The current SARS-CoV-2-induced pandemic has raised a
number of public health policy and scientific queries, related to
the virus origin, transmission, activity, contamination,
pathophysiologic effects and treatment. As of May 3, 2021, almost
188 million cases had been confirmed, while 4.05 million deaths had
been registered under the cause of death: 'COVID-19'. Although this
may underline an apogee of the third phase of the pandemic in some
countries, or may have been the result of certain interventions.
Public health policy approaches, communication campaigns,
pharmacological approaches, surveillance, and prevention practices
have been suggested.
The highly varying symptomatology and the
unpredictable global progress of COVID-19 have triggered an
unprecedentedly intensive activity in biomedical research and
public policy decisions. Furthermore, although the pathophysiology
of the disease is being progressively clarified, its complexity
remains vast, and preventive care approaches or treatments,
although both have significantly improved, remain
unsatisfactory.
Notably, the extremely rare yet highly unpredictable
and occasionally lethal vaccination-induced thrombotic
thrombocytopenia (VITT) syndrome has emphasized the gaps in the
current knowledge of certain unsuspected pathophysiological
pathways. The VITT morbid entity is of particular importance given
the generally mild and to a certain extent expected vaccination
side-effects, namely chills, fever, diarrhea, fatigue, muscle pain,
headache and mildly increased blood coagulability (1,2).
As of April 2021, 16 vaccination options were available: Two RNA
vaccines [BNT162b2 (Comirnaty) by Pfizer-BioNTech, mRNA.1273
(Spikevax) by Moderna], seven conventional inactivated ones
(CoronaVac, Covaxin, BBIBP-CorV, WIBP-CorV, Minhai-Kangtai, QazVac,
CovIran Bakerat), five viral vector-employing ones (Covishield and
Vaxzevria by Oxford Astra-Zeneca, the Janssen COVID-19 vaccine by
Johnson & Johnson, the Sputnik V and Sputnik Light by the
Gamaleya Research Institute of Epidemiology and Microbiology in
Russia, and the AD5-nCOV-Convidencia by CanSino Biologics Inc.),
and two protein subunit vaccines (EpiVacCorona and RDB-dimer).
Vaccination programs have been implemented so as to reach 'herd
immunity', in every country. According to national health authority
reports, as of August 30, 2021, 5.27 billion doses had been
administered globally. This is equal to 39.7% of the population on
the planet (where, however, only 1.6% of individuals in the
low-income countries had received at least one dose), having been
fully vaccinated (3). As of
August 30, 2021, 55.15% of the Greek population had been fully
vaccinated (3).
The aim of the present study was to illustrate the
signaling pathways implicated in SARS-CoV-2 infection, including
those of the extremely rare, yet severe VITT syndrome.
Furthermore, the interactions among the retrieved
genes/proteins were investigated by employing the Search Tool for
Retrieval of Interacting Genes/Proteins (STRING) database v11.0
(6,7), a database containing both primary
and predicted, physical and functional association data among genes
or proteins. These data are collected from diverse resources, such
as documented pathway knowledge, high-throughput experimental
studies, cross-species extrapolated information, automated text
mining results, computationally predicted interactions, etc. The
confidence threshold value for displaying interactions was set to
'high' (i.e., 0.7). The interactions in the generated network were
manipulated and visualized through Cytoscape (http://www.cytoscape.org/) (8), a software platform for network
processing and statistical analyses; the Edge Betweenness mode was
used to detect the number of the shortest paths that pass-through a
given edge in the COVID-19 network.
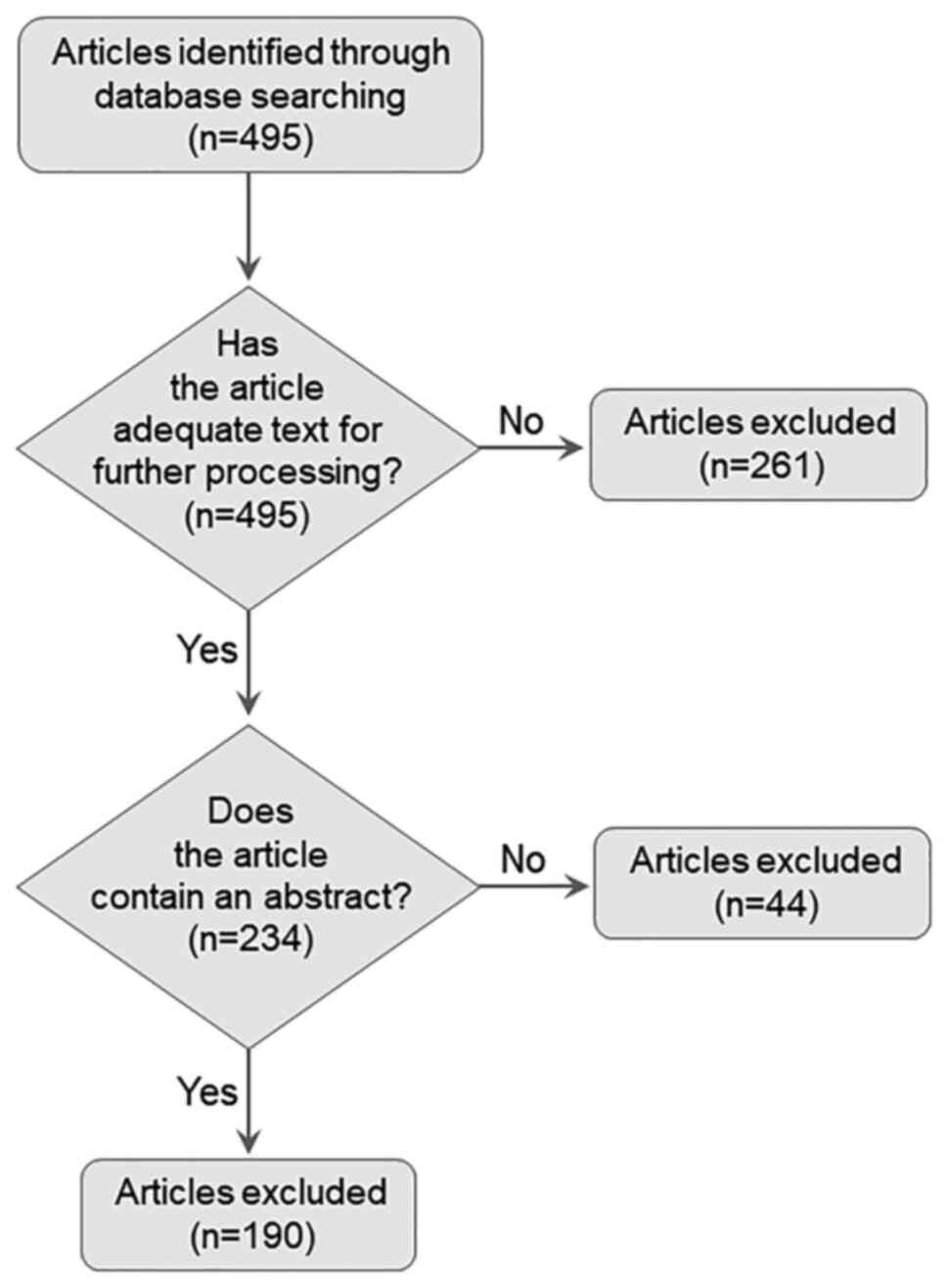
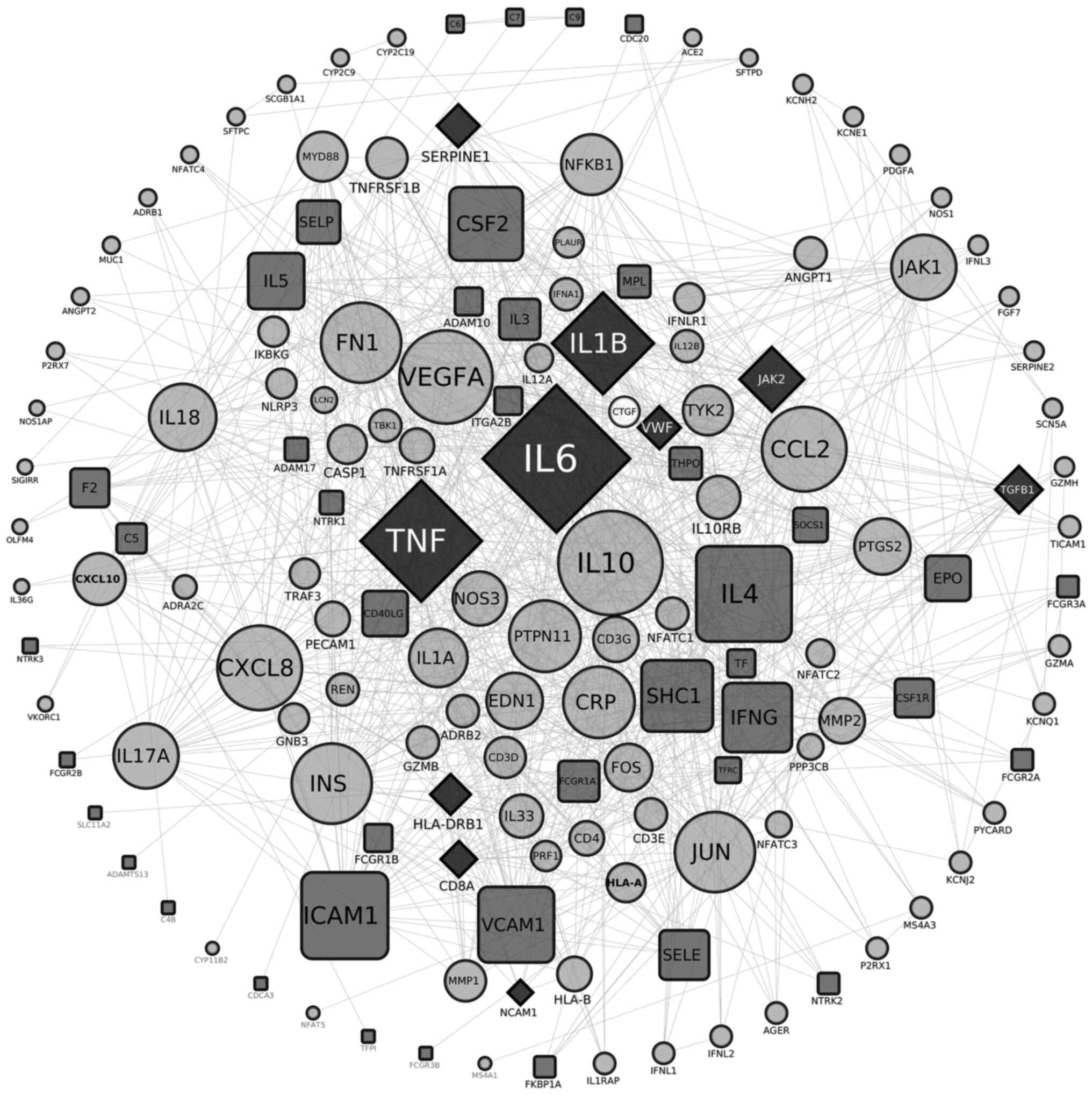
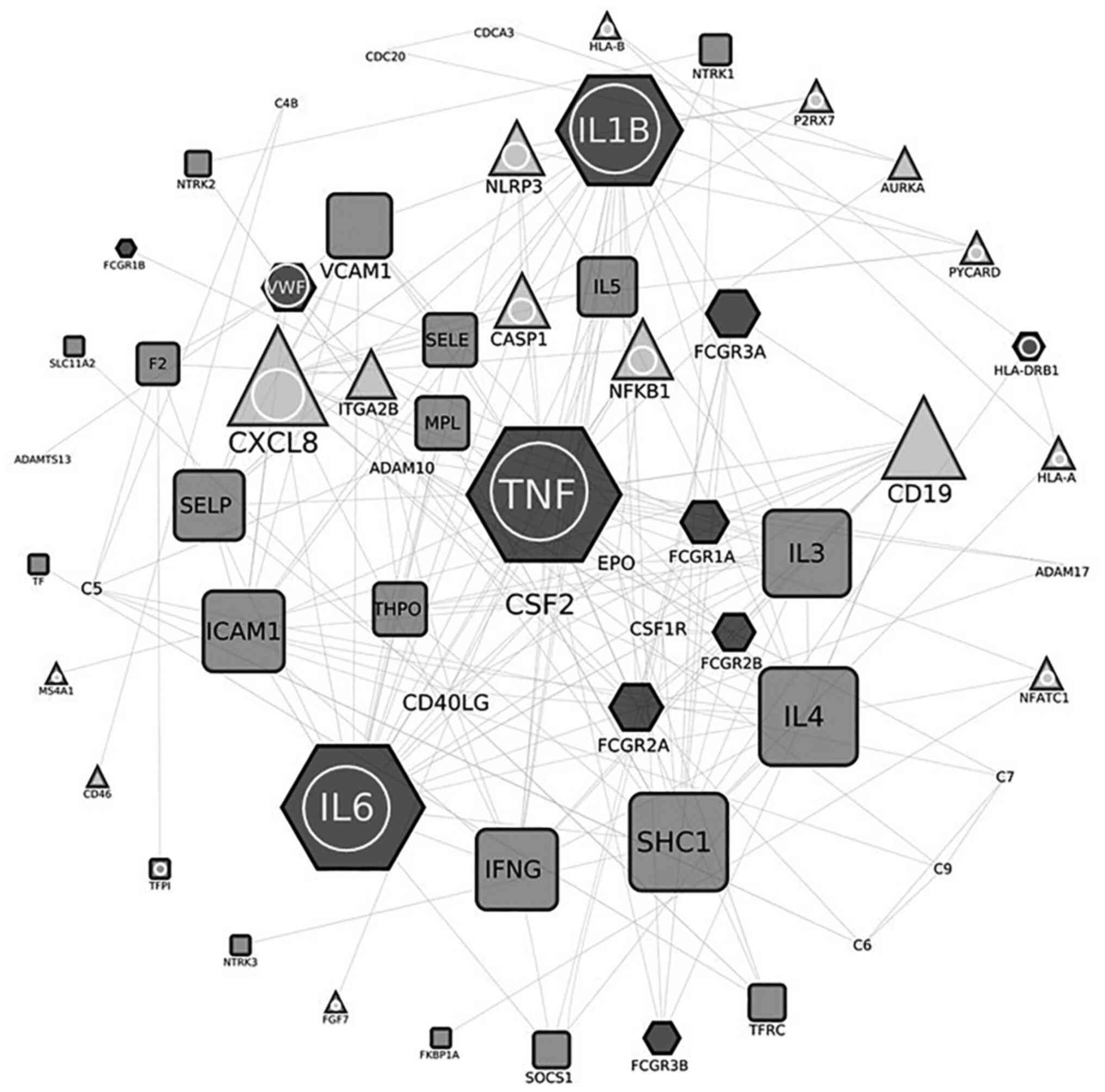
Subsequently, two interactomes were constructed: The
first one involving 119 nodes is described in Table I and illustrated in Fig. 3. Collectively, 119 nodes are
involved in COVID-19, while 57 are implicated in thrombocytopenia
[the latter profits from an unpublished work of ours (unpublished
data). Of these, 14 nodes were common in both entities (Figs. 3 and 4), namely AIM2, IFI16, NOD2, CD8A,
IL-1B, 1L-6, JAK2, NCAM1, HLA-DRB1, SERPINE1, TGFB1, TLR2, TNF and
VWF. The major hubs detected are displayed in the center of the
constructed circular network, while the less connected nodes are
shown at the periphery of the circle (Fig. 3). The thrombocytopenia-related
nodes are represented in square bullets, and the COVID-19-related
ones are presented in circles, whilst the common nodes are depicted
in rhomboids. The calculated average node degree of the entire
interactome was extremely high (11.9).
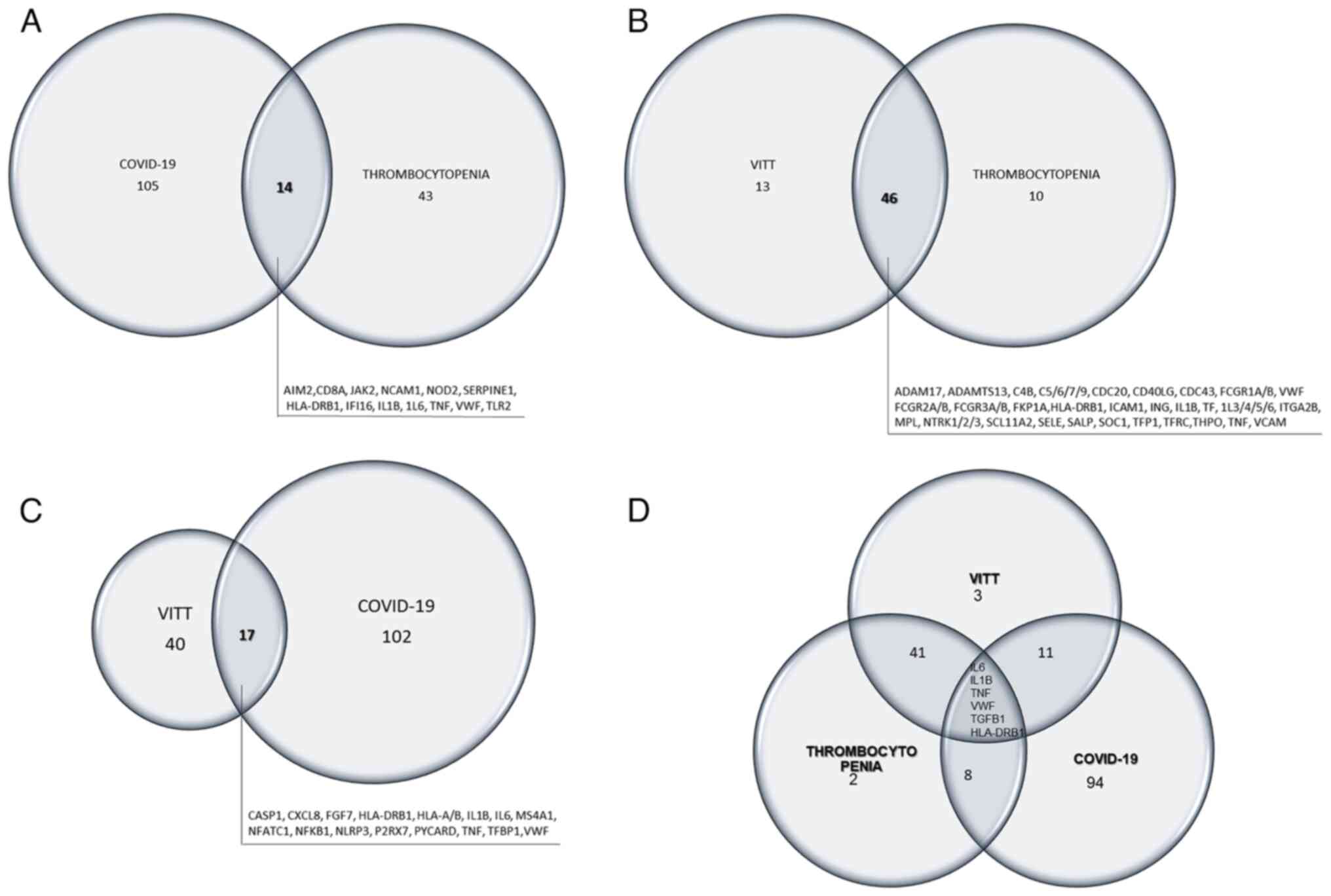
Venn diagrams were further created to illustrate the
nodes that are common between thrombocytopenia and COVID-19 or VITT
(Fig. 4A and B, respectively),
between COVID-19 and VITT (Fig.
4C), and amid the three morbid entities (Fig. 4D). The common nodes are listed in
each diagram in detail.
Epidemics were already identified as entities in
antiquity by Hippocrates and named by him in his Treatises 'On
Epidemics' (9,10). Viral epidemics were described
therein and in other works of the Hippocratic Corpus (11,12). On the other hand, Aristotle, the
ancient Greek physician and philosopher (4th century B.C.) wrote
that 'the creativeness of nature focuses on qualities rather
than quantities and description rather than measurements'
(13,14). This concept was rejected by
Newton's determinism and reductionism and was since forgotten,
until it was re-established by Wulff in 1999 (15). Indeed, subtle change in qualities
may trigger phase shift alterations with unpredictable
consequences, as the Chaos theory of dynamic systems recently
confirmed (16). According to
this concept, the systems theory was coined as representing a
rapid, cost and time-effective method of research (17). It may integrate basic,
preclinical and clinical research, and both human and animal
results to unravel new insights in complex and often unpredictable
issues. In the case of the COVID-19 pandemic, the urgency, and
certain ethical issues, make such an in silico approach a
sine qua non research method.
The human-to-human transmission of SARS-CoV-2 is
either mediated by respiratory droplets via sneezing/coughing or
even just breathing, while the disease demonstrates an incubation
period of 5-7 days (18). The
clinical outcomes range from asymptomatic to influenza-like, or to
even pneumonia and severe acute respiratory distress syndrome
(ARDS) (19), and thromboembolic
events (20,21), pointing to the lung tropism of
this virus. Dissimilarities in patients' profiles are attributed to
genetic and/or epigenetic variations and underlying pathologies.
Dissimilarities in severity may be attributed to the aforementioned
factors, but also to the size of the viral inoculum and/or viral
mutations.
Ariadne's thread appears to be the angiotensin I
converting enzyme 2 (ACE2), which clearly plays a crucial role.
SARS-CoV-2, via its spike S protein, a surface glycoprotein that
surrounds the spherical virus, is attached to ACE2 and this is
followed by entry into cells of the host (22-27). ACE2 is expressed in cells of a
number of human organs (including the skin, nasal and oral mucosa,
lung, nasopharynx, brain, lymph nodes, thymus, stomach, small
intestine, colon, bone marrow, spleen, liver and kidneys).
Additionally, its expression in lung alveoli (type 2 pneumonocytes)
and small intestine endothelium, as well as in the arterial and
other tissue smooth muscle epithelium (28), may trigger the release of
anaphylatoxin (29). There is
clinical evidence to confirm the aforementioned knowledge of
COVID-19 (29).
The network included dense interactions illustrating
clearly that SARS-CoV-2-specific T-cells are critical for the
extended damage caused by the 'cytokine storm' (or 'cytokine
release syndrome') (30,32) (Fig. 3). This excessive inflammatory
response may be lethal for some patients (29,33). Although the phenomenon may
manifest in other inflammatory conditions, including bacterial
sepsis, pneumonia, sterile inflammation, etc., the extent in the
secretion of several specific cytokines is different in
COVID-19-related storm (29). Of
note, COVID-19 infection has been associated with changes in the
blood coagulation mechanisms, with differing manifestations in
different patients, in distinct phases of the disease, and
independently of disease severity.
Autoimmune destruction of platelets, cytokine
release and high consumption of coagulation factors and platelets
have been observed in patients with SARS-CoV-2 infection
(Geronikolou et al, unpublished data) and initial
hypercoagulability (34).
Thromboembolic events increase by 31% in patients with COVID-19
admitted in intensive care units (35,36); the phenomenon may be interpreted
by the 'two way activation theory' (20,37), i.e., thrombogenesis via
inflammation-relevant pathways, with parallel occurrence of release
of VWF large polymers. The coagulation and platelet profiles of
patients with COVID-19 are then rather normal, unlike in patients
with sepsis where platelets are activated and consumed, with the
occurrence of thrombocytopenia (38). Only a few patients may then
survive, particularly of those with extensive disseminated
intravascular coagulation (38).
Thrombosis has been observed in situ in the lungs, as well
as in a systemic manner, in a similar fashion with classic sepsis
and acute respiratory distress syndrome. Reported thromboembolic
complications include mostly venous pulmonary embolism (38), aortic graft thrombosis, and
mesenteric ischemia; coronary and cerebral thrombosis cases have
been reported, although these are rare. The so-called 'COVID toe'
is a sign of thrombosis accompanied by arterial and venous clots,
urgent oxygen demand and multiple organ dysfunction (20,36,39).
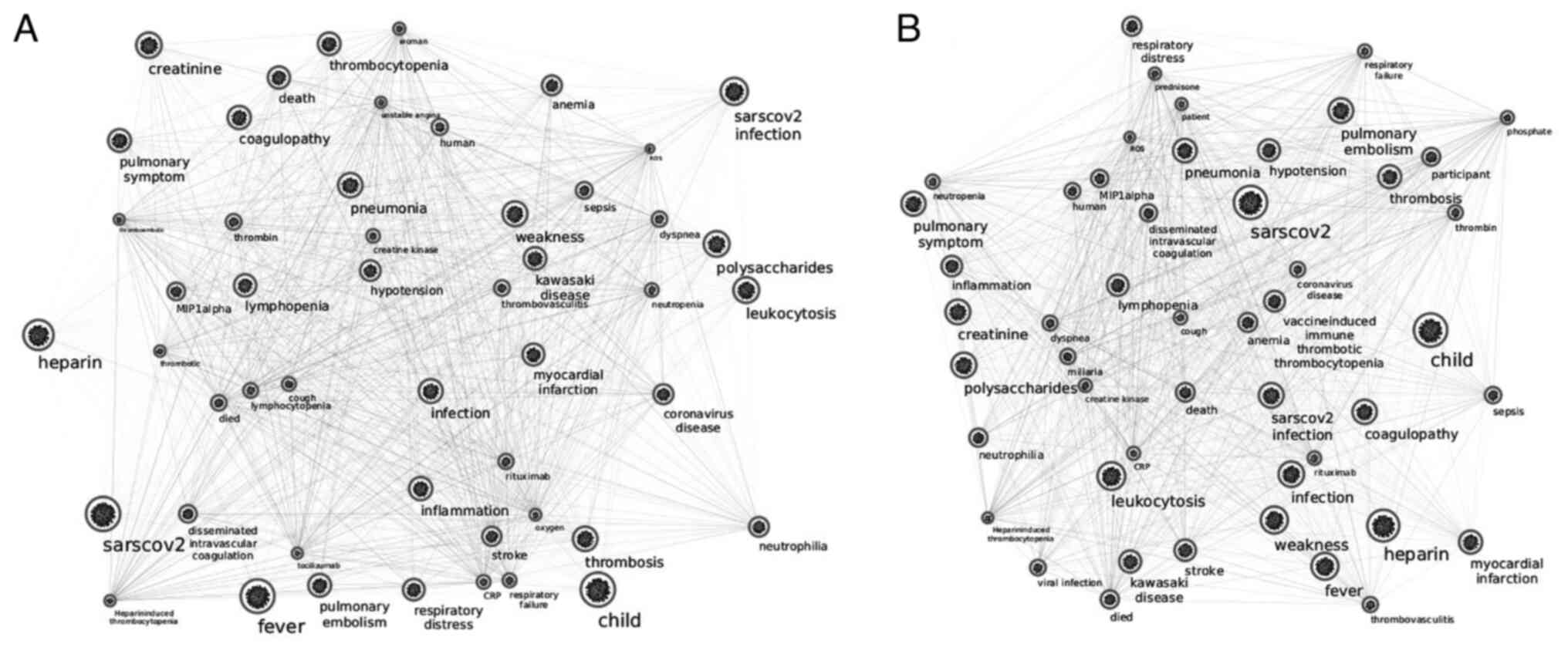
COVID-19 and thrombocytopenia interactomes share
only 14 nodes (AIM2, IFI16, TLR2, NOD2, NKAM1, IL-6, TNF, JAK2,
IL-1B, SERPINE1, HLA-DRB1, TGFB1, CD8A, and VWF) (Fig. 3), most of which serve as major
hubs (IL-6, TNF, JAK2, IL-1B, SERPINE1, TGFB1, CD8A and VWF) in the
herein presented interactome (Figs.
1 and 2).
Cytokines, such as IL-1B, 1L-6 and TNF contribute to
the so-called cytokine storm, as aforementioned. Moreover, JAK2 is
a kinase suspected to be implicated in thrombocytopenia via reduced
levels of thrombopoietin or via decreased expression levels of
their cognate receptors (cMpl receptors). JAK2 mutations (V617F)
that are present in the majority of patients with
myeloproliferative disease, may increase hematopoietic cell
sensitivity to erythropoietin and thrombopoietin. NKAM1 or CD56 is
a homophilic binding glycoprotein expressed on the surface of
neurons, glia cells and skeletal muscles. NKAM1 is a prototypic
marker of NK cells, also present in CD8+ T-cells. These
cell types exhibit diminished antiviral ability and cytotoxic
impairment during COVID-19 infection (24). CD8A1 is a cytotoxic marker for
T-cell populations. SERPINE1 or plasminogen activator inhibitor-1
is a protein encoded by the SERPINE1 gene, which
participates in both thrombosis and atherogenesis (40).
TGFB1 is a multifunctional peptide, with diverse
activities, including the control of cell growth, proliferation,
differentiation, and apoptosis. It can also down-regulate the
activity of immune cells via decreasing the expression levels of
cytokine receptors, such as that of IL-2. Several types of T-cells
secrete TGFB1, so as to inhibit cytotoxicity and the secretion of
certain cytokines, such as interferon-γ, TNF-α and various
interleukins, such as IL-6. This makes this molecule a potential
target of therapeutic value. On the other hand, the hemostatic VWF
is detected in blood plasma, endothelium and megakaryocytes, as
well as in subendothelial connective tissue. This factor appears to
be also increased and implicated in autoimmune diseases, such as
thrombotic thrombocytopenic purpura, as well as in stroke and
atrial fibrillation, due to the platelet clots that are potentially
formed when its levels are elevated.
Recent literature has further revealed that an HLA
class I and II molecule, that is, HLA-DRB1, which is common in
COVID-19 and in thrombocytopenia networks (Fig. 2), may play a role in the observed
COVID-19 individual and ethnic diversity in clinical severity
and/or response to therapy or vaccination (41-44). Of note, HLA-DRB1 is
interconnected with the lymphocyte function markers CD3D, CD3E,
CD3G, CD4, lymphocyte regulation positive FCGR1A, FCGR1B, HLA class
I and II molecules, such as HLA-A, HLA-B, similar to the NCAM1,
PTPN1, SHC1 and VCAM1 molecules that have been implicated in
thrombosis and atherosclerosis. NCAM1 is involved in cell-cell
adhesion in neural-muscle cells in the embryonic phase and later,
and more notably, in the responsiveness to viral infections (rabies
virus and papilloma virus) (45). PTPN1 is a potential therapeutic
target of obesity and type 2 diabetes mellitus as well (46); SHC1 is implicated in reactive
oxygen species regulation, thus, in the oxidative stress response
(47), while VCAM1 is directly
involved in thrombosis and atherogenesis and acute respiratory
syndrome (48-51).
Various coagulation mechanisms have been implicated
in VITT: High levels of D-dimers and low levels of fibrinogen have
been observed in patients (2,52,53). On the other hand, early reports
of VITT described a higher incidence of the syndrome in young
women, exhibiting both age-dependence and sexual dimorphism. VITT,
though very rare, is of utmost importance. Yet, in March, 2021, the
European Medicines Agency (EMA) issued a statement noting that the
thromboembolic events of VITT in vaccinated populations were not
higher than in general population (54). Subsequently, the 'risk vs.
benefit' equilibrium was weighed by the World Health Organization
(WHO), promoting the benefit of the vaccination vs. the extremely
low risk of thromboembolic risk of VITT in the general population
(55).
VITT is currently termed 'thrombosis with
thrombocytopenia syndrome (TTS)' by the Centers for Disease Control
and Prevention (CDC) and the US Food and Drug Administration (FDA)
(56), and is characterized by
arterial and venous thrombosis at unexpected sites (i.e., cerebral
venous sinuses, splanchnic vessels of variant severity and/or
positive platelet factor (PF) 4-heparin ELISA ('HIT' ELISA)
syndrome (52), exhibiting both
age dependence and sexual dimorphism (more frequent in individuals
<50 years old and of the female sex) (2). The laboratory and clinical features
of this syndrome are similar to those of the heparin-induced
thrombocytopenia (HIT) syndrome and/or the HIT-like autoimmune
thrombosis with thrombocytopenia syndrome (2,52,53), both of which have already been
reported following surgery, the uptake of certain pharmaceuticals,
or during some infections in patients that are not being treated
with heparin. The therapeutic suggestions of this recently coined
syndrome include early initiation of non-heparin anticoagulation,
high-dose IVIG, and/or prednisolone (57).
The genetic basis of the VITT syndrome appears to be
closely intertwined with that of the COVID-19 disease and, as such,
they share 16 nodes: CASP1, CXCL8, FGF7, HLA-A, HLA-B, IL1B, IL6,
MS4A1, NFATC1, NFKB1, NLP3, P2RX7, PYCARD, TNF, TFP1, VWF (Figs. 3Figure 4-5). The purpose of the vaccine is to
inhibit pathways that mediate this condition (52,58). More importantly, the relevant
research is ongoing with the extremely rare cases of this syndrome,
as VITT incidence is ~0.74-1 cases per 100,000 vaccinated subjects
(52). Of note, the
anti-COVID-19 vaccines do not cause illness and the two morbid
entities (COVID-19 and VITT) are by no means identical, with the
etiopathology of the latter being actually autoimmune, with
auto-antibodies against platelet factor 4. More explicitly,
COVID-19 network shares 14 nodes with thrombocytopenia (AIM2, CD8A,
HLA-DRB1, IFI16, IL1B, IL6, JAK2, NCAM1, NOD2, SERPINE1, TGFB1,
TLR2, TNF and VWF), while VITT (which is a type of
thrombocytopenia) shares 46 nodes with thrombocytopenia (Figs. 3Figure 4-5). Notably, SHC1, STXBP2, CDC20 and
ADAM10 are silenced in VITT, while AURKA, CD46, CD19 are uniquely
expressed following vaccination (apparently not expressed in common
thrombocytopenia or in COVID-19) (Figs. 3Figure 4-5). These molecules were not previously
identified as VITT-related and are, thus, a novel finding, at least
to the best of our knowledge.
It is known that the NLP3 inflammasome is implicated
in both COVID-19 and VITT, apart from its participation in other
inflammatory reactions (59). It
has also been previously demonstrated that acute thrombotic events
may manifest during hypoxia, as shown in COVID-19, due to an early
proinflammatory state in the venous milieu, mediated by a
HIF-induced NLP3 inflammasome complex (60,61). In the network constructed in the
present study, NLP3 connects with CASP1, IL-IB, IL17A, CXCL8, IL-6,
MYD88, NFKB1, P2RX7, PYCARD and TNF.
P2RX7 exhibits sexually dimorphic and contrasting
roles in the pathogenesis of thrombosis, depending on the pathogen
type, the severity of infection, the cell type infected and the
level of tissue activation (62). In the thrombocytopenia/
COVID-19/VITT cases, the viral load, the cell-type infected and the
infecting virus strain or certain vaccine types have been
associated with NLP3 hyperactivation, which in the presence of
comorbidities, such as liver, renal, gut or respiratory tract
illnesses, diabetes mellitus, previous infections, exposure to
pollutants, and/or lifestyle factors, such as smoking and obesity,
may upend the roles of P2RX7 and PYCARD to those of
tissue-damaging, or even lethal factors (62,63). More importantly, the persistent
neurological effects ('long-COVID-19') observed in a large
percentage of patients with COVID-19 may be explained via the
activation of these pathways. Thus, P2RX7 antagonists may be
promising therapeutics in the management of both VITT and
'long-COVID-19' (62,64), as P2RX7 receptor stimulation has
been implicated in lung damage, psychiatric disorders and
pathological inflammation (65,66). In the COVID-19 interactome, P2RX7
directly interacts with NLP3, CASP1 and P2RX1. On the contrary, in
the VITT network, P2RX7 directly interacts only with NLP3, IL1B and
CASP1. Accordingly, PYCARD interacts with NLP3, CASP1, IL1B, IL18
and IKBKG in COVID-19, and with NLP3, CASP1 and IL1B in the VITT
syndrome (Table II). The common
node in all possible combinations, as shown in Table II, is CASP1, a downstream event
of the NLP3 inflammasome; CASP1 activation promotes IL1B
production, which may be prevented by a pan-caspase inhibitor or by
glyburide treatment (67).
To this end, the present study investigated the
aforementioned issues through the construction of molecular
networks and the detection of at least one known
COVID/VITT/thrombocytopenia molecule that confirmed that
endothelial dysfunction and blood thrombosis are the key players of
both COVID-19 and VITT morbid entities. One limitation of the
present study is that it included only wild-type genes and their
products. To the best of our knowledge, however, this is the first
effort made at providing a comprehensive network map of the
molecules involved in the underlying mechanisms of COVID-19, long
COVID-19 and/or VITT pathophysiology.
In conclusion, the interactomes presented herein
revealed therapeutic and vaccination modification targets (i.e.,
SHC1, NCAM1, HLAs, CD8A, PTPN1, VWF and TBP1). It was also
demonstrated that: i) NCAM1 is involved in SARS-CoV-2 infection
responsiveness, apart from papilloma and rabies virus infections,
and may be responsible for relevant vaccination side effects; ii)
NLP3, P2RX7 and PYCARD contribution may help explain (partly or
mostly) VITT and/or 'long COVID-19 side-effects'; iii) furthermore,
the antagonism of these latter nodes should focus on potential
pharmacological targets in the context of SARS-CoV-2 infection
and/or vaccine immunization responsiveness. In conclusion, network
construction is a powerful tool, which may be used to elucidate the
physiology and pathophysiology of different states in clinical
investigation. The highly interconnected network presented herein
highlights the complexity of COVID-19/VITT pathophysiology, mapping
the key role of cytokines, enzymes and immune response markers
(lymphocyte regulators and human leucocyte antigens) that may be
potential drug or vaccine targets. It was constructed using
wild-type genes and gene products, revealing the body's
predisposition to COVID-19 infection or VITT. Of note, the COVID-19
and thrombocytopenia common nodes appear to be key players in the
natural history of the illness.
The datasets used and/or analyzed during the current
study are available throughout the manuscript.
SAG and MM were involved in the conceptualization of
the study. SAG was involved in the study methodology. SAG, AP and
MM were involved in data validation. SAG and AP was involved in
formal analysis and in the investigative aspects of the study. SAG
was involved in the provision of resources (study material). SAG,
IT and AP was involved in data curation. IT provided the software
used in this study. SAG, IT, GPC and AP were involved in the
interpretation of the data, and in the writing and preparation of
the original draft. SAG, AP, MM and GPC were involved in the
writing, reviewing and editing of the manuscript. MM and GPC
supervised the study. SAG and GPC were involved in project
administration. All authors confirm the authenticity of the raw
data and have read and agreed to the published version of the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
No funding was received.
|
1
|
World Health Organization (WHO):
Coronovirus disease (COVID-19): Vaccines safety. WHO; Geneva:
2021
|
|
2
|
Greinacher A, Thiele T, Warkentin TE,
Weisser K, Kyrle PA and Eichinger S: Thrombotic thrombocytopenia
after ChAdOx1 nCov-19 vaccination. N Engl J Med. 384:2092–2101.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mathieu E, Ritchie H, Ortiz-Ospina E,
Roser M, Hasell J, Appel C, Giattino C and Rodés-Guirao L: A global
database of COVID-19 vaccinations. Nat Hum Behav. 5:947–953. 2021.
View Article : Google Scholar
|
|
4
|
Wei CH, Allot A, Leaman R and Lu Z:
PubTator central: Automated concept annotation for biomedical full
text articles. Nucleic Acids Res. 47(W1): W587–W593. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen Q, Allot A and Lu Z: LitCovid: An
open database of COVID-19 literature. Nucleic Acids Res. 49(D1):
D1534–D1540. 2021. View Article : Google Scholar :
|
|
6
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1): D607–D613. 2019.
View Article : Google Scholar
|
|
7
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49(D1): D605–D612. 2021. View Article : Google Scholar
|
|
8
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hippocrates: Epidemics 2, 4-7. Smith
Wesley D: Loeb Classical Library 477. Harvard University Press;
Cambridge, MA: 1994
|
|
10
|
Jouanna J: Hippocrates. John Hopkins
University Press; Baltimore, MD: 1999
|
|
11
|
Mammas IN and Spandidos DA: Paediatric
virology in the Hippocratic corpus. Exp Ther Med. 12:541–549. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pappas G, Kiriaze IJ and Falagas ME:
Insights into infectious disease in the era of Hippocrates. Int J
Infect Dis. 12:347–350. 2008. View Article : Google Scholar
|
|
13
|
Misselbrook D: Aristotle, hume and the
goals of medicine. J Eval Clin Pract. 22:544–549. 2016. View Article : Google Scholar
|
|
14
|
Wulff HR: The concept of disease: From
Newton back to Aristotle. Lancet. 354(Suppl): SIV501999. View Article : Google Scholar
|
|
15
|
Wulff HR: The concept of disease: From
Newton back to Aristotle. Lancet. 54:3541999.
|
|
16
|
Lorenz EN: Deterministic nonperiodic flow.
J Atmos Sci. 20:130–141. 1963. View Article : Google Scholar
|
|
17
|
Barabási AL, Gulbahce N and Loscalzo J:
Network medicine: A network-based approach to human disease. Nat
Rev Genet. 12:56–68. 2011. View
Article : Google Scholar :
|
|
18
|
Li Q, Guan X, Wu P, Wang X, Zhou L, Tong
Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al: Early transmission
dynamics in Wuhan, China, of novel coronavirus-infected pneumonia.
N Engl J Med. 382:1199–1207. 2020. View Article : Google Scholar :
|
|
19
|
Raoult D, Zumla A, Locatelli F, Ippolito G
and Kroemer G: Coronavirus infections: Epidemiological, clinical
and immunological features and hypotheses. Cell Stress. 4:66–75.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mondal S, Quintili AL, Karamchandani K and
Bose S: Thromboembolic disease in COVID-19 patients: A brief
narrative review. J Intensive Care. 8:702020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu P, Zhou Q and Xu J: Mechanism of
thrombocytopenia in COVID-19 patients. Ann Hematol. 99:1205–1208.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li W, Moore MJ, Vasilieva N, Sui J, Wong
SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough
TC, et al: Angiotensin-converting enzyme 2 is a functional receptor
for the SARS coronavirus. Nature. 426:450–454. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ge XY, Li JL, Yang XL, Chmura AA, Zhu G,
Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, et al: Isolation and
characterization of a bat SARS-like coronavirus that uses the ACE2
receptor. Nature. 503:535–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mazzoni A, Salvati L, Maggi L, Capone M,
Vanni A, Spinicci M, Mencarini J, Caporale R, Peruzzi B, Antonelli
A, et al: Impaired immune cell cytotoxicity in severe COVID-19 is
IL-6 dependent. J Clin Invest. 130:4694–4703. 2020. View Article : Google Scholar :
|
|
25
|
Sama IE, Ravera A, Santema BT, van Goor H,
Ter Maaten JM, Cleland JGF, Rienstra M, Friedrich AW, Samani NJ, Ng
LL, et al: Circulating plasma concentrations of
angiotensin-converting enzyme 2 in men and women with heart failure
and effects of renin-angiotensin-aldosterone inhibitors. Eur Heart
J. 41:1810–1817. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Diaz JH: Hypothesis:
Angiotensin-converting enzyme inhibitors and angiotensin receptor
blockers may increase the risk of severe COVID-19. J Travel Med.
27:taaa0412020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH,
Nitsche A, et al: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
18:271–280.e8. 2020. View Article : Google Scholar
|
|
28
|
Hamming I, Timens W, Bulthuis ML, Lely AT,
Navis G and van Goor H: Tissue distribution of ACE2 protein, the
functional receptor for SARS coronavirus. A first step in
understanding SARS pathogenesis. J Pathol. 203:631–637. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao T, Hu M, Zhang X, Li H, Zhu L, Liu H,
Dong Q, Zhang Z, Wang Z, Hu Y, et al: Highly pathogenic coronavirus
N protein aggravates lung injury by MASP-2-mediated complement
over-activation. medRxiv. ppmedrxiv-20041962. 2020.
|
|
30
|
Cao X: COVID-19: Immunopathology and its
implications for therapy. Nat Rev Immunol. 20:269–270. 2020.
View Article : Google Scholar
|
|
31
|
Channappanavar R and Perlman S: Pathogenic
human coronavirus infections: Causes and consequences of cytokine
storm and immunopathology. Semin Immunopathol. 39:529–539. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao J, Zhao J and Perlman S: T cell
responses are required for protection from clinical disease and for
virus clearance in severe acute respiratory syndrome
coronavirus-infected mice. J Virol. 84:9318–9325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meduri GU, Kohler G, Headley S, Tolley E,
Stentz F and Postlethwaite A: Inflammatory cytokines in the BAL of
patients with ARDS. Persistent elevation over time predicts poor
outcome. Chest. 108:1303–1314. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang N, Li D, Wang X and Sun Z: Abnormal
coagulation parameters are associated with poor prognosis in
patients with novel coronavirus pneumonia. J Thromb Haemost.
18:844–847. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Helms J, Tacquard C, Severac F,
Leonard-Lorant I, Ohana M, Delabranche X, Merdji H, Clere-Jehl R,
Schenck M, Fagot Gandet F, et al: High risk of thrombosis in
patients with severe SARS-CoV-2 infection: A multicenter
prospective cohort study. Intensive Care Med. 46:1089–1098. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Klok FA, Kruip MJHA, van der Meer NJM,
Arbous MS, Gommers DAMPJ, Kant KM, Kaptein FHJ, van Paassen J,
Stals MAM, Huisman MV and Endeman H: Incidence of thrombotic
complications in critically ill ICU patients with COVID-19. Thromb
Res. 191:145–147. 2020. View Article : Google Scholar :
|
|
37
|
Chang JC: Hemostasis based on a novel
'two-path unifying theory' and classification of hemostatic
disorders. Blood Coagul Fibrinolysis. 29:573–584. 2018. View Article : Google Scholar
|
|
38
|
Chang JC: Sepsis and septic shock:
Endothelial molecular pathogenesis associated with vascular
microthrombotic disease. Thromb J. 17:102019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Seirafianpour F, Sodagar S, Pour Mohammad
A, Panahi P, Mozafarpoor S, Almasi S and Goodarzi A: Cutaneous
manifestations and considerations in COVID-19 pandemic: A
systematic review. Dermatol Ther. 33:e139862020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vaughan DE: PAI-1 and atherothrombosis. J
Thromb Haemost. 3:1879–1883. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Badary OA: Pharmacogenomics and COVID-19:
Clinical implications of human genome interactions with repurposed
drugs. Pharmacogenomics J. 21:275–284. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen MR, Kuo HC, Lee YJ, Chi H, Li SC, Lee
HC and Yang KD: Phenotype, susceptibility, autoimmunity, and
immunotherapy between Kawasaki disease and coronavirus disease-19
associated multisystem inflammatory syndrome in children. Front
Immunol. 12:6328902021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Romero-López JP, Carnalla-Cortés M,
Pacheco-Olvera DL, Ocampo-Godínez JM, Oliva-Ramírez J,
Moreno-Manjón J, Bernal-Alferes B, López-Olmedo N, García-Latorre
E, Domínguez-López ML, et al: A bioinformatic prediction of antigen
presentation from SARS-CoV-2 spike protein revealed a theoretical
correlation of HLA-DRB1*01 with COVID-19 fatality in Mexican
population: An ecological approach. J Med Virol. 93:2029–2038.
2021. View Article : Google Scholar
|
|
44
|
Anzurez A, Naka I, Miki S, Nakayama-Hosoya
K, Isshiki M, Watanabe Y, Nakamura-Hoshi M, Seki S, Matsumura T,
Takano T, et al: Association of HLA-DRB1*09:01 with severe
COVID-19. HLA. 98:37–42. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rotondo JC, Bosi S, Bassi C, Ferracin M,
Lanza G, Gafà R, Magri E, Selvatici R, Torresani S, Marci R, et al:
Gene expression changes in progression of cervical neoplasia
revealed by microarray analysis of cervical neoplastic
keratinocytes. J Cell Physiol. 230:806–812. 2015. View Article : Google Scholar
|
|
46
|
Combs AP: Recent advances in the discovery
of competitive protein tyrosine phosphatase 1B inhibitors for the
treatment of diabetes, obesity, and cancer. J Med Chem.
53:2333–2344. 2010. View Article : Google Scholar
|
|
47
|
Finkel T and Holbrook NJ: Oxidants,
oxidative stress and the biology of ageing. Nature. 408:239–247.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Choi YM, Kwon HS, Choi KM, Lee WY and Hong
EG: Short-term effects of beraprost sodium on the markers for
cardiovascular risk prediction in type 2 diabetic patients with
microalbuminuria. Endocrinol Metab (Seoul). 34:398–405. 2019.
View Article : Google Scholar
|
|
49
|
Nomura S, Taniura T, Shouzu A, Omoto S,
Suzuki M, Okuda Y and Ito T: Effects of sarpogrelate,
eicosapentaenoic acid and pitavastatin on arterioslcerosis
obliterans-related biomarkers in patients with type 2 diabetes
(SAREPITASO study). Vasc Health Risk Manag. 14:225–232. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng Y, Liu SQ, Sun Q, Xie JF, Xu JY, Li
Q, Pan C, Liu L and Huang YZ: Plasma microRNAs levels are different
between pulmonary and extrapulmonary ARDS patients: A clinical
observational study. Ann Intensive Care. 8:232018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Attia EF, Jolley SE, Crothers K, Schnapp
LM and Liles WC: Soluble vascular cell adhesion molecule-1
(sVCAM-1) is elevated in bronchoalveolar lavage fluid of patients
with acute respiratory distress syndrome. PLoS One.
11:e01496872016. View Article : Google Scholar :
|
|
52
|
Cines DB and Bussel JB: SARS-CoV-2
vaccine-induced immune thrombotic thrombocytopenia. N Engl J Med.
384:2254–2256. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Schultz NH, Sørvoll IH, Michelsen AE,
Munthe LA, Lund-Johansen F, Ahlen MT, Wiedmann M, Aamodt AH,
Skattør TH, Tjønnfjord GE and Holme PA: Thrombosis and
thrombocytopenia after ChAdOx1 nCoV-19 vaccination. N Engl J Med.
384:2124–2130. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
European Medicines Agency (EMA): COVID-19
Vaccine AstraZeneca: PRAC investigating cases of thromboembolic
events-vaccine's benefits currently still outweigh risks-update.
2021.
|
|
55
|
World Health Organization (WHO): Statement
of the WHO global advisory committee on vaccine safety (GACVS)
COVID-19 subcommittee on safety signals related to the AstraZeneca
COVID-19 vaccine. WHO; Geneva: 2021
|
|
56
|
Bussel JB, Connors JM, Cines DB, Dunbar
CE, Michaelis LC, Kreuziger LB, Lee AYY and Pabinger-Fasching I:
Thrombosis with thrombocytopenia syndrome (also termed
vaccine-induced thrombotic thrombocytopenia). American Society of
Haematology; Washington, DC: 2021
|
|
57
|
Thaler J, Ay C, Gleixner KV, Hauswirth AW,
Cacioppo F, Grafeneder J, Quehenberger P, Pabinger I and Knöbl P:
Successful treatment of vaccine-induced prothrombotic immune
thrombocytopenia (VIPIT). J Thromb Haemost. 19:1819–1822. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Smadja DM, Mentzer SJ, Fontenay M, Laffan
MA, Ackermann M, Helms J, Jonigk D, Chocron R, Pier GB, Gendron N,
et al: COVID-19 is a systemic vascular hemopathy: Insight for
mechanistic and clinical aspects. Angiogenesis. 24:755–788. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kashir J, Ambia AR, Shafqat A, Sajid MR,
AlKattan K and Yaqinuddin A: Scientific premise for the involvement
of neutrophil extracellular traps (NETs) in vaccine-induced
thrombotic thrombocytopenia (VITT). J Leukoc Biol. Sep 1–2021.Epub
ahead of prin. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gupta N, Sahu A, Prabhakar A, Chatterjee
T, Tyagi T, Kumari B, Khan N, Nair V, Bajaj N, Sharma M and Ashraf
MZ: Activation of NLRP3 inflammasome complex potentiates venous
thrombosis in response to hypoxia. Proc Natl Acad Sci USA.
114:4763–4768. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Salaro E, Rambaldi A, Falzoni S, Amoroso
FS, Franceschini A, Sarti AC, Bonora M, Cavazzini F, Rigolin GM,
Ciccone M, et al: Involvement of the P2X7-NLRP3 axis in leukemic
cell proliferation and death. Sci Rep. 6:262802016. View Article : Google Scholar :
|
|
62
|
Ribeiro DE, Oliveira-Giacomelli Á, Glaser
T, Arnaud-Sampaio VF, Andrejew R, Dieckmann L, Baranova J, Lameu C,
Ratajczak MZ and Ulrich H: Hyperactivation of P2X7 receptors as a
culprit of COVID-19 neuropathology. Mol Psychiatry. 26:1044–1059.
2021. View Article : Google Scholar
|
|
63
|
Savio LEB, de Andrade Mello P, da Silva CG
and Coutinho-Silva R: The P2X7 receptor in inflammatory diseases:
Angel or demon. Front Pharmacol. 9:522018. View Article : Google Scholar
|
|
64
|
Pacheco PAF and Faria RX: The potential
involvement of P2X7 receptor in COVID-19 pathogenesis: A new
therapeutic target? Scand J Immunol. 93:e129602021. View Article : Google Scholar
|
|
65
|
Ortiz GG, Pacheco-Moisés FP, Macías-Islas
M, Flores-Alvarado LJ, Mireles-Ramírez MA, González-Renovato ED and
Her nández-Nava r ro VE: Role of the blood-brain barrier in
multiple sclerosis. Arch Med Res. 45:687–697. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Di Virgilio F, Tang Y, Sarti AC and
Rossato M: A rationale for targeting the P2X7 receptor in
coronavirus disease 19. Br J Pharmacol. 177:4990–4994. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ferreira AC, Soares VC, de
Azevedo-Quintanilha IG, Dias SDSG, Fintelman-Rodrigues N,
Sacramento CQ, Mattos M, de Freitas CS, Temerozo JR, Teixeira L, et
al: SARS-CoV-2 engages inflammasome and pyroptosis in human primary
monocytes. Cell Death Discov. 7:432021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Moss ML and Bartsch JW: Therapeutic
benefits from targeting of ADAM family members. Biochemistry.
43:7227–7235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Souza JSM, Lisboa ABP, Santos TM, Andrade
MVS, Neves VBS, Teles-Souza J, Jesus HNR, Bezerra TG, Falcão VGO,
Oliveira RC and Del-Bem LE: The evolution of ADAM gene family in
eukaryotes. Genomics. 112:3108–3116. 2020. View Article : Google Scholar
|
|
70
|
Xu J, Xu X, Jiang L, Dua K, Hansbro PM and
Liu G: SARS-CoV-2 induces transcriptional signatures in human lung
epithelial cells that promote lung fibrosis. Respir Res.
21:1822020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Katneni UK, Alexaki A, Hunt RC, Schiller
T, DiCuccio M, Buehler PW, Ibla JC and Kimchi-Sarfaty C:
Coagulopathy and thrombosis as a result of severe COVID-19
infection: A microvascular focus. Thromb Haemost. 120:1668–1679.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tian J, Sun D, Xie Y, Liu K and Ma Y:
Network pharmacology-based study of the molecular mechanisms of
Qixuekang in treating COVID-19 during the recovery period. Int J
Clin Exp Pathol. 13:2677–2690. 2020.PubMed/NCBI
|
|
73
|
Boron WF and Boulpaep EL: Medical
physiology: A cellular and molecular approach. Saunders Elsevier;
Philadelphia, PA: 2012
|
|
74
|
Fitzpatrick D, Purves D and Augustine G:
Neuroscience. 3rd edition. Sinauer Associates, Inc; Sunderland, MA:
2004
|
|
75
|
Wang Q, Zhu W, Xiao G, Ding M, Chang J and
Liao H: Effect of AGER on the biological behavior of non-small cell
lung cancer H1299 cells. Mol Med Rep. 22:810–818. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Man SM, Karki R and Kanneganti TD: AIM2
inflammasome in infection, cancer, and autoimmunity: Role in DNA
sensing, inflammation, and innate immunity. Eur J Immunol.
46:269–280. 2016. View Article : Google Scholar
|
|
77
|
Bafunno V, Firinu D, D'Apolito M, Cordisco
G, Loffredo S, Leccese A, Bova M, Barca MP, Santacroce R, Cicardi
M, et al: Mutation of the angiopoietin-1 gene (ANGPT1) associates
with a new type of hereditary angioedema. J Allergy Clin Immunol.
141:1009–1017. 2018. View Article : Google Scholar
|
|
78
|
PubMed Gene database: ANGPT2 angiopoietin
2 [Homo sapiens (human)]. https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=ShowDetailView&TermToSearch=285
Accessed December 12, 2020.
|
|
79
|
Marumoto T, Honda S, Hara T, Nitta M,
Hirota T, Kohmura E and Saya H: Aurora-A kinase maintains the
fidelity of early and late mitotic events in HeLa cells. J Biol
Chem. 278:51786–51795. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li N, Zhang J, Liao D, Yang L, Wang Y and
Hou S: Association between C4, C4A, and C4B copy number variations
and susceptibility to autoimmune diseases: A meta-analysis. Sci
Rep. 7:426282017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Horiuchi T and Tsukamoto H:
Complement-targeted therapy: Development of C5- and C5a-targeted
inhibition. Inflamm Regen. 36:112016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hobart MJ, Fernie BA and DiScipio RG:
Structure of the human C7 gene and comparison with the C6, C8A,
C8B, and C9 genes. J Immunol. 154:5188–5194. 1995.PubMed/NCBI
|
|
83
|
Xia S, Zhang Z, Magupalli VG, Pablo JL,
Dong Y, Vora SM, Wang L, Fu TM, Jacobson MP, Greka A, et al:
Gasdermin D pore structure reveals preferential release of mature
interleukin-1. Nature. 593:607–611. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Cohen GM: Caspases: The executioners of
apoptosis. Biochem J. 326:1–16. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Avrutsky MI and Troy CM: Caspase-9: A
multimodal therapeutic target with diverse cellular expression in
human disease. Front Pharmacol. 12:7013012021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Singh S, Anshita D and Ravichandiran V:
MCP-1: Function, regulation, and involvement in disease. Int
Immunopharmacol. 101:1075982021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Coperchini F, Chiovato L, Ricci G, Croce
L, Magri F and Rotondi M: The cytokine storm in COVID-19: Further
advances in our understanding the role of specific chemokines
involved. Cytokine Growth Factor Rev. 58:82–91. 2021. View Article : Google Scholar :
|
|
88
|
Guan E, Wang J and Norcross MA:
Identification of human macrophage inflammatory proteins 1alpha and
1beta as a native secreted heterodimer. J Biol Chem.
276:12404–12409. 2001. View Article : Google Scholar
|
|
89
|
Charrier A and Brigstock DR: Regulation of
pancreatic function by connective tissue growth factor (CTGF,
CCN2). Cytokine Growth Factor Rev. 24:59–68. 2013. View Article : Google Scholar
|
|
90
|
Garcillán B, Fuentes P, Marin AV, Megino
RF, Chacon-Arguedas D, Mazariegos MS, Jiménez-Reinoso A, Muñoz-Ruiz
M, Laborda RG, Cárdenas PP, et al: CD3G or CD3D knockdown in
mature, but not immature, T lymphocytes similarly cripples the
human TCRαβ complex. Front Cell Dev Biol. 9:6084902021. View Article : Google Scholar
|
|
91
|
Heritable gene regulation in the CD4:CD8 T
cell lineage choice. Front Immunol. 8:2912017.PubMed/NCBI
|
|
92
|
Sharma P, Pandey AK and Bhattacharyya DK:
Determining crucial genes associated with COVID-19 based on COPD
findings✶,✶✶. Comput Biol Med. 128:1041262021.
View Article : Google Scholar
|
|
93
|
Zou M, Su X, Wang L, Yi X, Qiu Y, Yin X,
Zhou Z, Niu X, Wang L and Su M: The molecular mechanism of multiple
organ dysfunction and targeted intervention of COVID-19 based on
time-order transcriptomic analysis. Front Immunol. 12:7297762021.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jing Y, Luo L, Chen Y, Westerberg LS, Zhou
P, Xu Z, Herrada AA, Park CS, Kubo M, Mei H, et al: SARS-CoV-2
infection causes immunodeficiency in recovered patients by
downregulating CD19 expression in B cells via enhancing B-cell
metabolism. Signal Transduct Target Ther. 6:3452021. View Article : Google Scholar :
|
|
95
|
Badbaran A, Mailer RK, Dahlke C, Woens J,
Fathi A, Mellinghoff SC, Renné T, Addo MM, Riecken K and Fehse B:
Digital PCR to quantify ChAdOx1 nCoV-19 copies in blood and
tissues. Mol Ther Methods Clin Dev. 23:418–423. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Grewal IS and Flavell RA: CD40 and CD154
in cell-mediated immunity. Annu Rev Immunol. 16:111–135. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Riley-Vargas RC, Gill DB, Kemper C,
Liszewski MK and Atkinson JP: CD46: Expanding beyond complement
regulation. Trends Immunol. 25:496–503. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Lundstrom K, Barh D, Uhal BD, Takayama K,
Aljabali AAA, Abd El-Aziz TM, Lal A, Redwan EM, Adadi P, Chauhan G,
et al: COVID-19 vaccines and thrombosis-roadblock or dead-end
street? Biomolecules. 11:10202021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Chen J, Goyal N, Dai L, Lin Z, Del Valle
L, Zabaleta J, Liu J, Post SR, Foroozesh M and Qin Z: Developing
new ceramide analogs and identifying novel sphingolipid-controlled
genes against a virus-associated lymphoma. Blood. 136:2175–2187.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Dementyeva E, Kryukov F, Kubiczkova L,
Nemec P, Sevcikova S, Ihnatova I, Jarkovsky J, Minarik J,
Stefanikova Z, Kuglik P and Hajek R: Clinical implication of
centrosome amplification and expression of centrosomal functional
genes in multiple myeloma. J Transl Med. 11:772013. View Article : Google Scholar :
|
|
101
|
Martinez FO, Combes TW, Orsenigo F and
Gordon S: Monocyte activation in systemic Covid-19 infection: Assay
and rationale. EBioMedicine. 59:1029642020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Root RK and Dale DC: Granulocyte
colony-stimulating factor and granulocyte-macrophage
colony-stimulating factor: Comparisons and potential for use in the
treatment of infections in nonneutropenic patients. J Infect Dis.
179(Suppl 2): S342–S352. 1999. View
Article : Google Scholar
|
|
103
|
Zhang N, Zhao YD and Wang XM: CXCL10 an
important chemokine associated with cytokine storm in COVID-19
infected patients. Eur Rev Med Pharmacol Sci. 24:7497–7505.
2020.PubMed/NCBI
|
|
104
|
Bergamaschi C, Terpos E, Rosati M, Angel
M, Bear J, Stellas D, Karaliota S, Apostolakou F, Bagratuni T,
Patseas D, et al: Systemic IL-15, IFN-γ, and IP-10/CXCL10 signature
associated with effective immune response to SARS-CoV-2 in BNT162b2
mRNA vaccine recipients. Cell Rep. 36:1095042021. View Article : Google Scholar
|
|
105
|
Du HX, Zhu JQ, Chen J, Zhou HF, Yang JH
and Wan HT: Revealing the therapeutic targets and molecular
mechanisms of emodin-treated coronavirus disease 2019 via a
systematic study of network pharmacology. Aging (Albany NY).
13:14571–14589. 2021. View Article : Google Scholar
|
|
106
|
Lombardero M, Kovacs K and Scheithauer BW:
Erythropoietin: A hormone with multiple functions. Pathobiology.
78:41–53. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Petrović J, Pešić V and Lauschke VM:
Frequencies of clinically important CYP2C19 and CYP2D6 alleles are
graded across Europe. Eur J Hum Genet. 28:88–94. 2020. View Article : Google Scholar
|
|
108
|
Kell AM and Gale M Jr: RIG-I in RNA virus
recognition. Virology. 479-480:110–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Boron WF and Boulpaep EL: Medical
physiology: A cellular and molecular approach. 2nd edition.
Saunders Elsevier; Philadelphia, PA: 2009
|
|
110
|
Devreese KMJ: COVID-19-related laboratory
coagulation findings. Int J Lab Hematol. 43(Suppl 1): S36–S42.
2021. View Article : Google Scholar
|
|
111
|
Patel KR, Roberts JT and Barb AW: Multiple
variables at the leukocyte cell surface impact Fc γ
receptor-dependent mechanisms. Front Immunol. 10:2232019.
View Article : Google Scholar
|
|
112
|
Kelton JG, Smith JW, Santos AV, Murphy WG
and Horsewood P: Platelet IgG Fc receptor. Am J Hematol.
25:299–310. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Qiao J, Al-Tamimi M, Baker RI, Andrews RK
and Gardiner EE: The platelet Fc receptor, FcγRIIa. Immunol Rev.
268:241–252. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Fearon DT and Carroll MC: Regulation of B
lymphocyte responses to foreign and self-antigens by the CD19/CD21
complex. Annu Rev Immunol. 18:393–422. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Hartwig JH, Barkalow K, Azim A and
Italiano J: The elegant platelet: Signals controlling actin
assembly. Thromb Haemost. 82:392–398. 1999. View Article : Google Scholar
|
|
116
|
Viertlboeck BC, Schweinsberg S, Hanczaruk
MA, Schmitt R, Du Pasquier L, Herberg FW and Göbel TW: The chicken
leukocyte receptor complex encodes a primordial, activating,
high-affinity IgY Fc receptor. Proc Natl Acad Sci USA.
104:11718–11723. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Tan Y and Tang F: SARS-CoV-2-mediated
immune system activation and potential application in
immunotherapy. Med Res Rev. 41:1167–1194. 2021. View Article : Google Scholar
|
|
118
|
Hotchkiss KM, Clark NM and
Olivares-Navarrete R: Macrophage response to hydrophilic
biomaterials regulates MSC recruitment and T-helper cell
populations. Biomaterials. 182:202–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Springer S, Menzel LM and Zieger M: Google
trends provides a tool to monitor population concerns and
information needs during COVID-19 pandemic. Brain Behav Immun.
87:109–110. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Brockmeyer NH, Potthoff A, Kasper A,
Nabring C, Jöckel KH and Siffert W: GNB3 C825T polymorphism and
response to anti-retroviral combination therapy in HIV-1-infected
patients-a pilot study. Eur J Med Res. 10:489–494. 2005.PubMed/NCBI
|
|
121
|
Uddin MN, Akter R, Li M and Abdelrahman Z:
Expression of SARS-COV-2 cell receptor gene ACE2 is associated with
immunosuppression and metabolic reprogramming in lung
adenocarcinoma based on bioinformatics analyses of gene expression
profiles. Chem Biol Interact. 335:1093702021. View Article : Google Scholar
|
|
122
|
Bieberich F, Vazquez-Lombardi R, Yermanos
A, Ehling RA, Mason DM, Wagner B, Kapetanovic E, Di Roberto RB,
Weber CR, Savic M, et al: A single-cell atlas of lymphocyte
adaptive immune repertoires and transcriptomes reveals age-related
differences in convalescent COVID-19 patients. Front Immunol.
12:7010852021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Fricke-Galindo I and Falfán-Valencia R:
Genetics insight for COVID-19 susceptibility and severity: A
review. Front Immunol. 12:6221762021. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Jiang Z, Wei F, Zhang Y, Wang T, Gao W, Yu
S, Sun H, Pu J, Sun Y, Wang M, et al: IFI16 directly senses viral
RNA and enhances RIG-I transcription and activation to restrict
influenza virus infection. Nat Microbiol. 6:932–945. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Kennedy RB, Poland GA, Ovsyannikova IG,
Oberg AL, Asmann YW, Grill DE, Vierkant RA and Jacobson RM:
Impaired innate, humoral, and cellular immunity despite a take in
smallpox vaccine recipients. Vaccine. 34:3283–3290. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Kotenko SV: IFN-λs. Curr Opin Immunol.
23:583–590. 2011. View Article : Google Scholar :
|
|
127
|
Wu UI and Holland SM: Host susceptibility
to non-tuberculous mycobacterial infections. Lancet Infect Dis.
15:968–980. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Voloudakis G, Hoffman G, Venkatesh S, Lee
KM, Dobrindt K, Vicari JM, Zhang W, Beckmann ND, Jiang S, Hoagland
D, et al: IL10RB as a key regulator of COVID-19 host susceptibility
and severity. medRxiv. View Article : Google Scholar
|
|
129
|
Vecchié A, Bonaventura A, Toldo S, Dagna
L, Dinarello CA and Abbate A: IL-18 and infections: Is there a role
for targeted therapies. J Cell Physiol. 236:1638–1657. 2021.
View Article : Google Scholar
|
|
130
|
Peters VA, Joesting JJ and Freund GG: IL-1
receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav
Immun. 32:1–8. 2013. View Article : Google Scholar :
|
|
131
|
Bénard A, Jacobsen A, Brunner M, Krautz C,
Klösch B, Swierzy I, Naschberger E, Podolska MJ, Kouhestani D,
David P, et al: Interleukin-3 is a predictive marker for severity
and outcome during SARS-CoV-2 infections. Nat Commun. 12:11122021.
View Article : Google Scholar :
|
|
132
|
Zizzo G and Cohen PL: Imperfect storm: Is
interleukin-33 the Achilles heel of COVID-19? Lancet Rheumatol.
12:e779–e790. 2020. View Article : Google Scholar
|
|
133
|
Walsh PT and Fallon PG: The emergence of
the IL-36 cytokine family as novel targets for inflammatory
diseases. Ann NY Acad Sci. 1417:23–34. 2018. View Article : Google Scholar
|
|
134
|
Nussbaum JC, Van Dyken SJ, von Moltke J,
Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla
A, Liang HE and Locksley RM: Type 2 innate lymphoid cells control
eosinophil homeostasis. Nature. 502:245–248. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Coomes EA and Haghbayan H: Interleukin-6
in Covid-19: A systematic review and meta-analysis. Rev Med Virol.
30:1–9. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Das UN: Bioactive lipids in
COVID-19-further evidence. Arch Med Res. 52:107–120. 2021.
View Article : Google Scholar
|
|
137
|
Islam ABMMK, Khan MA, Ahmed R, Hossain MS,
Kabir SMT, Islam MS and Siddiki AMAMZ: Transcriptome of
nasopharyngeal samples from COVID-19 patients and a comparative
analysis with other SARS-CoV-2 infection models reveal disparate
host responses against SARS-CoV-2. J Transl Med. 19:322021.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
O'Brien JR: Shear-induced platelet
aggregation. Lancet. 335:711–713. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Langmuir P, Yeleswaram S, Smith P, Knorr B
and Squier P: Design of clinical trials evaluating ruxolitinib, a
JAK1/JAK2 inhibitor, for treatment of COVID-19-associated cytokine
storm. Dela J Public Health. 6:50–54. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Melman YF, Krummerman A and McDonald TV:
KCNE regulation of KvLQT1 channels: Structure-function correlates.
Trends Cardiovasc Med. 12:182–187. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Gouas L, Nicaud V, Berthet M, Forhan A,
Tiret L, Balkau B and Guicheney P; D.E.S.I.R. Study Group:
Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc
interval length in a healthy population. Eur J Hum Genet.
13:1213–1222. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Lazzerini PE, Acampa M, Laghi-Pasini F,
Bertolozzi I, Finizola F, Vanni F, Natale M, Bisogno S, Cevenini G,
Cartocci A, et al: Cardiac arrest risk during acute infections:
Systemic inflammation directly prolongs QTc interval via
cytokine-mediated effects on potassium channel expression. Circ
Arrhythm Electrophysiol. 13:e0086272020. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Szendrey M, Guo J, Li W, Yang T and Zhang
S: COVID-19 drugs chloroquine and hydroxychloroquine, but not
azithromycin and remdesivir, block hERG potassium channels. J
Pharmacol Exp Ther. 377:265–272. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Dahl SL, Woodworth JS, Lerche CJ, Cramer
EP, Nielsen PR, Moser C, Thomsen AR, Borregaard N and Cowland JB:
Lipocalin-2 functions as inhibitor of innate resistance to
mycobacterium tuberculosis. Front Immunol. 9:27172018. View Article : Google Scholar :
|
|
145
|
Vincenti MP and Brinckerhoff CE:
Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in
arthritis: Integration of complex signaling pathways for the
recruitment of gene-specific transcription factors. Arthritis Res.
4:157–164. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
146
|
Jabłońska-Trypuć A, Matejczyk M and
Rosochacki S: Matrix metalloproteinases (MMPs), the main
extracellular matrix (ECM) enzymes in collagen degradation, as a
target for anticancer drugs. J Enzyme Inhib Med Chem. 31(Suppl 1):
S177–S183. 2016. View Article : Google Scholar
|
|
147
|
Pisano TJ, Hakkinen I and Rybinnik I:
Large vessel occlusion secondary to COVID-19 hypercoagulability in
a young patient: A case report and literature review. J Stroke
Cerebrovasc Dis. 29:1053072020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Apostolidis SA, Kakara M, Painter MM, Goel
RR, Mathew D, Lenzi K, Rezk A, Patterson KR, Espinoza DA, Kadri JC,
et al: Cellular and humoral immune responses following SARS-CoV-2
mRNA vaccination in patients with multiple sclerosis on anti-CD20
therapy. Nat Med. 27:1990–2001. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Lu W, Liu X, Wang T, Liu F, Zhu A, Lin Y,
Luo J, Ye F, He J, Zhao J, et al: Elevated MUC1 and MUC5AC mucin
protein levels in airway mucus of critical ill COVID-19 patients. J
Med Virol. 93:582–584. 2021. View Article : Google Scholar
|
|
150
|
Conti P, Ronconi G, Caraffa A, Gallenga
CE, Ross R, Frydas I and Kritas SK: Induction of pro-inflammatory
cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19
(COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. J Biol Regul
Homeost Agents. 34:327–331. 2020.PubMed/NCBI
|
|
151
|
Morsy S: NCAM protein and SARS-COV-2
surface proteins: In-silico hypothetical evidence for the
immunopathogenesis of Guillain-Barré syndrome. Med Hypotheses.
145:1103422020. View Article : Google Scholar
|
|
152
|
Root-Bernstein R: Innate receptor
activation patterns involving TLR and NLR synergisms in COVID-19,
ALI/ARDS and sepsis cytokine storms: A review and model making
novel predictions and therapeutic suggestions. Int J Mol Sci.
22:21082021. View Article : Google Scholar :
|
|
153
|
Watanabe T, Kitani A, Murray PJ and
Strober W: NOD2 is a negative regulator of Toll-like receptor
2-mediated T helper type 1 responses. Nat Immunol. 5:800–808. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Esposito E and Cuzzocrea S: The role of
nitric oxide synthases in lung inflammation. Curr Opin Investig
Drugs. 8:899–909. 2007.PubMed/NCBI
|
|
155
|
Gamkrelidze M, Intskirveli N, Vardosanidze
K, Goliadze L, Chikhladze KH and Ratiani L: Myocardial dysfunction
during septic shock (review). Georgian Med News. 237:40–46.
2014.
|
|
156
|
Zang X, Li S, Zhao Y, Chen K, Wang X, Song
W, Ma J, Tu X, Xia Y, Zhang S and Gao C: Systematic meta-analysis
of the association between a common NOS1AP genetic polymorphism,
the QTc interval, and sudden death. Int Heart J. 60:1083–1090.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Guan SP, Seet RCS and Kennedy BK: Does
eNOS derived nitric oxide protect the young from severe COVID-19
complications. Ageing Res Rev. 64:1012012020. View Article : Google Scholar
|
|
158
|
Thom SR, Fisher D, Xu YA, Garner S and
Ischiropoulos H: Role of nitric oxide-derived oxidants in vascular
injury from carbon monoxide in the rat. Am J Physiol.
276:H984–H992. 1999.
|
|
159
|
Valent A, Danglot G and Bernheim A:
Mapping of the tyrosine kinase receptors trkA (NTRK1), trkB (NTRK2)
and trkC(NTRK3) to human chromosomes 1q22, 9q22 and 15q25 by
fluorescence in situ hybridization. Eur J Hum Genet. 5:102–104.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Liu W, Chen L, Zhu J and Rodgers GP: The
glycoprotein hGC-1 binds to cadherin and lectins. Exp Cell Res.
312:1785–1797. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Hennigs JK, Lüneburg N, Stage A, Schmitz
M, Körbelin J, Harbaum L, Matuszcak C, Mienert J, Bokemeyer C,
Böger RH, et al: The P2-receptor-mediated Ca2+
signalosome of the human pulmonary endothelium-implications for
pulmonary arterial hypertension. Purinergic Signal. 15:299–311.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Russo MV and McGavern DB: Immune
surveillance of the CNS following infection and injury. Trends
Immunol. 36:637–650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Tuuminen R, Nykänen A, Keränen MA, Krebs
R, Alitalo K, Koskinen PK and Lemström KB: The effect of
platelet-derived growth factor ligands in rat cardiac allograft
vasculopathy and fibrosis. Transplant Proc. 38:3271–3273. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Blum E, Margalit R, Levy L, Getter T,
Lahav R, Zilber S, Bradfield P, Imhof BA, Alpert E and Gruzman A: A
Potent leukocyte transmigration blocker: GT-73 showed a protective
effect against LPS-induced ARDS in mice. Molecules. 26:45832021.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Rovina N, Akinosoglou K, Eugen-Olsen J,
Hayek S, Reiser J and Giamarellos-Bourboulis EJ: Soluble urokinase
plasminogen activator receptor (suPAR) as an early predictor of
severe respiratory failure in patients with COVID-19 pneumonia.
Crit Care. 24:1872020. View Article : Google Scholar
|
|
166
|
Kumar S, Jain A, Choi SW, da Silva GPD,
Allers L, Mudd MH, Peters RS, Anonsen JH, Rusten TE, Lazarou M and
Deretic V: Mammalian Atg8 proteins and the autophagy factor IRGM
control mTOR and TFEB at a regulatory node critical for responses
to pathogens. Nat Cell Biol. 22:973–985. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Hoxha M: What about COVID-19 and
arachidonic acid pathway. Eur J Clin Pharmacol. 76:1501–1504. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Keikha M, Ghazvini K, Eslami M, Yousefi B,
Casseb J, Yousefi M and Karbalaei M: Molecular targeting of PD-1
signaling pathway as a novel therapeutic approach in HTLV-1
infection. Microb Pathog. 144:1041982020. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Coggeshall KM: Negative signaling in
health and disease. Immunol Res. 19:47–64. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
de Souza JG, Starobinas N and Ibañez O:
Unknown/enigmatic functions of extracellular ASC. Immunology.
163:377–388. 2021. View Article : Google Scholar
|
|
171
|
Larabi A, Barnich N and Nguyen HTT: New
insights into the interplay between autophagy, gut microbiota and
inflammatory responses in IBD. Autophagy. 16:38–51. 2020.
View Article : Google Scholar :
|
|
172
|
Brown MJ: Renin: Friend or foe? Heart.
93:1026–1033. 2007. View Article : Google Scholar
|
|
173
|
Amraei R and Rahimi N: COVID-19,
renin-angiotensin system and endothelial dysfunction. Cells.
9:16522020. View Article : Google Scholar :
|
|
174
|
Yanatori I, Yasui Y, Noguchi Y and Kishi
F: Inhibition of iron uptake by ferristatin II is exerted through
internalization of DMT1 at the plasma membrane. Cell Biol Int.
39:427–434. 2015. View Article : Google Scholar
|
|
175
|
Denham NC, Pearman CM, Ding WY, Waktare J,
Gupta D, Snowdon R, Hall M, Cooper R, Modi S, Todd D and Mahida S:
Systematic re-evaluation of SCN5A variants associated with Brugada
syndrome. J Cardiovasc Electrophysiol. 30:118–127. 2019. View Article : Google Scholar
|
|
176
|
Smadja DM, Guerin CL, Chocron R, Yatim N,
Boussier J, Gendron N, Khider L, Hadjadj J, Goudot G, Debuc B, et
al: Angiopoietin-2 as a marker of endothelial activation is a good
predictor factor for intensive care unit admission of COVID-19
patients. Angiogenesis. 23:611–620. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Bongiovanni D, Klug M, Lazareva O,
Weidlich S, Biasi M, Ursu S, Warth S, Buske C, Lukas M, Spinner CD,
et al: SARS-CoV-2 infection is associated with a pro-thrombotic
platelet phenotype. Cell Death Dis. 12:502021. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Wu D and Yang XO: Dysregulation of
pulmonary responses in severe COVID-19. Viruses. 13:9572021.
View Article : Google Scholar :
|
|
179
|
Katzen J and Beers MF: Contributions of
alveolar epithelial cell quality control to pulmonary fibrosis. J
Clin Invest. 130:5088–5099. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Nandy D, Sharma N and Senapati S:
Systematic review and meta-analysis confirms significant
contribution of surfactant protein D in chronic obstructive
pulmonary disease. Front Genet. 10:3392019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Di Lisa F, Kaludercic N, Carpi A, Menabò R
and Giorgio M: Mitochondria and vascular pathology. Pharmacol Rep.
61:123–130. 2009. View Article : Google Scholar
|
|
182
|
Dinarello CA, Nold-Petry C, Nold M, Fujita
M, Li S, Kim S and Bufler P: Suppression of innate inflammation and
immunity by interleukin-37. Eur J Immunol. 46:1067–1081. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Montalbetti N, Simonin A, Kovacs G and
Hediger MA: Mammalian iron transporters: families SLC11 and SLC40.
Mol Aspects Med. 34:270–287. 2013. View Article : Google Scholar
|
|
184
|
Schulert GS, Blum SA and Cron RQ: Host
genetics of pediatric SARS-CoV-2 COVID-19 and multisystem
inflammatory syndrome in children. Curr Opin Pediatr. 33:549–555.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Pachlopnik Schmid J and de Saint Basile G:
Angeborene hämophagozytische lymphohistiozytose (HLH). Klin
Padiatr. 222:345–350. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Shi JH, Xie X and Sun SC: TBK1 as a
regulator of autoimmunity and antitumor immunity. Cell Mol Immunol.
15:743–745. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Zimecki M, Actor JK and Kruzel ML: The
potential for Lactoferrin to reduce SARS-CoV-2 induced cytokine
storm. Int Immunopharmacol. 95:1075712021. View Article : Google Scholar :
|
|
188
|
Chabot PR, Raiola L, Lussier-Price M,
Morse T, Arseneault G, Archambault J and Omichinski JG: Structural
and functional characterization of a complex between the acidic
transactivation domain of EBNA2 and the Tfb1/p62 subunit of TFIIH.
PLoS Pathog. 10:e10040422014. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Speeckaert MM, Speeckaert R and Delanghe
JR: Biological and clinical aspects of soluble transferrin
receptor. Crit Rev Clin Lab Sci. 47:213–228. 2010. View Article : Google Scholar
|
|
190
|
Bg S, Gosavi S, Ananda Rao A, Shastry S,
Raj SC, Sharma A, Suresh A and Noubade R: Neutrophil-to-lymphocyte,
lymphocyte-to-monocyte, and platelet-to-lymphocyte ratios:
Prognostic significance in COVID-19. Cureus.
13:e126222021.PubMed/NCBI
|
|
191
|
Campbell GR, To RK, Hanna J and Spector
SA: SARS-CoV-2, SARS-CoV-1, and HIV-1 derived ssRNA sequences
activate the NLRP3 inflammasome in human macrophages through a
non-classical pathway. iScience. 24:1022952021. View Article : Google Scholar :
|
|
192
|
Borrello S, Nicolò C, Delogu G, Pandolfi F
and Ria F: TLR2: a crossroads between infections and autoimmunity.
Int J Immunopathol Pharmacol. 24:549–556. 2011. View Article : Google Scholar
|
|
193
|
Zheng M, Karki R, Williams EP, Yang D,
Fitzpatrick E, Vogel P, Jonsson CB and Kanneganti TD: TLR2 senses
the SARS-CoV-2 envelope protein to produce inflammatory cytokines.
Nat Immunol. 22:829–838. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Khan S, Shafiei M, Longoria C, Schoggins
JW, Savani R and Zaki H: SARS-CoV-2 spike protein induces
inflammation via TLR2-dependent activation of the NF-κB pathway.
Elife. 10:e685632021. View Article : Google Scholar
|
|
195
|
Sohn KM, Lee SG, Kim HJ, Cheon S, Jeong H,
Lee J, Kim IS, Silwal P, Kim YJ, Paik S, et al: COVID-19 patients
upregulate toll-like receptor 4-mediated inflammatory signaling
that mimics bacterial sepsis. J Korean Med Sci. 35:e3432020.
View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Guven-Maiorov E, Keskin O, Gursoy A,
VanWaes C, Chen Z, Tsai CJ and Nussinov R: TRAF3 signaling:
Competitive binding and evolvability of adaptive viral molecular
mimicry. Biochim Biophys Acta. 1860:2646–2655. 2016. View Article : Google Scholar
|
|
197
|
Callaway E: The quest to find genes that
drive severe COVID. Nature. 595:346–348. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Kaur S, Tripathi DM and Yadav A: The
enigma of endothelium in COVID-19. Front Physiol. 11:9892020.
View Article : Google Scholar :
|
|
199
|
Rovas A, Osiaevi I, Buscher K, Sackarnd J,
Tepasse PR, Fobker M, Kühn J, Braune S, Göbel U, Thölking G, et al:
Microvascular dysfunction in COVID-19: The MYSTIC study.
Angiogenesis. 24:145–157. 2021. View Article : Google Scholar
|
|
200
|
Holcomb D, Alexaki A, Hernandez N, Hunt R,
Laurie K, Kames J, Hamasaki-Katagiri N, Komar AA, DiCuccio M and
Kimchi-Sarfaty C: Gene variants of coagulation related proteins
that interact with SARS-CoV-2. PLoS Comput Biol. 17:e10088052021.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Shahidi M: Thrombosis and von Willebrand
factor. Adv Exp Med Biol. 906:285–306. 2017. View Article : Google Scholar
|