEDV)-left ventricular end-systolic volume
/LVEDV) ×100; left ventricular fractional shortening (LVFS) (%) =
left ventricular internal-diastolic diameter (LVIDD)-left
ventricular internal-systolic diameter)/LVIDD ×100.
The mice were sacrificed one week following the
treatment and the cardiac tissue were quickly collected and fixed
in 4% formaldehyde for seven days at room temperature. Tissues were
embedded in paraffin and sliced into 4 µm thick sections.
Next, the sections were mounted on normal glass slides and stained
with hematoxylin and eosin (H&E) for 2 min at room temperature
and Masson's Trichrome (MT) for 10 min at room temperature. Four
hearts were analyzed in each group and five distinct fields were
examined in each slide, resulting in the evaluation of 20 fields in
total from each group. Each slide was examined using an optical
microscope and quantified with ImageJ 1.50i software (National
Institutes of Health, Bethesda, MD, USA).
Statistical analysis
The data are presented as the mean ± standard
deviation (n=4). The comparisons between the experimental groups
were conducted using either an unpaired two-tailed Student's
t-test, or a one-way analysis of variance with a
Student-Newman-Keuls post hoc test. Statistical analyses were
performed using SPSS version 23.0 (SPSS, Inc., Chicago, IL, USA)
and GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Characterization of human peripheral
blood-derived exosomes
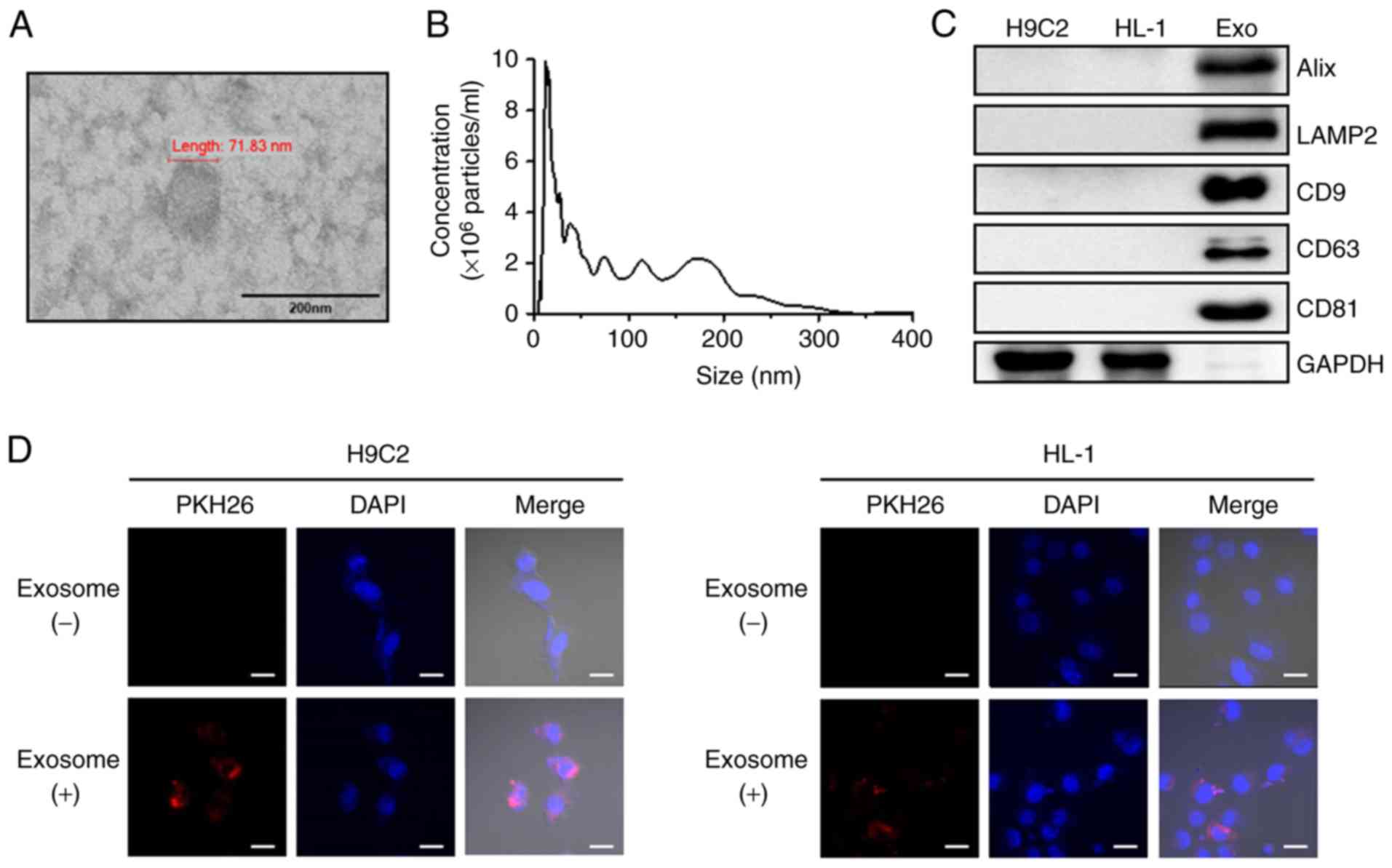
Exosomes isolated from human peripheral blood were
examined by TEM, NTA and western blot analysis. TEM analysis
revealed that the isolated exosomes had a typical round morphology
(Fig. 1A) and NTA revealed that
the isolated exosomes had a mean particle size distribution of 104
nm (Fig. 1B). Consistent with
these results, western blot analysis demonstrated the presence of
known exosome markers, including Alix, LAMP2, CD9, CD63 and CD81,
in the exosomes. The exosomes revealed high expression of markers,
but GAPDH was absent (Fig. 1C).
The detection of shape, size and protein markers of typical
exosomes indicated that the exosomes were successfully isolated
from human peripheral blood. To further investigate whether human
peripheral blood-derived exosomes were taken up by cardiac cell
lines, the exosomes with were labelled with PKH26, a red florescent
dye. After 24 h treatment of H9C2 and HL-1 cells with labeled
exosomes, a positive PKH26 signal was validated using confocal
microscopy (Fig. 1D). These
observations suggest that human peripheral blood-derived exosomes
were effectively absorbed in the two cardiac cell lines.
Exosome-mediated delivery of miRNA in
vitro
In order to determine the miRNA delivery potential
of human peripheral blood derived-exosomes in vitro,
exosomes were loaded with either miR-21 mimic or inhibitor using
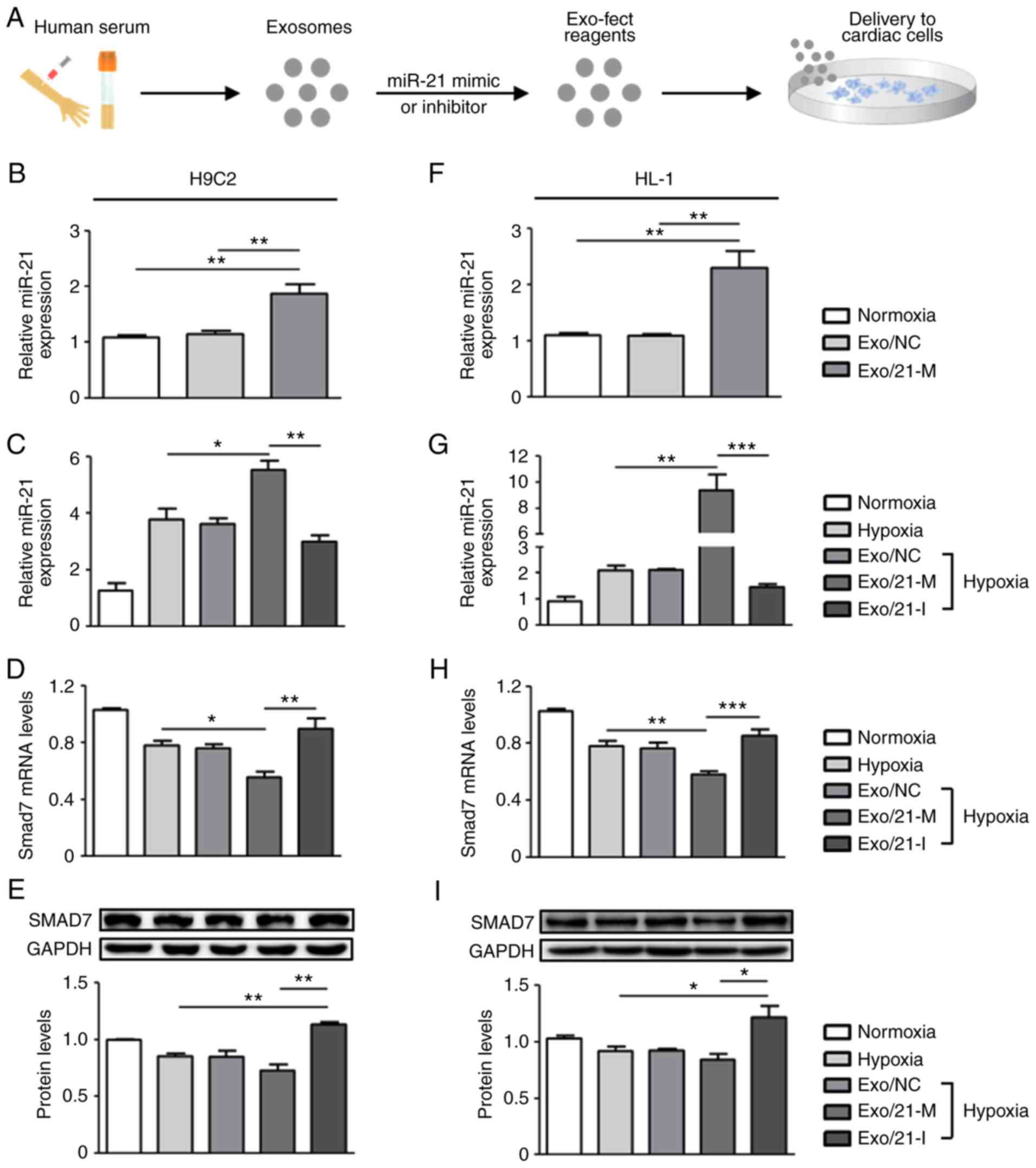
the Exo-Fect™ Exosome Transfection reagent (Fig. 2A). Following treatments with
miR-21 mimic- or inhibitor-loaded exosomes in H9C2 and HL-1 cells,
RT-qPCR was performed to measure the levels of miR-21. Under
normoxic conditions, miR-21 expression levels in H9C2 and HL-1
cells were significantly upregulated following treatment with
miR-21 mimic-loaded exosomes compared with normoxic controls
(P<0.01; Fig. 2B and F). Next,
it was detected that miR-21 expression was upregulated in hypoxic
cardiac cells compared with normoxic controls and that miR-21
expression was regulated by the presence of its mimic and inhibitor
when compared to the hypoxic controls. Under the conditions of
hypoxic H9C2 cells, miR-21 mimic and inhibitor significantly
upregulated and downregulated miR-21 expression compared with
hypoxic controls, respectively (P<0.05; Fig. 2C). Similarly, in hypoxic HL-1
cells, miR-21 mimic-loaded exosomes significantly increased miR-21
expression compared with the normoxic and hypoxic controls
(P<0.01), whereas miR-21 inhibitor-loaded exosomes significantly
reduced miR-21 expression (P<0.005; Fig. 2G). Collectively, these
observations suggest that human peripheral blood-derived exosomes
effectively delivered miR-21 mimic or inhibitor to the two cardiac
cell lines under all tested conditions.
It is well known that miRNAs exert their functions
via the suppression of their target gene expression by binding to
the 3′-untranslated region of target mRNAs (25,26); thus, the present study next
examined the mRNA and protein expression levels of Smad7, a direct
target of miR-21, previously implicated in fibrosis regulation
(19,27). It was observed that treatment with
miR-21 mimic-loaded exosomes in H9C2 cells significantly
downregulated the mRNA and protein expression levels of Smad7
compared with hypoxic controls and that the expression of Smad7 was
significantly upregulated in cells treated with miR-21
inhibitor-loaded exosomes (P<0.05; Fig. 2D and E). Similarly, in HL-1 cells,
hypoxia induced the significantly reduced expression levels of
Smad7 mRNA and protein compared with normoxic controls, which were
downregulated by miR-21 mimic- and upregulated by miR-21
inhibitor-loaded exosomes (P<0.05; Fig. 2H and I). Altogether, these results
demonstrate that miR-21 mimic- or inhibitor-loaded exosomes
were effectively delivered into cardiac cells and exhibited
therapeutic potential by regulating target genes associated with
physiopathological mechanisms.
Regulation of functional target using
miRNA-loaded exosomes
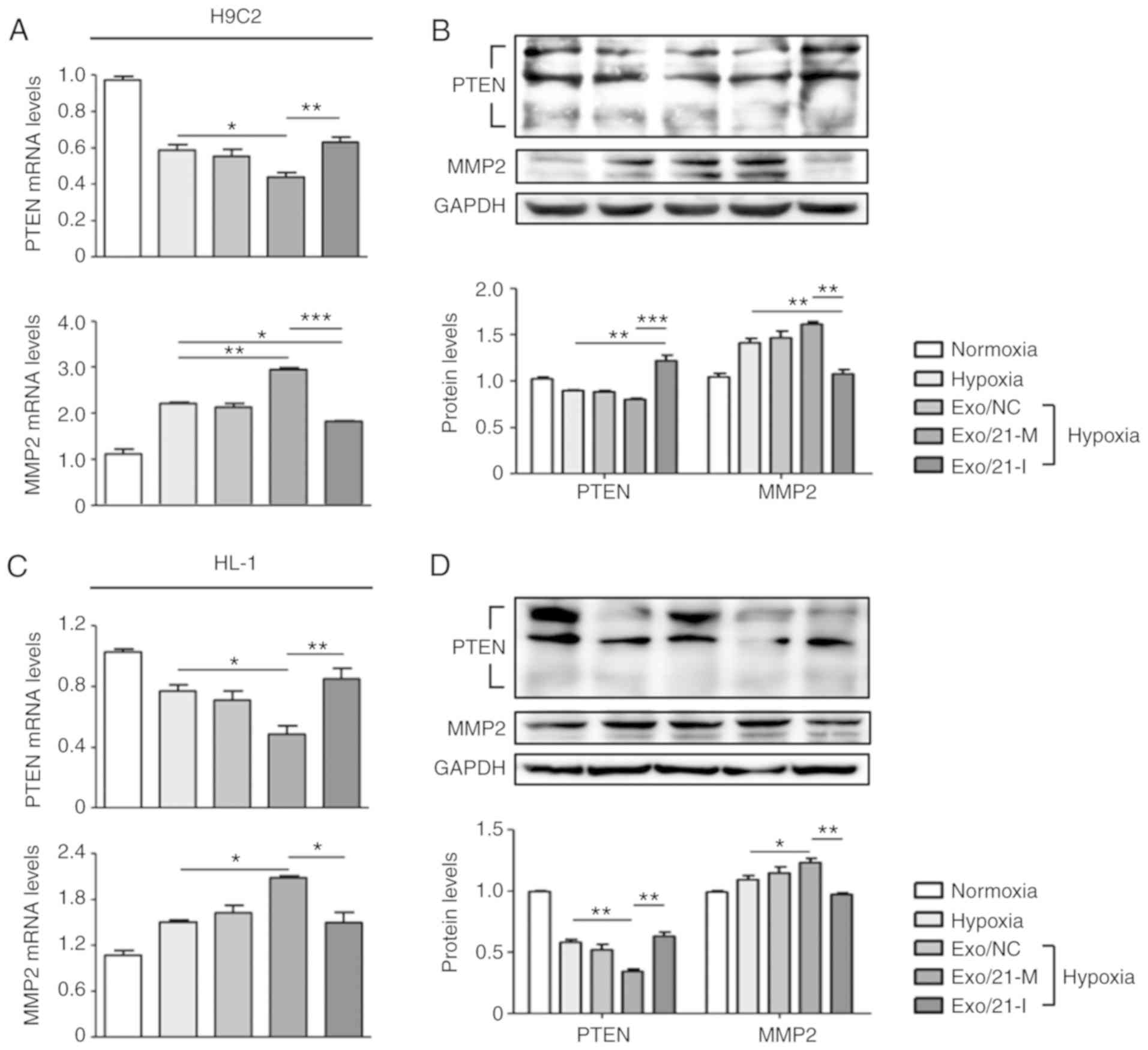
Furthermore, the present study assessed the
expression of PTEN, a well-known direct target of miR-21 (28), in order to validate the efficiency
of exosomes as a delivery system. PTEN is expressed in various cell
types, including cardiomyocytes, fibroblasts, endothelial cells and
vascular smooth muscle cells, where it modulates cell
survival/apoptosis, hypertrophy, contractility, fibrosis and
metabolism (28,29). Hence, PTEN is a crucial molecule
involved in the development of numerous cardiovascular diseases
(30). It was observed that
miR-21 mimic-loaded exosomes significantly suppressed PTEN
expression compared with hypoxic controls, whereas miR-21
inhibitor-loaded exosomes significantly promoted PTEN expression
compared with H9C2 and HL-1 cells treated with miR-21 mimic-loaded
exosomes (P<0.05; Fig. 3A-D).
In addition, previous reports have demonstrated that
miR-21-regulated expression of MMP-2 is modulated via a PTEN
pathway and results in cardiac fibrosis (18,31). In the two cell lines, miR-21
mimic-loaded exosomes significantly increased MMP-2 expression
compared with hypoxic controls, whereas miR-21 inhibitor-loaded
exosomes significantly reduced MMP-2 expression compared to cells
treated with miR-21 mimic-loaded exosomes (P<0.05; Fig. 3A-D Visualization and in vivo
tracking of the exosomes of murine melanoma B16-BL6 cells in mice
after intravenous injection).
Exosome-mediated delivery of miRNA in
vivo
C57BL/6 mice were subjected to MI to evaluate miR-21
expression levels in the infarcted, border and remote zones. MiR-21
expression levels were significantly upregulated in the MI mouse
model, compared with non-MI mice, particularly in the infarcted
zone of the heart (P<0.05; Fig.
4A). Next, the expression levels of Smad7, PTEN and MMP2 were
assessed using RT-qPCR and western blot analysis to determine
whether miR-21 mimic- or inhibitor-loaded exosomes were delivered
efficiently in vivo. Similar to the results of in
vitro experiment, it was observed that the mRNA and protein
expression levels of Smad7, PTEN and MMP2 were significantly
altered following treatment with miR-21 mimic-loaded exosomes when
compared with mice treated with inhibitor-loaded
exosomes(P<0.05; Fig. 4B and
C).
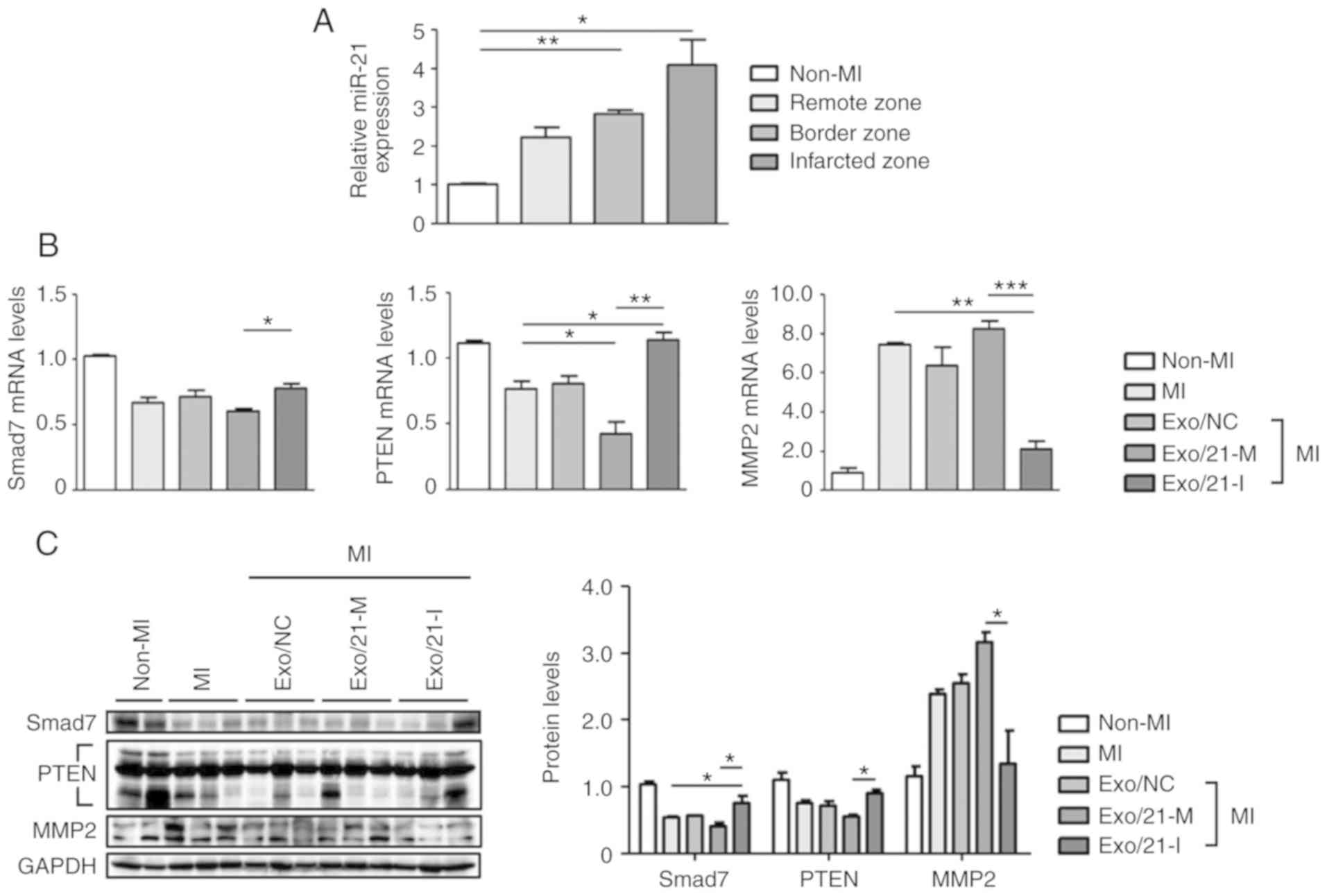 | Figure 4Delivery of miR-21 mimic- or
inhibitor-loaded exosomes to MI mouse model. (A) RT-qPCR was used
to determine miR-21 levels in different regions (infarcted, border
zone and remote zone) of non-MI and MI mice. Relative miR-21 levels
were normalized against U6. (B) Following an injection of miR-21
mimic- or inhibitor-loaded exosomes into the MI mouse model, Smad7,
PTEN and MMP2 mRNA expression levels were determined usong RT-qPCR.
(C) Smad7, PTEN and MMP2 protein levels were detected using western
blot analysis. *P<0.05, **P<0.01 and
***P<0.005 with comparisons shown by lines. MI,
myocardial infarction; miR, microRNA; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; Smad7, SMAD
family member 7; PTEN, phosphatase and tensin homolog; MMP2, matrix
metalloproteinase 2; Exo/NC, negative control miRNA-loaded exosome
injection; Exo/21-M, miR-21 mimic-loaded exosome injection;
Exo/21-I, miR-21 inhibitor-loaded exosome injection. |
Therapeutic potential of miRNA-loaded
exosomes against cardiac fibrosis
Finally, the present study investigated the
potential of human peripheral blood-derived exosomes to deliver
miRNA for the treatment of cardiac fibrosis following MI. The
echocardiography analysis revealed worse cardiac function in mice
treated with miR-21 mimic-loaded exosomes compared with mice
treated with miR-21 inhibitor-loaded exosomes, as the former group
of mice exhibited significantly decreased LVEF and LVFS compared
with the latter group of mice (P<0.05; Fig. 5A). As observed following the
H&E and MT staining, myocardial disorders and fibrosis were
significantly enhanced in MI mice compared with non-MI mice
(P<0.05). Histological examination of the hearts of mice treated
with miR-21 mimic-loaded exosomes revealed the dilatation of the
left ventricular chambers with thinning of the walls. However, the
histological changes were reduced following treatment with miR-21
inhibitor-loaded exosomes. In addition, cardiac fibrosis at the
infarcted and border zones increased in mice treated with miR-21
mimic-loaded exosomes compared with MI mice, and this effect was
reversed by treatment with miR-21 inhibitor-loaded exosomes
(Fig. 5B and C).
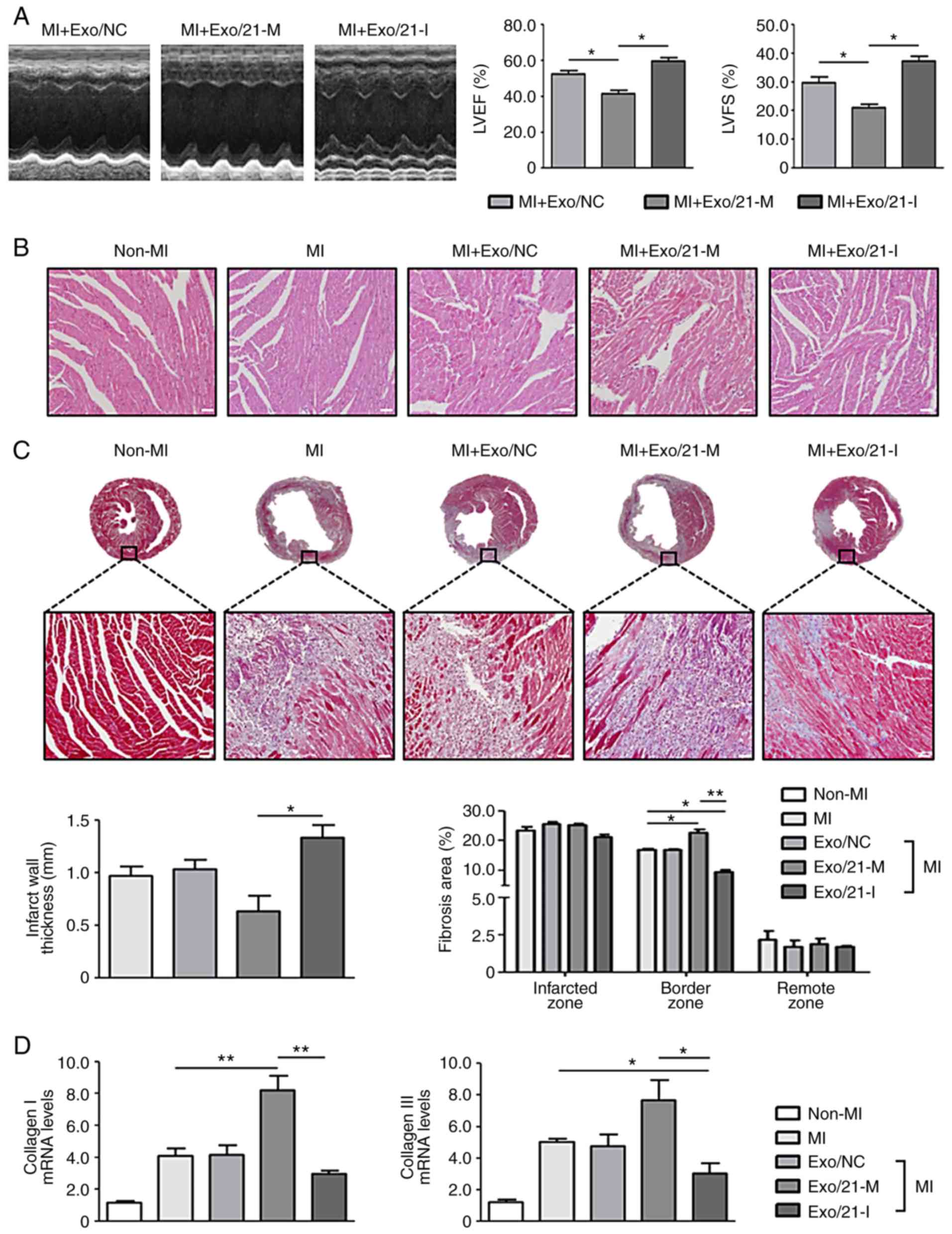 | Figure 5Effects of miR-21 mimic- or
inhibitor-loaded exosomes on cardiac fibrosis in mice. (A)
Representative images of M-mode echocardiography demonstrating the
LV wall motion of the infarcted hearts, and the mean data of LVEF
and LVFS. *P<0.05 with comparisons shown by lines.
(B) Representative haemotoxylin and eosin staining in the border
zone of the infarcted hearts (scale bar, 50 µm;
magnification, ×200). Pink staining indicates cardiac myocytes;
blue staining indicates nuclei. (C) Representative Masson's
Trichrome staining of mouse heart sections 1 week after MI (scale
bar, 50 µm; magnification: Top row, ×2; bottom row, ×200).
Blue staining indicates scar tissue; red staining indicates viable
myocardium. Bar graphs exhibit the infarct wall thickness and the
percentage of the fibrotic area (n=4 mice in each group). (D)
Collagen I and III mRNA expression levels in the infarcted hearts
were measured using reverse transcription-quantitative polymerase
chain reaction. *P<0.05 and **P<0.01
with comparisons shown by lines. miR, microRNA; MI, myocardial
infarction; Exo/NC, negative control miRNA-loaded exosome
injection; Exo/21-M, miR-21 mimic-loaded exosome injection;
Exo/21-I, miR-21 inhibitor-loaded exosome injection; LVEF, left
ventricular ejection fraction; LVFS, left ventricular fractional
shortening. |
Furthermore, the expression levels of myocardial
fibrosis markers, collagen I and III, were significantly
upregulated in MI mice compared with non-MI mice. Mice treated with
miR-21 mimic- or inhibitor-loaded exosomes significantly
upregulated and downregulated collagen I and III expression levels,
respectively (P<0.05; Fig.
5D). Collectively, these results revealed that human peripheral
blood-derived exosomes successfully delivered miR-21 mimics or
inhibitors and modulated cardiac fibrosis in vivo. Thus, the
regulation of mRNAs using miRNA-loaded exosomes derived from the
human peripheral blood may represent an effective therapeutic
approach against cardiac fibrosis following MI.
Discussion
MI is a public health problem and a leading cause of
mortality globally (32). The
development of impaired cardiac function and cardiac fibrosis due
to MI serves a key function in regulating cardiac dysfunction
(6,33). Currently, no efficient therapies
are available that may inhibit the progression of cardiac fibrosis
in infarcted hearts (8). Thus, in
the present study, it was examined whether miRNA delivery using
human peripheral blood-derived exosomes may function as an
effective therapeutic approach against MI by modulating cardiac
fibrosis.
To date, a number of studies have suggested that
extracellular vesicles (EVs) from in vitro cultured-cells or
serum possess cardioprotective effects due to miRNAs, already
present in the EVs (34,35); however, these studies have focused
on the function of EVs as protective vesicles as opposed to drug
carriers. Notably, the present study, to the best of our knowledge,
provides the first compelling evidence that human peripheral
blood-derived exosomes may be successfully used as a delivery
vehicle for miRNA.
miRNAs comprise a broad class of small non-coding
RNAs that control the expression of complementary target mRNAs
(36). Dysregulation of miRNAs by
a number of mechanisms has been observed in various diseases
including cardiac diseases (20,37). Previously, miR-21 has been
revealed to serve notable functions in cardiovascular disorders
(28). Furthermore, a number of
potential target genes involved in miR-21-mediated cardiovascular
effects have also been identified (28). For instance, miR-21 regulates the
extracellular regulated kinase-mitogen activated pathway kinase
signaling pathway in cardiac fibroblasts, which impacts cardiac
structure and function (20).
Furthermore, miR-21 serves a critical function in cardiac
fibroblast activation and cardiac fibrosis following MI via the
tumor growth factor-β/Smad7 signaling pathway (19). In addition, modulation of the
miR-21-regulated expression of MMP-2 via the PTEN pathway regulates
cardiac fibrosis (18,31). Altogether, these results confirm
that miR-21 may function as a therapeutic target for MI; thus the
present study examined miR-21 loaded into human peripheral
blood-derived exosomes.
Exosomes are small membrane-bound vesicles (30-100
nm) secreted by most cell types. Exosomes serve a major function in
cell-to-cell communication that may affect neighboring cells and
distant parts of the body by transferring molecules, including
proteins, mRNAs and miRNAs (9,38,39). The natural biocompatibility of
exosomes with mammalian cells suggests that they may overcome the
majority of cellular barriers and drug delivery hurdles, including
RNase susceptibility, endosomal accumulation, phagocytosis,
multidrug resistance, cytotoxicity and immunogenicity (40,41). However, exosome safety and
quantity may constitute major issues in their application in
clinical practice (14). As human
peripheral blood has been used extensively for transfusions and is
readily available, exosomes isolated from human peripheral blood
may circumvent these issues. Furthermore, considering the systemic
administration limitations of exosomes, including accumulation in
the liver, spleen and lungs or rapid clearance by macrophages
(42-44), direct injection or additional
genetic modification of peripheral blood-derived exosome, as
suggested by Kim et al (45), may increase the efficacy of their
delivery to the heart by preventing non-specific delivery to other
organs, thus increasing their therapeutic efficacy in clinical
settings.
In conclusion, these results suggest that human
peripheral blood-derived exosomes function as efficient vehicles
for the delivery of miRNAs, which in turn may potentially be used
for the treatment of cardiac diseases. This approach may be further
applied to therapeutic tools for various diseases, including
cancer, by delivering miRNAs other than miR-21 or diverse molecules
with functionality.
Funding
The present study was supported by research grants
from the Basic Science Research Program through the National
Research Foundation of Korea (grant no. 2017R1A2B3003303), the
Ministry of Education, Science and Technology (grant no.
NRF-2017R1A2B3003303) and the Korean Healthcare Technology R&D
project funded by the Ministry of Health and Welfare (grant no.
HI16C0058).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JK, HyelimP and BJ conceived and designed the
experiments. JK and HyewonP performed the experiments. JK, HK and
DM analyzed the data. JK, NY and BJ wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was ethically approved by the
Institutional Review Board of Severance Hospital and adhered to the
principles of the Declaration of Helsinki. All animal experiments
were ethically approved by the Institutional Animal Care and Use
Committee of Yonsei University College of Medicine (approval
reference no. 2011-0136).
Patient consent for publication
Written informed consent was obtained from the
subjects for participation in the present study study.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
LVEDV
|
left ventricular end-diastolic
volume
|
|
LVIDD
|
left ventricular internal-diastolic
diameter
|
Acknowledgments
Not applicable.
References
|
1
|
Thygesen K, Alpert JS, Jaffe AS, Simoons
ML, Chaitman BR and White HD: Third universal definition of
myocardial infarction. J Am Coll Cardiol. 60:1581–1598. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Swynghedauw B: Molecular mechanisms of
myocardial remodeling. Physiol Rev. 79:215–262. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nian M, Lee P, Khaper N and Liu P:
Inflammatory cytokines and postmyocardial infarction remodeling.
Circ Res. 94:1543–1553. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun M, Dawood F, Wen WH, Chen M, Dixon I,
Kirshenbaum LA and Liu PP: Excessive tumor necrosis factor
activation after infarction contributes to susceptibility of
myocardial rupture and left ventricular dysfunction. Circulation.
110:3221–3228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Regula KM and Kirshenbaum LA: Apoptosis of
ventricular myocytes: A means to an end. J Mol Cell Cardiol.
38:3–13. 2005. View Article : Google Scholar
|
|
6
|
van den Borne SW, Diez J, Blankesteijn WM,
Verjans J, Hofstra L and Narula J: Myocardial remodeling after
infarction: The role of myofibroblasts. Nat Rev Cardiol. 7:30–37.
2010. View Article : Google Scholar
|
|
7
|
Zamilpa R and Lindsey ML: Extracellular
matrix turnover and signaling during cardiac remodeling following
MI: Causes and consequences. J Mol Cell Cardiol. 48:558–563. 2010.
View Article : Google Scholar :
|
|
8
|
Fan Z and Guan J: Antifibrotic therapies
to control cardiac fibrosis. Biomater Res. 20:132016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Urbanelli L, Magini A, Buratta S, Brozzi
A, Sagini K, Polchi A, Tancini B and Emiliani C: Signaling pathways
in exosomes biogenesis, secretion and fate. Genes (Basel).
4:152–170. 2013. View Article : Google Scholar
|
|
10
|
Lin J, Li J, Huang B, Liu J, Chen X, Chen
XM, Xu YM, Huang LF and Wang XZ: Exosomes: Novel biomarkers for
clinical diagnosis. Scientific World Journal. 2015:6570862015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ha D, Yang N and Nadithe V: Exosomes as
therapeutic drug carriers and delivery vehicles across biological
membranes: Current perspectives and future challenges. Acta Pharm
Sin B. 6:287–296. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Batrakova EV and Kim MS: Development and
regulation of exosome-based therapy products. Wiley Interdiscip Rev
Nanomed Nanobiotechnol. 8:744–757. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Batrakova EV and Kim MS: Using exosomes,
naturally-equipped nanocarriers, for drug delivery. J Control
Release. 219:396–405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boon RA and Dimmeler S: MicroRNAs in
myocardial infarction. Nat Rev Cardiol. 12:135–142. 2015.
View Article : Google Scholar
|
|
16
|
Sun T, Dong YH, Du W, Shi CY, Wang K,
Tariq MA, Wang JX and Li PF: The role of microRNAs in myocardial
infarction: From molecular mechanism to clinical application. Int J
Mol Sci. 18:2017. View Article : Google Scholar
|
|
17
|
Wang J, Huang W, Xu R, Nie Y, Cao X, Meng
J, Xu X, Hu S and Zheng Z: MicroRNA-24 regulates cardiac fibrosis
after myocardial infarction. J Cell Mol Med. 16:2150–2160. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Roy S, Khanna S, Hussain SR, Biswas S,
Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ and Sen CK: MicroRNA
expression in response to murine myocardial infarction: miR-21
regulates fibroblast metalloprotease-2 via phosphatase and tensin
homologue. Cardiovasc Res. 82:21–29. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan J, Chen H, Ge D, Xu Y, Xu H, Yang Y,
Gu M, Zhou Y, Zhu J, Ge T, et al: Mir-21 promotes cardiac fibrosis
after myocardial infarction via targeting Smad7. Cell Physiol
Biochem. 42:2207–2219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thum T, Gross C, Fiedler J, Fischer T,
Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et
al: MicroRNA-21 contributes to myocardial disease by stimulating
MAP kinase signalling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Park H, Ku SH, Park H, Hong J, Kim D, Choi
BR, Pak HN, Lee MH, Mok H, Jeong JH, et al: RAGE siRNA-mediated
gene silencing provides cardioprotection against ventricular
arrhythmias in acute ischemia and reperfusion. J Control Release.
217:315–326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bala S, Csak T, Momen-Heravi F, Lippai D,
Kodys K, Catalano D, Satishchandran A, Ambros V and Szabo G:
Biodistribution and function of extracellular miRNA-155 in mice.
Sci Rep. 5:107212015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
National Research Council (US) Institute
for Laboratory Animal Research: Guide for the Care and Use of
Laboratory Animals. National Academies Press; Washington, DC:
1996
|
|
25
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang B, Hao J, Jones SC, Yee MS, Roth JC
and Dixon IM: Decreased Smad 7 expression contributes to cardiac
fibrosis in the infarcted rat heart. Am J Physiol Heart Circ
Physiol. 282:H1685–H1696. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng Y and Zhang C: MicroRNA-21 in
cardiovascular disease. J Cardiovasc Transl Res. 3:251–255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oudit GY and Penninger JM: Cardiac
regulation by phosphoinositide 3-kinases and PTEN. Cardiovasc Res.
82:250–260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oudit GY, Sun H, Kerfant BG, Crackower MA,
Penninger JM and Backx PH: The role of phosphoinositide-3 kinase
and PTEN in cardiovascular physiology and disease. J Mol Cell
Cardiol. 37:449–471. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tao H, Zhang JG, Qin RH, Dai C, Shi P,
Yang JJ, Deng ZY and Shi KH: LncRNA GAS5 controls cardiac
fibroblast activation and fibrosis by targeting miR-21 via
PTEN/MMP-2 signaling pathway. Toxicology. 386:11–18. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chi JS and Kloner RA: Stress and
myocardial infarction. Heart. 89:475–476. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Travers JG, Kamal FA, Robbins J, Yutzey KE
and Blaxall BC: Cardiac fibrosis: The fibroblast awakens. Circ Res.
118:1021–1040. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Gu H, Li Z, Xie Y, Yao Y, Zhu J, Xu Y,
Dai J, Zhong Q, Zhu CH, et al: Serum-derived extracellular vesicles
protect against acute myocardial infarction by regulating
miR-21/PDCD4 signaling pathway. Front Physiol. 9:3482018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Khan M, Nickoloff E, Abramova T, Johnson
J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VN,
Benedict C, et al: Embryonic stem cell-derived exosomes promote
endogenous repair mechanisms and enhance cardiac function following
myocardial infarction. Circ Res. 117:52–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Catalanotto C, Cogoni C and Zardo G:
MicroRNA in control of gene expression: An overview of nuclear
functions. Int J Mol Sci. 17:2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
He He L, Lim X, de Stanchina LP, Xuan E,
Liang Z, Xue Y, Zender W, Magnus L, Ridzon JD, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Théry C: Exosomes: Secreted vesicles and
intercellular communications. F100. Biol Rep. 3:152011.
|
|
39
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Syn NL, Wang L, Chow EK, Lim CT and Goh
BC: Exosomes in cancer nanomedicine and immunotherapy: Prospects
and challenges. Trends Biotechnol. 35:665–676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Usman WM, Pham TC, Kwok YY, Vu LT, Ma V,
Peng B, Chan YS, Wei L, Chin SM, Azad A, et al: Efficient RNA drug
delivery using red blood cell extracellular vesicles. Nat Commun.
9:23592018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Charoenviriyakul C, Takahashi Y, Morishita
M, Matsumoto A, Nishikawa M and Takakura Y: Cell type-specific and
common characteristics of exosomes derived from mouse cell lines:
Yield, physicochemical properties, and pharmacokinetics. Eur J
Pharm Sci. 96:316–322. 2017. View Article : Google Scholar
|
|
43
|
Smyth T, Kullberg M, Malik N, Smith-Jones
P, Graner MW and Anchordoquy TJ: Biodistribution and delivery
efficiency of unmodified tumor-derived exosomes. J Control Release.
199:145–155. 2015. View Article : Google Scholar :
|
|
44
|
Takahashi Y, Nishikawa M, Shinotsuka H,
Matsui Y, Ohara S, Imai T and Takakura Y: Visualization and in vivo
tracking of the exosomes of murine melanoma B16-BL6 cells in mice
after intravenous injection. J Biotechnol. 165:77–84. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim H, Yun N, Mun D, Kang JY, Lee SH, Park
H, Park H and Joung B: Cardiac-specific delivery by cardiac
tissue-targeting peptide-expressing exosomes. Biochem Biophys Res
Commun. 499:803–808. 2018. View Article : Google Scholar : PubMed/NCBI
|