Introduction
MiRNAs are short non-coding RNAs of 16 to 25
nucleotides in length. Longer precursor transcripts with hairpin
structures are first synthesized by RNA polymerase II and mature
miRNAs are generated after processing of the precursors by Drosha
and Dicer ribonucleases (1).
Depending on the degree of homology to their target sequence,
miRNAs induce translational repression or cleavage of mRNAs. A
single miRNA can target hundreds to a thousand or more mRNAs
(2), rendering it challenging to
attribute distinct functions to specific miRNAs. Perhaps as a
result of this complexity, in spite of the evolutionary
conservation of miRNAs, their function in physiology and disease
remains rather enigmatic. Our research group has demonstrated that
exogenous plant miRNAs in food can regulate the expression of
target genes in mammals which suggested that miRNA may have a
powerful ability to regulate gene expressions (3). Aberrations or corruptions of miRNA
functions may lead to deregulated cell proliferation,
tumorigenesis, and ultimately, cancer. Moreover, signaling pathways
of the core components of cancer cells also take part in the
post-transcriptional regulation of miRNAs (4).
Cancer is characterized by abnormal cell
proliferation that undergoes rapid, uncoordinated cell growth.
Malignant cancer, in contrast to benign cancer, is further
hallmarked by aggressive neoplasms that have the ability to invade
and annihilate adjacent tissues and metastasize to more distant and
sometimes specific tissues. Genes involved in cancer, be it
inceptionally or during the later invasive or metastasis stages,
are generally classified into oncogenes (OG) or tumor suppressor
(TS) genes. During the last decade, a unique set of cancer
regulator miRNAs have emerged and these are divided into oncomiRs
and anti-oncomiRs. OncomiRs and anti-oncomiRs negatively regulate
tumors uppressor genes and oncogenes, respectively.
Differential miRNA expression profiles of cancerous
and normal tissues have revealed signatures that facilitate
identifying and monitoring cancers (5). Researchers are now trying to use
these miRNA signatures therapeutically to support diagnosis,
prognosis or treatment of cancer. Instead of detailing the whole
body of identified oncomiRs and anti-oncomiRs (6,7), the
focus is on illustrating mechanistically how the miRNA pathway is
involved, or affected by, cancer at the hand of representative
examples (8).
A biological pathway, which is a series of actions
among molecules in a cell that leads to a certain product or a
change in a cell, can trigger the assembly of new molecules or turn
genes on and off, or spur a cell to move. Identifying what genes,
proteins and other molecules are involved in a biological pathway
can provide clues on what goes wrong when a disease strikes. The
most common pathways are involved in metabolism, the regulation of
genes and the transmission of signals. Deregulation of miRNAs are
involved in the process of cell proliferation activation, apoptosis
signaling pathway inactivation, as well as other genetic changes,
which together lead to cancer pathogenesis. Hitherto, accumulating
evidence has demonstrated that miRNAs are involved in mediating
several cancer-related pathways linked to programmed cell death
(PCD), indicating that miRNAs may function as the key regulators in
apoptosis and autophagy of cancer (9). Recent projects that deciphered the
different expressions of miRNAs in a certain cancer by RT-qPCR
after Solexa or Microarry screening only have found an array of
different mutations of miRNAs in different samples, then the
pathway involved by the most different expressed miRNAs can be
figured out. The problem is that these kinds of methods only could
find one miRNA-related pathway in a certain cancer. miRNAs commonly
involved in several cancers would function as key regulators of
many more cancer-related pathways, which may be a link between the
cancer-related pathway research and function research of
miRNAs.
The development of pathway strategies for the
analysis of cancer raises the question of whether one can use these
approaches to characterize and treat human cancer. Identifying the
molecular causes of cancer represented a major breakthrough in the
history of medicine, moving the discipline from pattern recognition
and therapeutic strategies based on syndromic pathophysiology to
molecular mechanism and evidence-based therapies derived from
clinical trials designed on the basis of molecular mechanism
(10).
In this study, we summarized 11 common CA-miRNAs
from previous observations. Many of these CA-miRNAs were located
near genomic breakpoints (11).
For example, miR-15/16-1 cluster is located within a 30-kb region
of chromosome 13q14 and that both genes are deleted or
downregulated in the majority (approximately 68%) of B cell chronic
lymphocytic leukemia (CLL) cases (12). On the other hand, one cluster of
microRNAs, the miR-17-92 polycistron, is located in a region of DNA
that is amplified in human B cell lymphomas (13). Upregulated expression of the mature
miRNAs from miR-17-92 cluster, has been confirmed in a wide range
of tumor-derived cell lines (14).
Protein class, molecular functions, biological processes and
canonical pathways involved by the targets of each CA-miRNA as well
as 5 main canonical pathways participated by certain CA-miRNAs,
were identified and analyzed, which may offer significant treatment
clues for the clinical therapy of cancer.
Materials and methods
Targets analysis of each CA-miRNAs
MiRNA targeting is mostly achieved through specific
base-pairing interactions between the 5′ end (‘seed’ region) of the
miRNA and sites within coding and untranslated regions (UTRs) of
mRNAs; target sites in the 3′UTR lead to more effective mRNA
destabilization (15). The
expression of a single target gene of a certain miRNA may not
provide enough information on the role of that miRNA in the
analyzed pathophysiological process (16). Therefore, we used a widely-used and
web-based software Targetscan (http://www.targetscan.org) to generate lists of
possible gene targets of each CA-miRNA. Then we input the targeted
genes into another web server Panther (http://www.pantherdb.org/) which is designed for gene
function cluster and we gained the protein class from the panther
analysis. After that, we clustered the same function class of
protein with top ten classes.
The web-based functional annotation tool Database
for Annotation, Visualization and Integrated Discovery (DAVID) v6.7
(http://david.abcc.ncifcrf.gov/tools.jsp) has key
components for disease analysis, gene ontology analysis and pathway
analysis (17).
Pathway mapping of cancer-associated
miRNA targets
The signaling pathways and processes that these gene
targets are involved in were explored using the systems biology
tool KEGG Mapper (http://www.genome.jp/kegg/tool/map_pathway2.html).
This KEGG database, containing 291 known pathways on molecular
interactions and reaction networks, pathways and processes
(18), allows the user to
visualize known biological systems within their data.
Results and Discussion
Cancer-associated miRNAs (CA-miRNAs)
Functional studies performed in cancer cell lines or
mouse models with various malignancies through overexpression or
knockdown of miRNAs have supported a role for some of these miRNAs
in carcinogenesis (15). Based on
previous experimental data, 11 CA-miRNAs are summarized as the most
common cancer-associated miRNAs (Table
I). The miRNA dysregulation could drive tumorigenesis, although
the roles miRNAs can adopt as tumor suppressors or oncogenes.
 | Table IPrediction of each CA-miRNA
target. |
Table I
Prediction of each CA-miRNA
target.
| CA-miRNAs | No. of predicted
target genes |
|---|
| let-7 family | 1,072 |
| miR-9 | 1,237 |
| miR-15a/16-1
cluster | 1,273 |
| miR-17-92
family | 2,656 |
| miR-21 | 164 |
| miR-26a | 186 |
| miR-34a/b/c | 852 |
| miR-155 | 440 |
| miR-200/141
family | 744 |
| miR-205 | 416 |
| miR-206 | 102 |
Let-7 is an anti-oncomiR and conserved in many
cancers. It functions as a post-transcriptional gatekeeper for cell
proliferation process. For example, let-7 family negatively
regulates RAS, a lung cancer oncogene involved in cancerous cells
by disturbing cell cycle progression (19,20).
Let-7 family (let-7a to i) display a striking upregulation in
differentiating SK-3rd cells and a high level of expression in the
parental SK-BR-3 cells that have not been enriched for breast
T-ICs13 (21).
MiR-9 has been strongly suggested to act as a
putative tumor suppressor gene in recurrent ovarian cancer
(22). In addition, it can be
affected by epigenetic inactivation due to aberrant
hypermethylation which is an early and frequent event in breast
cancer development (23). The
rescued expression of miR-9 could also promote medulloblastoma cell
growth arrest and apoptosis while targeting the proliferative
truncated TrkC isoform (24). On
the other hand, some research groups have reported that the level
of miR-9 is upregulated in breast cancer cells by directly
targeting CDH1, the E-cadherin-encoding mRNA, which could lead to
increased cell motility and invasiveness. MiR-9-mediated E-cadherin
downregulation results in the activation of β-catenin signaling
pathway, which could contribute to upregulated expression of the
gene encoding vascular endothelial growth factor (VEGF); this would
in turn lead to an increase of tumor angiogenesis. Certain miRNA
may mediate c-Myc induced mammary carcinogenesis (25). At the same time, expression of
miR-9 could also be activated by MYC and MYCN, both of which
directly bind to the mir-9-3 locus (26), which seem to be an obvious evidence
of feedback loop regulation.
MiR-15a and miR-16-1 act as putative tumor
suppressors by targeting the oncogene BCL2. These miRNAs form a
cluster at the chromosomal region of 13q14, which is frequently
deleted in cancer. The miR-15a and miR-16-1 cluster targets CCND1
(encoding cyclin D1) and WNT3A, which promotes several tumorigenic
features such as survival, proliferation and invasion. Deletion of
miR-15a and miR-16-1 genes results in loss of apoptosis. For
advanced prostate tumors, the level of miR-15a and miR-16 is
significantly decreased, whereas the expression of BCL2, CCND1 and
WNT3A is inversely upregulated (27). In chronic lymphocytic leukemia
(CLL), miR-15a and miR-16-1 are mostly lost or downregulated in the
majority of cases (28).
Overexpression of miRNAs encoded by the miR-17-92
cluster and its paralogs in multiple malignancies are known to act
as oncogenes. Expression of these miRNAs promotes cell
proliferation, suppresses apoptosis of cancer cells and induces
tumor angiogenesis (29). Analysis
of human medulloblastomas (MBs) demonstrated that 3 miR-17-92
cluster miRNAs (miR-92, miR-19a and miR-20) were overexpressed in
human MBs with a constitutively activated Sonic Hedgehog (SHH)
signaling pathway, but not found in other forms of the disease
(30).
MiR-21 expression is not only activated in multiple
types of cancers, such as breast, liver, brain, prostate and
myometrial cancers but also in different kinds of diseases, such as
cardiovascular disease. MiR-21 regulates a plethora of target
proteins which are involved in cellular survival, apoptosis and
cell invasiveness. MiR-21 regulation is complex due to a promoter
that is target for various transcription factors and hormones. The
consistent miR-21 overexpression under pathophysiological
conditions points to miR-21 as a valuable tool for new therapeutic
strategies. The overexpression of certain oncogenic miRNAs (miR-21
and miR-155) and the loss of several tumor suppressor miRNAs
(miR-206, miR-17-5p, miR-200, let-7 and miR-34) have been observed
in many breast cancers. The gene networks orchestrated by these
miRNAs are still largely unknown, although key targets have been
identified that may contribute to the disease phenotype (31). MiR-155 was more highly expressed in
activated B cell-like (ABC)-type than germinal center B cell-like
(GCB)-type cell lines, which are two subtypes of diffuse large B
cell lymphoma (DLBCL) (32).
MiR-26a is frequently amplified at the DNA level in
human glioma, most often in association with monoallelic PTEN loss
(30). Ectopic expression of
miR-26a influenced cell cycle progression by targeting the bona
fide oncogene EZH2, a Polycomb protein and global regulator of gene
expression (33). Its expression
in liver cancer cells in vitro can induce cell cycle arrest,
which may associate with direct targeting of cyclins D2 and E2
(34). A significant decrease in
miR-26a was detected in growing anaplastic carcinomas (ATC) in
comparison to normal thyroid tissue (35). MiR-26a, CDK4 and CENTG1 together
comprise a functionally integrated oncomir/oncogene DNA cluster
that promotes aggressiveness in human cancers by cooperatively
targeting the RB1, PI3K/AKT and JNK pathways (36).
The miRNA-34 family comprises three members:
miRNA-34a, miR-34b and miR-34c. MiR-34a is generated from a larger
transcriptional unit on chromosome 1p36; and both of miR-34b and
miR-34c are generated through the processing of a bicistronic
transcript from chromosome 11q23 (termed miR-34bc). The miR-34
family members have also been identified as promising prognostic
markers in non-small cell lung cancer (NSCLC); the family is
downmodulated in tumors compared with normal tissue. Restoration of
miR-34 expression in the pancreatic cancer cells by either
transfection of miR-34 mimics or infection with lentivirus
significantly inhibited clonogenic cell growth and invasion,
induced apoptosis and G1 and G2/M arrest in the cell cycle, and
sensitized the cells to chemotherapy and radiation (37). The likely growth inhibition
mechanism of miR-34a, as a tumor suppressor gene in human
neuroblastoma, is through cell cycle arrest followed by apoptosis.
BCL2 and MYCN were identified as miR-34a targets and likely
mediators of the tumor suppressor phenotypic effect (38). In HepG2 cells, ectopic expression
of miR-34a potently inhibited tumor cell migration and invasion in
a c-Met-dependent manner. It directly targeted c-Met and caused
reduction of both mRNA and protein levels of c-Met; thus, decreased
c-Met-induced phosphorylation of extracellular signal-regulated
kinases 1 and 2 (ERK1/2) (39).
From a large-scale miRnome analysis on lung, breast,
stomach, prostate, colon, and pancreatic tumors, some miRNAs have
been found with well characterized cancer association, which
include miR-155, miR-17-5p, miR-21, miR-92 and miR-106a (40). What is more, high miR-155 and low
let-7a-2 expression correlated with poor survival has been found in
lung cancer by univariate analysis as well as multivariate analysis
for miR-155 (41).
Downregulation of miR-141 and miR-200c in renal
clear cell carcinomas (CCCs) might be involved in suppression of
CDH1/E-cadherin transcription via upregulating ZFHX1B (42). The expression of miR-141 was also
found to be substantially reduced in several human gastric cancer
cell lines such as MGC-803, HGC-27, SGC-7901 and BGC-823 cells.
MiR-141 may be involved in the development of gastric cancer
through its inhibitory effect on cell proliferation (43). Members of the miR-200 family appear
to control the epithelial-to-mesenchymal transition (EMT) process,
as well as the sensitivity to EGFR therapy in bladder cancer cells.
Structural analysis of EGFR TK domain provides insights into EGFR
targeted therapies (44). The
expression of miR-200 is sufficient to restore EGFR dependency at
least in some of the mesenchymal bladder cancer cells. The targets
of miR-200 include ERRFI-1, which is a novel regulator of
EGFR-independent growth (45). On
the contrary, in ovarian cancer miR-200 family members are
expressed at low or negligible levels in normal ovarian surface
cells and substantially increase in expression, whereas expression
of ZEB1 and ZEB2 shows the opposite pattern (46).
MiR-205 exerts a tumor-suppressive effect in human
prostate by counteracting EMT process and reducing cell
migration/invasion, at least in part through the downregulation of
protein kinase Cε (47). MiR-200
and miR-205 loci are repressive chromatin marks, in muscle invasive
bladder tumors and undifferentiated bladder cell lines, which have
been found specifically silenced and gain promoter hypermethylation
(48). As a new oncosuppressor
gene in breast cancer, miR-205 is able to interfere with the
proliferative pathway mediated by kinase-inactive member HER
receptor family (49). However,
compared with normal tissues, the levels of miR-205, together with
miR-21 and miR-203 were found to be significantly up-modulated in
OVCAR3 cells which were demethylated with 5-aza-2′-deoxycytidine
(50). This suggested that miR-205
also has a role as an oncomiRNA.
As the product of MET proto-oncogene, Met
tyrosine-kinase receptor has been found overexpressed in human
rhabdomyosarcoma (RMS) cell lines and involved in RMS pathogenesis.
Upon the presence of miR-206, Met tyrosine-kinase receptor was
down-regulated in murine satellite cells in the onset of normal
myogenesis (51). Since there was
no evidence of miR-206 activation in serum derivate RMS cell lines,
miR-206 was suggested as tumor suppressor and has been identified
to be involved in breast cancer metastasis (52).
Thus, let-7 family (let-7a to i), miR-15a/16-1
cluster, miR-34a/b/c and miR-206 are classified as anti-oncomiRs or
TS (tumor suppressor), while miR-17-92 family (miR-17, miR-18a/b,
miR-20a/b, miR-106a/b, miR-93, miR-19a/b, miR-25, miR-92a and
miR-363), miR-21 and miR-155 play the roles as oncomiRs or OG
(oncogenes). What is more, miR-9, miR-26a, miR-200/141 family and
miR-205 possess two kinds of effects which are either anti-oncomiRs
or oncomiRs.
Predictions and protein classifications
of CA-miRNA targets
The miRNAs have the capacity to ‘tune’ the
expression of a target gene to a precise level (53). Therefore, investigation of target
gene is one of the keys for understanding miRNAs. Each CA-miRNA or
miRNA cluster has the ability to target between 102 and 2,656 mRNAs
of predicted genes (Table I) and
in addition some 3′UTRs of the mRNAs potentially have multiple
complementary sites for a given miRNA. To add to the complexity, in
fact this set of miRNAs may be a subset of the total number of
miRNAs that play a role in cancer. Moreover, this result was
consistent with the hypothesis that oncomiRs and anti-oncomiRs
mainly interact with tumor suppressor genes and oncogenes,
respectively (54).
A total of 5,001 unique targeted genes have been
found that are 11 identified CA-miRNAs related. Of all the targeted
genes, 1,850 items are from targets of anti-oncomiRs, so these
genes are more likely upregulated in cancer cells. Accordingly,
2,971 downregulated targeted genes are discovered from that of
oncomiRs and 2,204 ones from that of miRNAs which could act as
either oncomiRs or anti-oncomiRs. Moreover, 12 common genes (BNC2,
BRWD3, DCUN1D3, GATAD2B, KCNA1, KPNA4, PIK3R1, PURB, RBMS3, SATB1,
SOCS6 and TGFBR2) were found in the targets of oncomiRs and with
one gene the C5orf41, which was identified as a target of miRNAs
either as oncomiRs or anti-oncomiRs. These common target genes can
be potential cancer treatment targets.
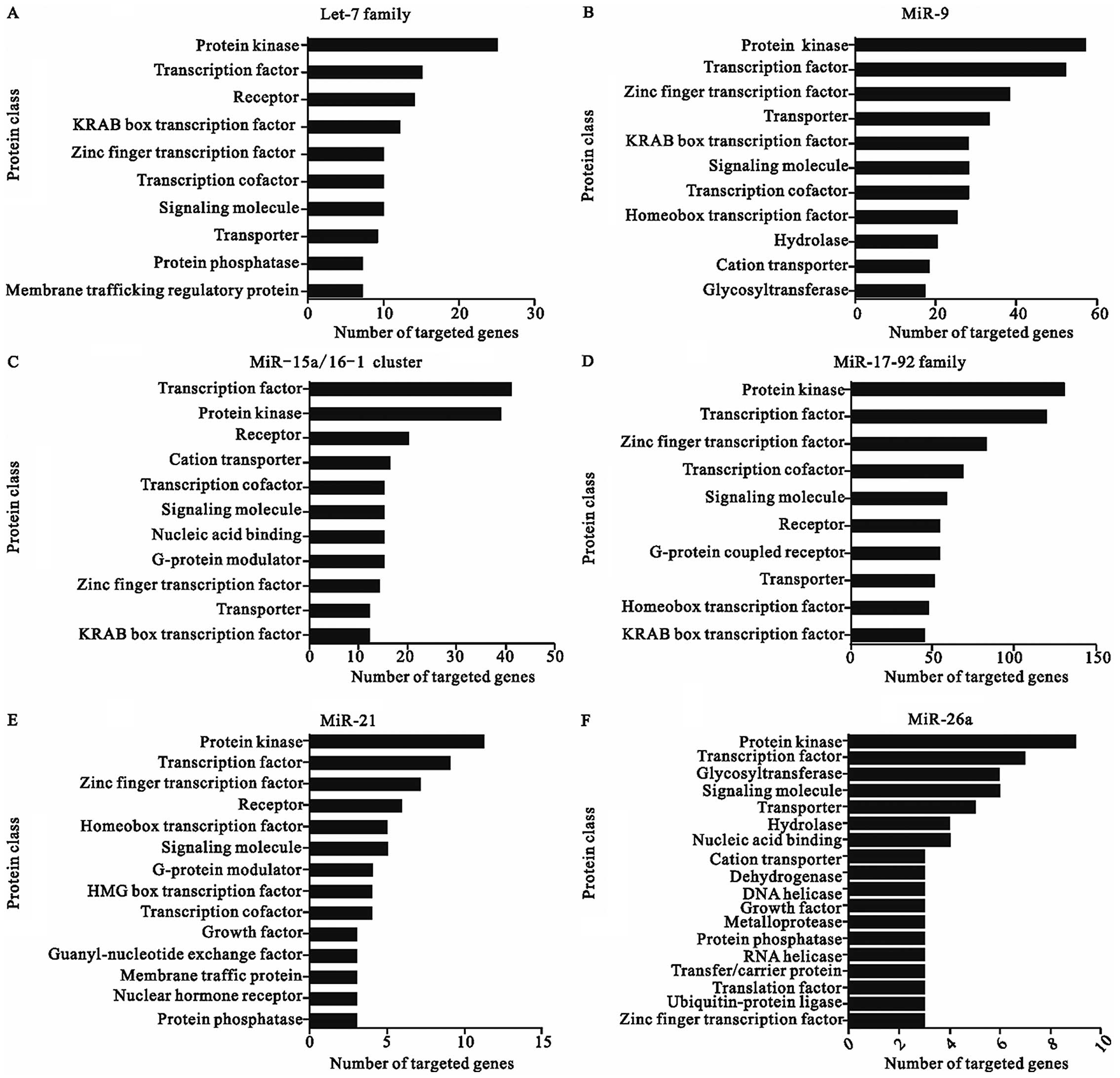
The major classes of potential targets of each
CA-miRNAs are shown in Fig. 1.
These potential miRNA targets belong to a great many of gene
families which play various roles during physiological and
pathological processes. Although the different seed region
sequences of these miRNAs leads to the diversity of different
targets, the protein class of all the targeted genes seemed
similar, suggesting that CA-miRNAs might function in the
post-transcriptional level mainly through manipulating the
expression of transcription factors and protein kinases. As a
recently recognized part of that regulation, miRNA-mediated events
seem to ensure the preciseness and fidelity of dynamic and
spatially restricted gene expression. Most of these targeted genes
could express the transcription factors which controlled the
expression of nearly all genes. Besides, another crucial part of
the predicted targets were diverse sorts of enzymes such as protein
kinase, which may participate in various signaling pathways. A
protein kinase which modifies other proteins by chemically adding
phosphate groups to them (phosphorylation) is one of the largest
and most influential of gene families: constituting some 2% of the
proteome, they regulate almost all biochemical pathways and may
phosphorylate up to 30% of the proteome. Interestingly, beside the
CA-miRNAs, other miRNAs also seem to have the same most common
targeted genes, which may be a general character of miRNA
regulation (55).
Over-represented protein classes of target genes are
transcription factors, protein kinases, receptors, components of
the miRNA machinery, and other proteins involved in translational
regulation, as well as components of the ubiquitin machinery, which
is crucial in the maintenance of normal cell life and this result
also represented novel feedback loops in gene regulation similarly
to previous research (56). As
miRNAs seem to play a role as more refined regulators of gene
expression, the minor percent of the targets contain diverse
proteins such as cytokine, protease, immunoglobulin superfamily
cell adhesion molecule, ATP-binding cassette (ABC) transporter, and
actin family cytoskeletal protein.
The largest number of genes are targeted by the
miR-17-92 cluster, which is not only because this miRNA cluster
contain more family members but also commit more dominant functions
except transcription factors and protein kinases such as signaling
molecule, G-protein coupled receptor, and hydrolase. On the other
hand, as a less significant regulator, miR-206 targets the minimum
amount of genes, which may function as G-protein modulator,
dehydrogenase, and hydrolase. In agreement with our results, some
research groups have found that miRNA oncogenes and tumor
suppressors clearly show different patterns in function,
evolutionary rate, expression, chromosome distribution, molecule
size, free energy, transcription factors and targets (57).
Molecular function, biological process
and signal pathway analysis of targets related to each
CA-miRNA
We scored the list of genes for each miRNA against
molecular function, biological process and canonical pathways. For
each miRNA, the top five rank for each analysis is shown in
Tables II and III. The targets for the CA-miRNAs were
most prominently predicted to function in regulation of
transcription, which is the dominant process of controlling gene
expression. Carcinogenesis in humans is a multistep process and
that these steps reflect genetic alterations that drive the
progressive transformation of normal human cells into highly
malignant derivatives. Similarly, pathways in cancer and MAPK
signaling pathway were the most-observed overlaps between the
pathways for these CA-miRNAs, suggesting that these miRNAs may
probably regulate carcinogenesis mainly through the two pathways.
The results indicated that those miRs are closely associated with
cancers, consistent with current observations described in cancer
associated miRNAs section.
 | Table IIMolecular function and biological
process analysis of each CA-miRNA. |
Table II
Molecular function and biological
process analysis of each CA-miRNA.
| CA-miRNAs | Molecular function
and biological process | % Regulated by
CA-miRNAs | P-value |
|---|
| let-7 family | Regulation of
transcription | 20.6% | 2.80E-04 |
| Transcription | 14.4% | 5.20E-02 |
| Regulation of
transcription, DNA-dependent | 13.7% | 7.90E-03 |
| Regulation of RNA
metabolic process | 13.7% | 1.20E-02 |
| Phosphorus
metabolic process | 10.2% | 9.40E-05 |
| miR-9 | Regulation of
transcription | 19.30% | 1.50E-06 |
| Transcription | 15.20% | 1.40E-04 |
| Regulation of RNA
metabolic process | 14.00% | 6.70E-06 |
| Regulation of
transcription, DNA-dependent | 13.40% | 3.40E-05 |
| Intracellular
signaling cascade | 8.90% | 7.60E-03 |
| miR-15a/16-1
cluster | Regulation of
transcription | 18.90% | 4.70E-04 |
| Transcription | 14.80% | 7.00E-03 |
| Regulation of RNA
metabolic process | 12.00% | 5.30E-02 |
| Intracellular
signaling cascade | 9.00% | 2.50E-02 |
| Phosphate metabolic
process | 8.90% | 1.10E-04 |
| miR-17-92
family | Regulation of
transcription | 21.00% | 1.20E-24 |
| Transcription | 16.70% | 1.20E-17 |
| Regulation of RNA
metabolic process | 14.30% | 3.00E-14 |
| Regulation of
transcription, DNA-dependent | 13.90% | 2.50E-13 |
| Intracellular
signaling cascade | 10.10% | 3.80E-11 |
| miR-21 | Regulation of
transcription | 23.30% | 2.90E-03 |
| Regulation of RNA
metabolic process | 18.40% | 1.70E-03 |
| Transcription | 17.80% | 2.40E-02 |
| Regulation of
transcription, DNA-dependent | 17.20% | 5.00E-03 |
| Positive regulation
of macromolecule metabolic process | 14.70% | 2.80E-06 |
| miR-26a | Transcription | 15.10% | 9.60E-02 |
| Phosphorus
metabolic process | 11.40% | 1.60E-03 |
| Phosphate metabolic
process | 11.40% | 1.60E-03 |
|
Phosphorylation | 9.20% | 6.10E-03 |
| Protein amino acid
phosphorylation | 8.60% | 2.70E-03 |
| miR-34a/b/c | Regulation of
transcription | 19.20% | 4.80E-05 |
| Transcription | 15.30% | 6.30E-04 |
| Regulation of RNA
metabolic process | 13.50% | 7.30E-04 |
| Regulation of
transcription, DNA-dependent | 13.10% | 1.10E-03 |
| Positive regulation
of macromolecule metabolic process | 7.40% | 5.10E-04 |
| miR-155 | Regulation of
transcription | 29.30% | 1.30E-14 |
| Transcription | 23.80% | 1.20E-11 |
| Regulation of
transcription, DNA-dependent | 19.00% | 4.50E-08 |
| Regulation of RNA
metabolic process | 19.00% | 1.20E-07 |
| Intracellular
signaling cascade | 12.40% | 2.00E-04 |
| miR-200/141
family | Regulation of
transcription | 21.50% | 1.90E-07 |
| Transcription | 18.10% | 3.50E-07 |
| Regulation of RNA
metabolic process | 16.40% | 1.00E-07 |
| Regulation of
transcription, DNA-dependent | 15.40% | 1.90E-06 |
| Intracellular
signaling cascade | 9.80% | 4.50E-03 |
| miR-205 | Regulation of
transcription | 22.50% | 6.90E-06 |
| Transcription | 18.60% | 3.10E-05 |
| Regulation of
transcription, DNA-dependent | 16.20% | 5.50E-05 |
| Regulation of RNA
metabolic process | 16.20% | 1.10E-04 |
| Positive regulation
of macromolecule metabolic process | 11.80% | 1.60E-08 |
| miR-206 | Transcription | 21.60% | 9.00E-03 |
| Regulation of
transcription | 21.60% | 7.20E-02 |
| Intracellular
signaling cascade | 12.40% | 8.50E-02 |
| Protein
localization | 11.30% | 2.30E-02 |
| Protein
transport | 9.30% | 6.00E-02 |
 | Table IIICanonical pathway analysis of each
CA-miRNA. |
Table III
Canonical pathway analysis of each
CA-miRNA.
| CA-miRNAs | Canonical
pathways | % Regulated by
CA-miRNAs | P-value |
|---|
| let-7 family | Pathways in
cancer | 4.20% | 9.00E-04 |
| MAPK signaling
pathway | 3.70% | 9.60E-04 |
| p53 signaling
pathway | 2.00% | 4.30E-04 |
| mTOR signaling
pathway | 1.20% | 2.10E-02 |
| miR-9 | Pathways in
cancer | 3.40% | 2.60E-05 |
| MAPK signaling
pathway | 2.60% | 5.40E-04 |
| Focal adhesion | 2.50% | 1.60E-05 |
| Endocytosis | 2.10% | 1.70E-04 |
| Regulation of actin
cytoskeleton | 2.10% | 1.80E-03 |
| miR-15a/16-1
cluster | Pathways in
cancer | 4.00% | 2.30E-08 |
| MAPK signaling
pathway | 2.90% | 2.00E-06 |
| Neurotrophin
signaling pathway | 2.30% | 7.00E-05 |
| Insulin signaling
pathway | 1.70% | 1.40E-02 |
| p53 signaling
pathway | 1.10% | 1.50E-02 |
| miR-17-92
family | Pathways in
cancer | 2.80% | 1.60E-05 |
| MAPK signaling
pathway | 2.70% | 1.90E-08 |
| Endocytosis | 2.20% | 8.00E-10 |
| Regulation of actin
cytoskeleton | 1.80% | 3.70E-04 |
| Focal adhesion | 1.80% | 1.40E-04 |
| miR-21 | MAPK signaling
pathway | 6.70% | 1.20E-04 |
| Pathways in
cancer | 5.50% | 9.50E-03 |
| Cytokine-cytokine
receptor interaction | 4.90% | 9.50E-03 |
| Jak-STAT signaling
pathway | 4.30% | 2.80E-03 |
| Pancreatic
cancer | 3.70% | 4.80E-04 |
| miR-26a | Wnt signaling
pathway | 2.70% | 1.90E-02 |
| miR-34a/b/c | Pathways in
cancer | 3.40% | 1.10E-03 |
| MAPK signaling
pathway | 2.40% | 2.60E-02 |
| Focal adhesion | 2.30% | 3.10E-03 |
| Endocytosis | 1.90% | 1.60E-02 |
| Regulation of actin
cytoskeleton | 1.90% | 5.20E-02 |
| miR-155 | Pathways in
cancer | 5.50% | 5.20E-06 |
| MAPK signaling
pathway | 4.30% | 1.10E-04 |
| T cell receptor
signaling pathway | 3.70% | 5.50E-08 |
| Neurotrophin
signaling pathway | 3.00% | 5.60E-05 |
| B cell receptor
signaling pathway | 2.50% | 1.50E-05 |
| miR-200/141
family | Pathways in
cancer | 3.60% | 2.70E-04 |
| MAPK signaling
pathway | 2.80% | 3.30E-03 |
| Wnt signaling
pathway | 1.90% | 3.00E-03 |
| Axon guidance | 1.50% | 1.80E-02 |
| Chronic myeloid
leukemia | 1.40% | 1.40E-03 |
| miR-205 | Tight junction | 2.70% | 8.80E-04 |
| Endocytosis | 2.50% | 2.40E-02 |
| Wnt signaling
pathway | 2.20% | 2.10E-02 |
| Ubiquitin mediated
proteolysis | 2.00% | 3.60E-02 |
| Adherens
junction | 1.50% | 3.00E-02 |
| miR-206 | Regulation of actin
cytoskeleton | 4.10% | 5.70E-02 |
Pathway mapping of cancer-associated
miRNA targets
The top five pathways regulated by the CA-miRNAs are
shown in Table IV. It inferred
that these 11 CA-miRNAs participate in carcinogenesis mainly
through the five pathways concerned with morphological changes,
intercellular communication and invasion of cancer cells. Three out
of these top 5 pathways (pathways in cancer, endocytosis and
regulation of actin cytoskeleton) are reported in the biological
pathway analysis for prostate cancer (58). Admittedly, there are definitely
other cancer-related pathways not emerged in the Rank Five list. It
may be because these targeted genes mainly occupied a large
percentage in the five pathways and they are still closely
connected with other cancer-related pathways.
 | Table IVThe top five pathways regulated by
all 11 CA-miRNAs. |
Table IV
The top five pathways regulated by
all 11 CA-miRNAs.
| Pathway DB | Name | Hits | Total | Percent |
|---|
| KEGG | Pathways in
cancer | 141 | 343 | 41.11% |
| MAPK signaling
pathway | 125 | 284 | 44.01% |
| Endocytosis | 103 | 240 | 42.92% |
| HTLV-I
infection | 93 | 198 | 46.97% |
| Regulation of actin
cytoskeleton | 93 | 228 | 40.79% |
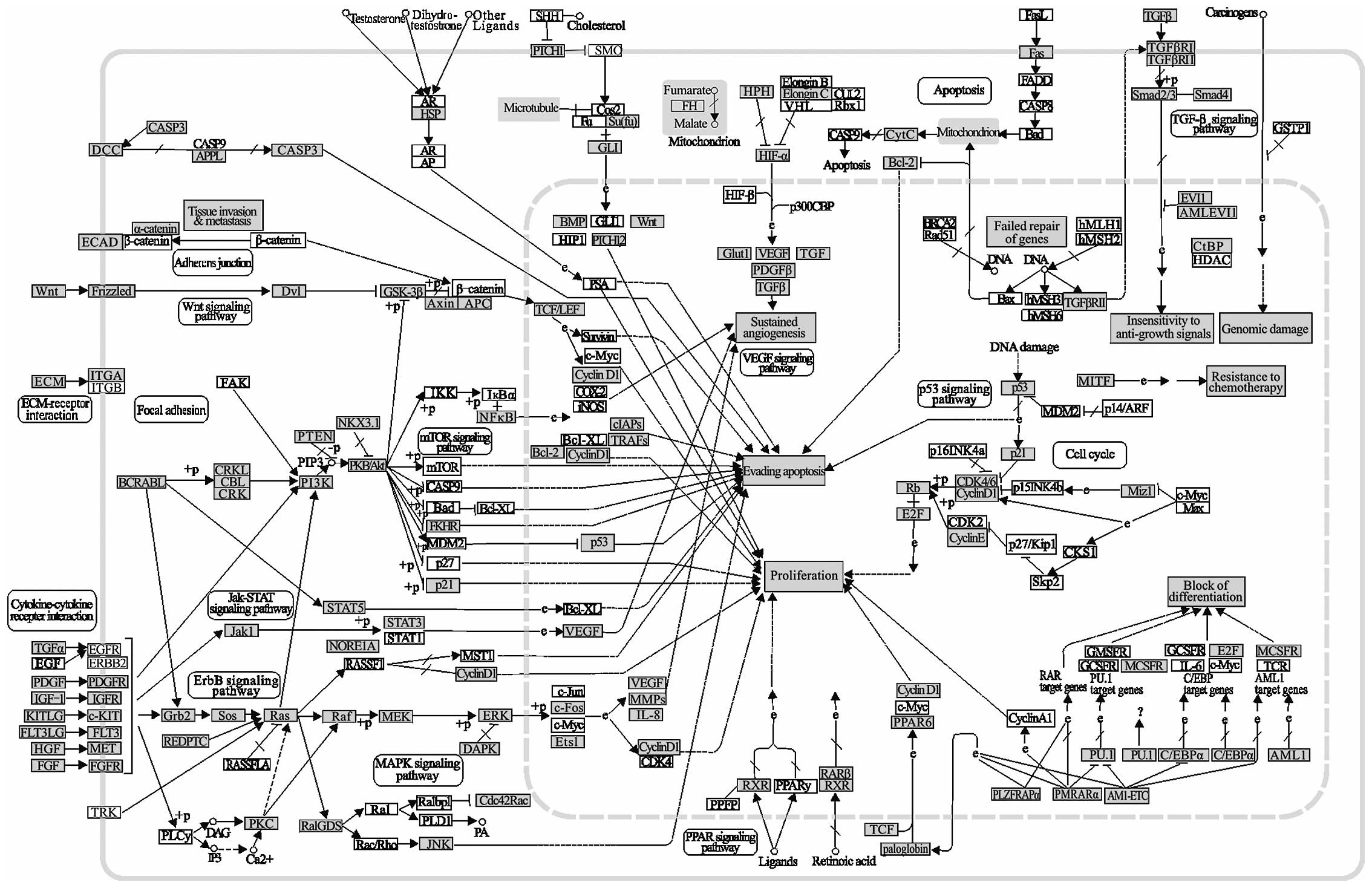
Pathways in cancer are highly saturated with gene
targets of the CA-miRNAs (Fig. 2).
This kind of pathway clearly demonstrated the acquisition of
biological capabilities such as block of differentiation,
resistance to apoptosis, unlimited replicative potential, sustained
angiogenesis, tissue invasion and metastasis for the transformation
from normal cells into highly malignant tumor cells. The effects of
alterations on many oncogenes and tumor suppressor genes are
complex due to the high number of changes in the interactions of
the biological pathways involved. However, more common
abnormalities in oncogenes and tumor suppressor genes regulated by
CA-miRNAs can be potential therapeutic targets. Indeed, miRNA
targeted genes in grey such as Wnt, STAT3, p21, P53, BCL-2, Fas,
TGF-β, and Rb are directly related with cancer (Fig. 2).
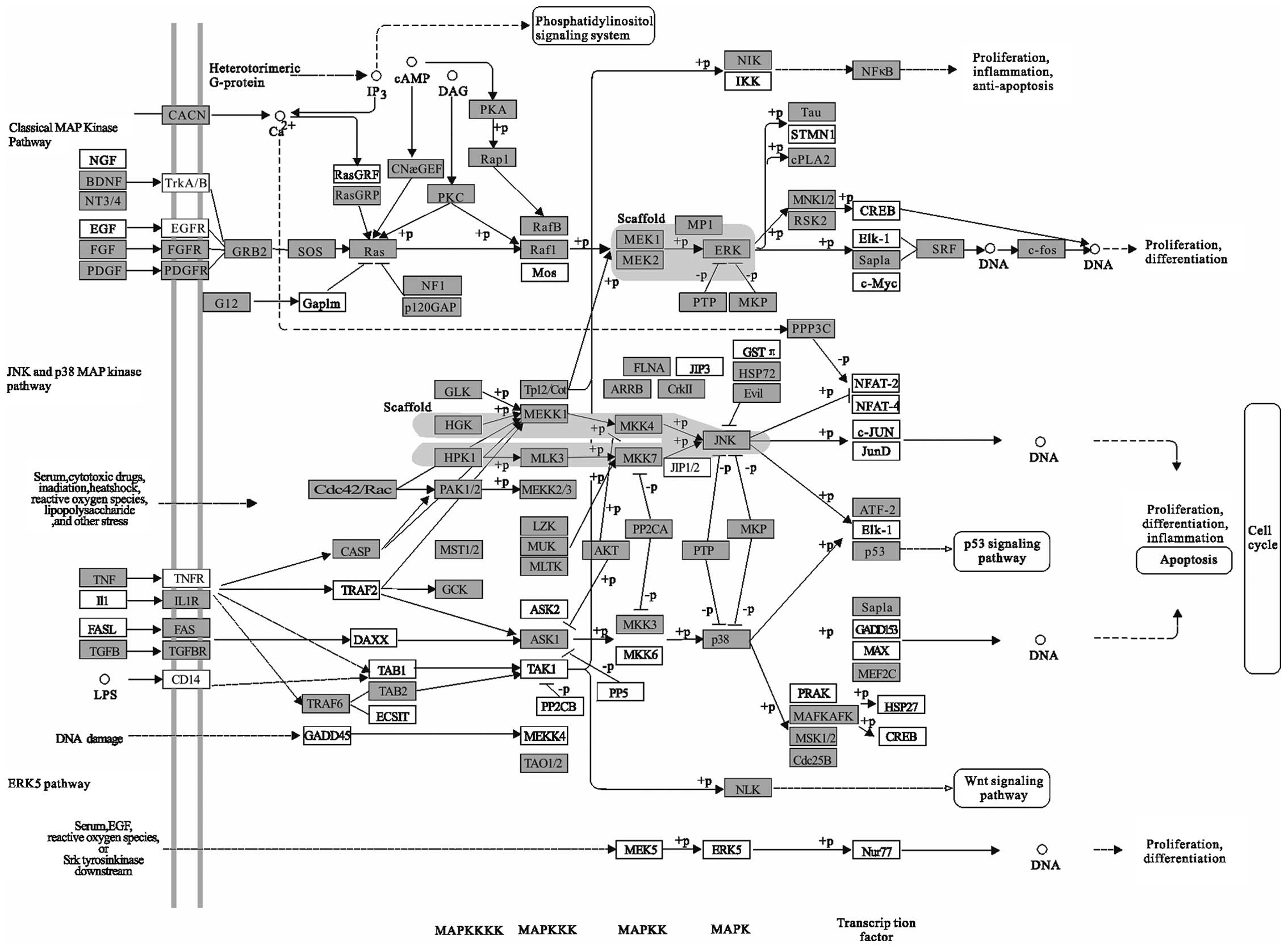
Mitogen-activated protein kinase (MAPK) pathway
functions as integrating signals that affect proliferation,
differentiation, survival and migration (Fig. 3). MAPK signaling is tightly
regulated so that optimal biological activities are achieved and
health is maintained. This pathway activation is a frequent event
in human cancer and is often the result of activating mutations in
the BRAF and RAS oncogenes. There are three main sub-families of
MAPK pathways in humans (classical MAPK pathways, JNK and p38 MAPK
pathway and ERK5 pathway), whose functions are regulated by
activators, inactivators, substrates and scaffolds, which together
form delicate signaling cascades in response to different
extracellular or intracellular stimulation. Unscheduled
proliferation is a hallmark of cancer, and the JNK and p38 MAPK
pathways regulate cell cycle progression at different transition
points by both transcription-dependent and
transcription-independent mechanisms. Members of MAP kinase (MAPK)
family are evolutionarily conserved regulators that mediate signal
transduction and play essential roles in various physiological
processes. Consistent with the importance of these events in
tumorigenesis, MAPK signaling is closely associated with cancers in
humans. The MAPKs are activated by mitogens and were found to be
upregulated in human tumors; this finding has led to the
development of inhibitors of this pathway for cancer therapeutics.
Studies in mouse models have been essential to better understand
how these MAPKs control cancer development, and these models are
expected to provide new strategies for the design of improved
therapeutic approaches (59).
Small-molecule inhibitors designed to target various steps of this
pathway have entered clinical trials (60). Pharmacological inhibition of the
kinase JNK blocked induction of oncomiR miR-155 in response to
either polyriboinosinic:polyribocytidylic acid or TNF-α, suggesting
that miR-155-inducing signals use the JNK pathway (61). In addition, miR-141 and miR-200a
target p38α and modulate the oxidative stress response (62). These previous results indicate that
miRNAs may be a promising clinical treatment for cancer through the
MAPK pathway.
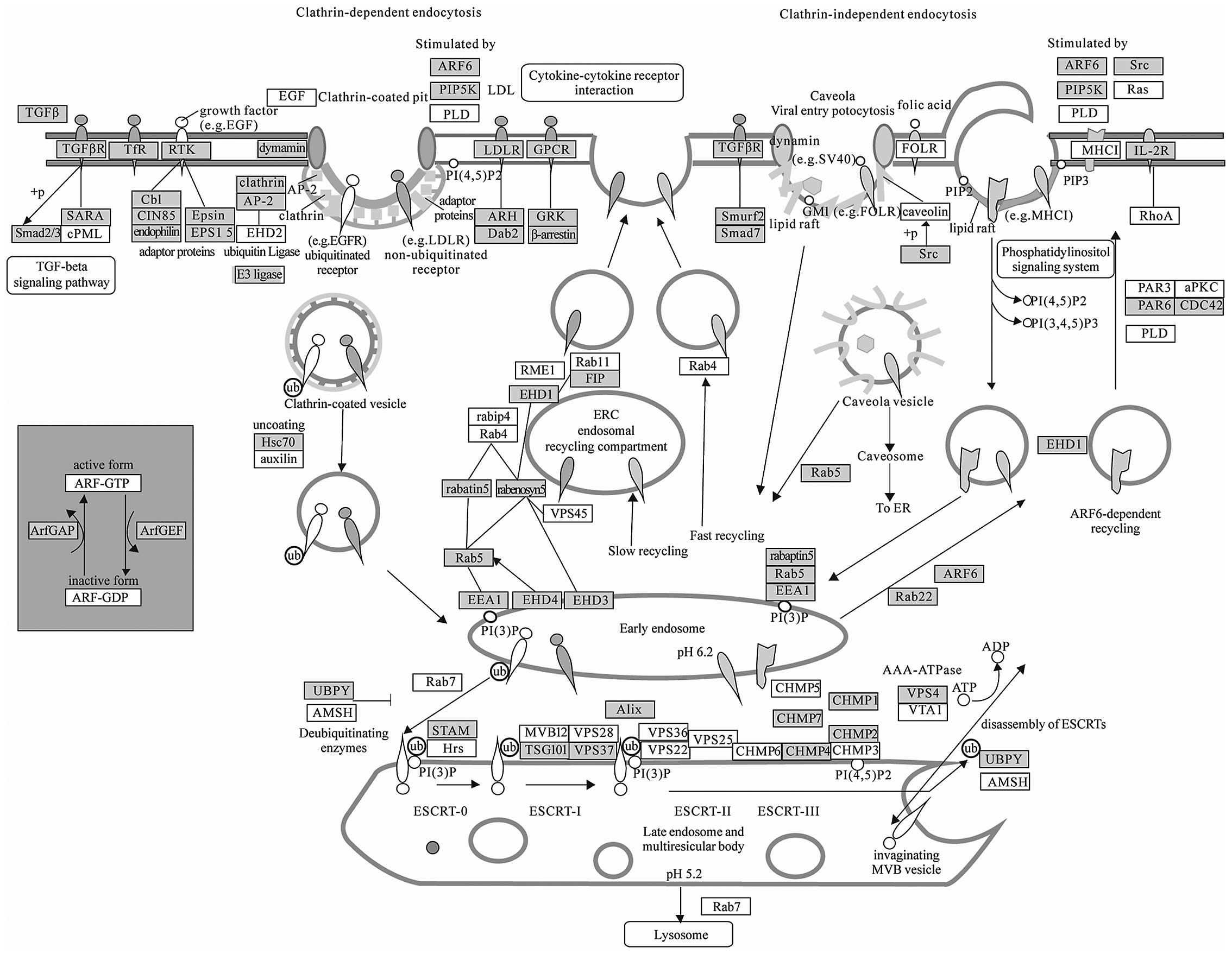
Endocytosis (Fig.
4) has been regarded as a long-term mechanism of signal
attenuation via receptor clearance from the cell surface.
Additional and quite unexpected functions for endocytosis have
emerged, which, together with its attenuation function, project a
central role for this process in cellular homeostasis and control
of proliferation (63). Subversion
of endocytic control is thus predicted to play a causative role in
hyper-proliferative conditions, first and foremost cancer (63). Recently, research has revealed that
microenvironment around tumor cells contains various circulating
miRNAs secreted by microvesicles or exosomes, which may enter into
other surrounded cells by endocytosis (64,65).
Therefore, this pathway cluster analysis suggests that endocytosis
needs to be paid more attention as a novel mechanism of
intercellular communication of tumor cells.
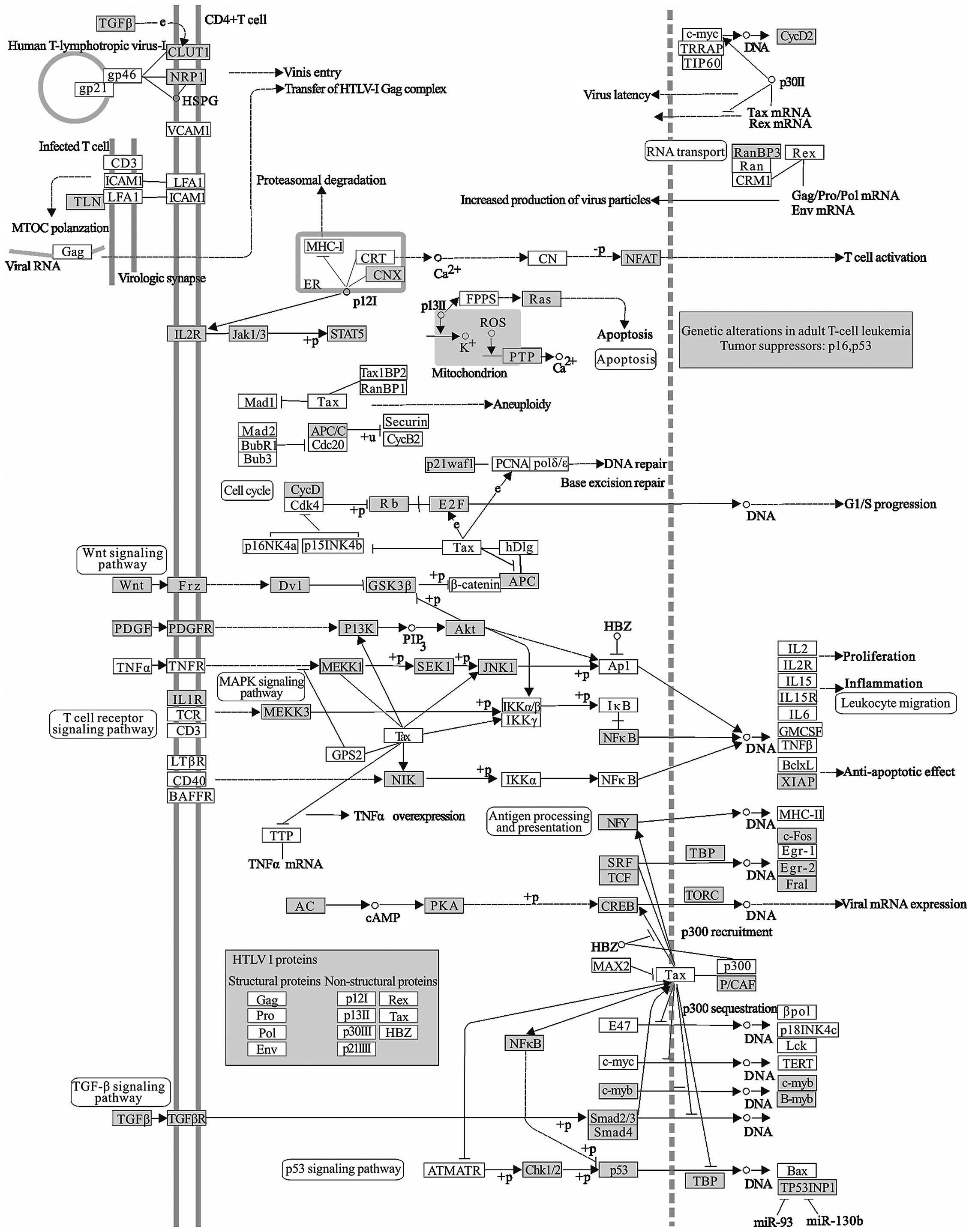
The human T cell leukemia virus type I (HTLV-I)
infection (Fig. 5) is associated
with adult T cell leukemia/lymphoma (ATL). ATL is a highly
aggressive neoplastic disease of CD4 positive T lymphocyte, which
is featured by the pleomorphic tumor cells with hyper-segmented
nuclei, called ‘flower cell’ (66). HTLV-I, as an oncogenic retrovirus,
encodes an oncogenic protein, Tax, which interferes with several
signaling pathways related to anti-apoptosis or cell proliferation.
The ability of Tax to both transcriptionally regulate cellular gene
expression and to functionally inactivate proteins involved in cell
cycle progression and DNA repair provide the basis for Tax-mediated
transformation and leukemogenesis (67). The modulation of the signaling by
Tax involves its binding to transcription factors like CREB/ATF,
NF-κB, SRF and NFAT.
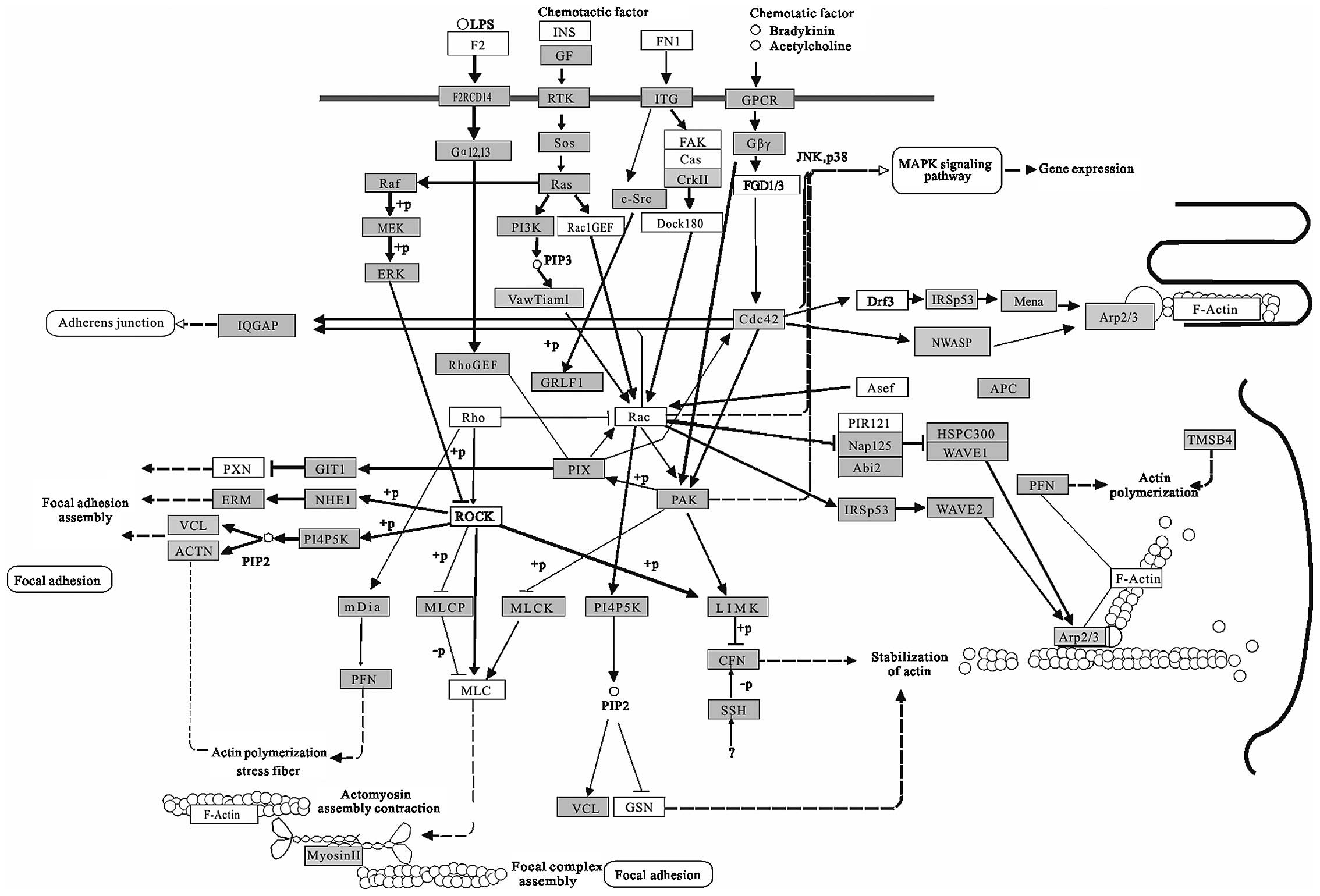
Regulation of actin cytoskeleton (Fig. 6) may be involved in morphological
changes of cancer cells. Several studies have demonstrated that
molecules that link migratory signals to the actin cytoskeleton are
upregulated in invasive and metastatic cancer cells (68). Aberrant regulation of cell
migration drives progression of cancer invasion and metastasis
(69-71). This pathway seems to be an overlap
with those participated in senescent cells, which also undergo
changes in morphology, becoming large and flattened (72). Beside this pathway, many
cancer-related pathways overlap with that involved in senescence
such as inflammatory pathway, IGF pathway, p16–p21 pathway and p53
pathway. In terms of the pathway, cancer and senescence seem to
share many same biological processes and cancer is defined as a
typical aging related disease (73). For the ability of targeting many
different genes, miRNAs provide a mechanism through which
widespread alternations could be induced.
In this study, we have used pathway mapping and
theoretical gene target identification to create a biological
framework by which to test the relevance of miRNAs in cancer
induction. The identification of CA-miRNAs related pathways with
the ability to regulate a complex pathological process such as
cancer, can be better using bioinformatic techniques followed by
experimental validation.
Acknowledgements
This project is supported by grants
from National Natural Science Foundation of China (31000323,
31070672), Specialized Research Fund for the Doctoral Program of
Higher Education of China (20100091120023) and Fundamental Research
Funds for the Central Universities (1095020823).
References
|
1.
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Zhang L, Hou D, Chen X, et al: Exogenous
plant MIR168a specifically targets mammalian LDLRAP1: evidence of
cross-kingdom regulation by microRNA. Cell Res. 22:107–126. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Ma J and Huang Y: Post-transcriptional
regulation of miRNA biogenesis and functions. Front Biol. 5:32–40.
2010. View Article : Google Scholar
|
|
5.
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
7.
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar
|
|
8.
|
Kwak PB, Iwasaki S and Tomari Y: The
microRNA pathway and cancer. Cancer Sci. 101:2309–2315. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Ming M, Zhao X, Zhao Z, Liu B and Bao J:
MicroRNA regulation of programmed cell death pathways in cancer.
Curr Chem Biol. 6:53–59. 2012.
|
|
10.
|
Loscalzo J, Kohane I and Barabasi AL:
Human disease classification in the postgenomic era: a complex
systems approach to human pathobiology. Mol Syst Biol. 3:1242007.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Calin GA, Dumitru CD, Shimizu M, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Tagawa H and Seto M: A microRNA cluster as
a target of genomic amplification in malignant lymphoma. Leukemia.
19:2013–2016. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar
|
|
16.
|
Pei Y, Wang Z, Fei F, Shao Z, Huang W and
Zhang X: Bioinformatics study indicates possible microRNA-regulated
pathways in the differentiation of breast cancer. Chin Sci Bull.
55:927–936. 2010. View Article : Google Scholar
|
|
17.
|
Huang da W, Sherman BT, Tan Q, et al: The
DAVID gene functional classification tool: a novel biological
module-centric algorithm to functionally analyze large gene lists.
Genome Biol. 8:R1832007.PubMed/NCBI
|
|
18.
|
Kanehisa M, Araki M, Goto S, et al: KEGG
for linking genomes to life and the environment. Nucleic Acids Res.
36:D480–D484. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Johnson CD, Esquela-Kerscher A, Stefani G,
et al: The let-7 microRNA represses cell proliferation pathways in
human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Adams BD, Guttilla IK and White BA:
Involvement of microRNAs in breast cancer. Semin Reprod Med.
26:522–536. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Laios A, O’Toole S, Flavin R, et al:
Potential role of miR-9 and miR-223 in recurrent ovarian cancer.
Mol Cancer. 7:352008. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Lehmann U, Hasemeier B, Christgen M, et
al: Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human
breast cancer. J Pathol. 214:17–24. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Ferretti E, De Smaele E, Po A, et al:
MicroRNA profiling in human medulloblastoma. Int J Cancer.
124:568–577. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Sun Y, Wu J, Wu SH, et al: Expression
profile of microRNAs in c-Myc induced mouse mammary tumors. Breast
Cancer Res Treat. 118:185–196. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Ma L, Young J, Prabhala H, et al: miR-9, a
MYC/MYCN-activated microRNA, regulates E-cadherin and cancer
metastasis. Nat Cell Biol. 12:247–256. 2010.PubMed/NCBI
|
|
27.
|
Bonci D, Coppola V, Musumeci M, et al: The
miR-15a-miR-16-1 cluster controls prostate cancer by targeting
multiple oncogenic activities. Nat Med. 14:1271–1277. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Calin GA, Cimmino A, Fabbri M, et al:
MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl
Acad Sci USA. 105:5166–5171. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Mendell JT: miRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Uziel T, Karginov FV, Xie S, et al: The
miR-17∼92 cluster collaborates with the Sonic Hedgehog pathway in
medulloblastoma. Proc Natl Acad Sci USA. 106:2812–2817. 2009.
|
|
31.
|
O’Day E and Lal A: MicroRNAs and their
target gene networks in breast cancer. Breast Cancer Res.
12:2012010.
|
|
32.
|
Lawrie CH, Soneji S, Marafioti T, et al:
MicroRNA expression distinguishes between germinal center B
cell-like and activated B cell-like subtypes of diffuse large B
cell lymphoma. Int J Cancer. 121:1156–1161. 2007. View Article : Google Scholar
|
|
33.
|
Sander S, Bullinger L, Klapproth K, et al:
MYC stimulates EZH2 expression by repression of its negative
regulator miR-26a. Blood. 112:4202–4212. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Kota J, Chivukula RR, O’Donnell KA, et al:
Therapeutic microRNA delivery suppresses tumorigenesis in a murine
liver cancer model. Cell. 137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Visone R, Pallante P, Vecchione A, et al:
Specific microRNAs are downregulated in human thyroid anaplastic
carcinomas. Oncogene. 26:7590–7595. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Kim H, Huang W, Jiang X, Pennicooke B,
Park PJ and Johnson MD: Integrative genome analysis reveals an
oncomir/oncogene cluster regulating glioblastoma survivorship. Proc
Natl Acad Sci USA. 107:2183–2188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Gallardo E, Navarro A, Vinolas N, et al:
miR-34a as a prognostic marker of relapse in surgically resected
non-small-cell lung cancer. Carcinogenesis. 30:1903–1909. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Cole KA, Attiyeh EF, Mosse YP, et al: A
functional screen identifies miR-34a as a candidate neuroblastoma
tumor suppressor gene. Mol Cancer Res. 6:735–742. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Li N, Fu H, Tie Y, et al: miR-34a inhibits
migration and invasion by down-regulation of c-Met expression in
human hepatocellular carcinoma cells. Cancer Lett. 275:44–53. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Nakada C, Matsuura K, Tsukamoto Y, et al:
Genome-wide microRNA expression profiling in renal cell carcinoma:
significant down-regulation of miR-141 and miR-200c. J Pathol.
216:418–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Du Y, Xu Y, Ding L, et al: Down-regulation
of miR-141 in gastric cancer and its involvement in cell growth. J
Gastroenterol. 44:556–561. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Nie W, Tang L, Zhang H, et al: Structural
analysis of the EGFR TK domain and potential implications for EGFR
targeted therapy. Int J Oncol. 40:1763–1769. 2012.PubMed/NCBI
|
|
45.
|
Adam L, Zhong M, Choi W, et al: miR-200
expression regulates epithelial-to-mesenchymal transition in
bladder cancer cells and reverses resistance to epidermal growth
factor receptor therapy. Clin Cancer Res. 15:5060–5072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Bendoraite A, Knouf EC, Garg KS, et al:
Regulation of miR-200 family microRNAs and ZEB transcription
factors in ovarian cancer: Evidence supporting a
mesothelial-to-epithelial transition. Gynecol Oncol. 116:117–125.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Gandellini P, Folini M, Longoni N, et al:
miR-205 exerts tumor-suppressive functions in human prostate
through down-regulation of protein kinase Cepsilon. Cancer Res.
69:2287–2295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Wiklund ED, Bramsen JB, Hulf T, et al:
Coordinated epigenetic repression of the miR-200 family and miR-205
in invasive bladder cancer. Int J Cancer. 128:1327–1334. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Iorio MV, Casalini P, Piovan C, et al:
microRNA-205 regulates HER3 in human breast cancer. Cancer Res.
69:2195–2200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Iorio MV, Visone R, Di Leva G, et al:
MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Taulli R, Bersani F, Foglizzo V, et al:
The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma
growth in xenotransplanted mice by promoting myogenic
differentiation. J Clin Invest. 119:2366–2378. 2009.PubMed/NCBI
|
|
52.
|
Negrini M and Calin GA: Breast cancer
metastasis: a microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Hobert O: miRNAs play a tune. Cell.
131:22–24. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Hammond SM: MicroRNAs as oncogenes. Curr
Opin Genet Dev. 16:4–9. 2006. View Article : Google Scholar
|
|
55.
|
Yang H, Zhang HY, Zhu L, Zhang CY and Li
DH: Identification and characterization of microRNAs in Macaca
fascicularis by EST analysis. Comp Funct Genom. Jul
5–2012.(Epub ahead of print).
|
|
56.
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar
|
|
57.
|
Wang D, Qiu C, Zhang H, Wang J, Cui Q and
Yin Y: Human microRNA oncogenes and tumor suppressors show
significantly different biological patterns: from functions to
targets. PLoS One. 5:e130672010. View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Yifei T, Jiajia C, Cheng L, Kaipia A and
Bairong S: MicroRNA expression analysis reveals significant
biological pathways in human prostate cancer. Systems Biology
(ISB). 2011 IEEE International Conference on 203–210.
|
|
59.
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Sebolt-Leopold JS and Herrera R: Targeting
the mitogen-activated protein kinase cascade to treat cancer. Nat
Rev Cancer. 4:937–947. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
O’Connell RM, Taganov KD, Boldin MP, Cheng
G and Baltimore D: MicroRNA-155 is induced during the macrophage
inflammatory response. Proc Natl Acad Sci USA. 104:1604–1609.
2007.PubMed/NCBI
|
|
62.
|
Mateescu B, Batista L, Cardon M, et al:
miR-141 and miR-200a act on ovarian tumorigenesis by controlling
oxidative stress response. Nat Med. 17:1627–1635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Polo S, Pece S and Di Fiore PP:
Endocytosis and cancer. Curr Opin Cell Biol. 16:156–161. 2004.
View Article : Google Scholar
|
|
64.
|
Al-Nedawi K, Meehan B, Micallef J, et al:
Intercellular transfer of the oncogenic receptor EGFRvIII by
microvesicles derived from tumour cells. Nat Cell Biol. 10:619–624.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
65.
|
Simons M and Raposo G: Exosomes -
vesicular carriers for intercellular communication. Curr Opin Cell
Biol. 21:575–581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66.
|
Yasunaga J and Matsuoka M: HTLV-I and
leukemogenesis. Uirusu. 56:241–249. 2006.(In Japanese).
|
|
67.
|
Gatza ML, Watt JC and Marriott SJ:
Cellular transformation by the HTLV-I Tax protein, a
jack-of-all-trades. Oncogene. 22:5141–5149. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
68.
|
Yamaguchi H and Condeelis J: Regulation of
the actin cytoskeleton in cancer cell migration and invasion.
Biochim Biophys Acta. 1773:642–652. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Condeelis J, Singer RH and Segall JE: The
Great Escape: when cancer cells hijack the genes for chemotaxis and
motility. Annu Rev Cell Dev Biol. 21:6952005. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Sahai E: Mechanisms of cancer cell
invasion. Curr Opin Genet Dev. 15:87–96. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Yamaguchi H, Wyckoff J and Condeelis J:
Cell migration in tumors. Curr Opin Cell Biol. 17:559–564. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Tominaga K, Olgun A, Smith JR and
Pereira-Smith OM: Genetics of cellular senescence. Mech Ageing Dev.
123:927–936. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
Burkle A, Caselli G, Franceschi C, et al:
Pathophysiology of ageing, longevity and age related diseases.
Immun Ageing. 4:42007. View Article : Google Scholar : PubMed/NCBI
|