Introduction
Epithelial cell adhesion molecule (EpCAM) is a
transmembrane glycoprotein that is involved in several biological
processes including cell adhesion, proliferation and
differentiation (1,2). It has been demonstrated that
EpCAM−/− mice exhibit impaired cell junctions in the
intestines, contributing to severe defects and phonotypes similar
to human congenital tufting enteropathy (2-4).
EpCAM expression is detected in various tissues from early-stage
embryos to adults, and in tumors. Huang et al (1) determined the expression pattern,
functions and underlying mechanisms of EpCAMs, revealing that the
expression pattern of liver EpCAM is particularly complex. EpCAM
has been reported to be expressed in hepatoblasts during embryonic
development and in adult liver cholangiocytes, but not in
hepatocytes (5,6). Previous studies have also identified
EpCAM as surface marker of stem cells in normal adult livers
(7,8). Furthermore, EpCAM mutant zebrafish
exhibit impaired hepatic development due to the activation of the
Wnt signaling pathway in the endoderm (9). However, whether EpCAM mutations
affect the development of mammalian livers has yet to be
elucidated.
The abnormal expression of EpCAM has been associated
with different liver diseases. Matsumoto et al (10) reported that proliferating ductal
cells (PDCs) expressing EpCAM may lead to the development of
hepatocellular carcinoma (HCC) in inflamed livers, indicating the
stem/progenitor cell origin of hepatocarcinogenesis. This protein
is also highly expressed in premalignant hepatic tissues and
EpCAM-positive cells may serve as HCC cancer stem cells (11-13). Mani et al (14) demonstrated that EpCAM underwent
regulated intramembrane proteolysis in hepatitis B virus
(HBV)-replicating cells. It was also revealed that the activation
of Wnt signaling may lead to HBV-associated HCC. It has been
reported that bile acids induce the expression of long non-coding
RNA H19, and the activation of hepatic H19RNA may promote
cholestatic liver fibrosis in mice via the ZEB1/EpCAM signaling
pathway in mice (15). Although
the expression pattern of EpCAM has been studied, the functions and
mechanisms of EpCAM in liver development and disease remain
unclear.
Circular RNAs (circRNAs), a novel type of non-coding
RNAs (ncRNAs) with a covalently closed-loop structure generated by
back splicing, play important roles in several biological
processes, including proliferation, apoptosis, development and
aging (16,17). circRNAs may also serve as
potential clinical biomarkers for various diseases, including
rheumatoid arthritis, breast cancer, cardiovascular diseases and
osteoarthritis (18-21). It has been demonstrated that a
number of circRNAs are differentially expressed between HCC and
normal liver tissues, which may be closely associated with the
development and prognosis of HCC (22). The altered expression of circRNAs
has also been implicated in hepatic steatosis and non-alcoholic
fatty liver disease (23,24).
The aforementioned studies indicate that the
expression of circRNAs may be involved in the functions of EpCAM
and that circRNAs may serve as biomarkers during liver development
and associated diseases. In the current study, the effects of EpCAM
on the expression profiles of circRNAs in the livers of
EpCAM−/− mice were investigated by high-throughput
sequencing. In addition, circRNA-microRNA (miRNA or miR)-mRNA
networks were assessed by target analysis. The present study
identified several novel circRNAs in the livers of
EpCAM−/− mice. These results may therefore enhance our
understanding of the functions of EpCAM during liver development
and in associated diseases.
Materials and methods
Generation of EpCAM−/−
mice
All animal experiments were approved by the
Committee on the Laboratory Animal Care and Use of Guangdong
Pharmaceutical University. Stable genetic EpCAM−/− mice
were obtained using the clusters of regularly inter-spaced short
palindromic repeats (CRISPRs)/CRISPR-associated protein 9 (Cas9)
gene editing technology. The method was as follows: The GGG CGA TCC
AGA ACA ACG AT sequence in exon 2 of EpCAM was identified as guide
RNA (gRNA) by Vector NTI11 software (version 11.5.1, Invitrogen)
(Fig. 1A). The gRNA and Cas9 gene
constructed in vitro were transfected into the fertilized
ova of C57BL/6 mice via electrotransfection, which was subsequently
transplanted into the oviducts of surrogate 10-week old ICR (n=12;
body weight, 30-34 g) female mice. Fertilized ova were obtained by
mating 10-15-week old C57BL/6 males (n=10; body weight, 20-30 g)
with 6-week-old C57BL/6 females (n=29; body weight, 18-20 g). F0
mice with successful gene knockout, confirmed by gene sequencing,
were mated with wild-type (WT) 8-12-week-old C57BL/6 mice (n=32;
body weight, 20-29 g). F1 hybrid mice were then screened out.
Female and male mice with identical knockout sequences were
selected for breeding F2 mice. Heterozygous F2 mice with stable
genetic characteristics were then selected for breeding and
conservation. Homozygous mice with the knockout gene were selected
for further analysis. All the mice were kept in an SPF mouse
facility, at 22°C, 60-65% humidity, with free access to food and
water. All mice were from Hunan SJA Laboratory Animal Co. Ltd.
Animal tissue collection for circRNA
sequencing
WT and EpCAM−/− mice were sacrificed on
the first day after birth, after which murine livers were weighed,
frozen in liquid nitrogen, and stored at -80°C for preservation.
The livers of every 3 mice (at least 200 mg) were collected as 1
sample. The control group (WT) and the EpCAM−/− group
had 3 samples each.
Hematoxylin and eosin (H&E) and
immunofluorescence staining
The mouse tissues were fixed in 4% paraformaldehyde
at 4°C overnight for H&E (Leagene Biotechnology) and
immunofluorescence staining H&E staining was performed on
4-µm-thick sections, stained with hematoxylin for 3 min and eosin
for 20 sec at room temperature. Immunofluorescence staining was
performed on frozen sections. Tissues were embedded in optimal
cutting temperature compound (OCT) (Sakura Finetek) after fixing,
and 7-µm-thick sections were mounted on the slides using the frozen
section machine (LEICA CM1860). The sections were boiled in 10 mM
citric acid (Merck) at pH 6.0 for 5 min, and then were exposed to
1% bovine serum albumin (BSA; Sigma) in phosphate-buffered saline
(PBS; HyClone) containing 0.1% Tween-20 to block non-specific
sites. Anti-rabbit EpCAM antibody (cat. no. ab71916; Abcam) was
incubated with the sections at 4°C overnight at a dilution of
1:200, and AlexFluor 488 secondary antibody (cat. no. A21206;
Thermo fisher Scientific, Inc.) was incubated with the sections at
room temperature for 1 h at a dilution of 1:1,000. Images of
H&E staining were captured using the PerkinElmer Automated
Quantitative Pathology System, and the images of immunofluorescence
staining were analyzed using an Olympus confocal microscope
(FV3000).
RNA extraction and library
construction
circRNA isolation, library construction, circRNA
sequencing and bioinformatics analysis were performed by the Gene
Denovo Biotechnology Co. Total RNA was extracted using TRIzol
reagent (Life Technologies) and, ribosomal RNA (rRNA) [Ribo-Zero
Gold (Human/Mouse/Rat) kit, Illumina] was removed to retain mRNA
and ncRNA. Enriched mRNAs and ncRNAs were separated into short
fragments and reverse transcribed into cDNA (NEB#7490 kit, New
England Biolabs) with random primers. Second-strand cDNA was then
synthesized (NEB#7490 kit, New England Biolabs) using DNA
polymerase I, RNaseH and dNTPs (NEB#7490 kit, New England Biolabs).
The cDNA fragments were purified using the QiaQuick PCR extraction
kit (Qiagen), end repaired, poly(A) added, and ligated into
Illumina sequencing adapters (NEB#7490 kit, New England Biolabs).
Uracil-N-Glycosylase (NEB#7490 kit, New England Biolabs) was then
used to digest the second-strand cDNA. Digested products were size
selected via agarose gel electrophoresis (Sigma), PCR-amplified,
and sequenced using Illumina HiSeqTM 4000 (Illumina).
circRNA sequencing analysis
Reads were further filtered to remove those of low
quality reads containing adapters or reads with >10% of unknown
nucleotides. Reads were then mapped to rRNA of mice (Genome version
GRCm38.p5, http://asia.ensembl.org/info/about/species.html) and
the reference genome was established using Bowtie2 (version 2.2.8,
http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)
and TopHat2 (version 2.1.1, http://ccb.jhu.edu/software/tophat/index.shtml)
software (25,26), respectively. Mapped reads were
abandoned and unmapped reads were collected for circRNA
identification. Each end of the unmapped reads (20 bp) was
extracted and aligned to the reference genome to find unique anchor
positions within splice site. Anchor reads that aligned in the
reversed orientation (head-to tail) were then analyzed via
Find_circ software (version 1) (27) to identify circRNAs. The type,
chromosome distribution and length distribution of the identified
circRNAs were then further analyzed. The functions of circRNA
source genes were also analyzed via functional enrichment analysis
as shown below.
Analysis of differentially expressed
circRNAs and database annotation
Differentially expressed circRNAs were identified
using the edgeR package (http://www.r-project.org/). circRNAs were then blasted
against the circBase for annotation and those that could not be
annotated were defined as novel circRNAs. A fold change ≥2 and
P-value <0.05 in the comparison between different groups were
considered to indicate statistically significant differences.
Gene ontology (GO) and kyoto encyclopedia
of genes and genomes (KEGG) pathway analysis
GO is a gene functional classification system that
defines the properties and functions of genes in different
organisms. GO has 3 ontologies: Molecular function, cellular
component and biological process. All the source genes were mapped
to GO terms in the GO database (http://www.geneontology.org/) and gene numbers were
calculated for every term, with FDR ≤0.05 as a threshold.
KEGG pathway analysis (https://www.kegg.jp/) identifies significantly
enriched pathways in source genes compared with the whole genome,
which helps to further understand the functions of genes. The
calculating formula is the same as GO analysis, and pathways with
FDR ≤0.05 were defined as significantly enriched.
Integrated analysis of
circRNAs-miRNAs-mRNAs
StarBase (v2.0) software was used to predict the
target miRNAs of known circRNAs. Mireap (https://sourceforge.net/projects/mireap/), Miranda
(v3.3a, http://miranda.org.uk/) and TargetScan
(v7.0, http://www.targetscan.org) were also
used to predict the target miRNAs of novel circRNAs. The
circRNA-miRNA-mRNA pathway regulatory network was analyzed via
miRTarBase (v6.1, http://mirtarbase.mbc.nctu.edu.tw/php/index.php), and
the resulting correlation was visualized by Cytoscape (https://cytoscape.org/).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the liver samples using
TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.), and
subjected to reverse transcription using the PrimeScript™ RT
Reagent kit (Takara). All primers of development and metabolism
associated genes are listed in Table
SI [produced by Sangon Biotech (Shanghai) Co., Ltd.]. qPCR was
performed using the SYBR Premix Ex Taq kit (Takara) with the
PikoReal PCR system (Thermo Fisher Scientific, Inc.,) under the
following conditions: 95°C for 30 sec, followed by 40 cycles at
95°C for 5 sec, 60°C for 20 sec and 65°C for 15 sec. GAPDH was used
as a reference gene.
To confirm the expression of circRNAs, qPCR was
performed under the following sequential conditions: 95°C for 5
min, 40 cycles at 95°C for 10 sec and 60°C for 34 sec. All the
primers of circRNAs were listed in Table SII (produced by Geneseed
Biotechnology Co., Ltd.). β-actin was used as the reference gene.
PCR products were tested via electrophoresis on 2% agarose gels and
bands were extracted and used for Sanger sequencing.
Statistical analysis
Statistical differences were determined using SPSS
23.0 software. An independent-sample t-test was used and the data
were presented as the means ± SD. P<0.05 was considered to
indicate a statistically significant difference.
Results
Generation of EpCAM knockout mice
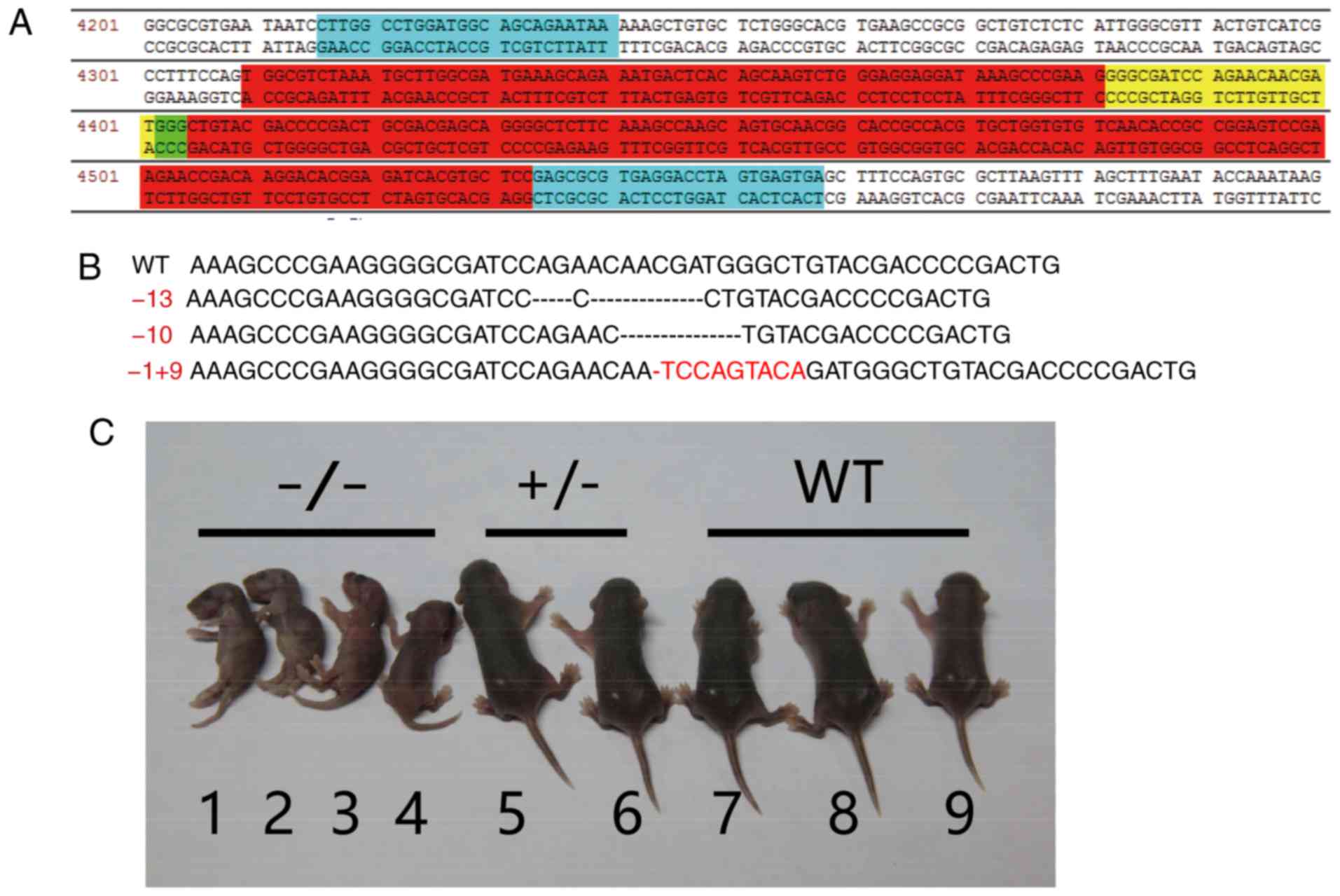
Three types of EpCAM gene knockout mice with
termination codon mutations were obtained and named -13 bp, -10 bp,
-1+9 bp mutant, respectively (Fig.
1B). The phenotypes of these mutants were identical. The -1+9
bp mutant mice were selected for further analysis. The phenotype of
the EpCAM knockout mice obtained via CRISPR/Cas9 was identical to
that obtained by the traditional gene targeting technique of our
previous study (2). As presented
in Fig. 1C, the
EpCAM−/− mice generated via CRISPR/Cas9 gene editing
were smaller than the heterozygous and WT mice at day 4 after birth
(Fig. 1C). The 89.36% (42/47) of
the mutant mice died within 1 week after birth, and all the mutant
mice died within 12 days. This result was similar to previous
studies, and the mice died of intestinal erosion as EpCAM knockout
affects the intestinal tight junction (2-4).
Expression pattern of glycogen-related
genes in the livers of EpCAM KO mice
As shown in Fig.
2A, the expression level of EpCAM was significantly decreased
in the EpCAM knockout mice. The results of immunofluorescence
staining also demonstrated that EpCAM was primarily expressed in
the liver cholangiocytes of WT mice at E18.5 (Figs. 2B and S1A) and at P0 (Fig. S1C). H&E staining revealed no
marked differences in the livers of the EpCAM−/− and WT
mice at E18.5, although some EpCAM−/− mice exhibited
reduced cytoplasmic vacuolation when compared with WT mice
(Fig. 2C), indicating defects in
hepatic glycogen storage (28).
In addition, no marked differences were observed following H&E
staining in the livers of EpCAM−/− and WT mice at P0.
The EpCAM−/− murine livers at P4 exhibited marked
vacuolation compared with those of the WT mice Fig. S1D.
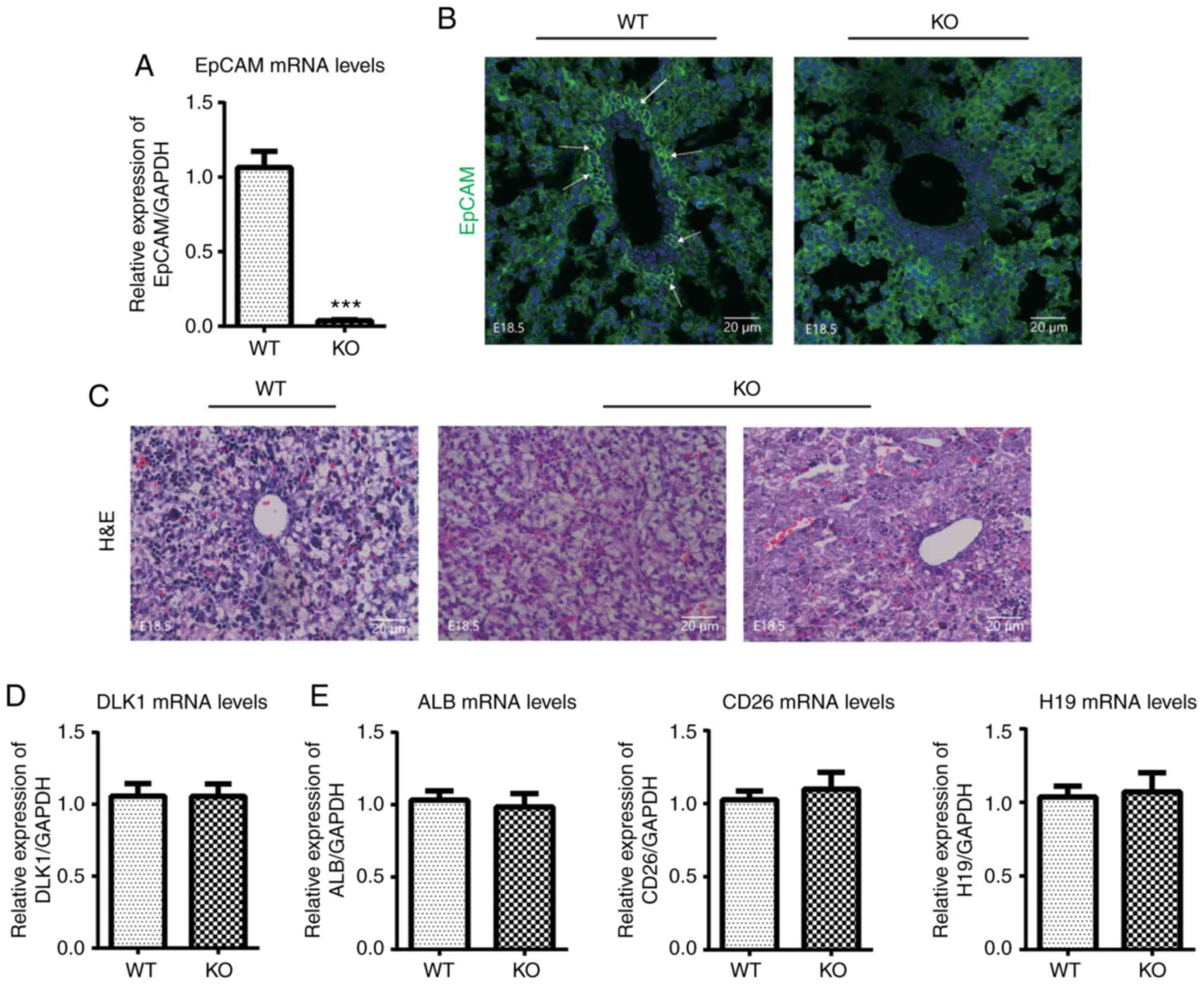 | Figure 2H&E staining and
immunofluorescence staining of EpCAM, and the expression of
development-associated genes at the mRNA level in the livers of
EpCAM−/− mice at embryonic day (E)18.5, as detected by
reverse transcription-quantitative PCR. (A) The expression of EpCAM
was significantly decreased in the livers of EpCAM−/−
mice at E18.5. (B) Immunofluorescence staining of EpCAM in the
livers of EpCAM−/− mice and WT mice at E18.5. (C)
H&E staining of EpCAM−/− murine livers and WT mice
of E18.5. The expression levels of (D) DLK1, (E) ALB, CD26 and H19,
(F) CD34 and Thy1, (G) Axin2 and Lgr5, (H) Hnf1β, CK19, CD133 and
Sox9 were determined. *P<0.05 and
***P<0.001, vs. WT mice. H&E, hematoxylin and
eosin; EpCAM, epithelial cell adhesion molecule; WT, wild-type;
DLK1, marker of hepatoblast delta-like 1 homologue; ALB, albumin;
Axin2, axis inhibition protein 2; Lgr5, leucine rich repeat
containing G-protein coupled receptor; Hnf1β, hepatocyte nuclear
factor 1β; CK19, cytokeratin 19; Sox9, SRY box 9. |
The expression levels of development associated
genes, including marker of hepatoblast delta-like 1 homologue
(DLK1; Fig. 2D), hepatocyte
albumin (ALB), CD26 and H19 (Fig.
2E), hepatic oval cell CD34 and thymus cell antigen 1 (Thy1)
(Fig. 2F), Wnt signal pathway
axis inhibition protein 2 (Axin2) and leucine rich repeat
containing G-protein coupled receptor 5 (Lgr5) (Fig. 2G) and cholangiocyte hepatocyte
nuclear factor 1β (Hnf1β), cytokeratin 19, CD133 and SRY box 9
(Sox9; Fig. 2H) were further
assessed by RT-qPCR. The results revealed that while the expression
of CD34 was increased in the knockout mice, the level of Axin2 was
significantly decreased. The expression of Hnf1β was also
increased, but not significantly (P=0.09).
Since some EpCAM−/− mice demonstrated
reduced cytoplasmic vacuolation of the liver, suggesting defects in
hepatic glycogen storage (28),
the expression of glycogen-associated genes was also determined
(Fig. 3). Glycogen phosphorylase
and glucose-6-phosphatase (G6PC) were determined to be associated
with glycogenolysis in the liver (29,30), and the expression of G6PC was
decreased in the knockout mice (P=0.08, Fig. 3A). The expression levels of two
glycogen synthesis-associated genes, UDP-glucose pyrophosphorylase
2 (Ugp2) and glucokinase (GCK) were significantly decreased
(P=0.08, Fig. 3B). The expression
levels of various genes that regulate glycogen catabolism,
including protein phosphatase 1 regulatory subunit 3B (PPP1R3B),
protein phosphatase 1 catalytic subunit α (PPC1CA), common β
subunit of phosphorylase kinase (PHKB), phosphorylase kinase
catalytic subunit, gamma 2 (PHKG2) remained unaltered (Fig. 3C). Additionally, no changes in the
expression of PAS domain containing serine/threonine kinase (PASK),
which regulates glycogen synthesis in the liver (31), were observed (Fig. 3D). The expression of solute
carrier family 2 (facilitated glucose transporter), member 2
[SLC2A2; also known as glucose transporter-2 (GLUT-2)], a gene
associated with disorders of glycogen storage (32), was significantly increased
(Fig. 3E).
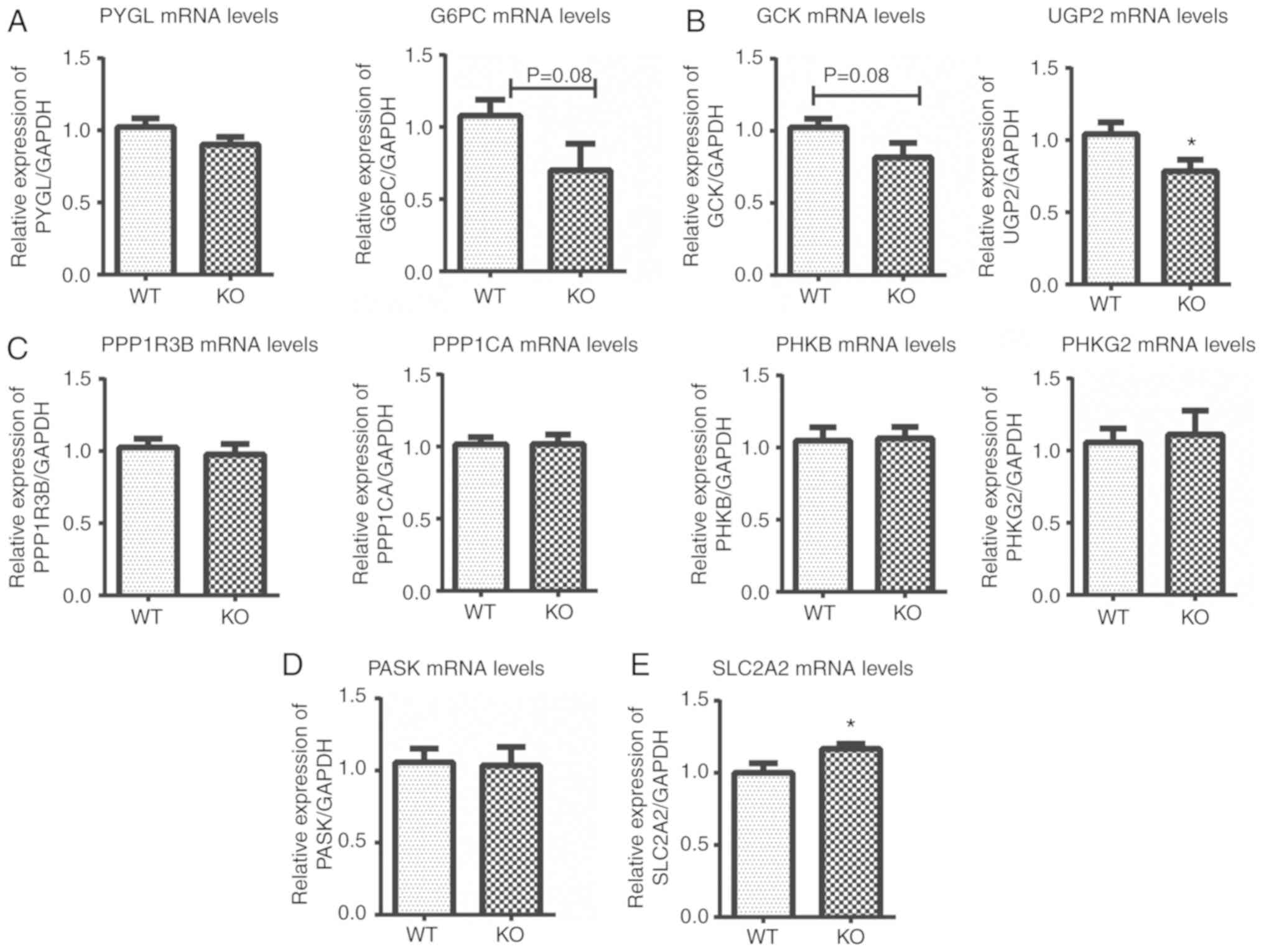 | Figure 3Expression of glycogen-associated
genes at the mRNA level in the livers of EpCAM−/− mice
at E18.5, as detected by reverse transcription-quantitative PCR.
The expression levels of (A) PYGL and G6PC, (B) GCK and UGP2, (C)
PPP1R3B, PPP1CA, PHKB and PHKG2, (D) PASK and (E) SLC2A2 were
determined. *P<0.05, vs. WT mice. EpCAM, epithelial
cell adhesion molecule; WT, wild-type; PYGL, glycogen
phosphorylase; G6PC, glucose-6-phosphatase; GCK, glucokinase; UPG2,
UDP-glucose pyrophosphorylase 2; PPP1R3B, regulatory subunit of
phosphoprotein phosphatase-1, PPC1CA, protein phosphatase 1
catalytic subunit α; PHKB, common β subunit of phosphorylase
kinase; PHKG2, γ subunit hepatic phosphorylase kinase; PASK, PAS
domain containing serine/threonine kinase; SLC2A2, solute carrier
family 2 member 2. |
Overview of circRNAs in the livers of
EpCAM−/− mice
According to the sequence data, 526,000,032 clean
reads (255,388,976 for EpCAM−/− mice and 270,611,056 for
WT mice) were generated. Following the removal of the low quality,
adapter-containing and poly-N containing reads, 518,617,322 high
quality clean reads (251,809,406 from EpCAM−/− mice and
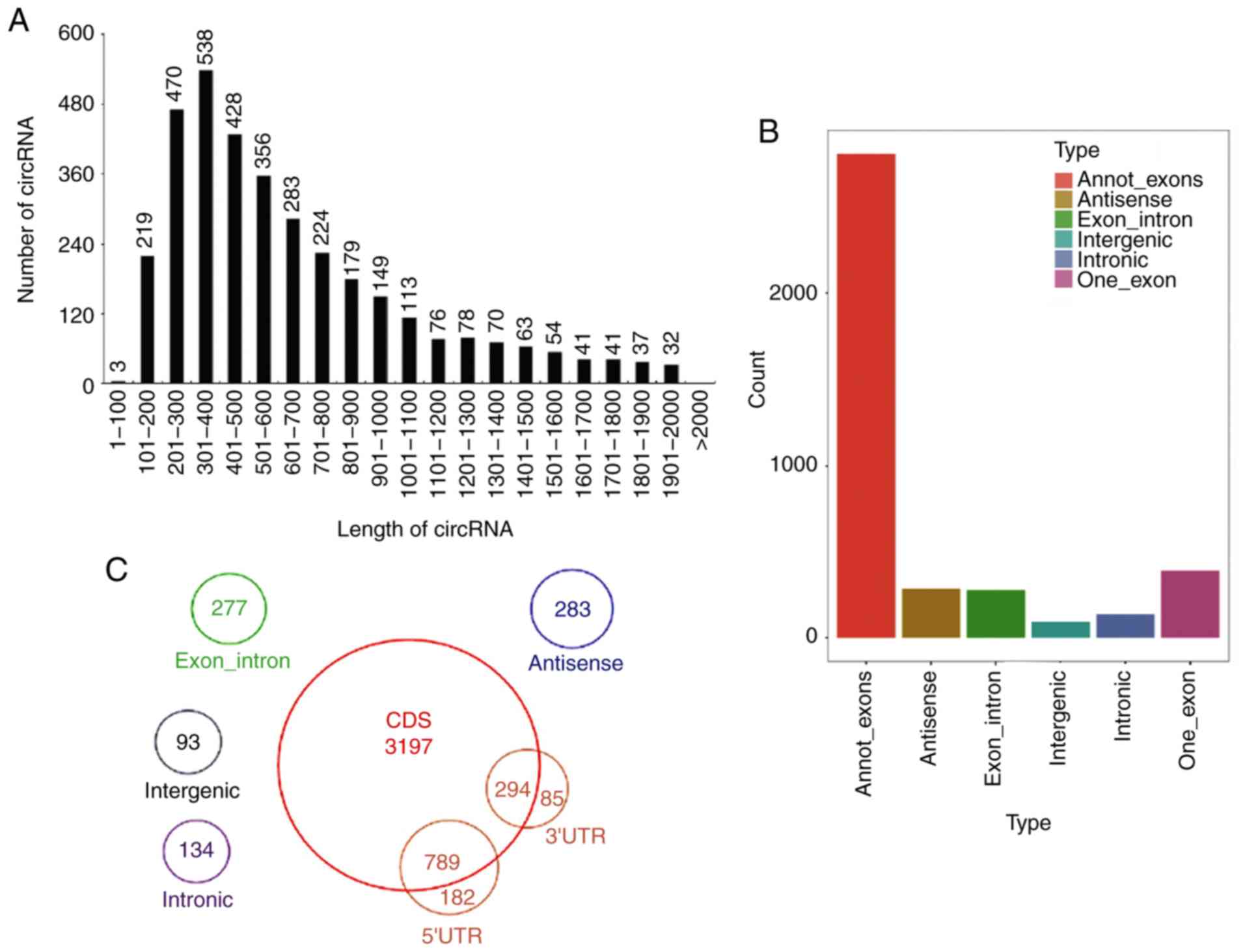
266,807,916 from WT mice) were obtained (Table SIII). A total of 3,984 circRNAs
were identified, including 292 known circRNAs and 3,692 novel
circRNAs. The majority of the circRNAs were ~300-400 nucleotides in
length (Fig. 4A). The length
distribution of known circRNAs and novel circRNAs are presented in
Fig. S2A and B, respectively.
Different types of circRNAs were identified, including annot_exons,
antisense, exon_intron, intergenic, intronic and one_exon circRNAs.
Among these, the most common type was annot_exon followed by
one_exon (Fig. 4B-C). The
different types of known and novel circRNAs are presented in
Fig. S2C and D, respectively, of
which, the first and second most common types were also annot_exon
and one_exon. In addition, the chromosome distributions of known
circRNAs and novel circRNAs are presented in Fig. S2E and F, respectively.
To elucidate the functions of circRNAs, all source
genes were mapped to terms in the GO database and compared with the
background. GO has 3 ontologies including biological process,
cellular component and molecular function. In the biological
process group, the majority of genes were associated with cellular
process, metabolic process and single-organism process. In the
cellular component group, the majority of genes were associated
with cell, cell part and organelle. Additionally, in the molecular
function group, the majority of genes were associated with binding
and catalytic activity (Fig.
4D).
KEGG assignments were used to classify the
functional annotations of source genes to further understand the
biological functions of circRNAs. The top 20 pathways are presented
in Fig. 4E and included ubiquitin
mediated proteolysis, thyroid hormone signaling, adherents
junction, cell cycle and cell senescence, regulation of actin
cytoskeleton, and insulin signaling pathway.
circRNA expression profiles in the livers
of EpCAM−/− mice
The differences in circRNA expression between the
livers of WT and EpCAM−/− mice were compared and those
with a log2 fold change >1 and P-value <0.05 were
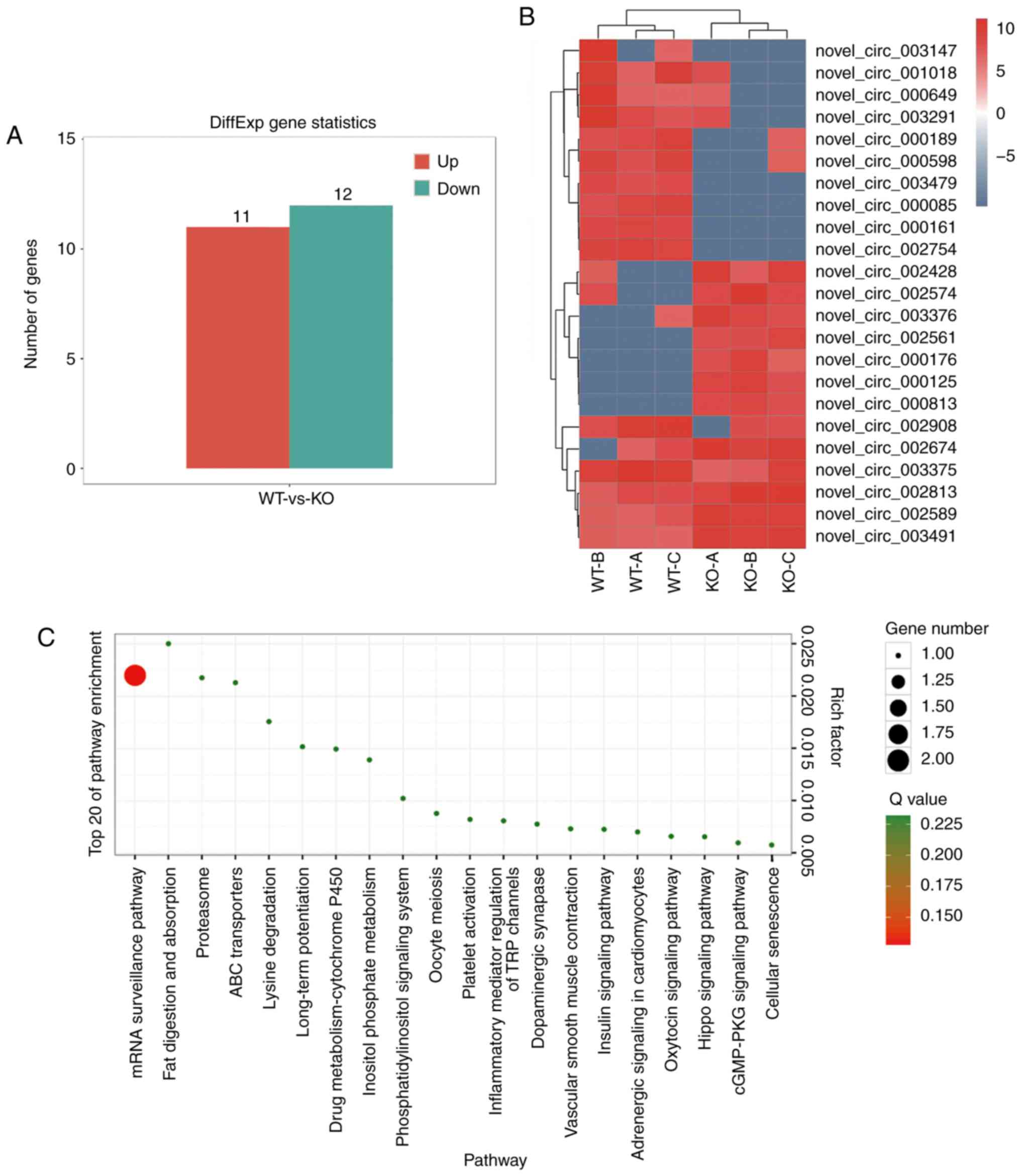
considered statistically significant. A total of 11 upregulated
circRNAs and 12 downregulated circRNAs were identified in the
livers of EpCAM−/− mice compared with the WT group
(Fig. 5A), including 6 known
circRNAs and 17 novel circRNAs. The basic information of the
upregulated and downregulated circRNAs and their predicted target
miRNAs is presented in Tables I
and II, respectively.
 | Table IBasic information and predicted
targets of up-regulated circRNAs. |
Table I
Basic information and predicted
targets of up-regulated circRNAs.
| circRNA ID | Location | Gene symbol | Targets |
|---|
|
novel_circ_000125 | chr 1 | Agap1 | mmu-miR-15b-5p;
mmu-miR-16-5p; mmu-miR-1907; mmu-miR-195a-5p; mmu-miR-195b |
|
novel_circ_000176 | chr 1 | Camsap2 | mmu-let-7a-2-3p;
mmu-miR-103-3p; mmu-miR-107-3p; mmu-miR-124-5p; mmu-miR-139-5p |
|
novel_circ_000813 | chr 12 | Psma6 | mmu-miR-1197-5p;
mmu-miR-154-3p; mmu-miR-1903; mmu-miR-1912-3p; mmu-miR-199a-3p |
|
novel_circ_002426 | chr 3 | Fmo5 | mmu-let-7a-5p;
mmu-let-7c-5p; mmu-let-7d-5p; mmu-let-7f-5p; mmu-let-7g-5p |
|
novel_circ_002561 | chr 4 | Hemgn | mmu-miR-103-3p;
mmu-miR-105; mmu-miR-107-5p; mmu-miR-1197-5p; mmu-miR-1198-5p |
|
novel_circ_002569 | chr 4 | Abca1 | mmu-let-7a-2-3p;
mmu-miR-103-1-5p; mmu-miR-103-2-5p; mmu-miR-1193-5p;
mmu-miR-135a-5p |
|
novel_circ_002574 | chr 4 | Tmem245 | mmu-miR-1192;
mmu-miR-1247-5p; mmu-miR-127-5p; mmu-miR-1306-5p;
mmu-miR-153-5p |
|
novel_circ_002674 | chr 4 | Zmym4 | mmu-miR-152-5p;
mmu-miR-1938; mmu-miR-1955-3p; mmu-miR-1958; mmu-miR-1964-5p |
|
novel_circ_002813 | chr 5 | Ppp1cb | mmu-miR-1264-3p;
mmu-miR-1298-3p; mmu-miR-1306-5p; mmu-miR-130b-5p;
mmu-miR-137-5p |
|
novel_circ_003376 | chr 7 | Pik3c2a | mmu-let-7f-2-3p;
mmu-miR-103-1-5p; mmu-miR-103-2-5p; mmu-miR-107-5p;
mmu-miR-1191a |
|
novel_circ_003491 | chr 8 | Nsd3 | mmu-miR-1224-3p;
mmu-miR-137-5p; mmu-miR-188-5p; mmu-miR-194-1-3p;
mmu-miR-214-5p |
 | Table IIBasic information and predicted
targets of down-regulated circRNAs. |
Table II
Basic information and predicted
targets of down-regulated circRNAs.
| circRNA ID | Location | Gene symbol | Targets |
|---|
|
novel_circ_000085 | chr 1 | Kansl1l | mmu-let-7a-2-3p;
mmu-let-7c-1-3p; mmu-let-7j; mmu-miR-101a-3p; mmu-miR-101c |
|
novel_circ_000161 | chr 1 | Tmcc2 | mmu-miR-1188-5p;
mmu-miR-1190; mmu-miR-1258-5p; mmu-miR-129-1-3p;
mmu-miR-129-2-3p |
|
novel_circ_000189 | chr 1 | Rnf2 | mmu-miR-1933-5p;
mmu-miR-19b-1-5p; mmu-miR-19b-2-5p; mmu-miR-302b-5p;
mmu-miR-302c-5p |
|
novel_circ_000596 | chr 11 | Smg6 | mmu-miR-105;
mmu-miR-107-5p; mmu-miR-1190; mmu-miR-1224-5p; mmu-miR-1231-5p |
|
novel_circ_000649 | chr 11 | Vezf1 | mmu-miR-106a-3p;
mmu-miR-1187; mmu-miR-1197-3p; mmu-miR-132-5p; mmu-miR-134-5p |
|
novel_circ_001018 | chr 13 | Klhl3 | mmu-miR-1251-3p;
mmu-miR-133a-3p; mmu-miR-133b-3p; mmu-miR-133c;
mmu-miR-135a-5p |
|
novel_circ_002754 | chr 4 | Rere | mmu-miR-181d-3p;
mmu-miR-1964-5p; mmu-miR-28a-5p; mmu-miR-666-3p;
mmu-miR-6929-3p |
|
novel_circ_002908 | chr 5 | Aff1 | mmu-miR-1197-3p;
mmu-miR-1198-5p; mmu-miR-137-3p; mmu-miR-145a-3p;
mmu-miR-145a-5p |
|
novel_circ_003147 | chr 6 | Slc41a3 | mmu-miR-1197-5p;
mmu-miR-1943-5p; mmu-miR-29a-3p; mmu-miR-29b-3p;
mmu-miR-29c-3p |
|
novel_circ_003291 | chr 7 | Gas2 | mmu-miR-124-3p;
mmu-miR-1904; mmu-miR-207; mmu-miR-210-5p; mmu-miR-24-3p |
|
novel_circ_003375 | chr 7 | Pik3c2a | mmu-miR-1898;
mmu-miR-1904; mmu-miR-216b-5p; mmu-miR-22-3p; mmu-miR-300-5p |
|
novel_circ_003479 | chr 8 | Efnb2 | mmu-let-7a-1-3p;
mmu-let-7c-2-3p; mmu-miR-105; mmu-miR-1192; mmu-miR-1224-3p |
The results of hierarchical clustering, which is one
of the most commonly used clustering techniques for gene expression
analysis, revealed the distinguishable circRNA expression profiles
between the WT and EpCAM −/− mice (Fig. 5B).
The results of KEGG pathway analysis demonstrated
that the top 20 pathways were the mRNA surveillance pathway, fat
digestion and absorption, insulin signaling, Hippo signaling and
cell senescence, as well as others (Fig. 5C).
The results of GO analysis determined that in the
biological process group, the majority of source genes of
differentially expressed circRNAs were associated with cellular
process, single-organism process, metabolic process and
developmental process. In the cellular component group, the
majority of genes were associated with cell, cell part, organelle
organelle part and membrane-enclosed lumen. Furthermore, in the
molecular function group, the majority of genes were associated
with binding and catalytic activity (Fig. 5D).
Validation of circRNA expression profiles
in the livers of EpCAM−/− mice
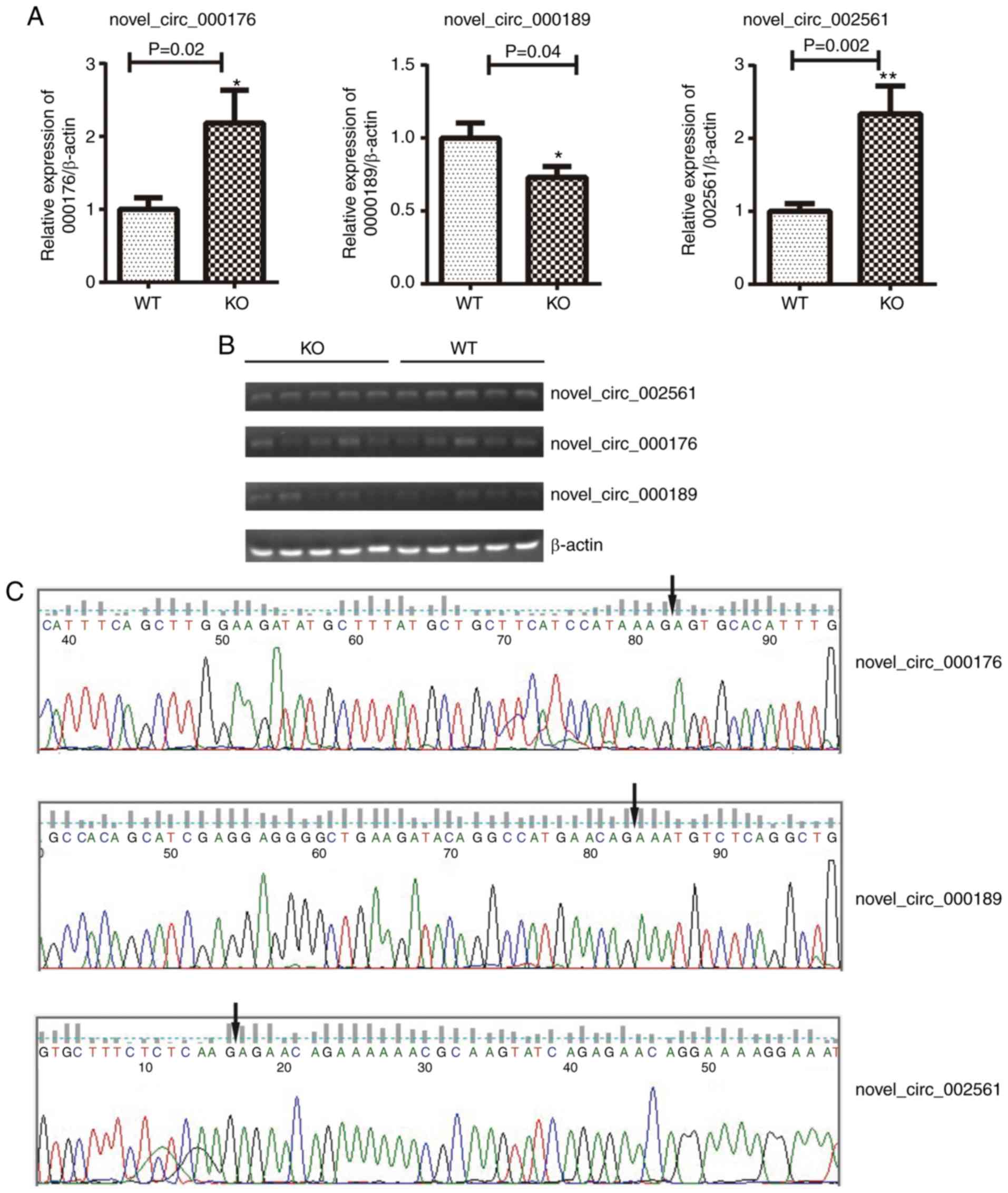
To validate circRNA profiles, 3 circRNAs were
randomly selected and amplified by RT-qPCR. The results revealed
that the expression levels of novel_circ_00176 and _circ_002561
were significantly increased in the livers of EpCAM−/−
mice compared with those of WT mice. Furthermore, the expression of
novel_circ_00189 was decreased in the livers of EpCAM−/−
mice (Fig. 6A). These results
were in agreement with the RNA-seq data. The PCR products were
subsequently analyzed using agarose gels to validate single DNA
amplifications (Fig. 6B). In
addition, the identity of 3 selected circRNAs were further
confirmed by Sanger sequencing (Fig.
6C).
Prediction of target association of
circRNA-miRNA-mRNA network
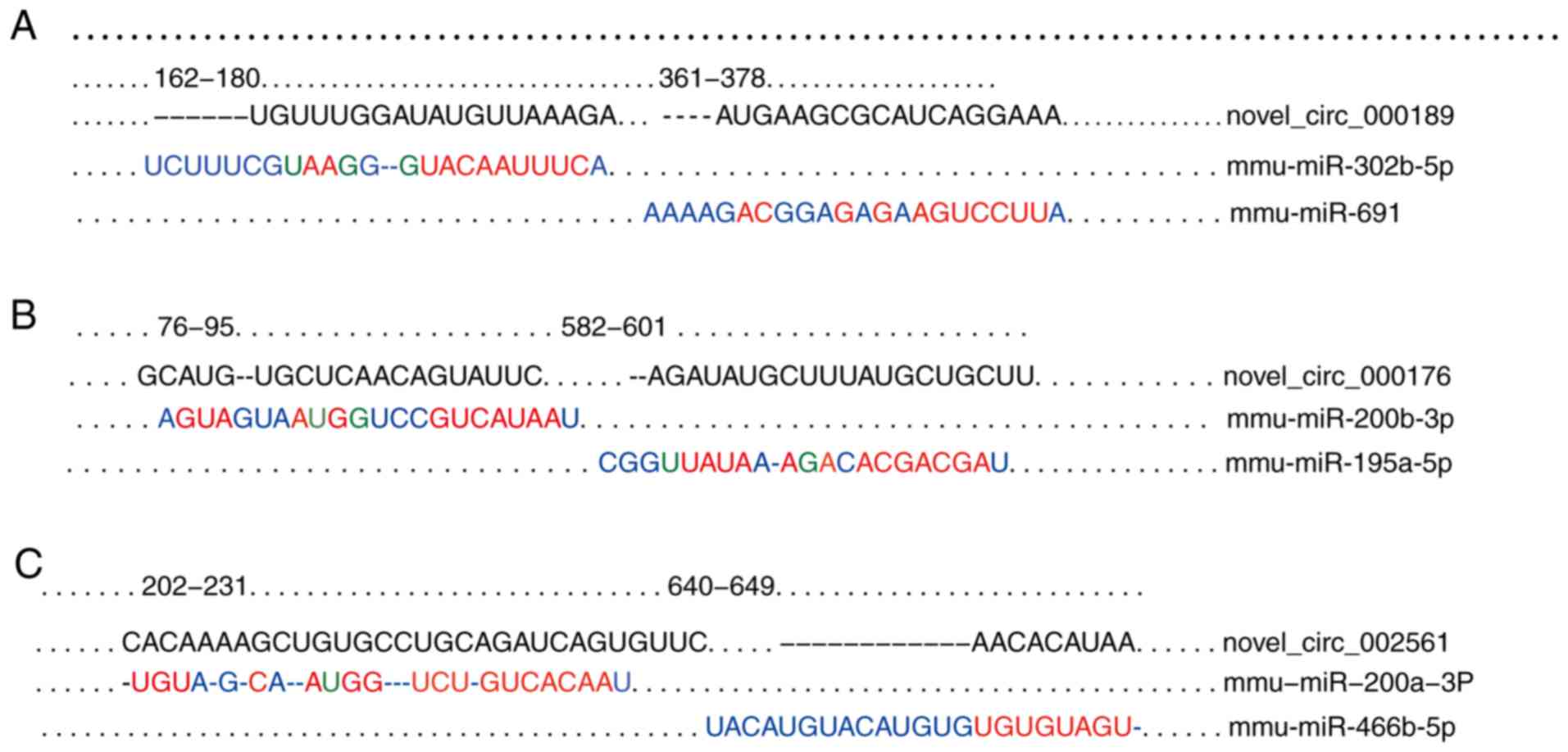
To confirm the functions of circRNAs as 'miRNAs
sponges', the mmu-miR-302b-5p and mmu-miR-691 binding sites of
novel_circ_000189; the mmu-miR-200b-3p and mmu-miR-195a-5p binding
sites of novel_circ_000176; and the mmu-miR-200a-3p and
mmu-miR-466b-5p binding sites of novel_circ_002561 are presented in
Fig. 7. All the established miRNA
binding sites of the 3 selected circRNAs are presented in Figs. S3-S5. The regulatory
circRNA-miRNA-mRNA networks of the 3 circRNAs were further
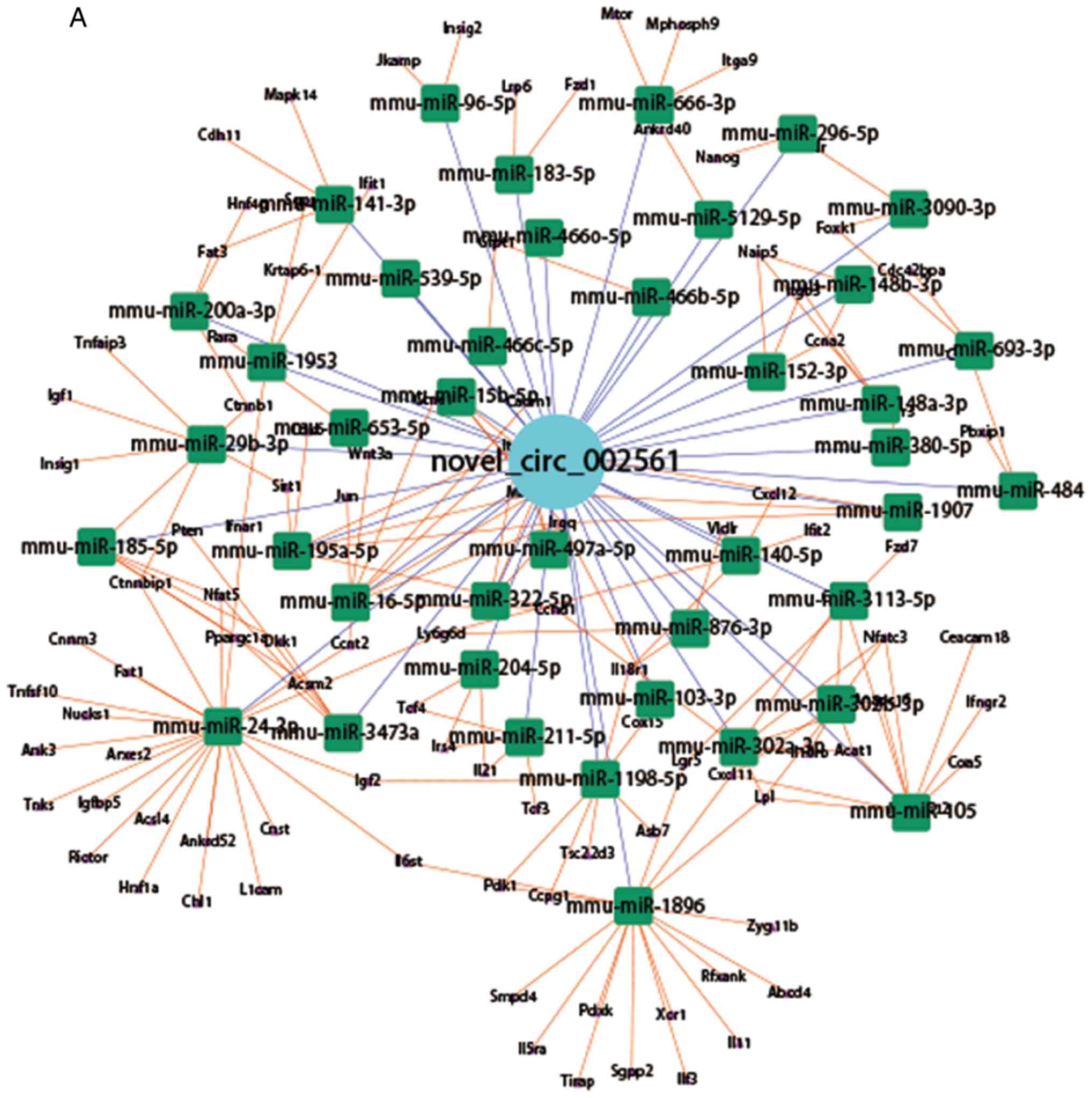
established. In novel_circ_002561, the target genes associated with
the Wnt signaling pathway [(Wnt family member 3a (Wnt3a), Lgr5,
transcription factor (Tcf)3 and Tcf4 and others], cell junctions
(cadherin 1, catenin beta-1 and cell adhesion molecule 1 and
others), cell cycle and division (cyclin E1, anaphase promoting
complex subunit 16, cyclin T2 and cyclin-dependent kinase 6 and
others), stem cell (Nanog), metabolism (insulin-induced gene 2,
hepatocyte nuclear factor 4 gamma, insulin-like growth factor 1 and
ATP binding cassette subfamily D and others) and inflammation
[interleukin (IL)11, IL6 signal transducer, IL5 receptor subunit
alpha and tumor necrosis factor alpha-induced protein 3] were
selected to establish the circRNA-miRNA-mRNA network. As presented
in Fig. 8A, novel_circ_002561 was
predicted to combine with 41 miRNAs to regulate the expression of
104 target genes. Some of the 41 miRNAs were mmu-miR-96-5p,
mmu-miR-666-3p, mmu-miR-183-5p, mmu-miR-141-3p, mmu-miR-296-5p and
mmu-miR-200a-3p. The networks of 2 further circRNAs,
novel_circ_000176 and novel_circ_000189 are presented in Fig. 8B and C, respectively. The network
of all the target genes of novel_circ_002561, novel_circ_000176 and
novel_ circ_000189 are presented in Fig. S6. The networks of the 6 known
differentially expressed circRNAs (novel_circ_000125,
novel_circ_000596, novel_circ_000649, novel_circ_002754,
novel_circ_002813 and novel_circ_003291) and 6 other randomly
selected novel differentially expressed circRNAs are presented in
Figs. S7-S8.
Discussion
EpCAM is a cell-cell adhesion molecule, which serves
important roles in cell signaling, proliferation, differentiation,
formation and maintenance of organ morphology (1). The phonotype of EpCAM knockout mice
primarily manifests the symptoms of intestinal defects, with
abnormalities in tight junctions and barrier functions of the
intestinal epithelium (2).
Recently, EpCAM was identified as a novel marker of cancer stem
cells in HCC. Additionally, EpCAM-positive HCC cells exhibited
hepatic cancer stem cell-like traits that could initiate highly
invasive HCC in severe combined immunodeficient mice (33).
In the current study, the results of
immunofluorescence staining demonstrated that EpCAM was mainly
expressed on the liver cholangiocytes of WT mice at E18.5 and P0.
However, this expression was completely eradicated in the livers of
EpCAM−/− mice, indicating that a successful knockout
mouse model was generated in the present study. RT-qPCR analysis of
genes associated with liver development revealed increased CD34
levels in the livers of EpCAM−/− mice, compared with WT
mice. CD34 is a marker of hepatic oval cells, resembling hepatic
progenitor cells (HPCs), located in the periportal region of the
liver (36). In response to liver
injury, HPCs become involved in the proliferation and
differentiation of liver cells (34). Since the expression of CD34
increased in the EpCAM knockout mice in the current study, this was
confirmed to resemble a type of liver injury. In addition, PDCs
have long been considered a type of putative liver stem/progenitor
cell, and EpCAM-positive PDCs with genetic alterations induced by
chemicals have been determined to develop into HCC, resembling
human cholangiolocellular carcinomas (CLCs). During this process,
the Wnt signaling pathway was demonstrated to be specifically
upregulated in the CLC components of PDC-derived HCC (10). The results of the present study
revealed that the expression of Axin2, an important component and
target gene of the Wnt signaling pathway, was significantly
decreased in the livers of EpCAM−/− mice. This result
indicated that EpCAM may interact with the Wnt signaling pathway to
play an important role in the liver development and liver diseases.
Furthermore, the present results revealed the distinct expression
of circRNAs in the livers of EpCAM knockout mice, such as
novel_circ_002561, which could regulate the transcription of Wnt
signaling related genes (Wnt3a, Lgr5, Tcf3 and Tcf4, and others).
It was therefore hypothesized that EpCAM may regulate liver
development via certain circRNAs.
circRNAs act as 'sponges' for microRNAs and RNA
binding proteins to regulate target gene expression (16,35,36). circRNAs have recently gained
interest due to their complex involvement in the regulation of
transcriptional processes their important roles in human diseases
and their potential to serve as biomarkers and potential clinical
targets (37). Although circRNAs
modulate transcription and interfere with splicing, the expression
pattern and functions of the majority of circRNAs remain largely
unexplored. Therefore, the study of circRNAs and their dynamic
expression patterns and complicated regulatory networks in
different biological processes and diseases will help to further
classify them (38). In the
present study, high-throughput sequencing identified 3,984
circRNAs, including 292 known circRNAs and 3,692 novel circRNAs in
the liver of WT and EpCAM−/− mice. The current study
also identified 11 upregulated and 12 downregulated circRNAs in the
livers of EpCAM−/− mice compared with the WT group.
Within these circRNAs, 6 were known circRNAs and 17 were novel
circRNAs. The results of GO analysis revealed that the majority of
source genes of the differentially expressed circRNAs were
associated with cellular process, single-organism process,
metabolic process and developmental process. These data were
consistent with the identification of development associated genes
via RT-qPCR. These results demonstrated the important role of EpCAM
in liver development.
H&E staining revealed reduced cytoplasmic
vacuolation of the liver, in EpCAM−/− mice, suggesting
defects in hepatic glycogen storage (28). The analysis of glycogen-associated
genes then identified two UDP-glucose pyrophosphorylase genes (UGP1
and UGP2), which are essential for sucrose and polysaccharide
synthesis (39). The results of
the current study also revealed that the expression of UGP2 was
decreased in the livers of EpCAM−/− mice. In addition,
the expression of SLC2A2 was increased in the EpCAM−/−
livers. SLC2A2 encodes the glucose transporter 2 (GLUT2), and a
defect may lead to neonatal diabetes, hepatomegaly and renal
Fanconi syndrome (40). The
results of the current study demonstrated that EpCAM affected
glycogen synthesis and genes associated with diseases of glycogen
storage. However, the underlying mechanisms warrant further
investigation in future studies. The results of KEGG pathway
analysis demonstrated that the top 20 pathways of differentially
expressed circRNA source genes included mRNA surveillance, fat
digestion and absorption and insulin signaling pathways. These
bioinformatics data were consistent with the results of RT-qPCR and
glycogen H&E staining, indicating that EpCAM may also be
involved in liver glycogen metabolism.
The results of H&E staining also revealed that
the livers of EpCAM −/− mice at P4 contained marked
vacuolation compared with the livers of WT mice.
EpCAM−/− mice also manifested intestinal barrier defects
and died shortly after birth as a result of intestinal erosion,
showing similar phenotypes with a previous study (2). It is hypothesized that
EpCAM−/− mice exhibiting intestinal defects may have
difficulties in digesting and absorbing food and energy, which
affected liver glycogen metabolism, causing vacuolation. This
hypothesis was supported by the data of the present study. However,
the underlying mechanisms require further investigation.
To further study the functions and mechanisms of
liver EpCAM, the target associations of the circRNA-miRNA- mRNA
networks was predicted. Based on experimental results, target genes
associated with the Wnt signaling pathway, cell junctions, cell
cycle and division, stem cells, metabolism and inflammation were
selected to establish the circRNA-miRNA-mRNA network. The results
identified Novel_circ_002561, located in chromosome 4, whose
expression was up-regulated in the livers of EpCAM−/−
mice. The circRNA-microRNA-mRNA network of novel_circ_002561, its
target genes and target microRNAs were also established. From
network analysis, the regulatory association was clearly
demonstrated, providing directions for further analysis.
In circRNAs profiling, 6 known circRNAs were
differentially expressed. Novel_circ_000125 (mm9_ circ_017649) and
novel_circ_002813 (mm9_circ_018676) were up-regulated in the livers
of EpCAM−/− mice, while novel_circ_000596
(mm9_circ_017983), novel_circ_000649 (m m9_ci rc_ 015947), novel_ci
rc_ 0 02754 (m m9_ circ_013483) and novel_circ_003291
(mm9_circ_003692) were down-regulated. As determined by Memczak
et al (27),
novel_circ_000125 and novel_circ_003291 were detected in the brains
and heads of mice, novel_circ_000596 was detected in the embryonic
stem cells (ES) and heads of mice, novel_circ_000649 was detected
in the heads of mice, novel_circ_002754 was detected in the ES and
brains of mice, and novel_circ_0002813 was detected in the ES,
brains and heads of mice. The current study determined that these
circRNAs were also expressed in the livers of WT mice at the P0
stage and that their expression was altered in the livers of
EpCAM−/− mice. However, their functions, mechanisms and
homologous human sequences remain obscure. The circRNAs determined
in the present study may provide useful information for further
study.
In conclusion, the current study revealed that the
expression of certain development and glycogen-associated genes was
altered in the livers of EpCAM−/− mice. The livers of
EpCAM−/− mice exhibited glycogen shortages at E18.5 and
vacuolation at P4. Based on RNA sequencing, a circRNA expression
profile of EpCAM−/− murine livers was established.
Several novel circRNAs were identified, and the circRNA-miRNA-mRNA
regulatory network was established. These results may provide
important information and direction for the future development of
novel targets for the treatment of liver disease.
Supplementary Materials
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 31671520 and 81803912), the
Science and Technology Project of Guangdong Province (grant nos.
2016B050501003 and 2017B050504005), the Scientific Research Project
of the Administration of Traditional Chinese Medicine of Guangdong
Province (grant no. 20182079), the Characteristic Innovation
Project (Natural Science) of the Education Department of Guangdong
Province and the 'Innovation Strong School Project' of Guangdong
Pharmaceutical University (grant no. 2017KTSCX102), and the Science
and Technology Project of Yue-Xiu District of Guangzhou (grant no.
2018-WS-011).
Availability of data and materials
The datasets generated and analyzed during the
current study are not publicly available due to the reason that
part of the dataset has not been analyzed and published yet, but
are available from the corresponding author on reasonable
request.
Authors' contributions
ZL, LL and JG designed the study and conceived the
report. YY analyzed and interpreted the results of RNA sequencing,
wrote the first draft of the manuscript and revised it critically.
SL, GC and LH established the mouse model. FY, YLi, LY, WL and YLe
performed the molecular experiments, including RT-qPCR and H&E
staining. YLe also created the figures. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were approved by the
Committee on the Laboratory Animal Care and Use of Guangdong
Pharmaceutical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to express their gratitude to
Professor Yaacov Ben-David (University of Toronto) for markedly
revising the text of the manuscript.
References
|
1
|
Huang L, Yang Y, Yang F, Liu S, Zhu Z, Lei
Z and Guo J: Functions of EpCAM in physiological processes and
diseases (Review). Int J Mol Med. 42:1771–1785. 2018.PubMed/NCBI
|
|
2
|
Lei Z, Maeda T, Tamura A, Nakamura T,
Yamazaki Y, Shiratori H, Yashiro K, Tsukita S and Hamada H: EpCAM
contributes to formation of functional tight junction in the
intestinal epithelium by recruiting claudin proteins. Dev Biol.
371:136–145. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guerra E, Lattanzio R, La Sorda R, Dini F,
Tiboni GM, Piantelli M and Alberti S: mTrop1/Epcam knockout mice
develop congenital tufting enteropathy through dysregulation of
intestinal E-cadherin/β-catenin. PLoS One. 7:pp. e493022012,
View Article : Google Scholar
|
|
4
|
Mueller JL, McGeough MD, Peña CA and
Sivagnanam M: Functional consequences of EpCam mutation in mice and
men. Am J Physiol Gastrointest Liver Physiol. 306:G278–G288. 2014.
View Article : Google Scholar :
|
|
5
|
Yousaf M, Tayyeb A and Ali G: Expression
profiling of adhesion proteins during prenatal and postnatal liver
development in rats. Stem Cells Cloning. 10:21–28. 2017.PubMed/NCBI
|
|
6
|
Tanaka M, Okabe M, Suzuki K, Kamiya Y,
Tsukahara Y, Saito S and Miyajima A: Mouse hepatoblasts at distinct
developmental stages are characterized by expression of EpCAM and
DLK1: Drastic change of EpCAM expression during liver development.
Mech Dev. 126:665–676. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Okabe M, Tsukahara Y, Tanaka M, Suzuki K,
Saito S, Kamiya Y, Tsujimura T, Nakamura K and Miyajima A:
Potential hepatic stem cells reside in EpCAM+ cells of normal and
injured mouse liver. Development. 136:1951–1960. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmelzer E, Zhang L, Bruce A, Wauthier E,
Ludlow J, Yao HL, Moss N, Melhem A, McClelland R, Turner W, et al:
Human hepatic stem cells from fetal and postnatal donors. J Exp
Med. 204:1973–1987. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu H, Ma J, Yang Y, Shi W and Luo L: EpCAM
is an endoderm-specific Wnt derepressor that licenses hepatic
development. Dev Cell. 24:543–553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matsumoto T, Takai A, Eso Y, Kinoshita K,
Manabe T, Seno H, Chiba T and Marusawa H: Proliferating
EpCAM-positive ductal cells in the inflamed liver give rise to
hepatocellular carcinoma. Cancer Res. 77:6131–6143. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamashita T, Forgues M, Wang W, Kim JW, Ye
Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, et al: EpCAM and
alpha-fetoprotein expression defines novel prognostic subtypes of
hepatocellular carcinoma. Cancer Res. 68:1451–1461. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamashita T, Ji J, Budhu A, Forgues M,
Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, et al:
EpCAM-positive hepatocellular carcinoma cells are tumor-initiating
cells with stem/progenitor cell features. Gastroenterology.
136:1012–1024. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji J, Yamashita T, Budhu A, Forgues M, Jia
HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al: Identification
of microRNA-181 by genome-wide screening as a critical player in
EpCAM-positive hepatic cancer stem cells. Hepatology. 50:472–480.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mani SK, Zhang H, Diab A, Pascuzzi PE,
Lefrançois L, Fares N, Bancel B, Merle P and Andrisani O:
EpCAM-regulated intra-membrane proteolysis induces a cancer stem
cell-like gene signature in hepatitis B virus-infected hepatocytes.
J Hepatol. 65:888–898. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song Y, Liu C, Liu X, Trottier J, Beaudoin
M, Zhang L, Pope C, Peng G, Barbier O, Zhong X, et al: H19 promotes
cholestatic liver fibrosis by preventing ZEB1-mediated inhibition
of epithelial cell adhesion molecule. Hepatology. 66:1183–1196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu C, Sun X, Li N, Wang W, Kuang D, Tong
P, Han Y and Dai J: CircRNAs in the tree shrew (Tupaia belangeri)
brain during postnatal development and aging. Aging (Albany NY).
10:833–852. 2018. View Article : Google Scholar
|
|
17
|
Maiese K: Disease onset and aging in the
world of circular RNAs. J Transl Sci. 2:327–329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo Q, Zhang L, Li X, Fu B, Deng Z, Qing
C, Su R, Xu J, Guo Y, Huang Z and Li J: Identification of circular
RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for
rheumatoid arthritis. Clin Exp Immunol. 194:118–124. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X and Fang L: Advances in circular
RNAs and their roles in breast cancer. J Exp Clin Cancer Re.
37:2062018. View Article : Google Scholar
|
|
20
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular functions and specific roles of circRNAs in the
cardiovascular system. Noncoding RNA Res. 3:75–98. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu CX and Sun S: An emerging role for
circular RNAs in osteoarthritis. Yonsei Med J. 59:349–355. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao R, Zou H and Liao W: Prospect of
circular RNA in hepatocellular carcinoma: A novel potential
biomarker and therapeutic target. Front Oncol. 8:3322018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo XY, He CX, Wang YQ, Sun C, Li GM, Su
Q, Pan Q and Fan JG: Circular RNA profiling and bioinformatic
modeling identify its regulatory role in hepatic steatosis. Biomed
Res Int. 2017:59361712017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo J, Zhou Y, Cheng Y, Fang W, Hu G, Wei
J, Lin Y, Man Y, Guo L, Sun M, et al: Metformin-induced changes of
the coding transcriptome and non-coding RNAs in the livers of
non-alcoholic fatty liver disease mice. Cell Physiol Biochem.
45:1487–1505. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Choi E, Zhang X, Xing C and Yu H: Mitotic
checkpoint regulators control insulin signaling and metabolic
homeostasis. Cell. 166:567–581. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stygar D, Andrare D, Bażanów B, Chełmecka
E, Sawczyn T, Skrzep-Poloczek B, Olszańska E, Karcz KW and Jochem
J: The impact of DJOS surgery, a high fat diet and a control diet
on the enzymes of glucose metabolism in the liver and muscles of
Sprague-Dawley rats. Front Physiol. 10:5712019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Adeva-Andany MM, González-Lucán M,
Donapetry-García C, Fernández-Fernández C and Ameneiros-Rodríguez
E: Glycogen metabolism in humans. BBA Clin. 5:85–100. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang DD, Zhang JG, Wang YZ, Liu Y, Liu GL
and Li XY: Per-Arnt-Sim Kinase (PASK): An emerging regulator of
mammalian glucose and lipid metabolism. Nutrients. 7:7437–7450.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hajiaghaalipour F, Khalilpourfarshbafi M
and Arya A: Modulation of glucose transporter protein by dietary
flavonoids in type 2 diabetes mellitus. Int J Biol Sci. 11:508–524.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Terris B, Cavard C and Perret C: EpCAM, a
new marker for cancer stem cells in hepatocellular carcinoma. J
Hepatol. 52:280–281. 2010. View Article : Google Scholar
|
|
34
|
Weiss TS and Dayoub R: Thy-1
(CD90)-positive hepatic progenitor cells, hepatoctyes, and
non-parenchymal liver cells isolated from human livers. Methods Mol
Biol. 1506:75–89. 2017. View Article : Google Scholar
|
|
35
|
Bose R and Ain R: Regulation of
transcription by circular RNAs. Adv Exp Med Biol. 1087:81–94. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Franz A, Rabien A, Stephan C, Ralla B,
Fuchs S, Jung K and Fendler A: Circular RNAs: A new class of
biomarkers as a rising interest in laboratory medicine. Clin Chem
Lab Med. 56:1992–2003. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Q, Yang ZL, Zou Q, Yuan Y, Li J,
Liang L, Zeng G and Chen S: SHP2 and UGP2 are biomarkers for
progression and poor prognosis of gallbladder cancer. Cancer
Invest. 34:255–264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Khandelwal P, Sinha A, Jain V, Houghton J,
Hari P and Bagga A: Fanconi syndrome and neonatal diabetes:
Phenotypic heterogeneity in patients with GLUT2 defects. CEN Case
Rep. 7:1–4. 2018. View Article : Google Scholar :
|