Introduction
Esophageal cancer (EC) is the eighth most frequent
type of cancer in the world and has a poor prognosis due to its
aggressiveness and poor survival rate. In 2003, >90% of cases of
EC were esophageal squamous cell cancer (ESCC) and esophageal
adenocarcinoma (ECA), and this proportion increased to 95% in 2017
(1,2). In patients with ECA, the nidus is
usually found in the distal esophagus, whereas the nidus in
patients with ESCC is more commonly distributed between the middle
and lower third of the esophagus (3,4).
ESCC is more frequent in developing countries, and risk factors,
including achalasia, smoking and alcohol use, are common to ESCC
(2). In terms of the clinical cT
category, ~20% patients with stage cT4a ESCC have a survival time
of 10 years, and <30% patients have a survival time of 5 years
(5). CDKN2A hypermethylation,
which is frequent in patients with ESCC, accounts for 40-62% of
cases, however, there is lack of valid evidence to confirm
methylated CDKN2A as a biomarker for ESCC (6).
MicroRNAs (miRNAs) are ~22 nt in length, and they
fall complementally in the 3′ untranslated region (3′UTR) in target
mRNAs, therefore, they have a critical function in animals and
plants (7). It has been reported
that >60% of human protein-coding genes are regulated by miRNAs,
and miRNAs originating from the diet or endogenous synthesis are
involved in physiology and pathology, including inflammation and
cancer (8,9). miRNAs can be upregulated or
downregulated in tumor tissues compared with normal tissues, and
they can be divided into tumor promoter and tumor suppressor miRNAs
(10). miRNA (miR)-15/16, miR-34
and the miR-200 family are reported to have inhibitory effects on
lung cancer; the overexpression of miR-15/16 inhibited non-small
cell lung cancer cell proliferation and invasion by regulating
TWIST1 (10,11); miR-17-92, miR-222/221 and miR-155
were found to promote the progression of breast cancer, and
miR-17-5p promoted breast cancer cell invasion and migration
through the inhibition of HMG box-containing protein 1 (10,12).
The miR-17-92 cluster is composed of six miRNAs
(miR-17, miR-18a, miR-19a, miR-19b, miR-20a and miR-92a-3p) and is
located on chr13q31.3 within the third intron of the
C13orf25/MIR17HG gene (13).
miR-92a-3p is considered as a tumor promotor, and a previous study
reported that miR-92a-3p acted as an oncomiR in colorectal cancer
cells via regulating the phosphatase and tensin homolog deleted on
chromosome 10 (PTEN)-mediated PI3K/AKT pathway (14). The serum levels of miR-92a-3p were
reported to be associated with ESCC (15). Therefore, miR-92a-3p was
considered a miRNA of interest and PTEN was considered a target
mRNA of interest in ESCC in the present study. The transfer of
miR-93-5p by exosomes promotes the proliferation of EC cells via
intercellular communication by targeting PTEN (16). PTEN has been reported to be
associated with tumors, including esophageal tumors. A novel
heterozygous mutation in the PTEN gene was reported to be
associated with an ovarian germ cell tumor complicated by growing
teratoma syndrome and overgrowth in a two-year-old female (17). The fibroblast growth factor
receptor 2-mediated phosphorylation of PTEN at tyrosine 240
contributes to the radioresistance of glioma (18). miR-21 suppresses anoikis through
targeting PTEN in human esophageal adenocarcinoma (19). Therefore, PTEN may be regulated
differently in EC. However, whether miR-92a-3p regulates PTEN in
ESCC remains unknown.
To investigate the role of the downregulation of
PTEN, it was hypothesized in the present study that miR-92a-3p
targeted PTEN to exert effects on the progression of ESCC. As the
modulation of miRNAs can be realized by miRNA mimics and miRNA
antagonists (20), the present
study used the miR-92a-3p mimic and miR-92a-3p inhibitor to
investigate the effect of miR-92a-3p on ESCC, according to cell
proliferation, migration and invasion, and examine effects of the
target gene of miR-92a-3p on ESCC cell proliferation, migration,
invasion and survival pathways. This study may provide a better
understanding of the mechanism underlying the occurrence of
ESCC.
Materials and methods
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
The samples used in the present study consisted of
23 EC tissues and adjacent tissues, which were collected from
patients with ESCC (14 males and 9 females, 43-65 years old) from
Shanxi Provincial People's Hospital, the HET-1A cell line (normal
esophageal cell line) and ESCC cell lines between 2016 and 2017.
The patients signed an informed consent form and the study was
approved by the Ethics Committee of Shanxi Provincial People's
Hospital (no. SP2016064926). RNAs were extracted from samples with
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) by centrifugation (Cence, Changsha, Hunan, China) at 8,000 × g
for 10 min at 4°C. The concentration of RNA was determined using a
Multiskan reader (Thermo Scientific, Waltham, MA, USA) at a
wavelength of 260 nm. A cDNA kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used to synthesize cDNA from the RNA
substrate, according to the manufacturer's instructions.
The deoxyribonucleotide primers, which were
synthesized by Sangon Biotech (Shanghai, China), used in the PCR
are listed in Table I. qPCR was
performed under the following conditions: Initial denaturation at
95°C for 90 sec, 45 cycles of denaturation at 95°C for 30 sec,
annealing at 50°C for 60 sec, extension at 72°C for 35 sec and a
final extension at 72°C for 10 min. The 2−∆∆Cq method
(21) was used to analyze
relative mRNA.
 | Table ISequences of primers used in
polymerase chain reaction. |
Table I
Sequences of primers used in
polymerase chain reaction.
| Primer name | Sequences
(5′-3′) |
|---|
| miR-92a-3p | F:
UAUUGCACUGUCCCGGCCUGU |
| R:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAGGCCG |
| U6 | F:
GTGCTCCCTGCTTCGGCAGCACATATAC |
| R:
AAAAATATGGAACGCTTCACGAATTTG |
Cell culture and cell transfection
HET-1A cells originate from the human esophagus and
were purchased from the Cancer Institute, Chinese Academy of
Medical Sciences (CICAMS; Beijing, China). The ESCC cell lines
(Eca-109, EC9706, KYSE-30, KYSE-150 and KYSE-220) and KYSE-510 cell
line originate from the esophagus of patients with ESCC and were
purchased from CICAMS. All cells were cultured with RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% 10,000
U/ml penicillin-10,000 µg/ml streptomycin (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C with 5% CO2 in an
incubator (Thermo Fisher Scientific, Inc.). The cells
(5×105) were inoculated into each well of 96-well
plates. The miR-92a-3p mimic, miR-92a-3p inhibitor, mimic control,
inhibitor control, pcDNA3.1 plasmid and PTEN-pcDNA3.1
overexpression plasmid were synthesized by Guangzhou RiboBio Co.,
Ltd. (Guangzhou, China). Depending on the group, 1 µg
PTEN-pcDNA3.1 overexpression plasmid (PTEN group) or pcDNA3.1
plasmid (NC group), 0.25 µg mimics (mimic group) or
inhibitor (inhibitor group) or mimics control (mimic control group)
or inhibitor control (inhibitor control group) were mixed (2:1)
with 1 µl lipofectamine (Invitrogen; Thermo Fisher
Scientific, Inc.) and free-FBS RPMI-1640 medium, and the mixed
solution was used to treat the cells for 4 h in an incubator at
37°C. Subsequently, the culture medium was replaced by the mixed
solution to culture the cells. Cells without any treatment were
used as a blank control (Blank group). The sequence were as
follows: Mimic, 3′-UGU CCG GCC CUG UUC ACG UUA U-5′; inhibitor,
3′-ACA GGC CGG GAC AAG UGC AAU A-5′; mimic control, 5′-UUU GUA CUA
CAC AAA AGU ACU G-3′; inhibitor control, 5′-CAG UAC UUU UGU GUA GUA
CAA A-3′.
Cell apoptosis and proliferation
assays
To analyze apoptosis, the cells (5×103
cells/well) were added into a 96-well plate. Insulin-like growth
factor 1 (IGF-1) was purchased from Bersee (Beijing, China). The
cells were resuspended with phosphate-buffered saline (PBS; Gibco;
Thermo Fisher Scientific, Inc.). Cell apoptosis were detected using
Annexin V FITC (5 µl) and propidium iodide (PI, 5 µl)
for 5 min at 37°C and analyzed using flow cytometry (Invitrogen;
Thermo Fisher Scientific, Inc.) and the machine system. The
experimental protocol was conducted following the manufacturer's
instructions.
For the proliferation assay, the cells were seeded
into a 96-well plate (Corning Incorporated, Corning, NY, USA) for
24 h, following which the reagents were used to treat the cells for
48 h. A cell counting kit-8 (CCK-8; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) was diluted with free-FBS RPMI-1640 medium at a
ratio of 1:9 and added to the cells and for 1 h in an incubator. A
Multiskan reader was used to determine the absorbance at a
wavelength of 450 nm.
Cell invasion and migration assays
Matrigel (100 µl; Sigma-Aldrich; Merck KGaA)
was diluted in free-FBS RPMI-1640 medium at a ratio of 1:8 and
added to the Transwell chamber (Corning Incorporated) in an
incubator for 6 h at 37°C to ensure solidification of the Matrigel.
The cells (1×106/ml) were resuspended with free-FBS
RPMI-1640 medium and were then added in top chamber of the
Transwell following treatment with the reagents for 48 h, and 20%
FBS RPMI-1640 medium was added to lower chamber of the Transwell.
The Transwell containing the cells was placed in an incubator at
37°C for 24 h. A methanol:acetone (1:1) solution (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China) was used to fix
the cells, and 4% crystal violet solution (Beijing Solarbio Science
& Technology Co., Ltd.) was used to stain the invaded cells.
Images were captured using Olympus DSX100 optical microscope
(Olympus Corporation, Tokyo, Japan).
The reagents were used to treat the cells for 48 h,
following which the cells were resuspended in 2% FBS culture
medium. The cells were added to a 35-mm dish, and a 200 µl
pipette tip (Sigma-Aldrich; Merck KGaA) was used to scratch the
cells. After 48 h, images of the scratches were captured using a
microscope. The shortened distances of the scratches were used to
determine the migration rates.
Western blot analysis
Total protein was extracted from the cells using
lysis solution (Invitrogen; Thermo Fisher Scientific, Inc.). The
protein concentration was determined with a BCA kit using a
Multiskan reader (Thermo Fisher Scientific, Inc.) at a wavelength
of 562 nm. The prestained protein ladder (Invitrogen, Carlsbad, CA,
USA) and protein samples (10 µg/lane) were separated by
SDS-PAGE on a 12% gel and were then transferred onto PVDF membranes
(Sigma-Aldrich; Merck KGaA) by cataphoresis. The protein membranes
were then stained with 1X ponceau solution (Beijing Solarbio
Science & Technology Co., Ltd.) and the blank spot of the
membranes were blocked by 5% bovine serum albumin solution (Beijing
Solarbio Science & Technology Co., Ltd.). Primary antibodies
were used to incubate the protein membranes for 12 h at 4°C. The
primary antibodies, from Abcam (Cambridge, MA, USA), were as
follows: Bcl-2 (cat. no. ab59348; 26 kDa), Bax (cat. no. ab32503;
21 kDa), cleaved caspase-3 (cat. no. ab2302; 17 kDa), PTEN (cat.
no. ab32199; 54 kDa), phosphorylated (p-)PI3K (cat. no. ab182651;
84 kDa), PI3K (cat. no. ab191606; 85 kDa), p-Akt (cat. no. ab38449;
56 kDa), Akt (cat. no. ab8805; 55 kDa; all 1:1,000) and GAPDH (cat.
no. ab8245; 36 kDa; 1:2,000). Secondary antibodies (cat. no.
ab6721; 1:5,000; Abcam) were used to incubate the protein membranes
for 2 h at room temperature. The protein membranes were stained
using an ECL kit (Sigma-Aldrich; Merck KGaA), and fluorescence was
determined using an imaging system (Tanon Science and Technology
Co., Ltd., Shanghai, China) and protein stains were analyzed using
the BandScan 5.0 system (Glyko, Inc., Novato, CA, USA).
Dual luciferase reporter assay
TargetScan 7.0 (www.targetscan.org) was used to predict the miR-92a-3p
target and its related sites. 293T cells (American Type Culture
Collection) were plated onto 12-well plates (5×105
cells/well) prior to cell transfection. Subsequently, either the
PTEN 3′UTR wild-type or PTEN 3′UTR mutant was cloned into the
psi-CHECK-2 vector (Promega Corporation, Madison, WI, USA). The
full length of PTEN 3′UTR was 5′-AGU UCU AGA AAU UUU GUG CAA UA-3′.
Subsequently, pGL-3 firefly luciferase reporters (1 µg per
well) were co-transfected with 50 nmol/l universal mimics control
(mimic control) or miR-92a-3p mimics and PTEN 3′UTR wild-type or
PTEN 3′UTR mutant using Lipofectamine 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. The cells transfected with mimics control and PTEN 3′UTR
wild-type were used as a control. Luciferase activity was
determined using a dual-luciferase reporter assay kit (Promega
Corporation).
Statistical analysis
For all experiments, three trials were performed in
triplicate. All values are expressed as the mean ± SD. The
differences between two groups were analyzed using Student's
t-test, or one-way ANOVA followed by Dunnett's t-test or Tukey post
hoc test using SPSS 22.0 (IBM, Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Effects of miR-92a-3p mimic and
miR-92a-3p inhibitor on ESCC cell proliferation
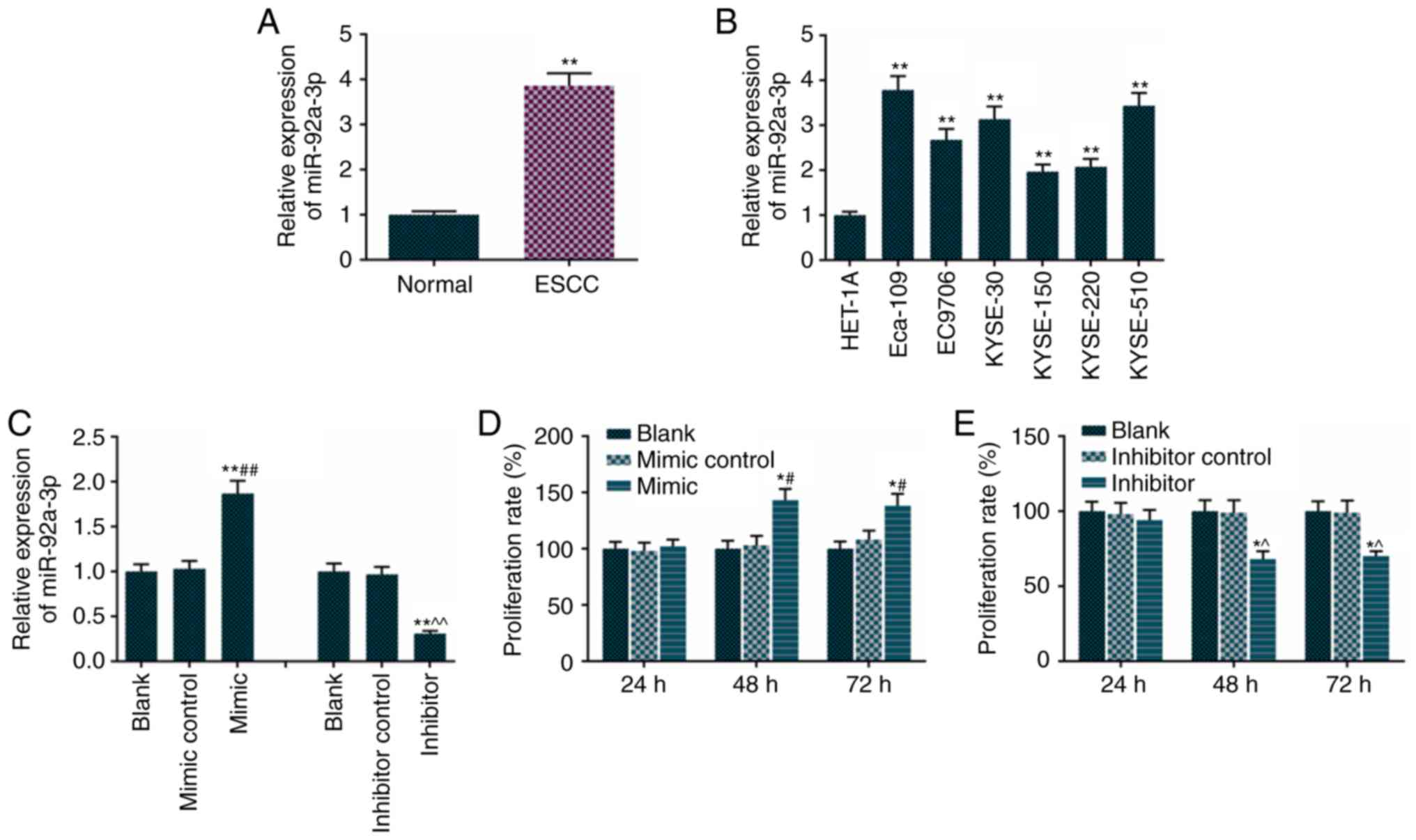
The results revealed that tissues from the patients
with ESCC had higher expression of miR-92a-3p, compared with normal
tissues (Fig. 1A). Therefore, the
relative expression of miR-92a-3p was detected in normal esophageal
and ESCC cell lines, including Eca-109, EC9706, KYSE-30, KYSE-150,
KYSE-220 and KYSE-510 cells. It was found that the ESCC cell lines
had higher levels of miR-92a-3p, compared with the normal esophagus
cells (Fig. 1B). The Eca-109 cell
line and ESCC cell line were used in subsequent experiments. The
miR-92a-3p mimic increased the levels of miR-92a-3p (Fig. 1C) and promoted Eca-109 cell
proliferation (Fig. 1D). However,
the miR-92a-3p inhibitor reduced Eca-109 cell proliferation
(Fig. 1E).
Effects of miR-92a-3p mimic and
miR-92a-3p inhibitor on Eca-109 cell apoptosis and the apoptosis
signaling pathway
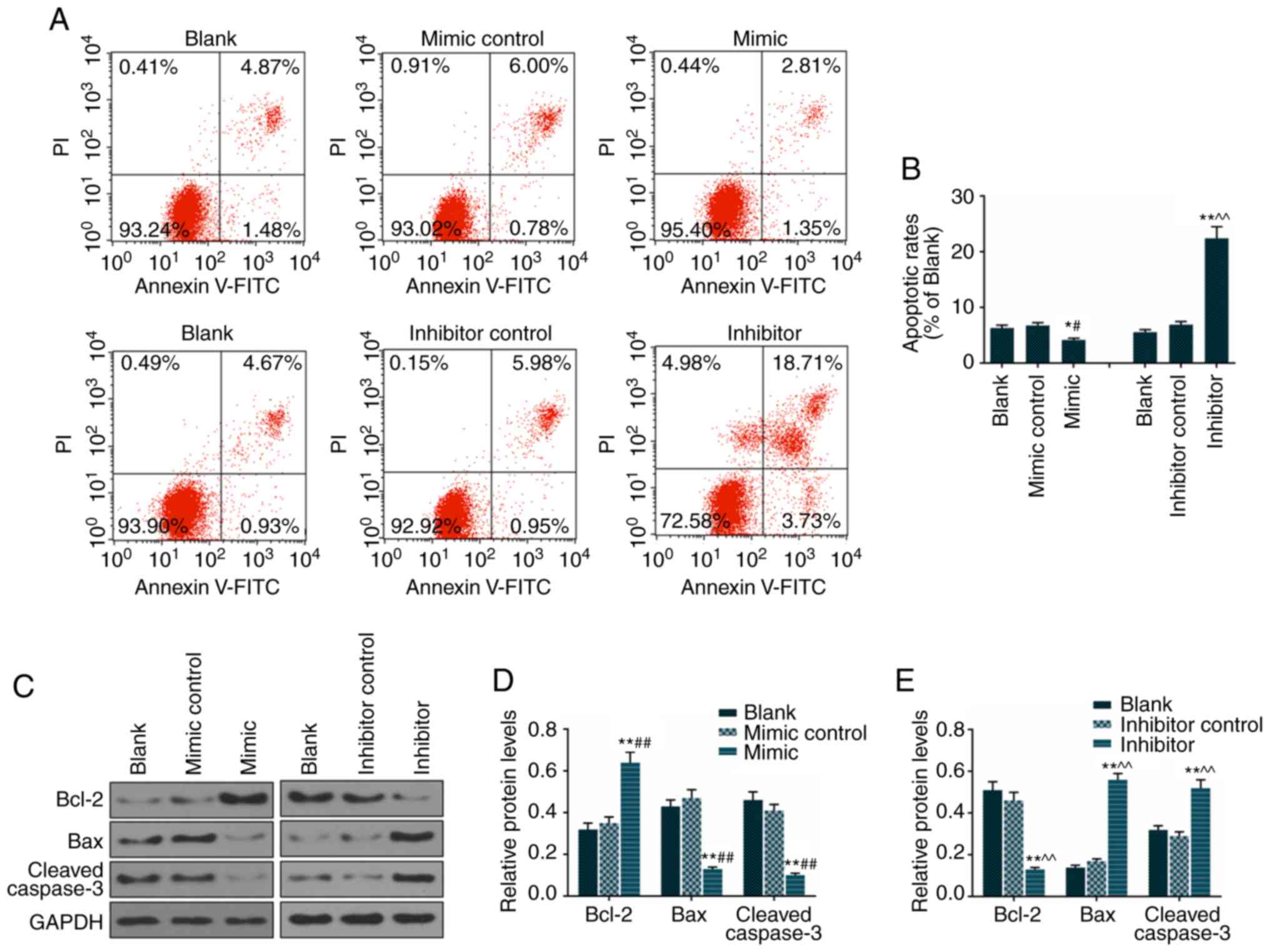
To examine the effect of miR-92a-3p on apoptosis, an
Annexin V/PI assay was used to determine apoptotic rates. The
apoptotic rates of Eca-109 cells transfected with miR-92a-3p mimic
(4.16%) decreased significantly, compared with rates in the blank
(6.35%) and mimic control (6.78%) groups. The apoptotic rates of
Eca-109 cells transfected with miR-92a-3p inhibitor (22.44%) were
increased significantly, compared with those in the blank (5.6%)
and inhibitor control (6.93%) groups. Therefore, the miR-92a-3p
mimic transfection decreased Eca-109 cell apoptosis and the
miR-92a-3p inhibitor increased Eca-109 cell apoptosis (Fig. 2A and B). The miR-92a-3p mimic was
found to decrease the protein levels of Bax and cleaved caspase-3
and increase the protein level of Bcl-2 (Fig. 2C and D). The miR-92a-3p inhibitor
promoted the protein expressions of Bax and cleaved caspase-3 and
decreased the level of Bcl-2 (Fig. 2C
and E).
Effects of miR-92a-3p mimic and
miR-92a-3p inhibitor on Eca-109 cell migration and invasion
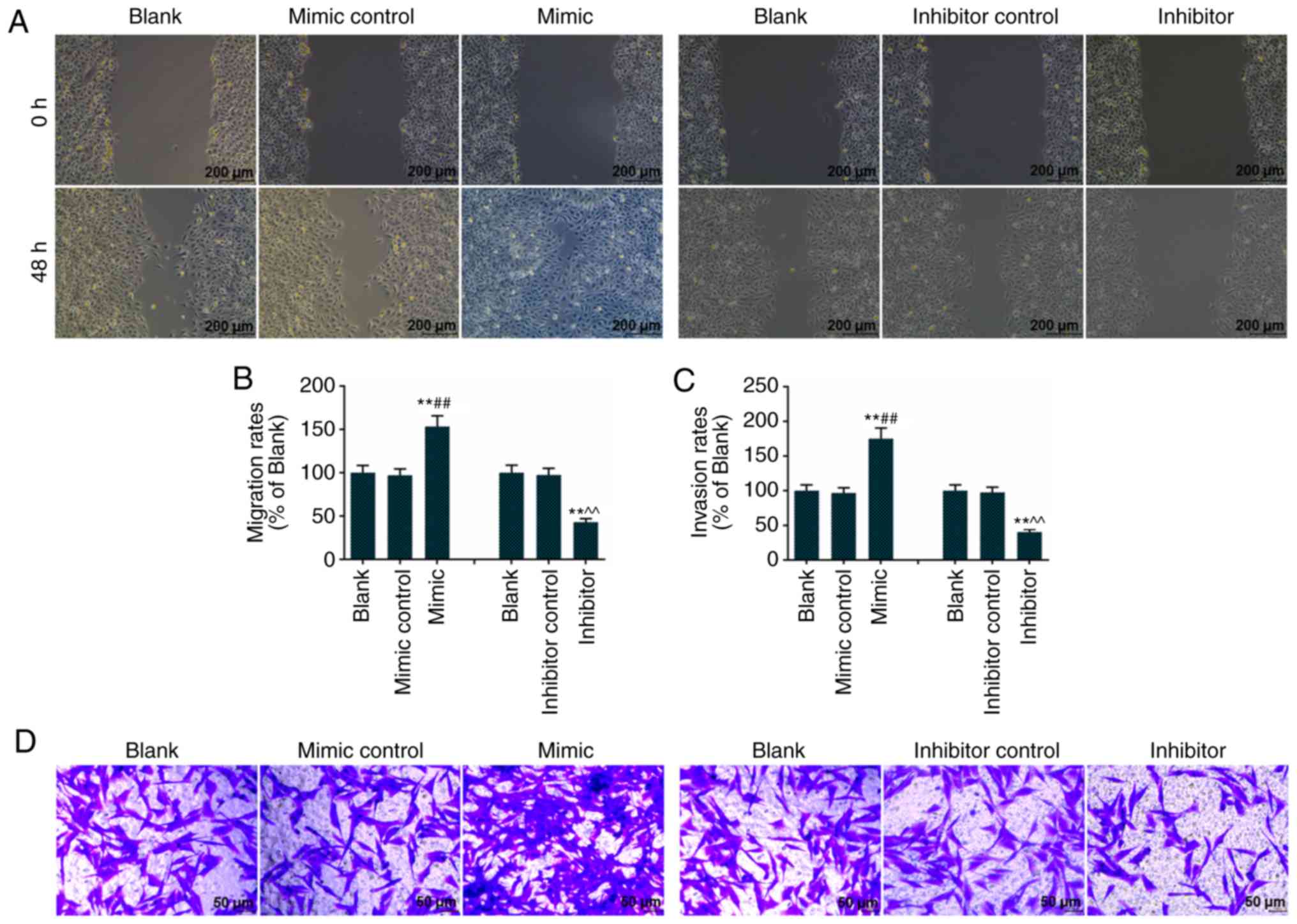
The miR-92a-3p mimic promoted Eca-109 cell
migration, as the Eca-109 cells treated with miR-92a-3p mimic
covered the plate in the scratch image captured at 48 h (Fig. 3A). The miR-92a-3p inhibitor
inhibited Eca-109 cell migration, as the scratch image captured at
48 h showed a longer scratch distance (Fig. 3A). Therefore, Eca-109 cells
treated with the miR-92a-3p mimic had the highest migration rate,
whereas Eca-109 cells treated with the miR-92a-3p inhibitor had the
lowest migration rate (Fig. 3B).
The miR-92a-3p mimic resulted in a higher invasion rate, and the
miR-92a-3p inhibitor resulted to a lower invasion rate in Eca-109
cells (Fig. 3C and D).
miR-92a-3p mimic inhibits the expression
of PTEN in Eca-109 cells
The miR-92a-3p mimic inhibited the luciferase
activity of the psi-CHECK-2 with PTEN 3′UTR in Eca-109 cells. The
alignment of designed miRNA mimic with the targeted section of PTEN
3′UTR is shown in Fig. 4A.
miR-92a-3p was significantly overexpressed in the mimic group,
compared with the blank and mimic control groups (Fig. 4B). The mutation of the PTEN 3′UTR
prevented the alignment of PTEN and miR-92a-3p. The luciferase
activity in the PTEN-MUT + mimic group was not changed compared
with the PTEN-WT + mimic control group, while the luciferase
activity in the PTEN-WT + mimic group was significantly lower
compared with PTEN-WT + mimic control group (Fig. 4C). The expression of PTEN was not
only inhibited in the mimic + NC group, compared with that in the
NC group (cells transfected with empty vector), but was also
inhibited in the mimic + PTEN group, compared with that in the PTEN
group. Therefore, the miR-92a-3p mimic inhibited the expression of
PTEN (Fig. 4D). The
overexpression of PTEN inhibited Eca-109 cell proliferation, and
the miR-92a-3p mimic accelerated proliferation of the PTEN-induced
Eca-109 cells (Fig. 4E).
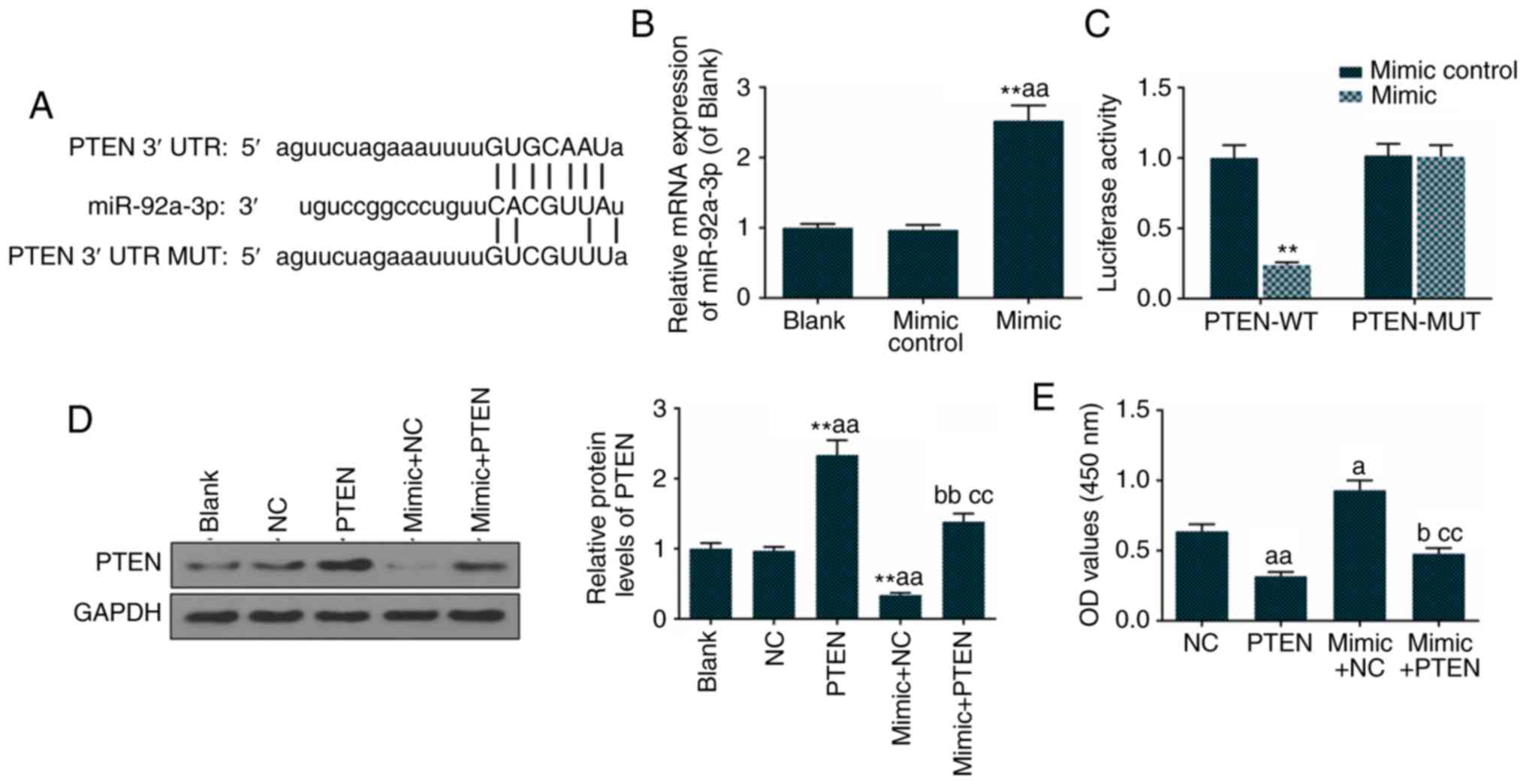 | Figure 4Overexpression of PTEN inhibits
Eca-109 proliferation. (A) TargetScan 7.2 predicted the target gene
of miR-92a-3p. (B) miR-92a-3p was overex-pressed in mimic group.
(C) A dual luciferase reporter assay was used to determine the
association between PTEN and miR-92a-3p. (D) PTEN, miR-92a-3p mimic
and their combination were transfected into Eca-109 cells for 48 h,
and the protein level of PTEN was analyzed by western blotting and
quantitation. (E) Cell proliferation was detected using a cell
counting kit-8. The values are presented as the mean ± SD.
Student's t-test was used to analyze differences between two
groups. **P<0.01 vs. blank group;
aP<0.05 and aaP<0.01 vs. NC group;
bP<0.05 and bbP<0.01 vs. PTEN group;
ccP<0.01 vs. mimic + NC group. miR, microRNA; PTEN,
phosphatase and tensin homolog deleted on chromosome 10; WT,
wild-type; MUT, mutant; 3′UTR, 3′ untranslated region; Blank,
untransfected cells; NC, cells transfected with empty vector; PTEN,
cells transfected with PTEN recombinant plasmid; mimic + NC, cells
transfected with mimics and empty vector; mimic + PTEN, cells
transfected with mimic and PTEN recombinant plasmid. |
miR-92a-3p downregulates PTEN and
inhibits its tumor suppressive function
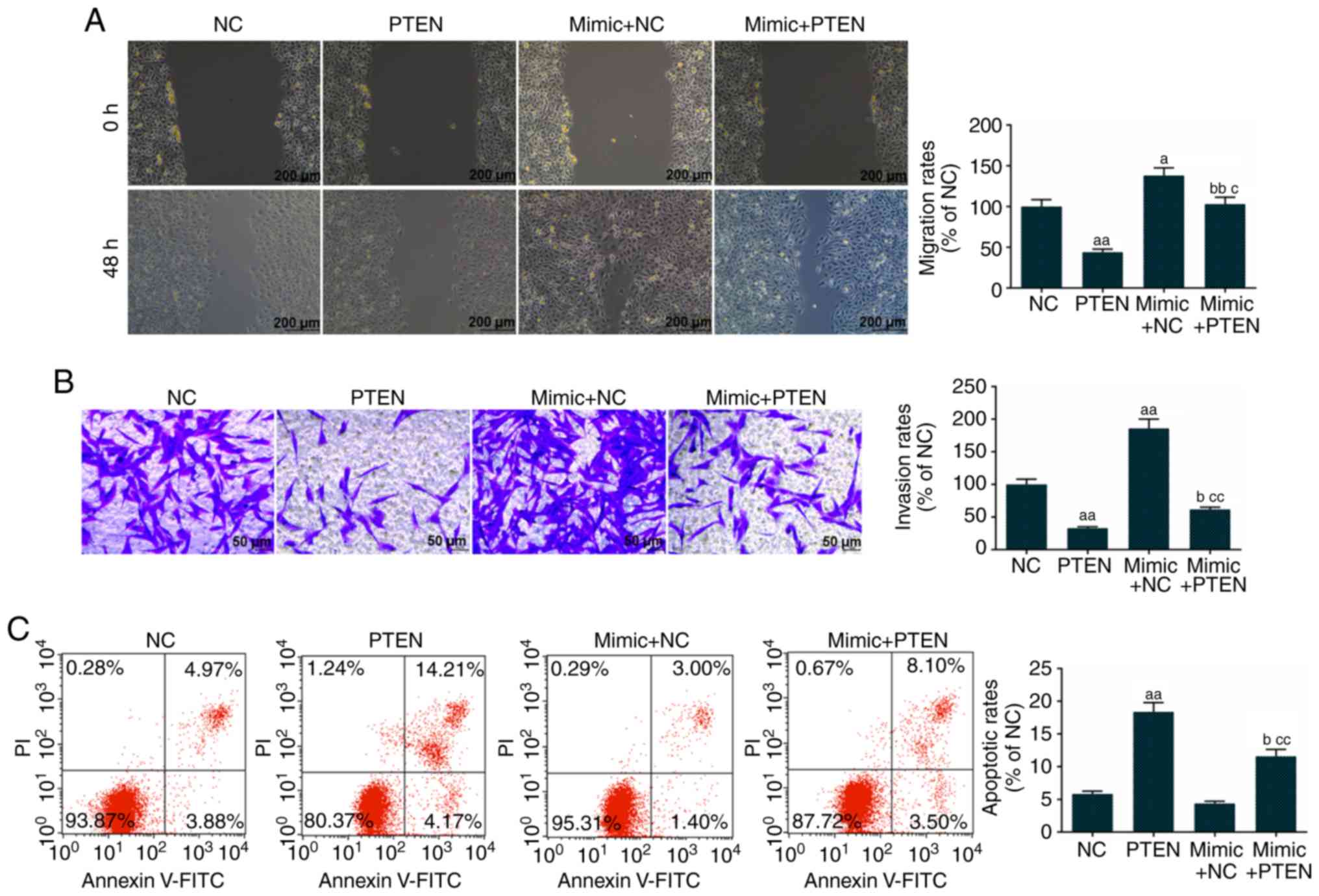
The overexpression of PTEN inhibited Eca-109 cell
migration, whereas the miR-92a-3p mimic promoted the migration of
PTEN-induced Eca-109 cells (Fig.
5A). The overexpression of PTEN decreased Eca-109 cell
invasion, however, the miR-92a-3p mimic increased the invasion of
PTEN-induced Eca-109 cells (Fig.
5B). The over-expression of PTEN promoted Eca-109 cell
apoptosis, whereas the miR-92a-3p mimic decreased the apoptotic
rate of the PTEN-induced Eca-109 cells (Fig. 5C).
miR-92a-3p mimic downregulates PTEN and
inhibits its PI3K and Akt phosphorylation-inhibiting function in
Eca-109 cells
The overexpression of PTEN decreased the levels of
p-PI3K and p-Akt in Eca-109 cells. The miR-92a-3p mimic promoted
the phosphorylation of PI3K and Akt in Eca-109 cells (Fig. 6A and B). The miR-92a-3p mimic
downregulated PTEN and inhibited its PI3K and Akt
phosphorylation-inhibiting function in Eca-109 cells. The results
showed that IGF-1 promoted cell proliferation, PTEN inhibited cell
proliferation through inactivation of the PI3K/Akt pathway, and the
over-expression of miR-92a-3p inhibited the function of PTEN
(Fig. 6C).
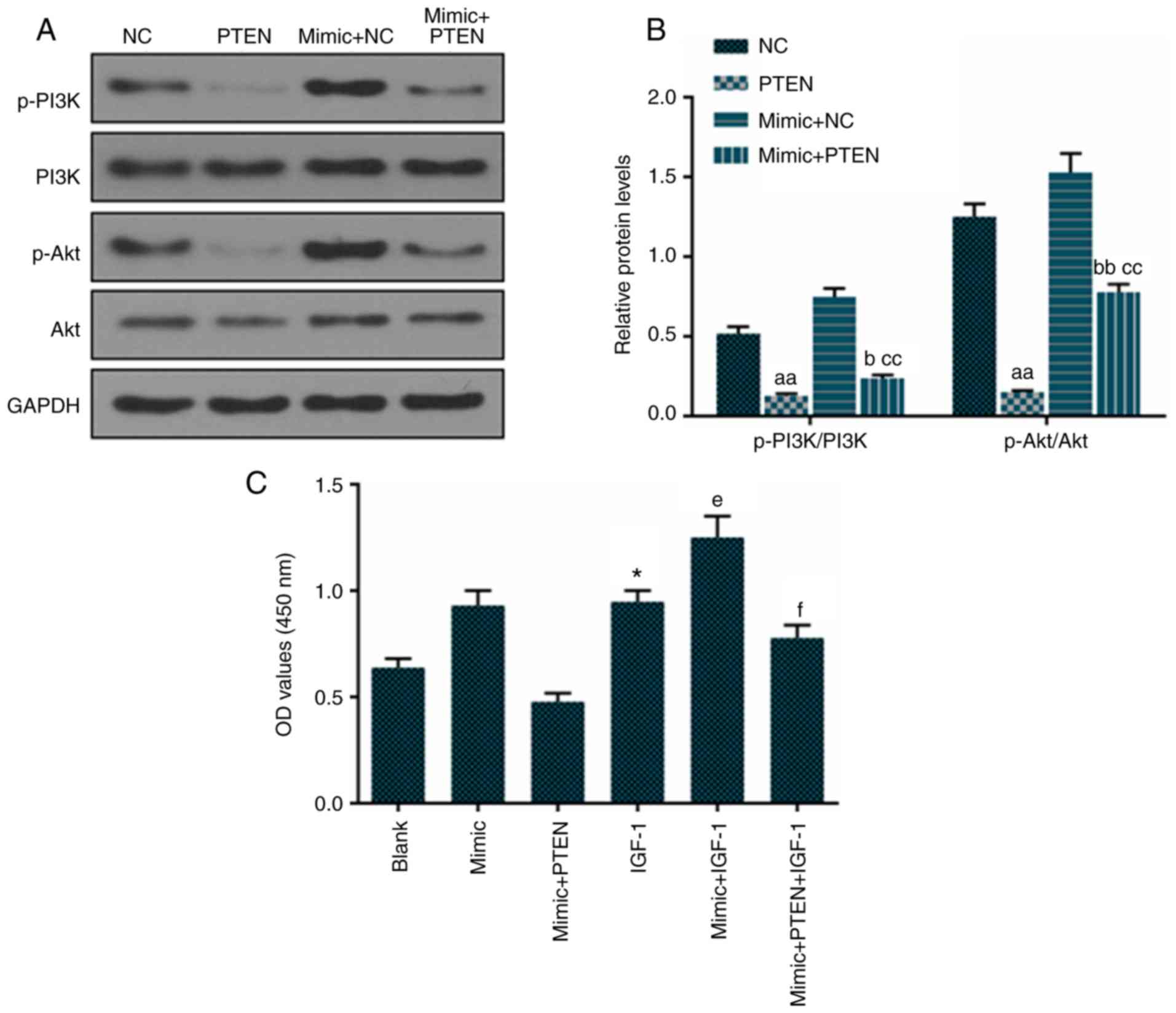 | Figure 6Overexpression of PTEN inhibits the
PI3K/Akt pathway in Eca-109 cells. PTEN, miR-92a-3p mimic and their
combination was transfected into Eca-109 cells for 48 h. (A)
Protein levels of p-PI3K, PI3K, p-Akt and Akt were measured by
western blotting and (B) quantification. (C) IGF-1, activator of
the PI3K/Akt pathway, alone and in combination was used to treat
Eca-109 cell for 48 h, and a cell counting kit-8 assay detected
cell proliferation. The values are presented as the mean ± SD.
Student's t-test was used to analyze differences between two
groups. *P<0.05 vs. Blank group,
aaP<0.01 vs. NC group, bP<0.05 and
bbP<0.01 vs. PTEN group, ccP<0.01 vs.
mimic + NC group; eP<0.05 vs. IGF-1 group;
fP<0.05 vs. mimic + IGF-1 group. PTEN, phosphatase
and tensin homolog deleted on chromosome 10; p-, phosphorylated;
NC, cells transfected with empty vector; IGF-1, insulin-like growth
factor 1. |
Discussion
The ESCC cells exhibited a higher expression of
miR-92a-3p, compared with that in normal esophageal cells (Fig. 1B), which was consistent with the
higher expression of miR-92a-3p in EC tissues from patients with
ESCC (Fig. 1A). The miR-92a-3p
mimic promoted the expression of miR-92a-3p in ESCC cells (Fig. 1C) and increased the proliferation
of ESCC cells (Fig. 1D). The
miR-92a-3p inhibitor restrained the expression of miR-92a-3p in
ESCC cells (Fig. 1C) and
inhibited the proliferation of ESCC cells (Fig. 1E). Therefore, the overexpression
of miR-92a-3p had a positive effect on ESCC cell proliferation.
Apoptosis is programmed cell death and serves a
crucial role in homeostasis of the body by eliminating injured
cells and abnormal cells. Apoptosis is also important in tissue
regulation and organogenesis, such as in the development of limbs
(22,23). The process of apoptosis includes
separation from cell group, condensation, fragmentation and
eventually phagocytosis through macrophages (24). Cell death originating from cell
apoptosis has been considered to be inevitable, although such an
assumption had not been demonstrated directly. However, Tang et
al demonstrated that cells dying from the execution stage of
apoptosis could be recovered following the removal of apoptotic
stimulation in 2012 (25). The
apoptotic pathway can be generally divided into extrinsic and
intrinsic pathways, and one intrinsic cell apoptotic pathway is the
release of cathepsins, which can promote Bid, activating Bax, and
resulting in permeabilization of the mitochondrial outer membrane
and release of cytochrome c (26). It is clear that the Bcl-2 gene
protects cells against apoptosis and that Bcl-2 can be inhibited by
the release of Bid (27,28). Caspases, a family of cysteine
proteases, can be stimulated when the apoptotic cell undergoes
autolytic cleavage (29). The
inhibition of miR-92a-3p promoted ESCC cell apoptosis and activated
the Bax/Bcl-2 and caspase-3 signaling pathways (Fig. 2). In addition, the overexpression
of miR-92a-3p inhibited apoptosis and inactivated the Bax/Bcl-2 and
caspase-3 signaling pathways (Fig.
2).
Cancer metastasis is a major cause of mortality in
patients with cancer. The first step of tumor metastasis involves
vascularization of the primary tumor through the secretion of
angiogenic factors, which increases tumor tissue stroma motility
and invasion (30). The invading
tumor cells penetrate blood vessels and enter the circulation
through lymphatic vessels (31).
Therefore, studies have been performed on cancer cell migration and
invasion in order to evaluate the function of drug or gene or on
the methods to inhibit migration and invasion (32-34). The overexpression of miR-92a-3p
increased ESCC cell migration and invasion, whereas the reduction
of miR-92a-3p inhibited ESCC cell migration and invasion (Fig. 3).
Zhang et al used TargetScan to predict
miR-100 target and its binding sites (35). The present study used TargetScan
7.0 to predict the miR-92a-3p target and its related sites, and it
was confirmed that the PTEN gene was a target gene of miR-92a-3p by
a dual luciferase report assay (Fig.
4A and B). The PTEN gene was found when chromosome 10q23 was
examined in 1997; the protein produced by the PTEN gene shared
sequence homology with cytoskeletal tensin, and mutations of PTEN
generally were detected in cancer (36,37). The overexpression of PTEN
inhibited ESCC cell proliferation (Fig. 4C-E). The miR-92a-3p mimic promoted
cell proliferation, which may only be attributed partially to the
downregulation of PTEN. The overexpression of miR-92a-3p also
inhibited apoptosis and promoted the migration and invasion of
PTEN-overexpressing ESCC cells (Fig.
5).
PTEN, which is characterized as a lipid and protein
phosphatase, negatively affects the PI3K/Akt pathway (38). The PI3K/Akt pathway is activated
in cancer, including renal carcinoma, prostate cancer and
hematologic malignancies (39-42). The results of the present study
supported that, in ESCC cells, the overexpression of PTEN inhibited
the PI3K/Akt pathway, which was promoted by miR-92a-3p (Fig. 6A and B). In order to investigate
that effect of the PI3K/Akt pathway on ESCC cell proliferation,
IGF-1, an activator of the PI3K/Akt pathway, was used (42). The results showed that IGF-1
promoted cell proliferation, PTEN inhibited cell proliferation
through inactivation of the PI3K/Akt pathway, and the
overexpression of miR-92a-3p inhibited the function of PTEN.
In conclusion, the present study supports the
hypothesis that the overexpression of miR-92a-3p promoted the
proliferation, migration and invasion and decreased the apoptosis
of ESCC cells. miR-92a-3p inhibited apoptosis via the Bax/Bcl-2 and
caspase-3 pathways and promoted proliferation, which may be
associated with the PI3K/Akt pathway. The effect of miR-92a-3p on
ESCC was realized by regulating PTEN. As target protectors are
designed against both the seed sequence and the flanking sequences
of a specific mRNA target, the phenotypic readout can be claimed to
be a result of specific targeting, rather than the net result of
miRNA downregulation of multiple targets. Further definitive
experiments, such as target-protector experiments, will be
performed in the future, and the effect of miR-92a-3p on tumor
formation in nude mice in vivo will also be
investigated.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during this study
are available from the corresponding author on reasonable
request.
Authors' contributions
XL and SG made substantial contributions to
conception and design. LM, QG and SZ contributed to data
acquisition, data analysis and interpretation. SZ, XL and SG
contributed to drafting the manuscript and/or critically revising
the manuscript for important intellectual content. All authors
approved the final version to be published. LM and QG agree to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in experiments involving
human participants were in accordance with the ethical standards of
the institutional and/or National Research Committee and with the
1964 Helsinki declaration and its later amendments or comparable
ethical standards. The patients signed an informed consent form and
the study was approved by the Ethics Committee of Shanxi Provincial
People's Hospital (no. SP2016064926).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
ESCC
|
esophageal squamous cell cancer
|
|
EC
|
esophageal cancer
|
|
ECA
|
esophageal adenocarcinoma
|
|
miRNA
|
microRNA
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
IGF-1
|
insulin-like growth factor 1
|
|
3′UTR
|
3′untranslated region
|
Acknowledgments
Not applicable.
References
|
1
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Short MW, Burgers KG and Fry VT:
Esophageal cancer. Am Fam Physician. 95:22–28. 2017.PubMed/NCBI
|
|
3
|
Daly JM, Fry WA, Little AG, Winchester DP,
McKee RF, Stewart AK and Fremgen AM: Esophageal cancer: Results of
an American college of surgeons patient care evaluation study. J Am
Coll Surg. 190:562–563. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siewert JR, Stein HJ, Feith M, Bruecher
BL, Bartels H and Fink U: Histologic tumor type is an independent
prognostic parameter in esophageal cancer: Lessons from more than
1,000 consecutive resections at a single center in the Western
world. Ann Surg. 234:360–369. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rice TW, Apperson-Hansen C, DiPaola LM,
Semple ME, Lerut TE, Orringer MB, Chen LQ, Hofstetter WL, Smithers
BM, Rusch VW, et al: Worldwide esophageal cancer collaboration:
Clinical staging data. Dis Esophagus. 29:707–714. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kaz AM and Grady WM: Epigenetic biomarkers
in esophageal cancer. Cancer Lett. 342:193–199. 2014. View Article : Google Scholar
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cui J, Zhou B, Ross SA and Zempleni J:
Nutrition, microRNAs, and human health. Adv Nutr. 8:105–112. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar :
|
|
11
|
Feng QQ, Dong ZQ, Zhou Y, Zhang H and Long
C: miR-16-1-3 ptargets TWIST1 to inhibit cell proliferation and
invasion in NSCLC. Bratisl Lek Listy. 119:60–65. 2018.
|
|
12
|
Li H, Bian C, Liao L, Li J and Zhao RC:
miR-17-5p promotes human breast cancer cell migration and invasion
through suppression of HBP1. Breast Cancer Res Treat. 126:565–575.
2011. View Article : Google Scholar
|
|
13
|
Cun J and Yang Q: Bioinformatics-based
interaction analysis of miR-92a-3p and key genes in
tamoxifen-resistant breast cancer cells. Biomed Pharmacother.
107:117–128. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ke TW, Wei PL, Yeh KT, Chen WT and Cheng
YW: miR-92a promotes cell metastasis of colorectal cancer through
PTEN-Mediated PI3K/AKT pathway. Ann Surg Oncol. 22:2649–2655. 2015.
View Article : Google Scholar
|
|
15
|
Shen Y, Ding Y, Ma Q, Zhao L, Guo X, Shao
Y, Niu C, He Y, Zhang F, Zheng D, et al: Identification of novel
circulating MicroRNA biomarkers for the diagnosis of esophageal
squamous cell carcinoma and squamous dysplasia. Cancer Epidemiol
Biomarkers Prev. Apr 15–2019, Epub ahead of print. View Article : Google Scholar
|
|
16
|
Liu MX, Liao J, Xie M, Gao ZK, Wang XH,
Zhang Y, Shang MH, Yin LH, Pu YP and Liu R: miR-93-5p transferred
by exosomes promotes the proliferation of esophageal cancer cells
via inter-cellular communication by targeting PTEN. Biomed Environ
Sci. 31:171–185. 2018.PubMed/NCBI
|
|
17
|
Tullius BP, Shankar SP, Cole S, Triano V,
Aradhya S, Huang EC, Sanchez T and Pawar A: Novel heterozygous
mutation in the PTEN gene associated with ovarian germ cell tumor
complicated by growing teratoma syndrome and overgrowth in a
two-year-old female. Pediatr Blood Cancer. e277882019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yan Y, Li Z, Zeng S, Wang X, Gong Z and Xu
Z: FGFR2-mediated phosphorylation of PTEN at tyrosine 240
contributes to the radioresistance of glioma. J Cell Commun Signal.
Apr 25–2019, Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao MY, Wang LM, Liu J, Huang X, Liu J
and Zhang YF: miR-21 Suppresses Anoikis through targeting PDCD4 and
PTEN in human esophageal adenocarcinoma. Curr Med Sci. 38:245–251.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tutar L, Tutar E and Tutar Y: MicroRNAs
and cancer; an overview. Curr Pharm Biotechnol. 15:430–437. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Jacobson MD, Weil M and Raff MC:
Programmed cell death in animal development. Cell. 88:347–354.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fuchs Y and Steller H: Programmed cell
death in animal development and disease. Cell. 147:742–758. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kerr JF, Wyllie AH and Currie AR:
Apoptosis: A basic biological phenomenon with wide-ranging
implications in tissue kinetics. Br J Cancer. 26:239–257. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang HL, Tang HM, Mak KH, Hu S, Wang SS,
Wong KM, Wong CS, Wu HY, Law HT, Liu K, et al: Cell survival, DNA
damage, and oncogenic transformation after a transient and
reversible apoptotic response. Mol Biol Cell. 23:2240–2252. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Joshi GN and Knecht DA: Multi-parametric
analysis of cell death pathways using live-cell microscopy. Curr
Protoc Toxicol. 58:Unit 4.40. 2013. View Article : Google Scholar
|
|
27
|
Dietrich JB: Apoptosis and anti-apoptosis
genes in the Bcl-2 family. Arch Physiol Biochem. 105:125–135.
1997.In French. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kvansakul M, Caria S and Hinds MG: The
Bcl-2 family in Host-Virus interactions. Viruses. 9:E2902017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Crowley LC and Waterhouse NJ: Detecting
cleaved caspase-3 in apoptotic cells by flow cytometry. Cold Spring
Harbor Protocols. Nov 1–2016, Epub ahead of print. View Article : Google Scholar
|
|
30
|
Han T, Kang D, Ji D, Wang X, Zhan W, Fu M,
Xin HB and Wang JB: How does cancer cell metabolism affect tumor
migration and invasion? Cell Adh Migr. 7:395–403. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Duff D and Long A: Roles for RACK1 in
cancer cell migration and invasion. Cell Signal. 35:250–255. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tungsukruthai S, Sritularak B and
Chanvorachote P: Cycloarto-biloxanthone inhibits migration and
invasion of lung cancer cells. Anticancer Res. 37:6311–6319.
2017.PubMed/NCBI
|
|
34
|
Yang X, Xu Y, Wang T, Shu D, Guo P,
Miskimins K and Qian SY: Inhibition of cancer migration and
invasion by knocking down delta-5-desaturase in COX-2 overexpressed
cancer cells. Redox Biol. 11:653–662. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang N, Fu H, Song L, Ding Y, Wang X,
Zhao C and Zhao Y, Jiao F and Zhao Y: MicroRNA-100 promotes
migration and invasion through mammalian target of rapamycin in
esophageal squamous cell carcinoma. Oncol Rep. 32:1409–1418. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast, and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Steck PA, Pershouse MA, Jasser SA, Yung
WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T,
et al: Identification of a candidate tumour suppressor gene, MMAC1,
at chromosome 10q23.3 that is mutated in multiple advanced cancers.
Nat Genet. 15:356–362. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Malaney P, Uversky VN and Davé V: PTEN
proteoforms in biology and disease. Cell Mol Life Sci.
74:2783–2794. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo H, German P, Bai S, Barnes S, Guo W,
Qi X, Lou H, Liang J, Jonasch E, Mills GB and Ding Z: The PI3K/AKT
pathway and renal cell carcinoma. J Genet Genomics. 42:343–353.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen H, Zhou L, Wu X, Li R, Wen J, Sha J
and Wen X: The PI3K/AKT pathway in the pathogenesis of prostate
cancer. Front Biosci (Landmark Ed). 21:1084–1091. 2016. View Article : Google Scholar
|
|
41
|
Mayer IA and Arteaga CL: The PI3K/AKT
pathway as a target for cancer treatment. Annu Rev Med. 67:11–28.
2016. View Article : Google Scholar
|
|
42
|
Yu M, Wang H, Xu Y, Yu D, Li D, Liu X and
Du W: Insulin-like growth factor-1 (IGF-1) promotes myoblast
proliferation and skeletal muscle growth of embryonic chickens via
the PI3K/Akt signalling pathway. Cell Biol Int. 39:910–922. 2015.
View Article : Google Scholar : PubMed/NCBI
|