Introduction
Neuroblastoma (NB) is one of the most common
pediatric solid tumors, accounting for 15% of all pediatric cancer
deaths. It originates from the sympathoadrenal lineage derived from
the neural crest. The clinical behavior is markedly heterogeneous
(1–3). Most tumors tend to grow aggressively
and often have a fatal outcome, but some tumors are favorable and
show spontaneous differentiation or regression. The stage of the
tumor at diagnosis, the age of the patient and the presence or
absence of MYCN amplification are the basic parameters used
for risk stratification to determine the management and treatment
of this disease. Recent progress in chemotherapy has dramatically
increased the survival rates of many pediatric cancers; however,
advanced stage NB with metastasis, especially those with genomic
amplification of the MYCN oncogene, are frequently resistant
to any therapy and the outcome for patients is still very poor
(1–3). Therefore, it is important to know the
mechanism of metastasis in NB in order to improve the treatment
results.
The epithelial-mesenchymal transition (EMT) is a
series of events during which epithelial cells lose many of their
epithelial characteristics and take on properties typical of
mesenchymal cells. EMT has an important role in the development of
many tissues during embryogenesis and similar cell changes are
recapitulated during pathological processes, such as fibrosis and
cancer. Numerous observations support the idea that EMT has a
central role in tumor progression and metastasis (4–7).
Cancer cells acquire mesenchymal gene expression patterns and
properties, resulting in reduced cell-cell adhesion and the
activation of proteolysis and motility. These activities promote
tumor invasion and metastasis. EMT is important in the progression
of tumor cells acquiring a more invasive, metastatic capacity. In
this study, we investigated the role of EMT in the progression of
NB in terms of invasiveness and metastasis.
Materials and methods
Tumor samples
Ninety-six primary NBs were obtained from the
Department of Pediatric Surgery, University of Tsukuba, and the
Division of Biochemistry, Chiba Cancer Center Research Institute,
Japan. Patients were aged between 0 months and 18 years at
diagnosis (median 16 months). The clinical characteristics of the
96 NBs are shown in Table I.
 | Table I.Tumor stages and MYCN amplification
of 96 neuroblastomas. |
Table I.
Tumor stages and MYCN amplification
of 96 neuroblastomas.
| Stage 1, 2 | Stage 4S | Stage 3 | Stage 4 | Total |
|---|
| MYCN | | | | | |
| Unamplified | 22 | 4 | 10 | 15 | 51 |
| Amplified | 2 | 2 | 11 | 30 | 45 |
| Total | 24 | 6 | 21 | 45 | 96 |
Cell lines
Six NB cell lines (SK-N-AS, SK-N-DZ, SK-N-SH, GOTO,
GANB and TGW) were used for invasion assays. SK-N-SH, SK-N-DZ and
SK-N-AS were kindly provided by Toru Sugimoto, Kyoto Prefectural
Medical University. TGW and GANB were provided by Chiba Cancer
Center. GOTO was purchased from American Type Culture Collection
(Manassas, VA, USA). These were maintained in Daigo’s medium
supplemented with 10% fetal bovine serum (BioWest, Nuaille, France)
at 37°C in a humidified 5% CO2 atmosphere.
RNA extraction and cDNA
transcription
Total-RNA was prepared from frozen tumor tissue by
the guanidine isothiocyanate-phenol method using Isogen (Wako
Junyaku Kogyo, Tokyo, Japan) according to the manufacturer’s
instructions. One microgram of each RNA was reverse transcribed to
cDNA with random hexamer primers and transcriptor reverse
transcriptase using the Transcriptor First Strand cDNA Synthesis
Kit (Roche, USA).
EMT assay
To examine the expression levels of the EMT-related
genes, we used an RT2 Profiler PCR Array for human EMT
(SA Biosciences) consisting of quantitative RT-PCR of 84
EMT-related genes. This array coated 96-well microtiter plates and
was performed using an ABI Prism 7700 Sequence Detection System
(Applied Biosystems, Foster City, CA, USA) according to the
following program: 95°C for 10 min, 43 cycles at 95°C for 15 sec
and then 60°C for 1 min.
Real-time quantitative RT-PCR
The expression levels of cardesmon 1 (CALD1),
epidermal growth factor (EGFR), desmoplakin (DSP),
secreted protein acidic and rich in cysteine (SPARC), zinc
finger E-box-binding homeobox 1 (ZEB1), zinc finger
E-box-binding homeobox 2 (ZEB2), fibronectin 1 (FN1),
vimentin (VIM), keratin 19 (KRT19), erythroblastic
leukemia viral oncogene homolog (ERBB3), regulator of
G-protein signaling 2 (RGS2), transcription factor 3
(TCF3) and TWIST1 were measured by the ABI Prism 7700
Sequence Detection System (Applied Biosystems) using Universal
ProbeLibrary (UPL)-based real-time quantitative RT-PCR (Roche
Diagnostics). UPL is based on only 165 short hydrolysis probes of
just 8–9 nucleotides, each of which is labeled at the 5′ end with
FAM and at the 3′ end with a dark quencher dye. Human ACTNB
(β-actin) was used as an internal control gene. The specific
primers used are shown in Table
II. The UPL probes used were nos. 52, 69, 78, 7, 77, 3, 68, 13,
33, 71, 37, 61, 35, 6 and 64 in UPL for CALD1, EGFR, DSP,
SNAIL2, SPARC, ZEB1, ZEB2, VIM, FN1, KRT19, ERBB3, RGS2, TCF3,
TWIST1 and human ACTNB, respectively. Each experiment
was carried out with each sample in triplicate and repeated twice.
The thermal cycling conditions were as follows: 50°C for 2 min,
95°C for 10 min, 40 cycles at 95°C for 15 sec and then 60°C for 1
min. Data from real-time PCR were calculated using the
ΔΔCt method as previously described (8).
 | Table II.Sequences of the primers used for
PCR. |
Table II.
Sequences of the primers used for
PCR.
| Gene | Forward primer | Reverse primer |
|---|
| CALD1 |
5′-GAGCGTCGCAGAGAACTTAGA-3′ |
5′-TCCTCTGGTAGGCGATTCTTT-3′ |
| EGFR |
5′-GCCTTGACTGAGGACAGCA-3′ |
5′-TTTGGGAACGGACTGGTTTA-3′ |
| DSP |
5′-CTTTGCGCCAATTCAATTAAG-3′ |
5′-CCAGTCCTGAGGTGTATGAGG-3′ |
| SNAIL2 |
5′-TGGTTGCTTCAAGGACACAT-3′ |
5′-GTTGCAGTGAGGGCAAGAA-3′ |
| SPARC |
5′-GTGCAGAGGAAACCGAAGAG-3′ |
5′-TGTTTGCAGTGGTGGTTCTG-3′ |
| ZEB1 |
5′-GGGAGGAGCAGTGAAAGAGA-3′ |
5′-TTTCTTGCCCTTCCTTTCTG-3′ |
| ZEB2 |
5′-AAGCCAGGGACAGATCAGC-3′ |
5′-CCACACTCTGTGCATTTGAACT-3′ |
| VIM |
5′-TACAGGAAGCTGCTGGAAGG-3′ |
5′-ACCAGAGGGAGTGAATCCAG-3′ |
| FN1 |
5′-GGAAAGTGTCCCTATCTCTGATACC-3′ |
5′-AATGTTGGTGAATCGCAGGT-3′ |
| KRT19 |
5′-GCCACTACTACACGACCATCC-3′ |
5′-CAAACTTGGTTCGGAAGTCAT-3′ |
| ERBB3 |
5′-CTGATCACCGGCCTCAAT-3′ |
5′-GGAAGACATTGAGCTTCTCTGG-3′ |
| RGS2 |
5′-GAAAAGGAAGCTCCAAAAGAGA-3′ |
5′-TTCTGGGCAGTTGTAAAGCA-3′ |
| TCF3 |
5′-CTCGGTCATCCTGAACTTGG-3′ |
5′-TCTCCAACCACACCTGACAC-3′ |
| TWIST1 |
5′-AAGGCATCACTATGGACTTTCTCT-3′ |
5′-GCCAGTTTGATCCCAGTATTTT-3′ |
| ACTNB |
5′-CCAACCGCGAGAAGATGA-3′ |
5′-CCAGAGGCGTACAGGGATAG-3′ |
Matrigel invasion assay
The invasive ability of NB cell lines was measured
using BD Falcon cell culture inserts with an 8-μm pore size
PET membrane and 24-well BD BioCoat Matrigel Invasion Chambers (BD
Biosciences, Bedford, MA, USA) according to the manufacturer’s
instructions. NB cell suspensions were adjusted to
1.0×105 cells per well on Matrigel invasion chamber
plates and non-matrigel coat invasion chamber (control inserts) and
cultured in routine medium in the absence or presence of FBS. After
incubation at 37°C under 5% CO2 for 72 h, the cells that
had invaded the chamber and migrated to the lower surface were
stained with Diff-Quik (Sysmex, Kobe, Japan) and manually counted
under a microscope. The invading cells were stained and counted in
5 random fields at ×100 magnification. The mean number of counted
cells was defined as the invasive ability. Each experiment was
repeated 3 times.
Statistical analysis
Survival analysis was performed according to the
Kaplan-Meier method and the log-rank test. Relative mRNA expression
levels were expressed as the mean ± SD. Student’s or Welch’s
t-tests were used to assess the significance of differences between
the groups. A p-value of <0.01 was considered statistically
significant. This study was approved by the institutional ethics
committee for human genome research of the University of Tsukuba
(no. 211).
Results
Analysis of EMT-related gene expression
in 11 NB tumors using EMT assay
Eleven NB tumors in various stages (Table III) were analyzed by EMT multiple
gene profiling microarray. The expressions of 84 EMT-related genes
were compared among the 11 tumors using the EMT assay. Seven genes
(CALD1, EGFR, DSP, SNAIL2, SPARC, ZEB1 and ZEB2) were
found to be differentially expressed between NBs with low stages
(stages 1 or 2) and those with high stages (stages 3 or 4). Five
genes (KRT19, ERBB3, RGS2, TCF3 and
TWIST1) were found to be differentially expressed between
MYCN-amplified and MYCN-non-amplified tumors. These
genes and two others highly expressed in NB tumors (VIM and
FN1) were further analyzed in 96 tumors using quantitative
PCR.
 | Table III.Characteristics of 11 neuroblastomas
used in EMT assay. |
Table III.
Characteristics of 11 neuroblastomas
used in EMT assay.
| Stage 1, 2 | Stage 4S | Stage 3, 4 | Total |
|---|
| MYCN | | | | |
| Unamplified | 1 | 1 | 1 | 3 |
| Amplified | 3 | 2 | 3 | 8 |
| Total | 4 | 3 | 4 | 11 |
Correlation of EMT-related gene
expression between low and high tumor stages
The expression levels of ERBB3, RGS2, TCF3,
CALD1, EGFR, DSP, SNAIL2, SPARC, ZEB1, ZEB2, VIM and FN1
did not show any significant differences between low- and
high-stage NB. In contrast, low expression of KRT19 was
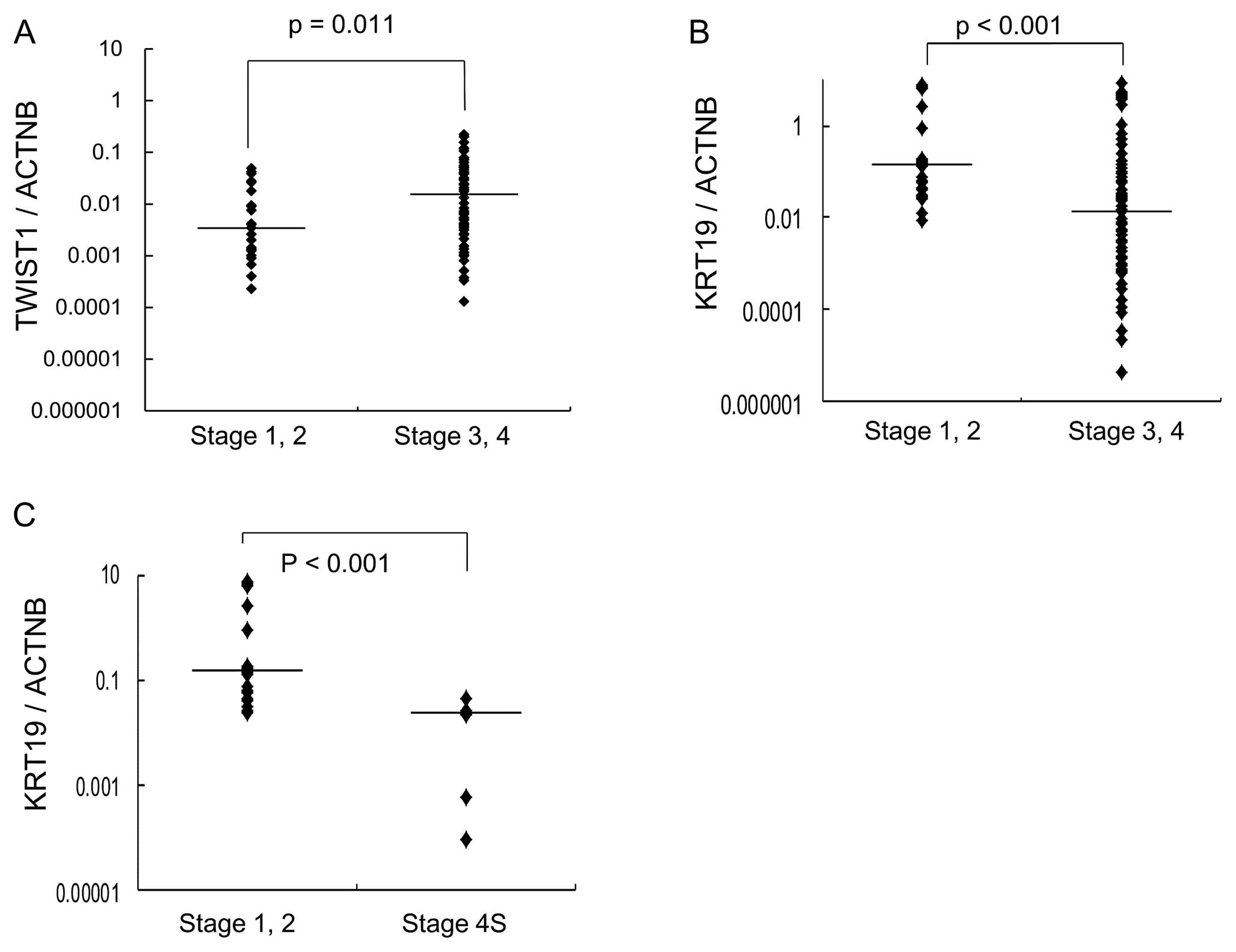
significantly associated with high stages of NB (Fig. 1B). TWIST1 was found to be
highly expressed in stage 3 or 4 NB (p=0.011) (Fig. 1A).
Correlation of EMT-related gene
expression with metastasis in localized primary tumors
Expression of these EMT-related genes was compared
between stage 1 or 2 localized NB and stage 4S NB. Expression of
KRT19 was significantly lower in stage 4S NB, which develops
metastasis in localized primary NB (Fig. 1C).
Correlation of EMT-related gene
expression with MYCN amplification
Expression of these EMT-related genes was compared
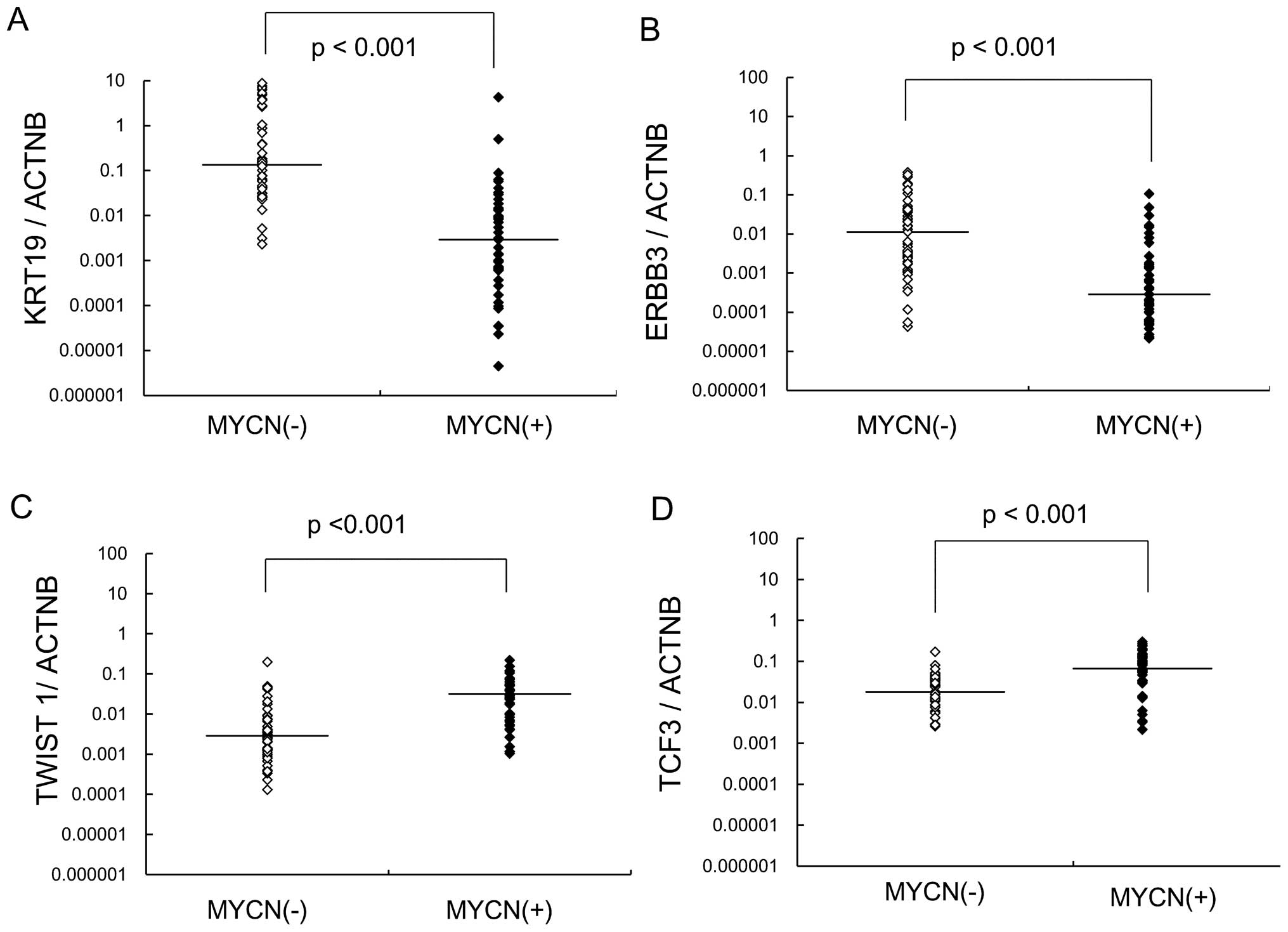
between NB with and without MYCN amplification. Expression
of KRT19 and ERBB3 was significantly decreased in NB
with MYCN amplification, while TCF3 and TWIST1
expression were increased (Fig.
2). MYCN-amplified NB showed significantly lower
expression of KRT19 and ERBB3 compared with
MYCN-unamplified NB.
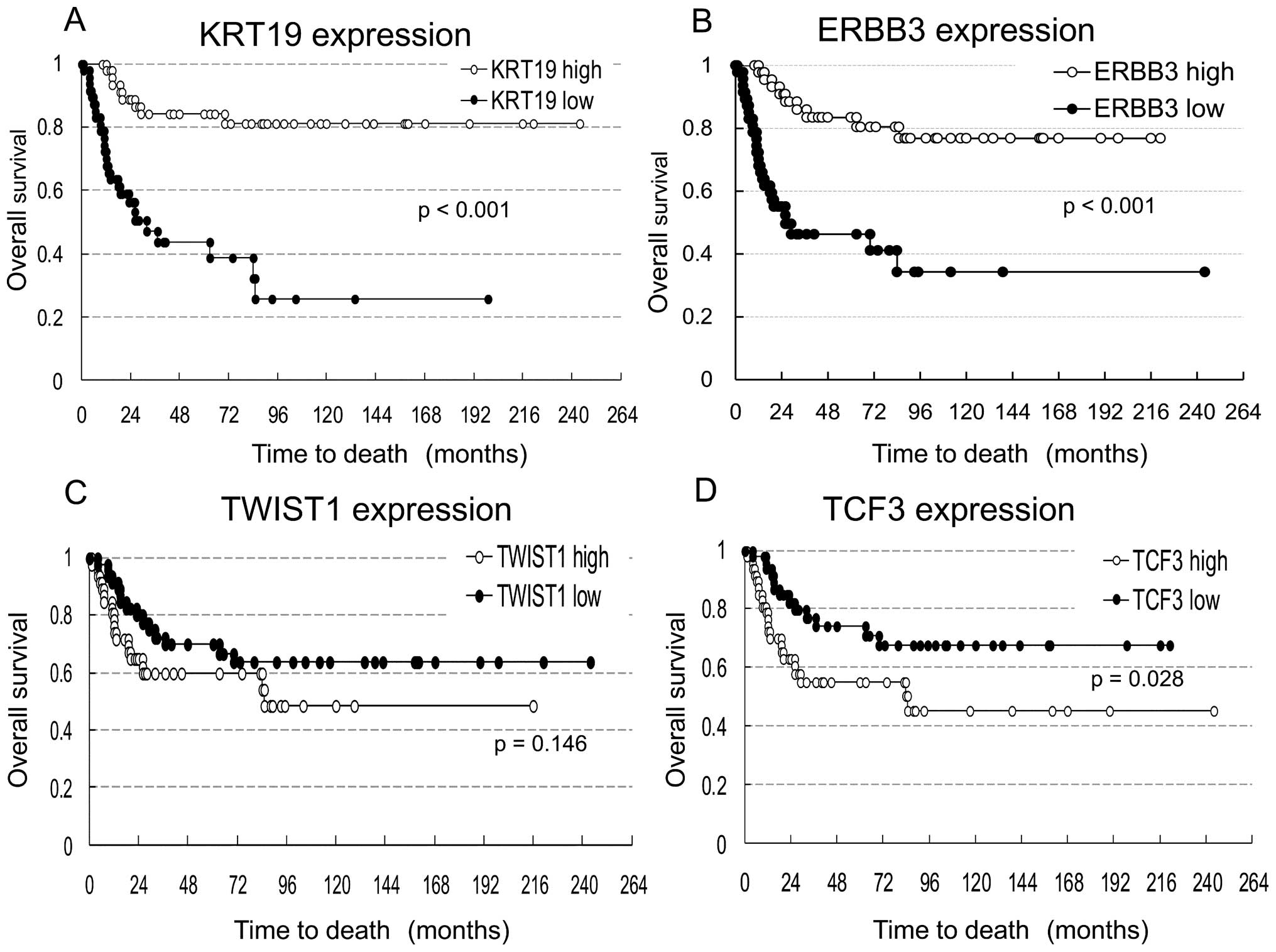
Overall survival rates for tumors with
VIM, FN1, KRT19, ERBB3, TCF3 and TWIST1 gene misregulation
Survival analysis was conducted in 94 NB tumors
excluding 2 in which NB was not the cause of death. These NBs were
divided into two groups: high expressers (47 NBs) and low
expressers (47 NBs) of 6 genes (VIM, FN1, KRT19, ERBB3, TCF3
and TWIST1). The median of log-transformed mRNA expression
level was used as the cut-off value. Kaplan-Meier survival curves
were compared for each gene between tumors with high and low
expression (Fig. 3). The graph
shows a trend toward increased survival for NB patients with
increased KRT19 or ERBB3 expression. Expression
levels of the other genes (VIM, FN1, TCF3 and
TWIST1) were not associated with patient survival.
The correlation of low KRT19 and ERBB3
expression with invasive ability in NB cell lines
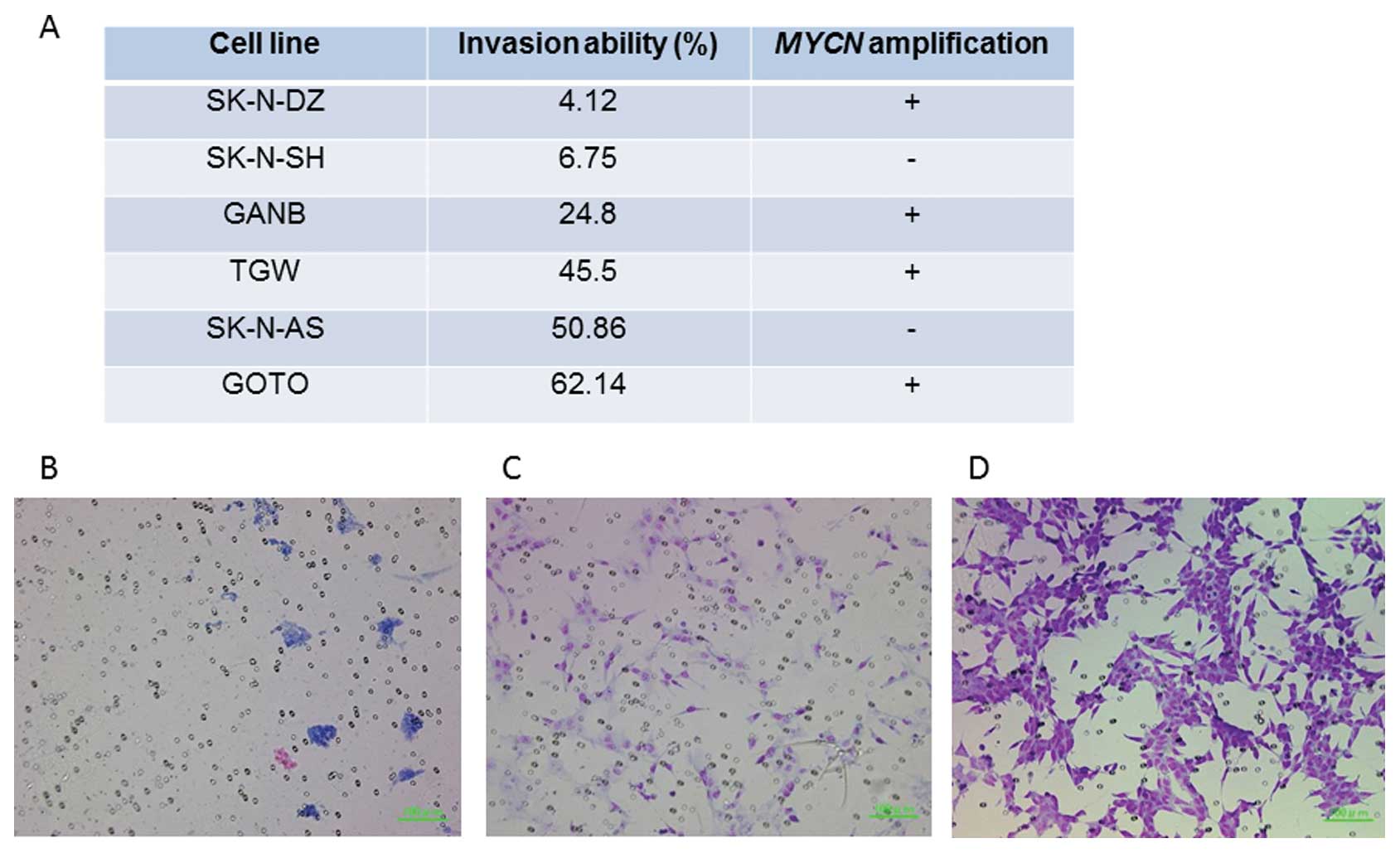
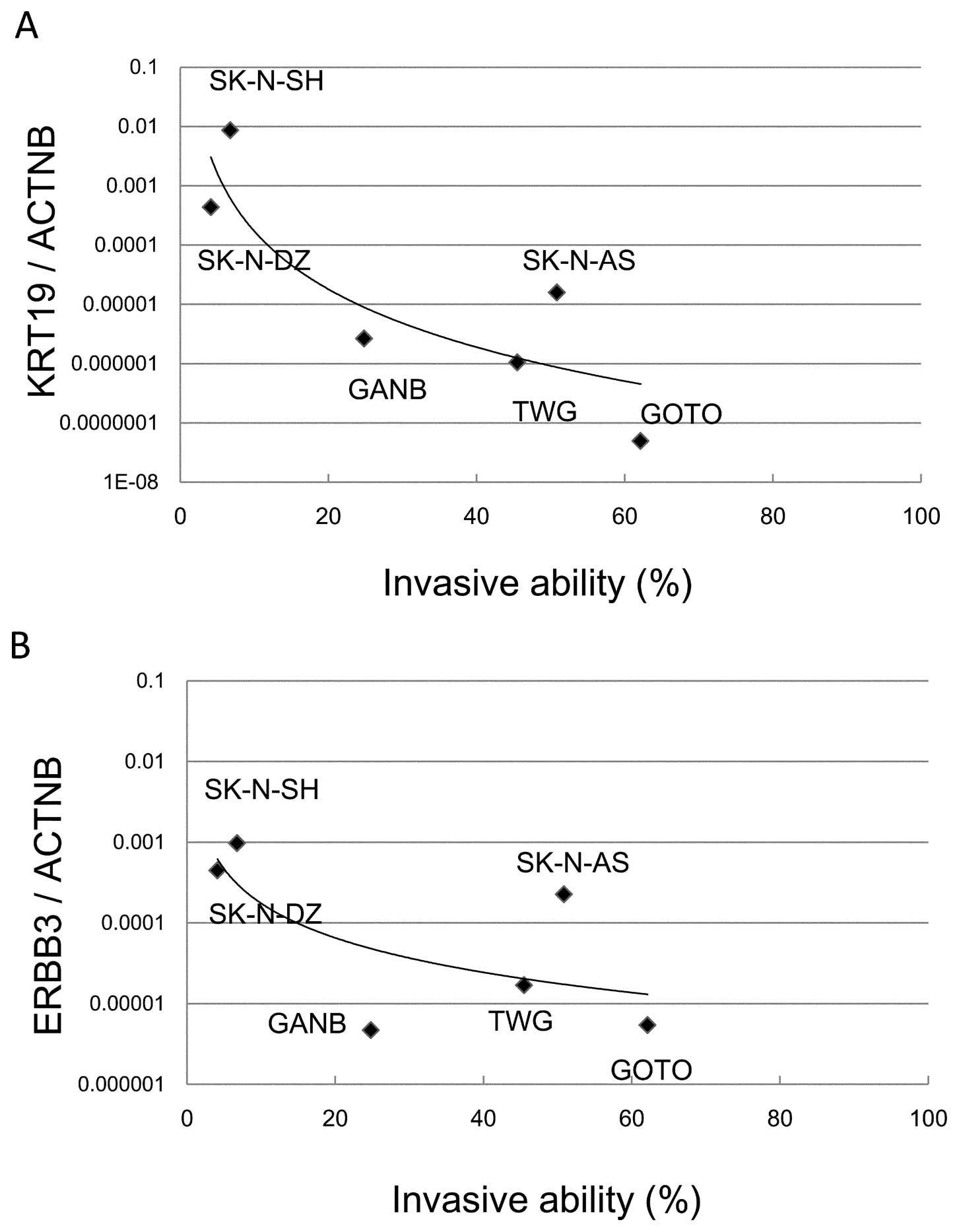
A Matrigel invasion assay demonstrated that two cell
lines (SK-N-SH and SK-N-DZ) showed significantly reduced invasive
ability (6.75 and 4.12%, respectively) while the 4 other cell lines
(GANB, TGW, SK-N-AS and GOTO) showed high invasive abilities (24.8,
45.5, 50.86 and 62.1%, respectively) (Fig. 4). The correlation of KRT19
and ERBB3 expression with invasive abilities was
investigated in the cell lines. The decreased expression of
KRT19 or ERBB3 was highly correlated with
invasiveness in NB cell lines (Fig. 5A
and B). SK-N-DZ, with high expression of KRT19 and
ERBB3 and MYCN amplification, had low invasive
ability; while SK-N-AS, with low expression of KRT19 and
MYCN non-amplification, showed high invasive ability.
Discussion
In this study, four EMT-related genes (KRT19,
ERBB3, TWIST1 and TCF3) were found to be
differentially expressed. Expression of KRT19 was
significantly decreased in high-stage NB compared to low-stage NB
(Fig. 1B). Downregulation of
KRT19 gene expression was highly associated with tumor
progression in NB. Furthermore, expression of KRT19 was
markedly decreased in NB with MYCN amplification. Decreased
expression of KRT19 was found to be significantly associated
with poor prognosis (Fig. 3).
Interestingly, expression of KRT19 was significantly
decreased in metastatic favorable stage 4S NB compared to localized
favorable stage 1 or 2 NB (Fig.
1C). These findings show that decreased expression of
KRT19 is strongly associated with the promotion of
metastasis in favorable NB. Keratin is an epithelial marker, and
downregulation of keratins is associated with EMT. Dysregulation of
keratin expression has long been recognized as a feature of
epithelial tumor progression (9).
A recent report also demonstrated that expression of KRT19
mRNA was significantly lower in tumors from patients that have died
from NB compared with patients with no evidence of disease, and
that low methylation of KRT19 was associated with a
favorable outcome (10).
Supporting this, our results demonstrated that low expression of
KRT19 was significantly associated with high tumor stages,
MYCN amplification and an unfavorable outcome in NB.
TWIST1 is a key regulator of embryogenesis
and is also known to be an EMT inducer. TWIST1 belongs to
the basic helix-loop-helix (bHLH) transcription factor family and
promotes EMT by repressing the expression of E-cadherin, which
leads to disassembly of adherens junctions and increased migratory
potential (11). The link between
TWIST1 expression and metastasis is clear and well
established (11,12). TWIST1 is known to be
overexpressed in MYCN-amplified NB tumors and cell lines and
is responsible for the inhibition of the ARF/p53 pathway involved
in the MYC-dependent apoptotic response. The cooperation of
TWIST1 and MYCN is thought to cause cell
transformation and malignant outgrowth (13,14).
In this study, TWIST1 was highly expressed in
MYCN-amplified NB as well as in high-stage NB. However, the
survival rates between patients with low and high expression of
TWIST1 were not significantly different (p= 0.146), so its
utility as a mesenchymal marker may be limited.
TCF3 (E12/E47) is a basic bHLH transcription
factor. A previous study implicated TCF3 as a repressor of
E-cadherin promoter activity and demonstrated its involvement in
the acquisition and maintenance of the mesenchymal phenotype
(15). In this study, high
expression of TCF3 was associated with MYCN
amplification in NB. Survival rates were not significantly
different between patients with high and low expression of
TCF3.
ERBB3 is a member of the epidermal growth
factor receptor (EGFR) family, which is composed of EGFR,
ERBB2 (HER2), ERBB3 (HER3) and
ERBB4 (HER4). Although ERBB3 lacks an active
tyrosine kinase domain, it can heterodimerize with other
ERBB receptors. Heterodimerization leads to the activation
of pathways which lead to cell proliferation or differentiation.
The role of EGFR in the proliferation of NB, and the utility of its
inhibitors in the treatment of NB, have all been well documented;
however, the data remain somewhat contradictory (16,17),
as other reports have demonstrated that exposure to EGF can induce
apoptosis in NB through the ERBB2 and ERBB3 receptors
(18–20). Richards et al reported that
non-EGFR ERBB family members (ERBB2, ERBB3 and
ERBB4) contributed to NB growth and survival, and that
pan-ERBB inhibition, rather than an EGFR specific inhibitor,
represents a potential therapeutic target (21). These findings suggest that
ERBB2, ERBB3 and ERBB4 play a significant role
in tumor progression of NB, but Gambini et al reported that
expression of ERBB2 was not related to tumor progression of
NB (22). Although a recent
immunohistological study suggested the significance of EGFR family
expression as a prognostic factor in NB, showing that EGFR
and HER2 expression is found in favorable NB and high
expression of HER4 is found in metastatic NB, the role of
HER family members in NB remains interrelated and complex (23). In our study, decreased expression
of ERBB3 was also correlated with MYCN-amplified NB
and poor survival rate. Several lines of evidence that provide
support for the pivotal role of ERBB3 in human
carcinogenesis have emerged in recent years (24). High expression of ERBB3 in
certain human cancers led early to the suggestion that it could be
a therapeutic target (25–28), but in some cancer cells the
mesenchymal phenotype was found to lose ERBB3 expression and
show resistance to EGFR inhibitors. Epithelial phenotype, however,
maintained ERBB3 expression (29,30).
The EMT might decrease the cellular dependency upon EGF signaling
by kinase switching; mesenchymal cells might acquire alternative
survival signals, thus becoming resistant to EGFR inhibitors
(30). Downregulation of
ERBB3 in NB might suggest similar kinase switching during
the EMT followed by tumor survival with the loss of EGF
dependency.
Next, we investigated the invasive abilities of six
NB cell lines using a Matrigel invasion assay to confirm the
association of tumor invasiveness with expression of KRT19
and ERBB3. While SK-N-DZ and SK-N-SH cell lines had a low
invasive ability (4.12 and 6.75%, respectively), the other cell
lines showed a high invasive ability (24.8–62.14%) (Fig. 4). Both cell lines with a low
invasive ability had low expression of KRT19 and
ERBB3 compared with the other cell lines (Fig. 5A and B). Interestingly, SK-N-DZ
showed a low invasive ability as expected from high expression
levels of KRT19 and ERBB3, although its MYCN
amplification should give it a high invasive ability. Thus,
although MYCN gene amplification is the most powerful
prognostic factor in NB, the expression levels of KRT19 or
ERBB3 might become another promising prognostic marker.
Acknowledgements
This study was supported by a
Grant-in-Aid for Challenging Exploratory Research from the Japan
Society for the Promotion of Science (JSPS) Grant 22659317 (to
HK).
References
|
1.
|
Brodeur GM: Neuroblastoma: biological
insights into a clinical enigma. Nat Rev Cancer. 3:203–216. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Maris JM: Recent advances in
neuroblastoma. N Engl J Med. 362:2202–2211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Maris JM: The biologic basis for
neuroblastoma heterogeneity and risk stratification. Curr Opin
Pediatr. 17:7–13. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: at the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: new insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Moll R, Franke WW, Schiller DL, Geiger B
and Krepler R: The catalog of human cytokeratins: patterns of
expression in normal epithelia, tumors and cultured cells. Cell.
31:11–24. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Carén H, Djos A, Nethander M, Sjöberg RM,
Kogner P, Enström C, Nilsson S and Martinsson T: Identification of
epigenetically regulated genes that predict patient outcome in
neuroblastoma. BMC Cancer. 11:662011.PubMed/NCBI
|
|
11.
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Karreth F and Tuveson DA: Twist induces an
epithelial-mesenchymal transition to facilitate tumor metastasis.
Cancer Biol Ther. 3:1058–1059. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Valsesia-Wittmann S, Magdeleine M,
Dupasquier S, Garin E, Jallas AC, Combaret V, Krause A, Leissner P
and Puisieux A: Oncogenic cooperation between H-Twist and N-Myc
overrides failsafe programs in cancer cells. Cancer Cell.
6:625–630. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Puisieux A, Valsesia-Wittmann S and
Ansieau S: A twist for survival and cancer progression. Br J
Cancer. 94:13–17. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Perez-Moreno MA, Locascio A, Rodrigo I,
Dhondt G, Portillo F, Nieto MA and Cano A: A new role for E12/E47
in the repression of E-cadherin expression and
epithelial-mesenchymal transitions. J Biol Chem. 276:27424–27431.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Ho R, Minturn JE, Hishiki T, Zhao H, Wang
Q, Cnaan A, Maris J, Evans AE and Brodeur GM: Proliferation of
human neuroblastomas mediated by the epidermal growth factor
receptor. Cancer Res. 65:9868–9875. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Tamura S, Hosoi H, Kuwahara Y, Kikuchi K,
Otabe O, Izumi M, Tsuchiya K, Iehara T, Gotoh T and Sugimoto T:
Induction of apoptosis by an inhibitor of EGFR in neuroblastoma
cells. Biochem Biophys Res Commun. 358:226–232. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Chiu B, Mirkin B and Madonna MB: Epidermal
growth factor can induce apoptosis in neuroblastoma. J Pediatr
Surg. 42:482–488. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Chiu B, Mirkin B and Madonna MB: Mitogenic
and apoptotic actions of epidermal growth factor on neuroblastoma
cells are concentration-dependent. J Surg Res. 135:209–212. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Chiu B, Mirkin B and Madonna MB: Novel
action of epidermal growth factor on caspase 3 and its potential as
a chemotherapeutic adjunct for neuroblastoma. J Pediatr Surg.
42:1389–1395. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Richards KN, Zweidler-McKay PA, Van Roy N,
Speleman F, Trevino J, Zage PE and Hughes DP: Signaling of ERBB
receptor tyrosine kinases promotes neuroblastoma growth in vitro
and in vivo. Cancer. 116:3233–3243. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Gambini C, Sementa AR, Boni L, Marino CE,
Croce M, Negri F, Pistoia V, Ferrini S and Corrias MV: Expression
of HER2/neu is uncommon in human neuroblastic tumors and is
unrelated to tumor progression. Cancer Immunol Immunother.
52:116–120. 2003.PubMed/NCBI
|
|
23.
|
Izycka-Swieszewska E, Wozniak A, Drozynska
E, Kot J, Grajkowska W, Klepacka T, Perek D, Koltan S, Bien E and
Limon J: Expression and significance of HER family receptors in
neuroblastic tumors. Clin Exp Metastasis. 28:271–282. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Sithanandam G and Anderson LM: The ERBB3
receptor in cancer and cancer gene therapy. Cancer Gene Therapy.
15:413–448. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Gullick WJ: The c-erbB3/HER3 receptor in
human cancer. Cancer Surv. 27:339–349. 1996.PubMed/NCBI
|
|
26.
|
Travis A, Pinder SE, Robertson JF, Bell
JA, Wencyk P, Gullick WJ, Nicholson RI, Poller DN, Blamey RW,
Elston CW and Ellis IO: C-erbB-3 in human breast carcinoma:
expression and relation to prognosis and established prognostic
indicators. Br J Cancer. 74:229–233. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Sergina NV, Rausch M, Wang D, Blair J,
Hann B, Shokat KM and Moasser MM: Escape from HER-family tyrosine
kinase inhibitor therapy by the kinase-inactive HER3. Nature.
445:437–441. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Baselga J and Swain SM: Novel anticancer
targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer.
9:463–475. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Fuchs BC, Fujii T, Dorfman JD, Goodwin JM,
Zhu AX, Lanuti M and Tanabe KK: Epithelial-to-mesenchymal
transition and integrin-linked kinase mediate sensitivity to
epidermal growth factor receptor inhibition in human hepatoma
cells. Cancer Res. 68:2391–2399. 2008. View Article : Google Scholar
|
|
30.
|
Thomson S, Petti F, Sujka-Kwok I, Epstein
D and Haley JD: Kinase switching in mesenchymal-like non-small cell
lung cancer lines contributes to EGFR inhibitor resistance through
pathway redundancy. Clin Exp Metastasis. 25:843–854. 2008.
View Article : Google Scholar : PubMed/NCBI
|