Introduction
The mixed-lineage leukemia (or myeloid-lymphoid
leukemia, MLL) gene on chromosome 11q23 is a common target for
chromosomal translocation associated with pediatric, adult and
therapy-related acute leukemias, and are generally associated with
aggressive disease and poor prognosis (1–4).
Acute lymphoblastic leukemia (ALL) carrying
11q23/MLL translocations represents 8–10% of total ALL and 60–70%
of infant (i.e. <1 year old) ALL (5,6). The
most common fusion rearrangement in ALL is the t(4;11)(q21;q23)
MLL-AF4 translocation, representing 5–10% of adult ALL, up to 5% of
pediatric ALL and >60% of infant ALL. The t(4;11)(q21;q23)
MLL-AF4 translocation is associated with high rates of early
treatment failure (7,8). Leukemic cells carrying the t(4;11)
(q21;q23) MLL-AF4 translocation may manifest some cyto-chemical and
ultrastructural features of monocytes, and thus t(4;11)(q21;q23)
MLL-AF4 leukemia has been classified as biphenotypic leukemia
(9). The t(11;19)(q23;p13.3)
MLL-ENL translocation represents 10% of the 11q23 cases in ALL and
5% in AML M4/M5 (8).
Acute myeloid leukemia (AML) with rearrangements of
11q23/MLL occurs in 10–15% of children (1–18 years old), 60–70% of
infants and 8–10% of adult patients (6,10).
The t(9;11)(p22;q23) MLL-AF9 translocation is the most frequent
rearrangement in AML subtype M5a, that represents approximately 15%
of AML in children >2 years old and approximately 50% of AML in
younger children (8). t(9;11)
(p22;q23) MLL-AF9 leukemia has a more favorable prognosis than
t(4;11)(q21;q23) MLL-AF4 leukemia (11), but the event-free and overall
survival rates in AMLs vary largely depending on the MLL fusion
partner gene involved, with t(9;11)(p22;q23) MLL-AF9 having the
most favorable outcome and t(6;11)(q27;q23) MLL-AF6 the poorest
(3).
In vivo bioluminescence imaging (BLI) has
become a powerful tool to evaluate cellular and molecular
physiologic and pathologic processes in living systems (12–15).
BLI has been used successfully in small animal models to study
several different types of tumor, such as prostate, breast, colon,
ovarian as well as lung cancer and is particularly appropriate for
haematopoietic tumors (16–22).
BLI is based on the activity of the enzyme luciferase, produced by
firefly Photinus pyralis, that acts as a bioluminescent
reporter gene. Luciferase catalyzes the oxidation of its substrate,
D-luciferin, in the presence of ATP, molecular oxygen and
Mg2+, conditions available only in viable cells, with
emission of light proportional to the expression of the enzyme
luciferase. Expression of luciferase may be accomplished in cells
through stable transfection of luciferase under the control of a
constitutive promoter (23). For
these features, BLI is cost-effective, does not need the use of
radioisotopes, is simple, sensitive and provides a strong light
signal with virtually no background noise because of the emission
of green light from firefly luciferase/luciferin reaction at
approximately 612 nm at body temperature. For in vivo BLI,
experimental animals, that do not physiologically express
luciferase, are first inoculated with cells that stably express
luciferase, then the substrate D-luciferin is intraperitoneally
(i.p.) administered to the animals, and light emission from the
luciferase-expressing cells is detected with an ultrasensitive
charge-coupled device (CCD) camera (24). The results can be presented as
qualitative pseudocolor images, representing the number of photons
emitted per area, or as quantitative photon counts. It is also
possible to establish a correlation between luminescence intensity
and cell expansion, since light emission increases as the
luciferase-expressing cells multiply in a linear relationship
(14). In vivo BLI is a
rapid and non-invasive method that permits whole-body studies of
experimental animals, preserving at the same time the wellness of
the animals, and that can be performed repeatedly to monitor the
leukemia growth kinetics and therapeutic efficacy using each animal
as its own control. Moreover, the high sensitivity of BLI enables
the early detection of very small amounts of luciferase-expressing
cells and of deep tumor, preceding the appearance of evident
symptoms and blood dissemination (25).
Small interfering RNAs (siRNAs) are double stranded
RNAs that mediate RNA interference (RNAi), an evolutionarily
conserved sequence-specific post-transcriptional gene silencing
(PTGS) mechanism, that consists of the degradation of complementary
mRNA or of the inhibition of mRNA translation (26,27).
siRNAs are derived from long double stranded RNA (dsRNA)
precursors, processed by RNase-III-like enzyme Dicer in 21
nucleotide siRNAs. siRNAs are subsequently rearranged into the RNA
induced silencing complex (RISC) that mediates mRNA-target
degradation. Alternatively, RNAi can be mediated by synthetic
siRNAs, introduced in the cell by delivery methods such as
electroporation, cationic liposomes, and nanoparticles. Preferred
RNAi targets are viral genes, oncogenes or aberrant
gain-of-function genes. RNAi has become the method of choice to
suppress gene expression in vitro and it is also emerging as
a powerful tool for in vivo research (28–31).
The success of the clinical use of oligonucleotide
therapeutics is subdued by their in vivo delivery systems,
that should overcome physiological barriers, such as degradation by
nucleases in serum and tissue, rapid excretion by glomerular
filtration and renal system, phagocytosis and then degradation by
circulating macrophages of the reticuloendothelial system,
difficulty to cross the capillary endothelial cells, trapping and
slow diffusion through the extracellular matrix. The challenge
in vivo is constituted by the evaluation and selection of
optimal delivery systems for siRNAs and other oligonucleotides to
overcome these barriers.
In this respect, several methods have been
evaluated: chemically modified oligonucleotides and different
delivery systems that can be complexed with siRNA or other
DNA-based oligonucleotides, such as cationic liposomes,
nanoparticles, cell penetrating peptides (CPP). Cationic
nanoparticles are interesting because of their proton sponge
effect: they fuse with the cellular membrane, facilitate
endocellular release of siRNAs, act as a buffer for the acid pH of
lisosomes and avoid lisosome fusion and siRNA pH-dependent
destruction. However, the optimization of the in vivo
delivery systems remains critical for a broad clinical success of
therapeutic gene targeting by oligonucleotides.
Here we report bioluminescent acute leukemia
xenograft mouse models of the most frequent and high risk
MLL-related acute leukemias (infant and adult MLL-AF9, MLL-ENL,
MLL-AF4). We transduced MLL-related cell lines with firefly
luciferase-expressing plasmid, then we inoculated
luciferase-expressing cells in NOD/SCID mice to generate in
vivo bioluminescent xenograft mouse models of leukemias. The
intensity of the BLI signals was investigated to explore the
correlation with the progression and distribution of leukemia in
the animal models for in vivo study of MLL-related
leukemias. To validate BLI for the detection of a therapeutic
response, systemic treatment with an anti-firefly
luciferase-targeting siRNA (siLuc) complexed with cationic
nanoparticles was administered to mice with MLL-AF4 acute leukemia.
The BLI signal showed a reduction after treatment with siLuc,
compared to the control mice and the mice treated with a control
siRNA.
Materials and methods
Cell lines
MLL-AF9 (THP-1 obtained from infant and MOLM-13 from
adult patients) (32,33) human acute monocytic leukemia cell
lines were cultured in RPMI-1640 (Gibco BRL), containing 10% fetal
bovine serum (FBS, EuroClone) and 1% penicillin/streptomycin (P/S,
Invitrogen). The MLL-AF4 (SEM) (9)
human acute byphenotypic leukemia cell line was cultured in
Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) containing
10% FBS and 1% P/S. The MLL-ENL (KOPN-8) (34) human B-cell precursor leukemia cell
line was cultured in RPMI-1640 containing 20% FBS and 1% P/S. A
retrovirus-packaging cell line for amphotropic retroviruses,
Phoenix A, was maintained in DMEM containing 10% FBS, and 1% P/S.
All cultures were maintained at 37°C and 5% CO2.
Transduction of leukemia cells
The luciferase retroviral expression plasmid
pMMP-Lucneo (kindly provided by Professor Andrew Kung, Harvard
Medical School, Boston, MA) was transfected using Lipofectamine
2000 (Invitrogen) into Phoenix cells to generate the amphotropic
retrovirus. Viral stock was collected 48 and 72 h post-transfection
and was used to transduce leukemia cell lines by spinoculation in
the presence of polybrene (hexadimethrine bromide, Sigma). Cells
were incubated for 48 h in growth medium and then selected for 15
days with 1 mg/ml of G418 (Calbiochem). Positive cell clones where
imaged using the light-tight chamber of a cooled CCD camera system
(LB981, Berthold Technologies) and were named THP-1-luc,
MOLM-13-luc, SEM-luc and KOPN-8-luc,
respectively.
In vitro bioluminescence
measurements
Cell clones with the highest light emission were
selected with the luciferase assay reagent (Promega). A cell
preparation (105 cells in 100 μl PBS) was plated into a
white 96-well cell culture microplate and luminescence signals were
measured following the addition of 100 μl of luciferase assay
reagent using a spectral scanning multimodal plate reader
(Varioskan Flash, Thermo Scientific). Luminescence measurements
were performed at room temperature (25°C) and emission spectra were
measured in the 560–670 nm range by collecting the light output
every 2 nm for 1 sec. Serial dilutions of selected clones were
plated in triplicate in a series starting from 105 cells
to assess the relationship between the viable cell count,
determined by a trypan blue exclusion assay, and BL signals for
each cell clone. After receiving 100 μl of luciferase assay system
substrate, the cells were imaged for 1 min with the same region of
interest (ROI) used to measure luminescent signals of cells in each
well using LB981 (Berthold Technologies). Linear regression
analysis was used to determine the correlation between
bioluminescence signal intensity and number of cells
(R2=0.98, p<0.001, n=3).
RT-PCR
RNA from each cell line was extracted using the
RNeasy kit (Qiagen), following the manufacturer’s protocol. Total
RNA was quantified with the NanoDrop2000 spectrophotometer (Thermo
Fisher Scientific). To generate first-strand cDNA, 1 μg of total
RNA was performed using Super Script II First Strand Synthesis Kit
(Invitrogen) on thermocycler PTC 225 (MJ Research). RNA was
incubated with random primers (50 ng/μl) and dNTPs (10 mM) for 5
min at 65°C, then for 1 min at 4°C. Then 15 mM MgCl2
containing buffer, DTT 0.1 M, and RNaseOUT™ Ribonuclease inhibitor
40 U/μl were added and the incubation was carried out for 2 min at
42°C. Finally, Superscript® II reverse transcriptase 50 U/μl was
added with the following program: 50 min at 42°C, 15 min at 70°C
and 30 min at 4°C. The resulting cDNA was subjected to PCR with a
mix of 0.2 μM of each sense and antisense primers, 0.2 mM dNTPs,
1.5 mM MgCl2 containing buffer (Eppendorf) and 1.25
units Taq polymerase (Eppendorf) with the following parameters:
95°C for 1 min; 39 cycles of 94°C for 30 sec, 60°C for 30 sec and
72°C for 1 min; 72°C for 7 min, 4°C for 10 min. The sequences of
the gene-specific primers were as follows: MLL-AF9 (THP-1)
5′-AGGACCGCCAAGAAAAGA-3′ (sense); 5′-CCTGGTCT GGGATGGTGTGAA-3′
(antisense), MLL-AF9 (MOLM-13) 5′-AGCACTCTCTCCAATGGC AATAGT-3′
(sense); 5′-AAG GACCTTGTTGCCTGGTCTG-3′ (antisense); MLL-AF4
5′-GCCCAAGTATCCCTGTA AAACA-3′ (sense); 5′-TAGGG
AAAGGAAACTTGGATGG-3′ (antisense); MLL-ENL 5′-CACCAGAATCAGGTCC
AGAGCA-3′ (sense); 5′-TCCTTGGCTGTGGTTCTGGGAT-3′ (antisense); human
GAPDH 5′-CCAATCTGATTCCACCAT GGC-3′ (sense);
5′-CTTGATTTTGGAGGGATCTCGC-3′ (antisense); murine GAPDH
5′-CCAAGGAGTAAGAAACC CTGGA-3′ (sense); 5′-GGCCCCTCCTGTTATTATGG-3′
(antisense). The amplification products were visualized on ethidium
bromide-stained agarose gels.
Animal models
Female 6 week-old severe combined immunodeficient
NOD/SCID mice were purchased from Charles River Laboratories. The
mice were separated into different groups for each type of cell
line and at least 10 mice for each cell line were used. Mice were
inoculated intravenously with 5×106 MOLM-13-luc,
SEM-luc or KOPN-8-luc cells, or 15×106
THP-1-luc cells. Mice were handled according to the
international guidelines (the Helsinki Declaration) and the
experiments were approved by the Bioethics Committee of Bologna
University.
In vivo bioluminescence imaging
Based on previous experiments on the kinetics of the
D-luciferin in vivo reaction (35), mice received an intraperitoneal
(i.p.) injection of 150 mg/kg D-luciferin (Gold Biotechnology) and
were placed in the LB981 (Berthold Technologies) under isoflurane
anaesthesia (Ugobasile). Photographic and luminescent images were
acquired starting 5 min after the D-luciferin injection, both in
prone and supine position, for 3 min. For all groups, leukemia
progression was monitored every 7 days until mice were sacrificed
due to the suffering from the extent of the leukemia dissemination.
The Winlight software version 2.9 (Berthold Technologies) was used
to analyze the BLI data. In vivo bioluminescence signals
were calculated as the sum of both prone and supine acquisitions
for each mouse after background subtraction (photon flux (ph/sec)
from a total body ROI of 500 mm2). The BL curves for
each xenograft leukemia model are reported as the means ± SEM of
the average of at least 10 mouse signals.
In vitro electroporation of small
interfering RNA duplexes
Small interfering RNA (siRNA) against firefly
luciferase gene (siLuc) and scrambled control (SCR) were purchased
from Sigma: siLuc sense 5′-CUUACGCUGAGUACUUCGATT-3′; antisense
5′-UCGAAGUACUCAGCGUAAGTT-3′; SCR sense
5′-UAGCGACUAAACACAUCAAdTdT-3′; antisense 5′-UUG
AUGUGUUUAGUCGCUAdTdT-3′.
Luciferase-expressing leukemia cell lines were
electroporated using the Ingenio Electroporation Kit (Mirus-Bio)
and the Nucleofector I Device (Lonza). A total of 1 million per
sample was harvested by centrifugation at 1000 rpm for 10 min.
Supernatant was removed and the cell pellet was resuspended in 105
μl of the electroporation reagent. siLuc or SCR siRNA (5 μg) were
mixed with cells and subsequently transferred into electroporation
cuvettes. MLL-AF4 acute biphenotypic leukemia SEM-luc cells
were electroporated using the V-01 program. Following
electroporation, 500 μl of prewarmed complete medium was
immediately added to the cells. Electroporated cells were seeded
into 6-well culture plates containing 1.5 ml of prewarmed medium
per well. Four hours after electroporation, 2 ml of prewarmed
complete medium per well was added, and 100 μl of cells in
triplicate were plated in 96-well culture plates for both
luciferase and viability cell assay at 24 and 48 h
post-transfection. Luciferase assay was performed as previously
described.
In vivo siRNA delivery
In vivo-jetPEI™ (optimized cationic linear
PEI-based transfection reagent for in vivo experiments) was
obtained from Polyplus-transfection (Illkirch). Fifty micrograms of
siLuc or SCR siRNA as negative control were diluted in 100 μl of 5%
glucose solution. In vivo-jetPEI was used at N/P ratio of
10; in vivo-jetPEI was diluted in 100 μl of 5% glucose
solution and mixed by vortexing for 10 sec. The in
vivo-jetPEI diluted solution was added to the nucleic acid
solution, mixed by vortexing for 10 sec and incubated for at least
15 min at RT before injecting into the mice. Treatments were
performed on MLL-AF4 acute biphenotypic leukemia SEM-luc
mice, starting 1 week after inoculation, and repeated every 48 h by
tail vein injection. Bioluminescence analysis was performed every
week as described above.
Organ collection
Blood samples were collected from the orbital sinus
with micro-haematocrit capillary tubes (Brand). A blood smear for
each sample was performed and stained with the May-Grünwald-Giemsa
stain. Briefly, few microliters of blood were used for blood
smears. The blood slides were stained for 3 min in the May-Grünwald
solution, washed with distilled water, stained for 7 min in the
Giemsa solution, then washed again with distilled water, and
finally let dry. Slides were observed at the optic microscope to
evaluate the leukemic infiltration. Bone marrow, spleen and liver
of dead mice were collected for molecular analysis. RNA from mouse
tissues were extracted with Trizol Reagent (Invitrogen), following
the manufacturer’s protocol. RT-PCR on mice RNA was performed as
previously described. The spleen of leukemic and control mice were
compared and photographed to evaluate the infiltration of leukemia
cells.
Results
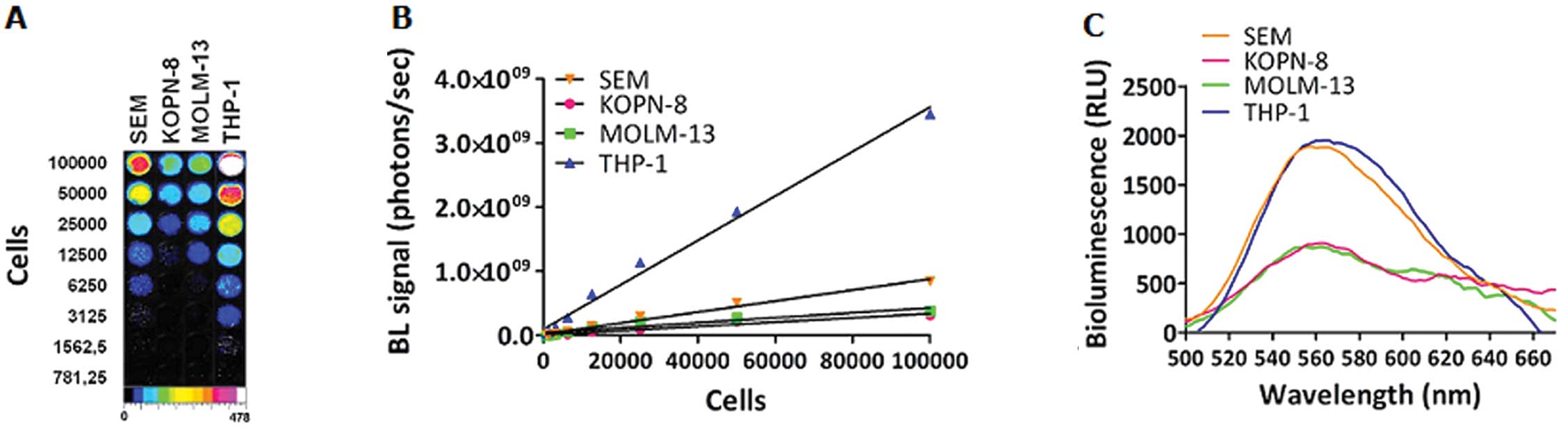
In vitro BLI imaging
We confirmed the linear relationship between cell
number and bioluminescence signals by the luciferase assay, that
appeared highly proportional to cell number in a linear range
(Fig. 1A and B). The correlation
coefficient was >0.99 for all cell clones. At the same plated
concentration, the highest bioluminescence signal was from
THP-1-luc cells, followed by SEM-luc; the
KOPN-8-luc and MOLM-13-luc cell lines showed the
lowest BL intensity. The same trend of bioluminescent signal was
noted in the emission spectra of intact cell clones (Fig. 1C). At an emission maximum of 562
nm, 105 cells of THP-1-luc and SEM-luc
emitted at about 2×103 RLU, MOLM-13-luc and
KOPN-8-luc at slightly <1×103. The luciferase
expression was not found to have any significant effects on the
expression of MLL fusion genes, proteins or proliferation rates of
the four cell lines.
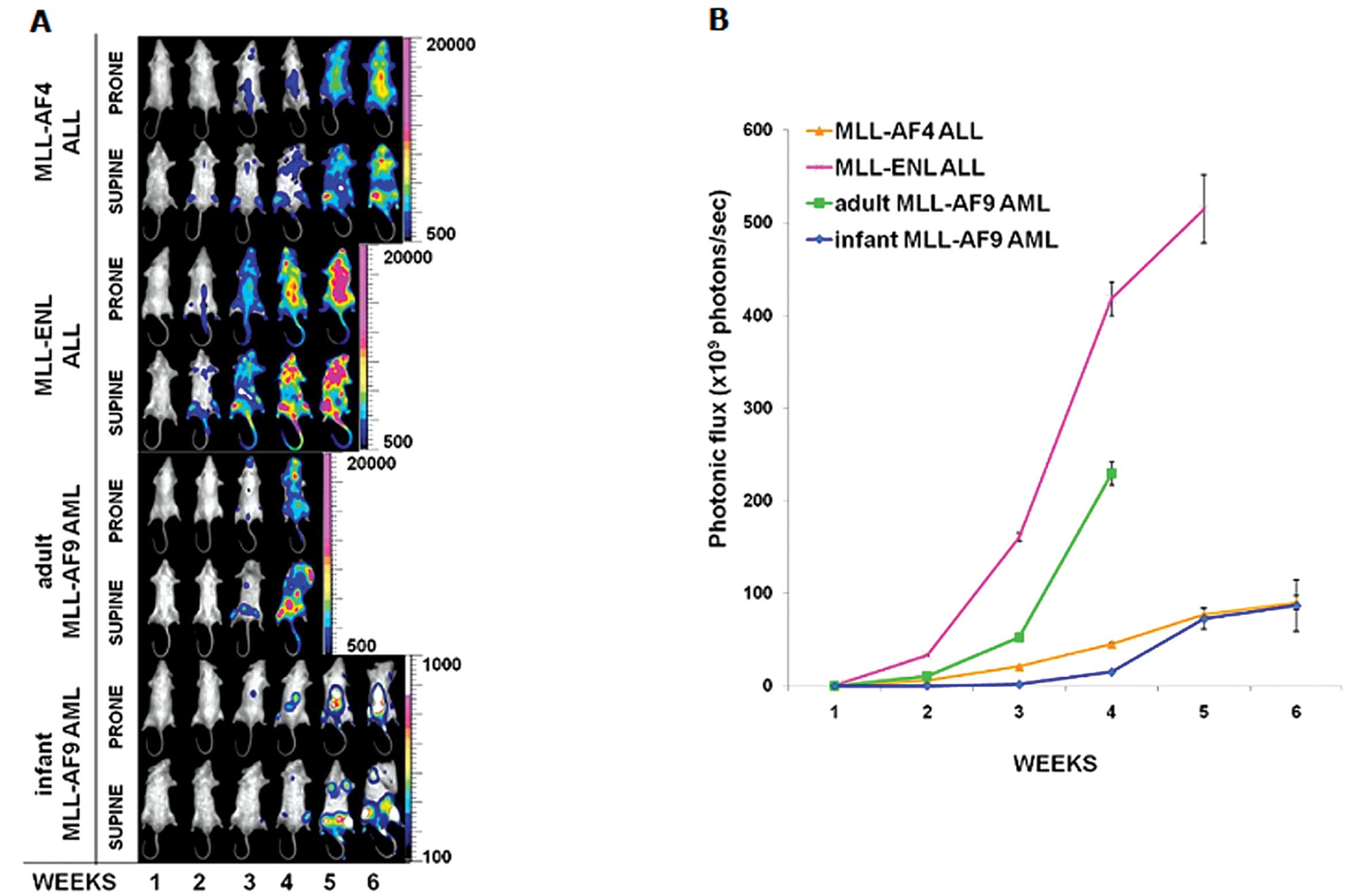
MLL-related acute leukemia progression
evaluated by BLI
All animals developed MLL-related acute leukemias as
detected by in vivo BLI. The four MLL-related acute leukemia
mouse models showed different progressions, due to the
inter-variability among the MLL fusion genes, and the
intra-variability between the infant and adult MLL-AF9 leukemias
(Fig. 2A and B). In three of the
four models, MLL-AF4 acute biphenotypic leukemia, MLL-ENL ALL and
adult MLL-AF9 AML, leukemia developed in the bone marrow and
progressed to a more disseminated disease resembling human
leukemia, whereas in the infant MLL-AF9 AML model leukemia
development and progression was restricted to the bone marrow. The
adult MLL-AF9 AML model presented a latency time of three weeks and
an end point of four weeks, which was the shortest time among the
four models. It also showed diffuse metastasis in the subcutaneous
and osseous planes, with sporadic infiltration of lymphatic
structures as lymph nodes. The infant MLL-AF9 AML model had the
slowest latency time; it developed leukemia at the third week, and
showed the lowest BL signal among the four models. Furthermore, the
infant MLL-AF9 AML model did not show dissemination of human
leukemia cells into the blood system. The MLL-ENL ALL model
revealed the shortest latency time among the four models, and the
BL signal was already evident at the second week after cell
inoculation. The end point of the MLL-ENL ALL mice was at the fifth
week, the BL signal of MLL-ENL ALL progression was also the highest
among the four models and spread in the whole body, due to
dissemination in various organs. The MLL-AF4 acute biphenotypic
leukemia model showed a latency time of two weeks and the end point
was six weeks. The MLL-AF4 acute biphenotypic leukemia mice also
developed alopecia on the head and back due to the infiltration of
the leukemic cells under the skin.
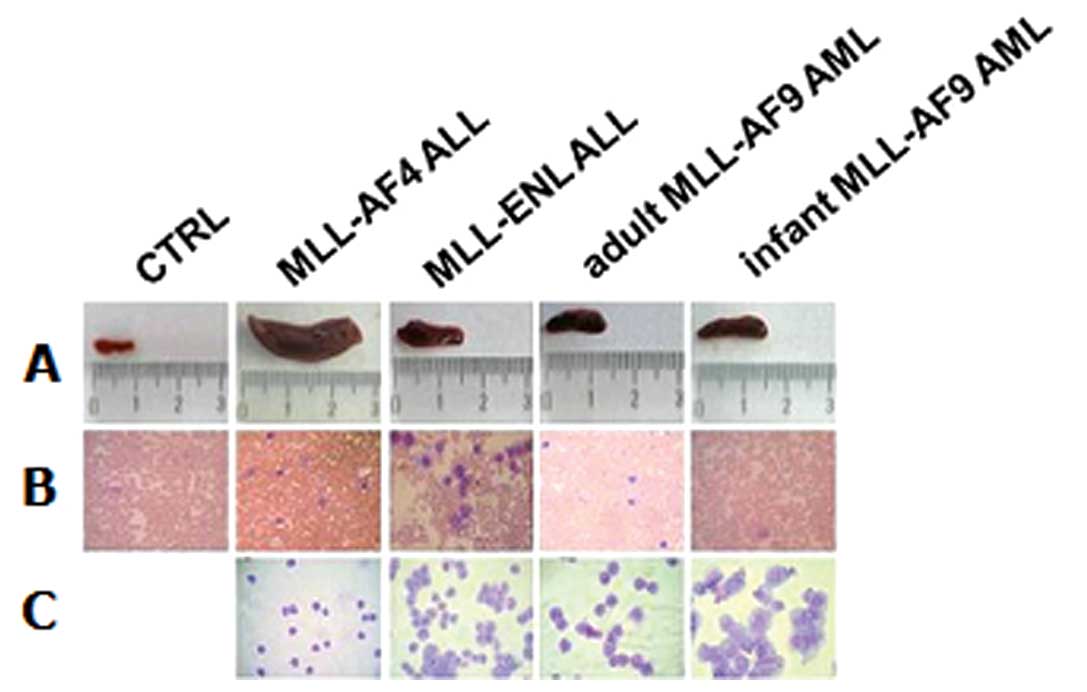
Clinical findings
To confirm the in vivo BLI results, clinical
macroscopic observations on organs were performed. Mouse spleens
were compared to evaluate the splenomegaly due to infiltration of
human leukemia cells (Fig. 3A).
The MLL-AF4 acute biphenotypic leukemia model showed a strong
infiltration into the spleen with evident splenomegaly. Peripheral
blood smears were performed at the end point of mice to evaluate
the blood infiltration of human leukemia cells and were compared to
peripheral blood smears of control mice (Fig. 3B) and to cell lines alone (Fig. 3C). MLL-AF4 biphenotypic leukemia,
MLL-ENL ALL and adult MLL-AF9 revealed dissemination of leukemia
cell lines into the blood system, except for the infant MLL-AF9 AML
cells, as we had expected.
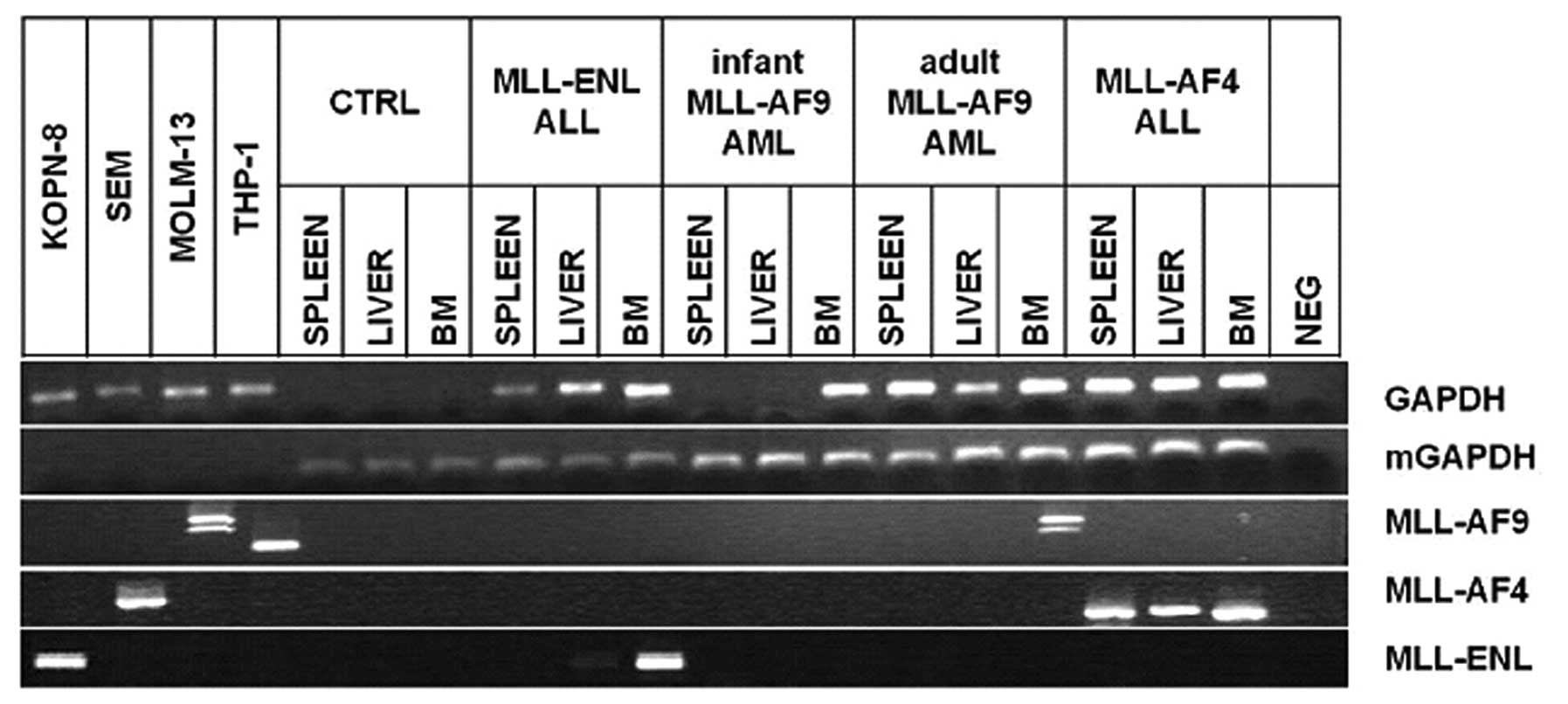
Tissue molecular analysis
Bone marrow, spleen and liver from mice were
analyzed by RT-PCR and were found differently positive for MLL
fusion gene expression (Fig. 4).
Human GAPDH housekeeping mRNA was found negative in samples from
control mice and positive in samples from leukemic mice, excepted
for the spleen and liver of mice inoculated with infant MLL-AF9 AML
cells, confirming the BL results; murine GAPDH (mGAPDH)
housekeeping mRNA was found positive in all mice samples and
negative in human leukemic cell lines, confirming the specificity
of the RNA extraction from mice. The MLL-AF9 mRNA was found
positive only in the bone marrow sample from adult MLL-AF9 AML
mice, showing the two previously described mRNA splicing isoforms
of MLL-AF9 in MOLM-13 cells (36).
The MLL-AF4 mRNA was found positive in all the tissues from MLL-AF4
acute biphenotypic leukemia mice. The MLL-ENL mRNA was found
clearly positive in the bone marrow and less clear but present in
the liver of the MLL-ENL ALL mice.
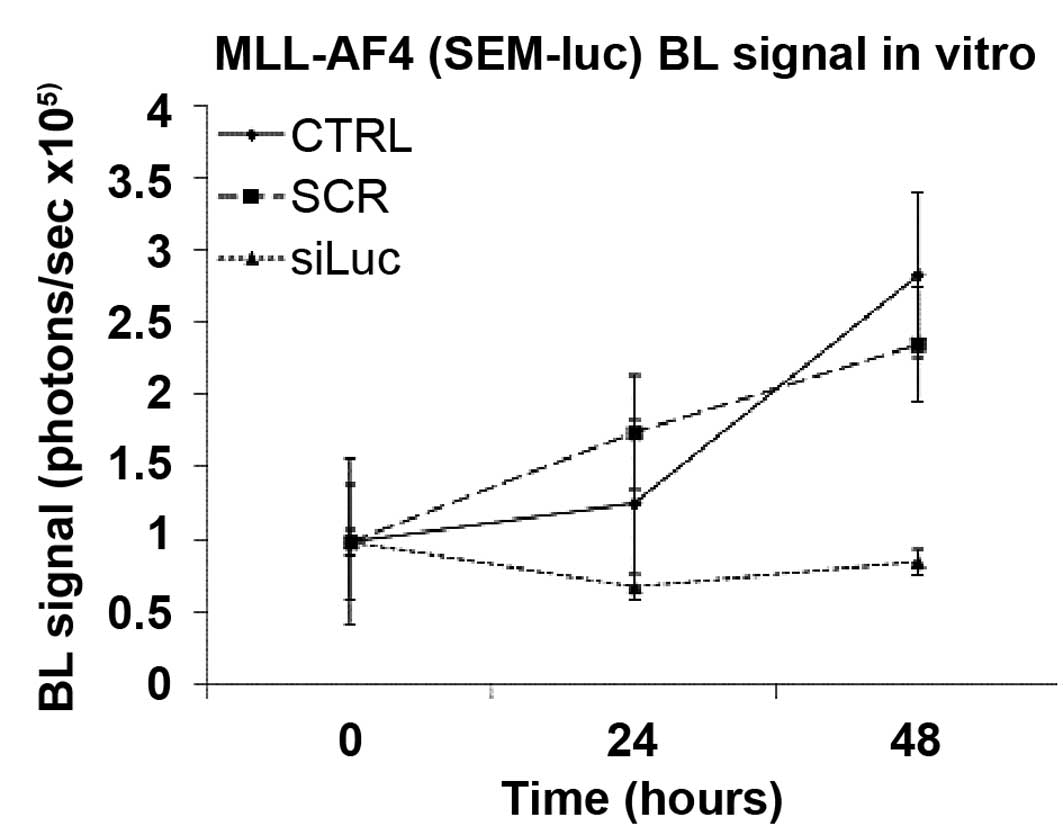
In vitro transfection of siRNA
We transiently transfected the MLL-AF4 acute
biphenotypic leukemia SEM-luc cell line with a siRNA against
luciferase mRNA (siLuc) and with a scrambled siRNA (siSCR). The
luminescence analysis showed in vitro reduction of
luminescence signal in the samples treated with siLuc compared to
the control samples or the samples treated with siSCR, at 24 and 48
h post-transfection (Fig. 5).
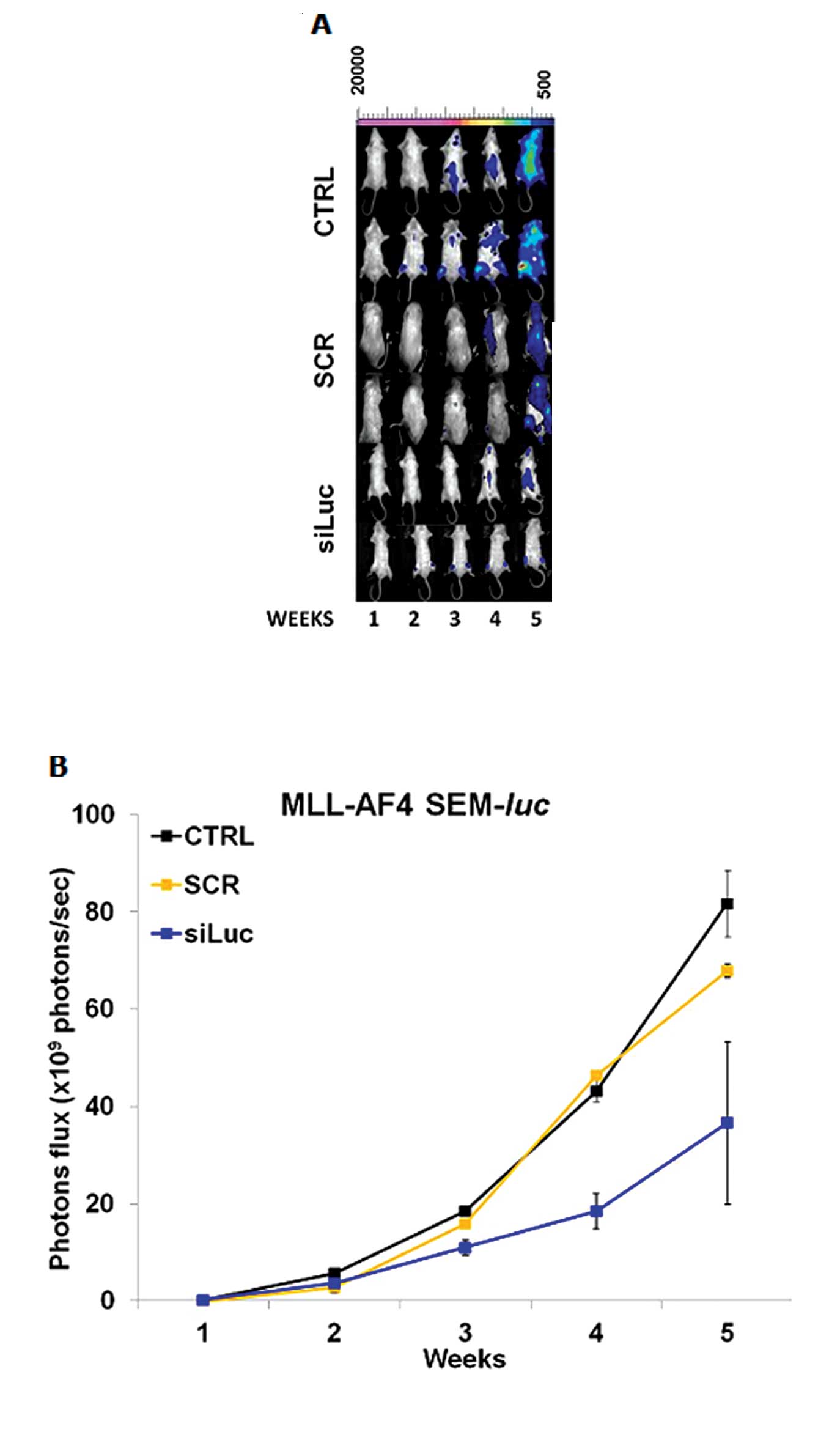
In vivo siRNA delivery
We treated MLL-AF4 acute biphenotypic leukemia
(SEM-luc) mice with siLuc and we monitored the BL signal
during the treatment. We compared the BL signal in treated mice
with siSCR-treated or untreated control mice (Fig. 6). We observed a statistically
significant difference between BL signals in siLuc and
siSCR-treated mice from the fourth week of treatment to the end
point, confirming the specificity of the luciferase gene
downregulation and the validity of the mouse model (Fig. 6A and B).
Discussion
Our bioluminescent mouse models of the most frequent
and high risk MLL-related acute leukemias provide a non-invasive,
sensitive, rapid and affordable method to study the different
behaviors of leukemia progression. The high sensitivity of in
vivo BLI enables the early detection of tumors, either
superficially or in deep tissues such as the bone marrow, preceding
the appearance of symptoms (25).
Moreover, our MLL-related acute leukemia mouse
models show the different course of infant and adult human MLL-AF9
AML, and the rapid aggressiveness of human MLL-ENL ALL and MLL-AF4
acute biphenotypic leukemia. Among the four models, three, MLL-AF4
acute biphenotypic leukemia, MLL-ENL ALL and adult MLL-AF9 AML,
developed leukemias in the bone marrow and progressed to a more
disseminated disease, and the results were confirmed by in
vivo BLI, clinical findings and RT-PCR molecular analysis on
tissues. Leukemia development and progression was restricted to the
bone marrow only in the infant MLL-AF9 AML model; this behavior is
possibly due to the lower infiltration trend of infant MLL-AF9 AML.
The two MLL-AF9 AML models showed a different course, probably due
to their different origin: the THP-1 cell line was established from
the peripheral blood of a 1-year-old boy with subtype M5a acute
monocytic leukemia (32), whereas
the MOLM-13 cell line was established from the peripheral blood of
a 20-year-old male patient at relapse of acute monocytic leukemia,
subtype M5a, which had evolved from myelodysplastic syndrome
(33). In vivo BLI allowed
us to distinguish the different courses of leukemia growth in
infant and adult MLL-AF9 AML models, and enabled the study of the
mechanism of action of the same MLL-AF9 oncogene in different
contexts, such as in infant or adult patients.
The parallelism between the results of in
vivo BLI and the clinical course of MLL-related acute leukemias
supports our proposed models as suitable tools for the study of
MLL-leukemias.
Moreover, our in vivo BLI models may be
complementary to transgenic mouse models of MLL-related acute
leukemias due to their capability to reveal different details of
disease progression and could be helpful for the leukemia research
community to elucidate the trafficking in vivo of the
different MLL-related acute leukemias.
The accurate definition of the leukemic evolution
from the early beginning to the final stages in these MLL-related
acute leukemia mouse models represents optimal characteristics for
the definition of preclinical treatment schedules in drug
development studies for MLL-related acute leukemias.
To confirm this hypothesis, we treated the MLL-AF4
acute biphenotypic leukemia bioluminescent mice with a specific
anti-firefly luciferase siRNA, complexed with
polyethylenimine-based nanoparticles to protect them from serum
nuclease degradation and to deliver the siRNA into the cells. We
chose the MLL-AF4 acute biphenotypic leukemia mouse model due to
the poor prognosis of MLL-AF4 acute biphenotypic leukemias in
pediatric patients and due to the widest window of latency and
progression in mice between all our leukemia mouse models. We
monitored the in vivo siRNA-mediated downregulation of
luciferase activity by in vivo BLI. We observed a
significant lower BL signal in treated mice compared to untreated
control mice and to siSCR-treated mice.
Finally, we propose our MLL-related acute leukemia
bioluminescent mouse model for in vivo siRNA therapeutic
treatments as a powerful tool to validate the efficacy of
much-needed new therapies for this aggressive group of
leukemias.
Acknowledgements
This work was supported by the CARISBO
Foundation, the del Monte Foundation of Bologna and Ravenna, AIRC
and AGEOP.
References
|
1
|
Krivtsov AV and Armstrong SA: MLL
translocations, histone modifications and leukaemia stem-cell
development. Nat Rev Cancer. 7:823–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bach C and Slany RK: Molecular pathology
of mixed-lineage leukemia. Future Oncol. 5:1271–1281. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thirman MJ, Gill HJ, Burnett RC, et al:
Rearrangement of the MLL gene in acute lymphoblastic and acute
myeloid leukemias with 11q23 chromosomal translocations. N Engl J
Med. 329:909–914. 1993. View Article : Google Scholar
|
|
4
|
Meyer C, Kowarz E, Hofmann J, et al: New
insights to the MLL recombinome of acute leukemias. Leukemia.
23:1490–1499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pui CH, Chessells JM, Camitta B, et al:
Clinical heterogeneity in childhood acute lymphoblastic leukemia
with 11q23 rearrangements. Leukemia. 17:700–706. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pui CH, Schrappe M, Ribeiro RC and
Niemeyer CM: Childhood and adolescent lymphoid and myeloid
leukemia. Hematology Am Soc Hematol Educ Program. 118–145.
2004.PubMed/NCBI
|
|
7
|
Hess JL: MLL: a histone methyltransferase
disrupted in leukemia. Trends Mol Med. 10:500–507. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pizzo PA and Poplack DG: Principles and
Practice of Pediatric Oncology. 5th Edition. Lippincott Williams
& Wilkins; Philadelphia, PA: 2006
|
|
9
|
Greil J, Gramatzki M, Burger R, et al: The
acute lymphoblastic leukaemia cell line SEM with t(4;11)
chromosomal rearrangement is biphenotypic and responsive to
interleukin-7. Br J Haematol. 86:275–283. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Balgobind BV, Raimondi SC, Harbott J, et
al: Novel prognostic subgroups in childhood 11q23/MLL-rearranged
acute myeloid leukemia: results of an international retrospective
study. Blood. 114:2489–2496. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Frascella E, Rondelli R, Pigazzi M, et al:
Clinical features of childhood acute myeloid leukaemia with
specific gene rearrangements. Leukemia. 18:1427–1429. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dothager RS, Flentie K, Moss B, Pan MH,
Kesarwala A and Piwnica-Worms D: Advances in bioluminescence
imaging of live animal models. Curr Opin Biotechnol. 20:45–53.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luker GD and Luker KE: Optical imaging:
current applications and future directions. J Nucl Med. 49:1–4.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klerk CP, Overmeer RM, Niers TM, et al:
Validity of bioluminescence measurements for noninvasive in vivo
imaging of tumor load in small animals. Biotechniques. 43:7–13.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Greer LF and Szalay AA: Imaging of light
emission from the expression of luciferases in living cells and
organisms: a review. Luminescence. 17:43–74. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Söling A and Rainov NG: Bioluminescence
imaging in vivo-application to cancer research. Expert Opin Biol
Ther. 3:1163–1172. 2003.Erratum in: Expert Opin Biol Ther 3: 1315,
2003.
|
|
17
|
Negrin RS and Contag CH: In vivo imaging
using bioluminescence: a tool for probing graft-versus-host
disease. Nat Rev Immunol. 6:484–490. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Inoue Y, Tojo A, Sekine R, et al: In vitro
validation of bioluminescent monitoring of disease progression and
therapeutic response in leukaemia model animals. Eur J Nucl Med Mol
Imaging. 33:557–565. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Inoue Y, Izawa K, Tojo A, et al:
Monitoring of disease progression by bioluminescence imaging and
magnetic resonance imaging in an animal model of hematologic
malignancy. Exp Hematol. 35:407–415. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Inoue Y, Izawa K, Kiryu S, Kobayashi S,
Tojo A and Ohtomo K: Bioluminescent evaluation of the therapeutic
effects of total body irradiation in a murine hematological
malignancy model. Exp Hematol. 36:1634–1641. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Armstrong SA, Kung AL, Mabon ME, et al:
Inhibition of FLT3 in MLL: Validation of a therapeutic target
identified by gene expression based classification. Cancer Cell.
3:173–183. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Edinger M, Cao YA, Verneris MR, Bachmann
MH, Contag CH and Negrin RS: Revealing lymphoma growth and the
efficacy of immune cell therapies using in vivo bioluminescence
imaging. Blood. 15:640–648. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Maes W, Deroose C, Reumers V, et al: In
vivo bioluminescence imaging in an experimental mouse model for
dendritic cell based immunotherapy against malignant glioma. J
Neurooncol. 91:127–139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Inoue Y, Kiryu S, Izawa K, Watanabe M,
Tojo A and Ohtomo K: Comparison of subcutaneous and intraperitoneal
injection of D-luciferin for in vivo bioluminescence
imaging. Eur J Nucl Med Mol Imaging. 36:771–779. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wetterwald A, van der Pluijm G, Que I, et
al: Optical imaging of cancer metastasis to bone marrow: a mouse
model of minimal residual disease. Am J Pathol. 160:1143–1153.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aronin N: Target selectivity in mRNA
silencing. Gene Ther. 13:509–516. 2006. View Article : Google Scholar
|
|
27
|
Meister G and Tuschl T: Mechanisms of gene
silencing by double-stranded RNA. Nature. 431:343–349. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tuschl T: RNA interference and small
interfering RNAs. Chembiochem. 2:239–245. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sledz CA and Williams BR: RNA interference
in biology and disease. Blood. 106:787–794. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cejka D, Losert D and Wacheck V: Short
interfering RNA (siRNA): tool or therapeutic? Clin Sci. 110:47–58.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Behlke MA: Progress towards in vivo use of
siRNAs. Mol Ther. 13:644–670. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsuchiya S, Yamabe M, Yamaguchi Y,
Kobayashi Y, Konno T and Tada K: Establishment and characterization
of a human acute monocytic leukemia cell line (THP-1). Int J
Cancer. 26:171–176. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Matsuo Y, MacLeod RA, Uphoff CC, et al:
Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and
MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9
fusion resulting from an occult chromosome insertion, ins(11;9)
(q23;p22p23). Leukemia. 11:1469–1477. 1997. View Article : Google Scholar
|
|
34
|
Matsuo Y and Drexler HG: Establishment and
characterization of human B cell precursor-leukemia cell lines.
Leuk Res. 22:567–579. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mezzanotte L, Fazzina R, Michelini E, et
al: In vivo bioluminescence imaging of murine xenograft cancer
models with a red-shifted thermostable luciferase. Mol Imaging
Biol. 12:406–414. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Montemurro L, Tonelli R, Fazzina R,
Martino V, Marino F and Pession A: Identification of two MLL-MLLT3
(alias MLL-AF9) chimeric transcripts in the MOLM-13 cell line.
Cancer Genet Cytogenet. 154:96–97. 2004. View Article : Google Scholar : PubMed/NCBI
|