Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed cancer in males and the second in females, with over 1.2
million new cancer cases and 608,700 deaths estimated to have
occurred in 2008 (1). In China,
CRC remains the fifth most common cancer type and the fourth most
common cause of cancer-related death (2). Moreover, the incidence of CRC is
increasing rapidly in recent years in China (3). The development of CRC from normal
epithelial cells to malignant carcinomas involves a multi-step
process with accumulation of both genetic and epigenetic changes,
leading to a temporal activation of oncogenes and inactivation of
tumor suppressor genes (4).
Despite research and resources dedicated to elucidating the
molecular mechanisms of CRC, the molecular pathogenesis of CRC is
complicated and remains unclear.
MiRNAs are a class of small non-coding 18- to
25-nucleotide-long RNAs that negatively regulate gene expression by
binding to the 3′-untranslated region (3′-UTR) of target messenger
(m) RNAs; this causes translational repression or degradation
(5). Up to October 2011, more than
1,400 human microRNAs have been reported (6,7).
Studies have demonstrated that microRNAs play a crucial role in
almost all cellular biological processes including development,
metabolism, survival, differentiation, proliferation, apoptosis and
the immune response (8–12). It may function as both oncomiR
(oncogene-like miRNA) and tumor suppressors (anti-oncomiR)
depending on the miRNAs and the tumor type (13–15).
MiRNAs have been demonstrated to play an important
role in the multistep processes of carcinogenesis. Specifically,
miR-21 is consistently overexpressed in diverse types of
malignancy, including breast cancer (16–18),
pancreatic cancer (16,19,20),
cholangiocarcinoma (21),
hepatocellular carcinoma (22),
gastric cancer (16), esophagus
cancer (23), colorectal cancer
(16,24), brain tumor (25), cervical cancer (26), ovarian cancer (27), prostate cancer (16), lung cancer (16), leukemia (28,29)
and osteosarcoma (30). The human
miR-21 gene is located on chromosome 17q23-1 overlapping with the
TMEM49 gene, a human homologue of rat Vacuole Membrane Protein 1
(VMP-1), and it has been shown to be implicated in multiple
malignancy-related processes including cell proliferation,
apoptosis, invasion and metastasis (16–44).
According to recent reports, several significant miR-21 targets
associated with malignancy have been described: phosphatase and
tensin homologue (PTEN) (21,22,31,32),
programmed cell death 4 (PDCD4) (23,33,34),
reversion-inducing cysteine-rich protein (RECK) (35,36),
maspin (37), tropomyosin 1 (TPM1)
(38), heterogeneous nuclear
ribonucleoprotein K (HNRPK) (39),
and TAp63 (39), Sprouty (SPRY1
and 2) (40,41), ras homolog gene family member B
(RhoB) (42), tissue inhibitor of
metalloproteinase 3 (TIMP3) (34,35,43,44).
However, the expression of miR-21 and its target
gene PTEN, as well as their relationship has not been established
in CRC. The role and relevant pathway of miR-21 in carcinogenesis
and development of CRC and whether its antagonism could be used as
a potential treatment for CRC remains illusive. In this study, we
analyzed the expression levels of miR-21 in samples of CRC tissues
and correlated them with clinicopathological features of CRC.
Subsequently, we used anti-miR-21 inhibitor (IN) anti-sense
oligonucleotide (ASO) to transiently knockdown miR-21 in HCT116
colon cancer cells in order to evaluate the role of this miRNA in
cell proliferation, apoptosis, cell cycle, invasion and migration,
and investigated the expression of PTEN, its target gene, as well
as its regulatory mechanism.
Materials and methods
Patients and tissue samples
CRC tissues were obtained from the 30 consecutive
patients who underwent primary surgical resection of CRC between
September 2008 and March 2009 at the First Affiliated Hospital of
Chongqing Medical University (Chongqing, China). No local or
systemic treatment was conducted in these patients before the
operation. A total of 10 adenomas as benign lesions with moderate
dysplasia were resected by endoscopic mucosal resection (EMR). Then
10 specimens of normal colorectal mucosal tissues were evaluated as
normal controls. Tissue samples were immediately frozen in liquid
nitrogen after resection and stored at −80°C until use. One section
of each sample was stained with hematoxylin and eosin (H&E) and
was used for histopathological evaluation. The study was approved
by the Research Ethics Committee of Chongqing Medical University,
China. Informed consent was obtained from all patients.
Cell culture
Human colorectal cancer cell lines Colo320, Lovo,
HCT116, HT29 and SW480 cell lines were purchased from Shanghai
Institute for Biochemistry, Chinese Academy of Sciences (Shanghai,
China). All cell lines were cultured in RPMI-1640 supplemented with
10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100
μg/ml streptomycin in humidified 5% CO2 at 37°C.
Trypsin (0.25%) solution was used to detach the cells from the
culture flask.
Oligonucleotide transfection
Anti-miR-21 inhibitor (5′-UCAA CAUCAGUCUGA UAA
GCUA-3′) and mismatched sequence negative control oligonucleotide
(5′-CAGUACUUUUGUGUA GUACAA-3′) were synthesized by GenePharma,
Shanghai, transfected into HCT116 cells (200 nmol/well) using
Oligofectamine reagent (Invitrogen, Carlsbad, CA, USA). RNA
oligonucleotides were transfected into cells using Lipofectamine
2000 (Invitrogen) according to the manufacturer’s protocol.
Transfection efficiency was evaluated by GFP expression in control
vector or real-time PCR. Medium was replaced 8 h later and cells
were collected for the next experiments 48 h post-transfection. The
transfection was performed in triplicates.
Plasmid construct
The 3′-UTR of the human PTEN gene was PCR amplified
from human genomic DNA (5′-GCTCTAGATC CCGCCCAAGCATGAAC-3′ and
5′-GCTCTAGACCATTTT ATAAATGTCATCATC-3′), and cloned into the
Xba1-site of pGL3-control vector (Promega, Madison, WI, USA), which
is designated pGL3-PTEN-wt after sequencing. Site-directed
mutagenesis of the miR-21 target-site in the PTEN 3′-UTR was
carried out using site-directed mutagenesis kit (Takara, Dalian,
China), with pGL3-PTEN-wt as a template, and named pGL3-PTEN-mut
(FW, 5′-TTCTCGCGATGATGTATACAGT TTTTTATG-3′, RV,
5′-CTTTTATGTAAACATCATAAG CTCA-3′).
Cell proliferation assay
Cells were seeded in 96-well plate at 4,000 cells
per well the day before transfection. The HCT116 cells were
transfected with miR-21 negative control (NC) and anti-miR-21
inhibitor (IN). Cell Counting Kit 8 (CCK8, Dojindo, Tokyo, Japan)
assay was used to measure the viable, proliferating cells at 24,
48, 72 and 96 h after transfection. The absorbance at 450 nm was
measured using a thermo spectrophotometer. Each cell group was
measured in triplicate.
Luciferase assay
Cells were seeded in 24-well plates 24 h before
transfection. HCT-116 cells were transiently transfected with
wild-type (pGL3-PTEN-wt) or mutant (pGL3-PTEN-mut) reporter plasmid
containing miR-21 potential binding sites in the presence or
absence of miR-21 using Lipofectamine 2000. Luciferase assays were
performed 36 h post-transfection using the Dual-luciferase assay
system (Promega, Madison, WI, USA) and they were normalized for
transfection efficiency with cotransfected Renilla luciferase. All
experiments were performed in triplicate.
Matrigel invasion assay
Twenty-four hours after transfection,
2×104 HCT116 cells were suspended in 0.25 ml of culture
medium with 1% FBS and plated in the top chamber with
matrigel-coated membrane (Becton Dickinson, USA). The cells were
incubated for 36 h, after which the cells that did not invade
through the pores were removed by a cotton swab. Cells on the lower
surface of the membrane were stained with H&E for
visualization, and counted under a light microscope in 5 random
fields with magnification, ×400.
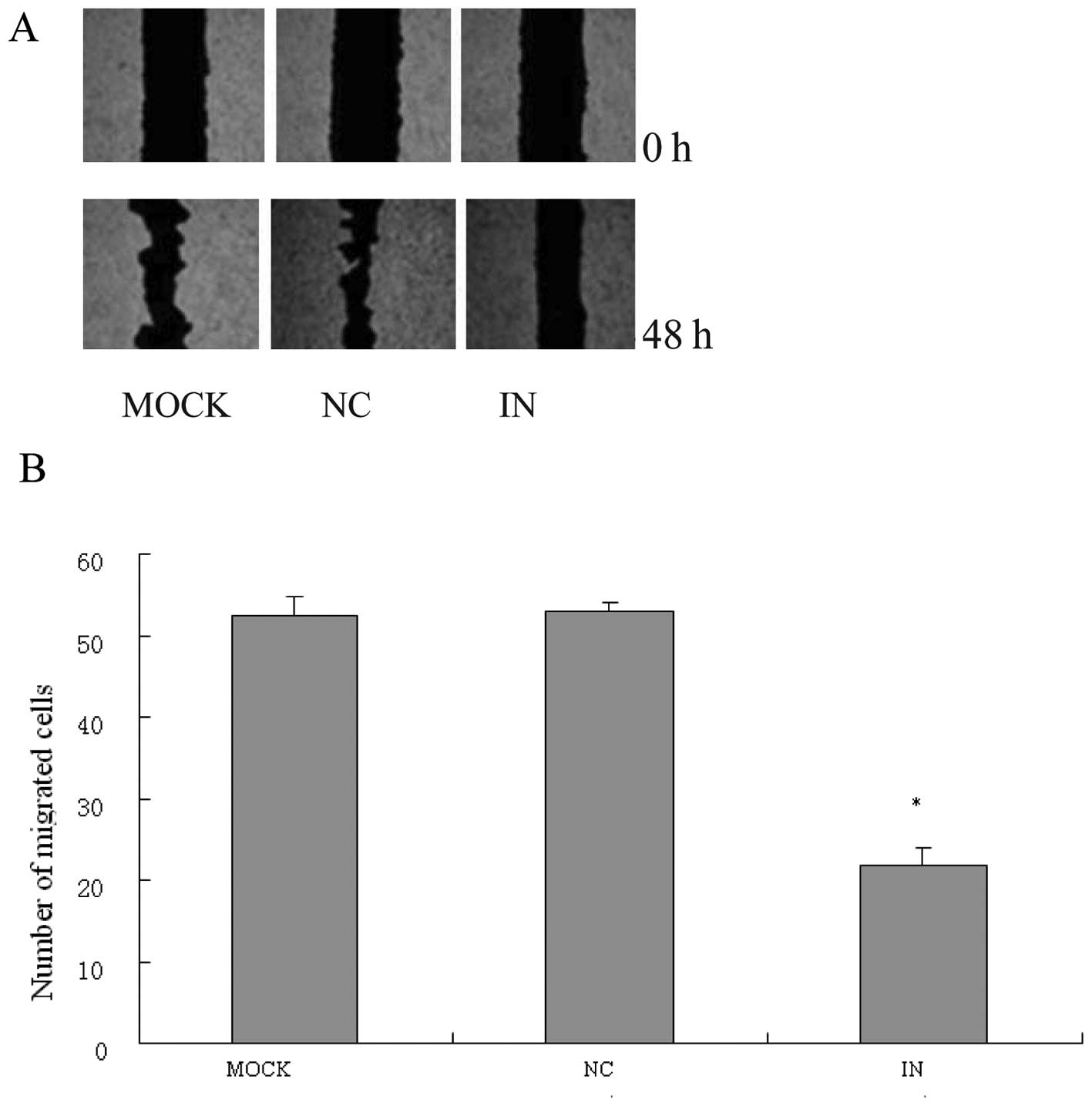
Scratch wound-healing motility assay
The scratch wound assay was performed as previously
described (36). When HCT116 cells
were seeded and grown to confluence, a scratch was set with a
pipette tip running though the dish and cultured under standard
conditions for 24 h. Plates were washed twice with fresh medium to
remove non-adherent cells and then photographed. The cell migration
was evaluated by counting cells that migrated from the wound edge.
Each cell group was measured in triplicate.
Colony formation assay
HCT116 cells were transfected with negative control
(50 nM) and anti-miR-21 inhibitor (50 nM) in 24-well plates at a
density of 5×105 cells/well. Cells were subsequently
trypsinized at 24 h and seeded in 6-well culture plates (500 cell
for each well) to form colonies. After 7 days, colonies were fixed
in 4% paraformaldehyde and stained in a 0.1% crystal violet
solution. Each cell group was measured in triplicate.
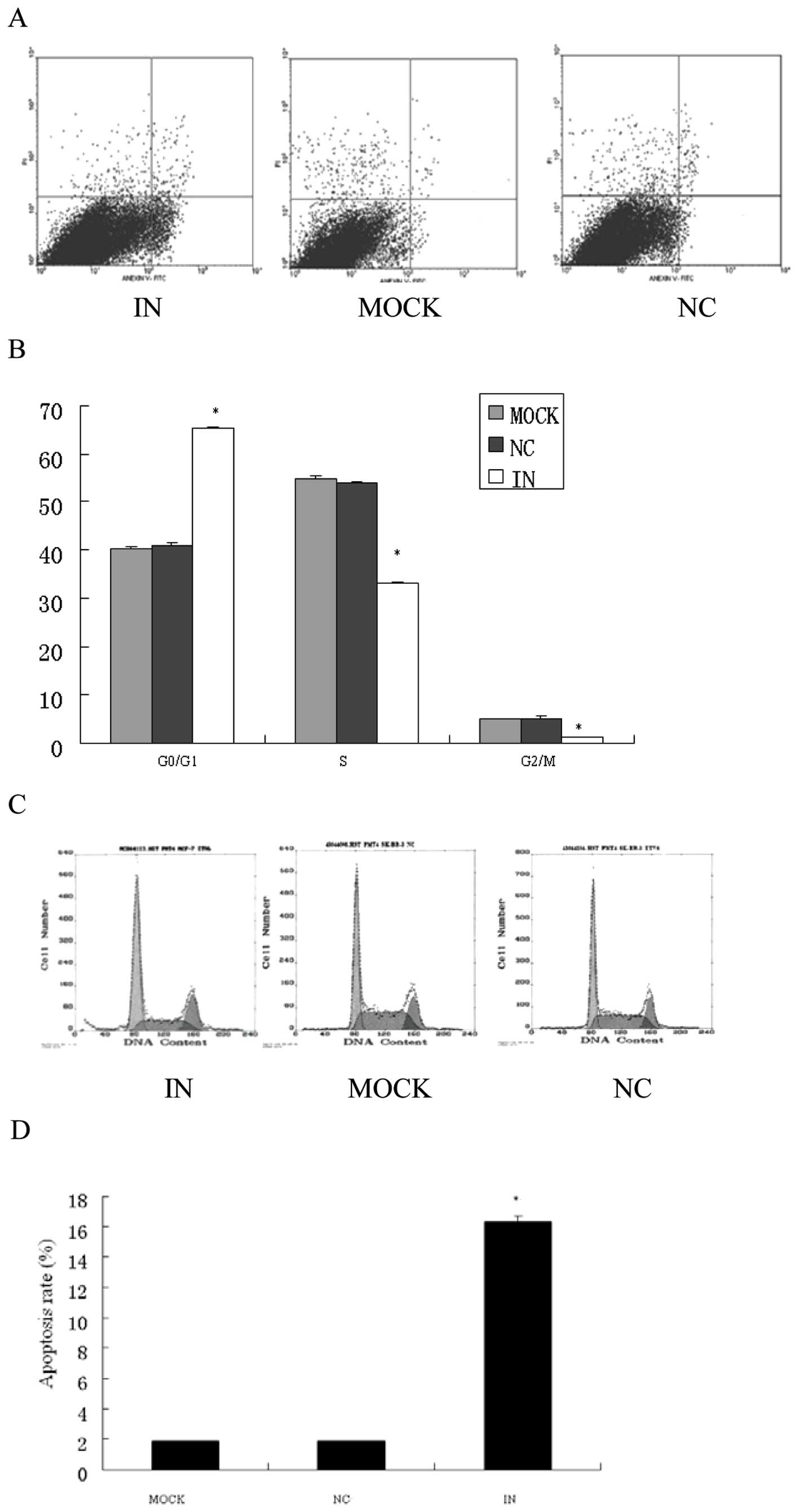
Cell cycle assay
For cell cycle analysis, parental and transfected
cells in the log phase of growth were stained with propidium iodide
(PI; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and examined
with a fluorescence-activated cell sorting (FACS) flow cytometer
(FCM) and DNA histograms were analyzed with modified software. Each
test was repeated in triplicate.
Apoptosis assay
For apoptosis assays, floating and adherent cells
were harvested 24 or 48 h after transfection, and then combined and
washed with PBS. Annexin V in combination with PI (Santa Cruz
Biotechnology) was added to the cells and samples were analyzed
within 30 min after staining. Quantification of fluorescence was
done by flow cytometry as described above. Each test was repeated
in triplicate.
RNA isolation and qRT-PCR
Total-RNA was extracted using TRIzol reagent
(Invitrogen) for both miR-21 and PTEN-mRNA analyses according to
the manufacturer’s instructions.
For detection of miR-21 expression, stem-loop RT-PCR
was performed as previously described (45). qPCR was carried out using
SYBR-Premix Ex Taq™ (Takara) according to the manufacturer’s
protocol. For detection of PTEN-mRNA expression, qPCR was performed
using Quantitect SYBR-Green PCR kit (Takara). β-actin was used to
normalize PTEN-mRNA expression levels.
Western blot analysis
Western blot analysis was performed according to
standard procedures. Total protein was isolated from HCT116 cells.
Protein concentrations were determined by BCA Protein Assay kit
(Pierce Biotechnology, Rockford, IL, USA). The membrane was first
incubated with antibody against PTEN, phosphorylated AKT (p-AKT),
PI3K p85α (Tyr458), p-mTOR and MMP-9 (Santa Cruz Biotechnology),
then with anti-β-actin antibody (Sigma, St. Louis, MO, USA) as a
loading control. Signals were detected by secondary antibodies
labeled with HRP and signal intensity was determined by Quantity
One software.
Immunohistochemistry
For immunohistochemistry, a mouse monoclonal
antibody (Zhongshan Technology, Beijing, China) were used as
primary antibodies for overnight incubation at 4°C. The sections
were subsequently treated with goat anti-mouse secondary antibody,
followed by further incubation with streptavidin-horseradish
peroxidase complex (Zhongshan Technology). Diaminobenzidine
(Zhongshan Technology) was used as a chromogen and sections were
lightly counterstained with hematoxylin. The proportion of PTEN
immunostaining tumor cells varied from 0 to 100%, and a four-grade
scoring system was used to evaluate the degree of immunostaining:
score 0, <5%; score 1, 5–25%; score 2, 25–50%; score 3, >50%
of tumor cells with positive immunostaining.
Statistical analysis
All values in the present study were reported as
mean ± standard deviation (SD) from three independent experiments.
Statistics was determined using ANOVA, χ2 test or
Student’s t-test using SPSS17.0 (Windows). The inverse correlation
of PTEN protein and miR-21 expression levels was examined by
Spearman correlation analysis. P-values <0.05 were considered
statistically significant.
Results
MiR-21 is overexpressed in CRC tissues
and clinicopathological characteristics
The results indicate that, among the 30 CRC samples
analyzed, the relative expression of miR-21
(2−ΔΔCt=4.956±1.892) was significantly upregulated
compared with the adenomas (1.662±0.496) and normal tissues
(1.024±0.043) (p<0.01, respectively). MiR-21 expression was
significantly higher in adenomas than it in the normal tissues
determined by post-hoc analyses (p<0.05). Moreover, the high
miR-21 group was significantly associated with invasion depth,
lymph node metastasis (present), poor differentiation and advanced
TNM stage (III, IV) (p<0.05, respectively). On the other hand,
no significant differences were observed regarding gender,
histological type, tumor size (p>0.05, respectively), implying
miR-21 might be involved in the development and metastasis of
cancer, and has a prognostic implication for CRC. This result is
consistent with the previous findings (46–53).
Low PTEN expression correlates with
clinicopathological variables
As shown in Table
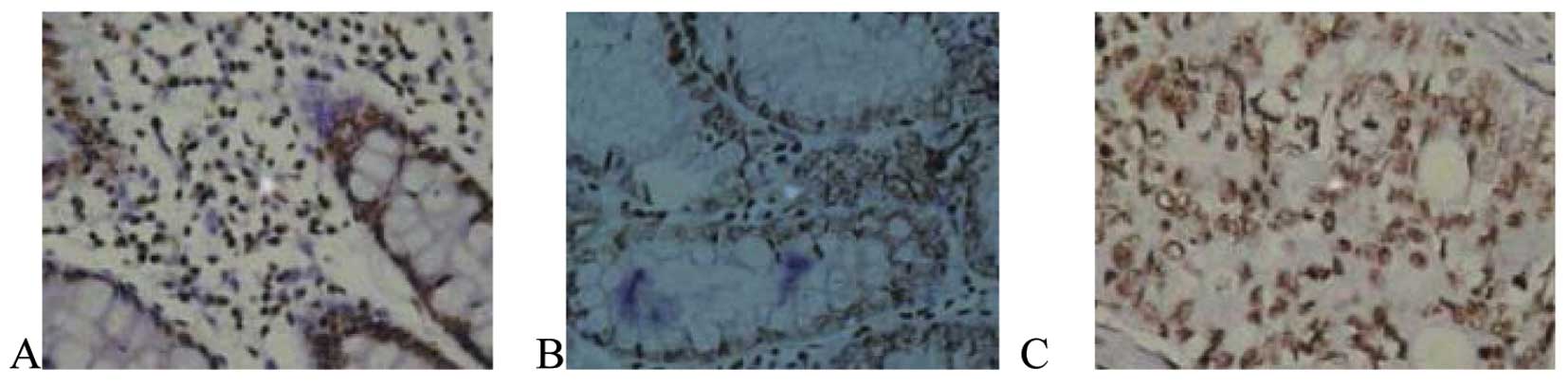
I, 14 of 30 (46.67%) CRC tissues were positive for PTEN, mainly
weakly positive and expressed in the cytoplasm (Fig. 1), which was distinctly lower than
that in adenomas and normal controls (p<0.01). Moreover, the low
PTEN was significantly associated with invasion depth, lymph node
metastasis (present), poor differentiation and advanced TNM stage
(III, IV) (p<0.05, p<0.05, p<0.05, p<0.01 and
p<0.05, respectively). On the other hand, no significant
differences were observed regarding gender, histological type
(p>0.05, respectively).
 | Table I.Correlation between miR-21
overexpression and PTEN in CRC. |
Table I.
Correlation between miR-21
overexpression and PTEN in CRC.
| MiR-21 (n=30) | PTEN
|
|---|
| Negative
(n=16) | Positive
(n=14) |
|---|
| 3.32±0.29
(n=13) | 4 (30.77) | 9 (69.23) |
| 6.12±1.86
(n=17) | 12 (70.59) | 5 (29.41) |
PTEN protein correlates inversely with
miR-21 in CRC cell lines and tissues
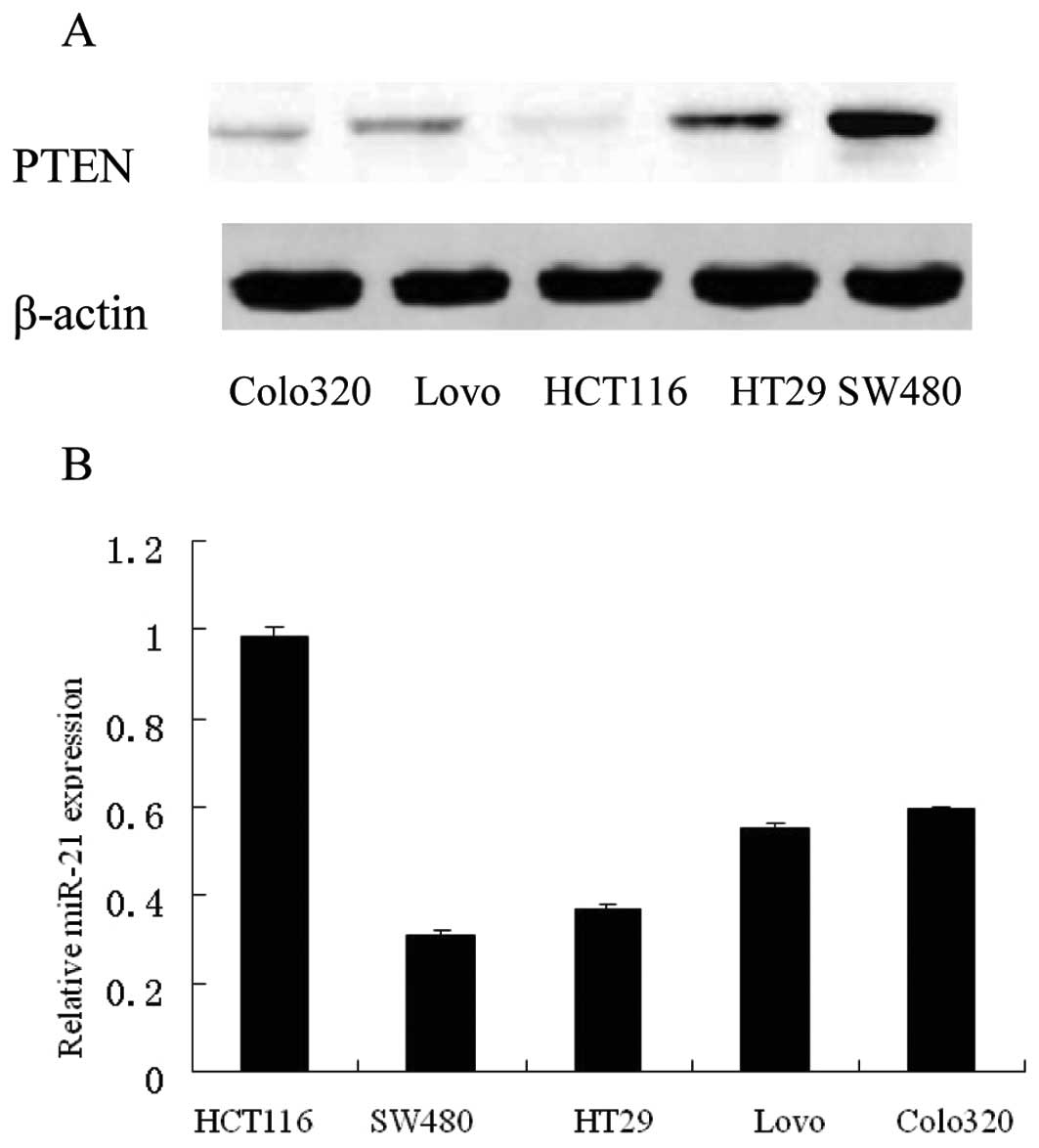
We determined expression levels of miR-21 and PTEN
(protein and mRNA) in 5 different colorectal cancer cell lines
(Fig. 2). In cell lines with high
endogenous miR-21 (for example, HCT116, Colo320, Fig. 2B) as measured by miRNA qRT-PCR, a
low amount of PTEN protein was observed (Fig. 2A and C), whereas cell lines with
low miR-21 (for example, SW480, HT29, Fig. 3B) showed high amounts of PTEN
protein (Fig. 2A and C). Across
all 5 cell lines tested, we found a significant inverse correlation
between miR-21 and PTEN protein levels (r=−0.972, p<0.01).
However, the differences in PTEN protein are higher than for
PTEN-mRNA among the cells (Fig. 2C and
D) (p=0.000 for PTEN protein, p=0.010 for PTEN mRNA). These
data suggest that miR-21 expression is inversely correlated with
PTEN expression in human colorectal cancer cells, and it is
possible that miR-21 negatively regulates PTEN
post-transcriptionally.
We then surveyed correlation between miR-21
expression and PTEN expression in CRC tissues, we found 9 cases
were positive for PTEN among the 13 cases of CRC tissues with
relatively low miR-21 expression (3.32±0.29), but only 5 cases were
positive for PTEN among the 17 cases of CRC tissues with relatively
high miR-21 expression (6.12±1.86). Spearman’s rank correlation
analysis showed that PTEN expression was negatively correlated with
miR-21 expression (r=−0.396, p<0.05) (Table I).
Anti-miR-21 inhibits CRC cell growth,
proliferation and migration, and induces apoptosis
HCT116 cell line was selected to investigate miR-21
functions and targets by using sequence-specific functional
inhibition of miR-21, because the cell line expressed higher levels
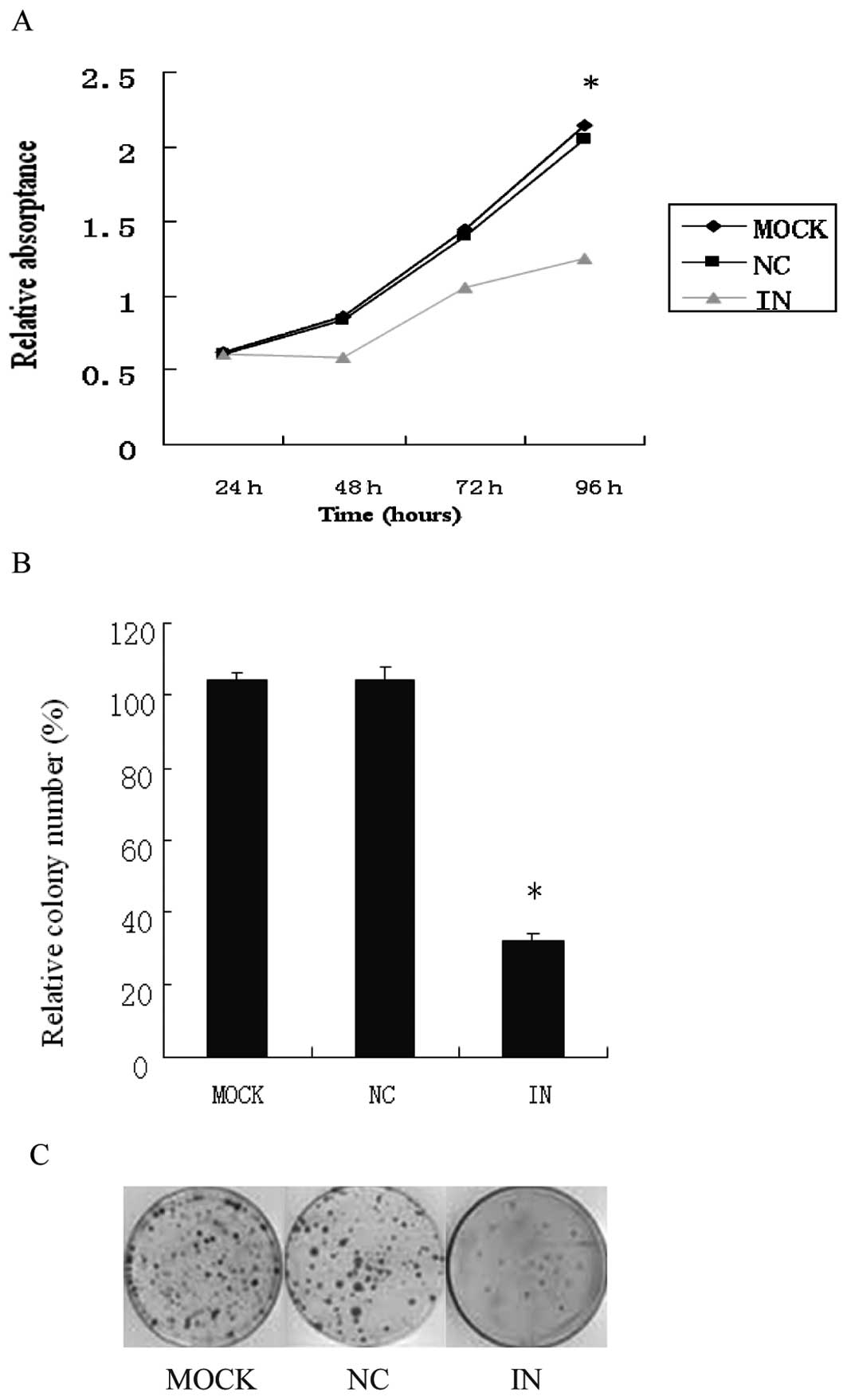
of miR-21. Knockdown of miR-21 with ASO reduced significantly
miR-21 levels in HCT116 cells (Fig.
3B). Anti-miR-21 inhibitor led to a significant decrease in
HCT116 cell growth and proliferation (p<0.01, Fig. 3A). As shown in Fig. 3B, the number of colonies from
HCT116 cells transfected with anti-miR-21 inhibitor was
significantly lower than that of negative control and MOCK group
(p=0.05). Furthermore, the size of the colonies from the cells
transfected with anti-miR-21 inhibitor was much smaller than those
of the control group (Fig. 3C).
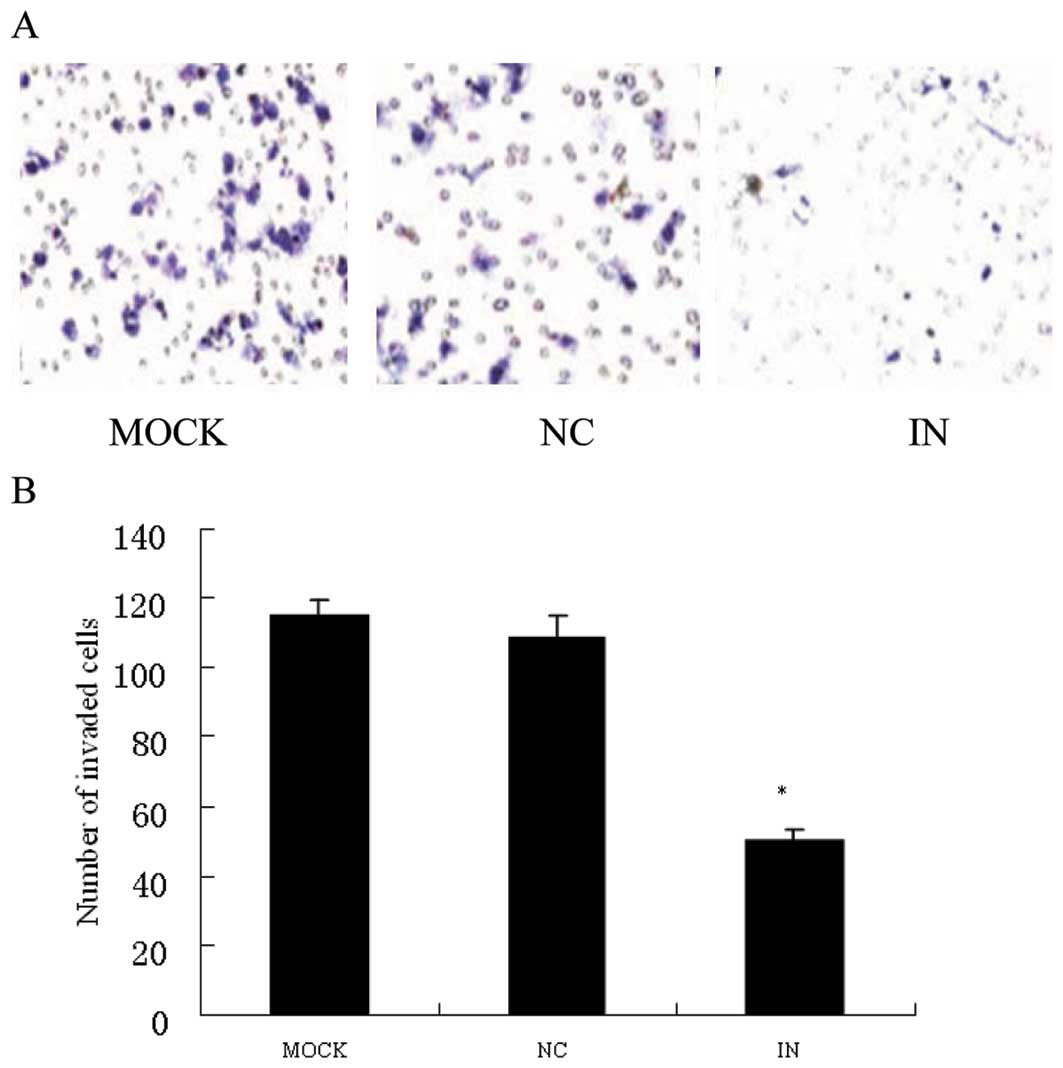
In vitro matrigel invasion assay showed that anti-miR-21
inhibitor significantly suppressed HCT116 cell invasion
(p<0.05), compared with anti-miR-21NC and MOCK group (Fig. 4A and B). Wound healing assay showed
that wound repair was delayed in anti-miR-21 inhibitor compared
with anti-miR-21NC and MOCK (Fig.
5A). Anti-miR-21 inhibitor significantly suppressed HCT116 cell
migration (p<0.05), compared with anti-miR-21NC at 24 h after
wound scratch (Fig. 5B). These
data demonstrate the tumorigenic properties of miR-21 in regulating
cell growth, proliferation, invasion and migration.
Induced apoptosis in vitro
To further analyze whether decreased viability was a
result of cell cycle arrest, the cell cycle distribution was
analyzed using FCM. At 48 h after transfection, FCM analysis showed
a 25% increase in G0/G1 phase cells and a 20%
decrease in S-phase cells and a 20% decrease in G2/M
phase cells in HCT116 cells transfected with a miR-21 inhibitor
(p<0.05, Fig. 6A and B). We
also analyzed the effect of miR-21 inhibitor on apoptosis by
conducting Annexin V and PI double staining. The rate of apoptosis
significantly increased in cells transfected with anti-miR-21
inhibitor (48 h after transfection) compared with blank and NC
control (p<0.05, Fig. 6C and
D). It suggested that miR-21 may function as a strong
antiapoptotic factor in human colorectal cancer cells.
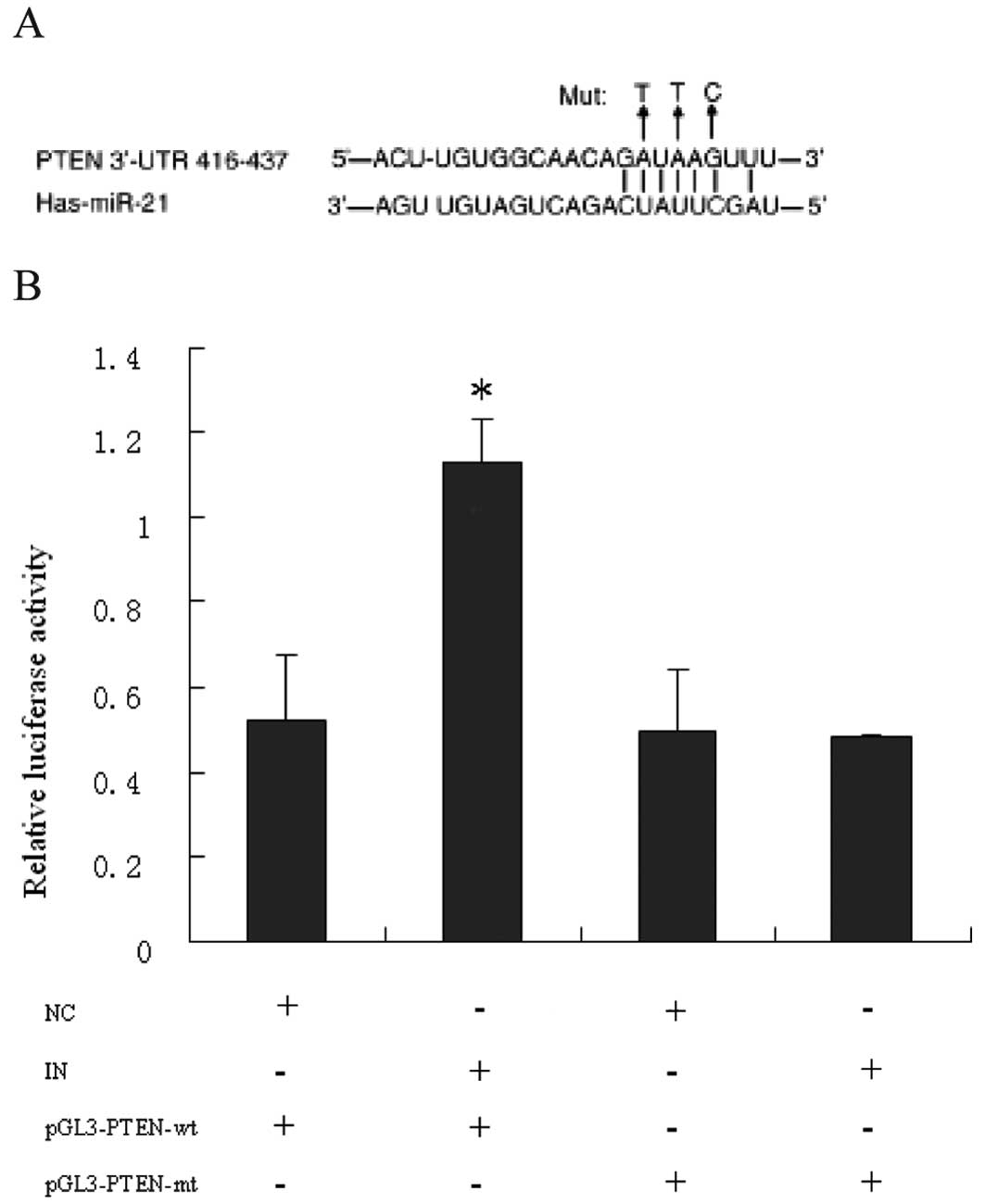
PTEN is a direct target of miR-21 in
CRC
To verify that PTEN may be a tumor-suppressor gene
in CRC and miR-21 may directly target PTEN, we constructed
luciferase-reporter plasmids that contain the wt or mutant 3′-UTR
segments of PTEN (Fig. 7A). The wt
or mutant reporter plasmid was cotransfected into HCT116 cells
along with miR-21 inhibitor or negative control (NC). Compared with
NC, miR-21 inhibitor significantly increased the relative
luciferase activity when cotransfected with the wt reporter
plasmid. However, the mutant reporter plasmid abolished miR-21
inhibitor-mediated increase in luciferase activity (Fig. 7B). These findings suggest that
miR-21 suppresses PTEN by direct binding to the 3′-UTR of PTEN.
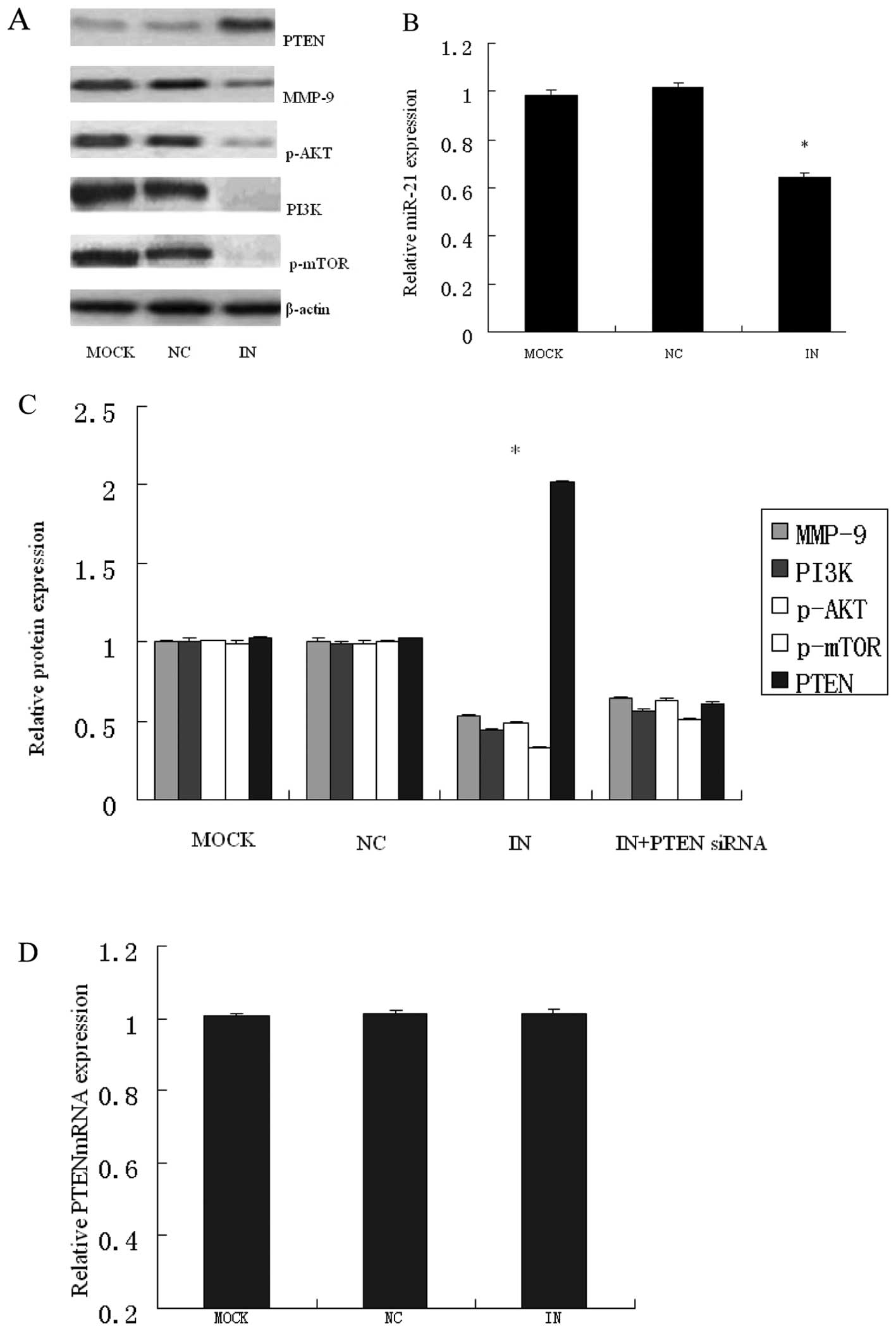
MiR-21 regulates expression of PTEN and
its downstream targets
We assessed whether the downregulation of miR-21
affected PTEN protein expression in HCT116 cell lines. HCT116 cells
were transiently transfected with miR-21 inhibitor, inhibitor
negative control or blank control culture medium. The decrease in
endogenous miR-21 levels with miR-21 inhibitor (Fig. 8B) significantly increased PTEN
protein expression (Fig. 8C and
A), but it almost caused no alternations in PTEN mRNA (Fig. 8D). These findings showed that
miR-21 inhibits PTEN protein production, rather than degrading its
mRNA. It further suggests that, in CRC, miR-21 regulates PTEN at
the level of protein translation. This finding strongly suggests
that PTEN is regulated by miR-21 in CRC at post-transcription
level.
As AKT was an important downstream target of PTEN
and expression of PTEN resulted in the reduced levels of
phosphorylated AKT (p-AKT), and PTEN suppressed the expression of
matrix metalloproteinase-2 (MMP-2) and MMP-9 through FAK
dephosphorylation (22,31), the levels of p-AKT, PI3K, p-mTOR
and MMP-9 were also examined at the same time. As shown in Fig. 8C, the expression of p-AKT, PI3K,
p-mTOR and MMP-9 was concomitantly decreased by miR-21 inhibitor.
In addition, siRNA to PTEN abrogated the reduction in p-Akt, PI3K,
p-mTOR and MMP-9 by anti-miR-21 (Fig.
8C). In combination, these studies define an important role for
PTEN as a mediator of the biological effects of miR-21 on cell
proliferation and invasion in human CRC.
Discussion
Although miRNA signatures for CRC cancer have been
established, the molecular mechanism of the mir-21-mediated
tumorigenesis, development and metastases of CRC needs to be
elucidated. In the present study, we identified miR-21
over-expression in adenomas and CRC. In vitro, miR-21
blockade with ASO inhibits growth and induces apoptosis in CRC cell
lines, which is mediated by retrieving PTEN expression. In
vivo, injection of miR-21 ASO into CRC xenografts implanted
subcutaneously in nude mice suppressed tumor growth.
MiR-21 is one of the most prominent miRNAs
implicated in human cancer. It has been proposed that miR-21 acts
as a potential oncogene. MiR-21 is upregulated in various cancer
types and has been associated with poor survival and poor
therapeutic outcome (46–48). Its expression is obviously
upregulated in diverse types of malignancies, including CRC. We
found the expression levels of miR-21 were significantly higher in
tumor tissues than the levels in adenomas and normal tissues
(p=0.000).
Moreover, overexpression of miR-21 was closely
correlated with advanced TNM clinical stage (p<0.05) and poor
differentiation (p<0.05), and lymph node metastases (p<0.05)
which are the main prognostic factors for CRC, implying miR-21
might be involved in the development and metastasis of cancer, and
has a prognostic implication for CRC. This result is consistent
with the previous findings (46–53).
We further demonstrated that knockdown of miR-21 with ASO could
significantly decrease cell proliferation, invasion and migration
abilities in HCT116 cells. Higher expression levels of miR-21 in
adenomas and carcinomas relative to normal surrounding colonic
tissue suggest that this represents an early cellular event in the
progression to cancer. These findings suggest that the aberrant
expression of miR-21 maybe a marker for early diagnosis of CRC,
directly affects invasive and metastatic potential of tumor cells
and is implicated in the progression of CRC.
However, it was contradicted by another study
(54), which discovered that the
level of miR-21 was underexpressed in a subset of
Cdc25A-overexpressing colon cancers. The reason may lie in the
amount and quality of tissue samples in the latter study or the
specific cell line and culture condition in the former study.
Folini et al(55) also
argued that the oncogenic properties of miR-21 could be cell and
tissue dependent and that the potential role of a given miRNA as a
therapeutic target should be contextualized with respect to the
disease. Cheng et al(56)
reported that inhibition of miR-21 significantly promotes
proliferation in HeLa cervical carcinoma cells, which may suggest
its role as a tumor suppressor. It has been considered that miR-21
may have completely different effects, depending on the cellular
environment or context (26,57).
It suggested that miRNAs could participate in many signal pathways
that have been proven to be important in tumorigenesis,
inflammation and cell growth, and miRNAs seem to be involved in a
complicated network of epigenetics of cancer.
Various studies have reported that miR-21 plays an
important role not only in tumor growth but also in the invasion
and metastasis by targeting multiple tumor suppressor genes
including PTEN, PDCD4, BCL-2, TPM1, and RECK (32–38).
PTEN (phosphatase and tensin homolog deleted on chromosome 10)
(58), also called MMAC1 (59) or TEP1 (60), is a tumor suppressor gene. It is
localized to chromosome 10q23.2 and is often lost in late-stage
human cancers, especially that of the prostate, brain and
endometrium. PTEN negatively regulates intracellular levels of
PI-(3,4,5)P3, most likely via direct dephosphorylation, suggesting
that it exerts its role as a tumor suppressor by negatively
regulating the PI-3 K/PKB/Akt signaling pathway (61). This pathway is known to play a key
role in numerous cellular functions including proliferation,
adhesion, angiogenesis, migration, invasion, metabolism, survival
and chemoresistance (62). Our
study shows that 46.67% of CRC samples were positive for PTEN
expression and 53.33% were negative that is consistent with
previous studies (63,64).
In the present study, PTEN expression showed a
negative correlation with the expression of miR-21 in CRC tissues
and cell lines. It was shown that the ability of HCT116 cell
proliferation and migration was inhibited significantly after
transfection with miR-21 inhibitor. Furthermore, we demonstrated
that inhibition of miR-21 can significantly increase PTEN
expression in HCT116 cell lines, suggesting CRC might enhance
malignant biological behavior by inactivating PTEN genetically and
upregulating miR-21. As shown in Fig.
7, luciferase activity of the wt, but not mutant, PTEN-3′-UTR
reporter was significantly increased in HCT116 cells transfected
with miR-21 inhibitor compared with negative control. Furthermore,
miR-21 expression inversely correlated with PTEN protein levels,
however, without obvious change in PTEN mRNA level and decreased
phosphorylated AKT expression. Thus, it can be demonstrated that
miR-21 negatively regulated the PTEN at the post-translational
level, performing as oncogene in the CRC.
Asangani et al(33) reported that miR-21 may target PDCD4
directly in human CRC RKO cells and demonstrated that miR-21
induced invasion, intravasation and metastasis. Taken together,
miR-21 may negatively regulate PDCD4 and PTEN, which in turn alter
focal adhesion kinase phosphorylation and expression of several
MMPs and thereby contribute to cancer cell migration and
invasion.
Although PTEN as a target gene of miR-21 has been
validated in HCC (22), breast
cancer (65) and non-small cell
lung cancer (31), and gastric
cancer (32), Hatley et
al(66) confirmed that PTEN
was not regulated by miR-21 in non-small cell lung cancer and
miR-21 high expression was not sufficient in the non-small cell
lung cancer tumorigenesis model. These findings further demonstrate
that miR-21, along with its known targets and a few associated
genes, forms a complex regulatory network and mechanism that plays
an important role in CRC formation and progression.
In conclusion, miR-21 is overexpressed in CRC, and
aberrant expression of miR-21 can alter multiple biological
processes of human CRC cells such as proliferation, apoptosis,
migration, and invasion, probably through regulating PTEN and other
critical target genes. Our data suggest that miR-21 can serve as a
biomarker for CRC, and can possibly become a potential therapeutic
target for CRC.
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Zhang YL, Zhang ZS, Wu BP and Zhou DY:
Early diagnosis for colorectal cancer in China. World J
Gastroenterol. 8:21–25. 2002.
|
|
3.
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
4.
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Chim SS, Shing TK, Hung EC, et al:
Detection and characterization of placental microRNAs in maternal
plasma. Clin Chem. 54:482–490. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Mayr C, Hemann MT and Bartel DP:
Disrupting the pairing between let-7 and Hmga2 enhances oncogenic
transformation. Science. 315:1576–1579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Giraldez AJ, Cinalli RM, Glasner ME, et
al: MicroRNAs regulate brain morphogenesis in zebrafish. Science.
308:833–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Chen CZ, Li L, Lodish Hf and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Hornstein E, Mansfield JH, Yekta S, et al:
The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Marson A, Levine SS, Cole MF, et al:
Connecting microRNA genes to the core transcriptional regulatory
circuitry of embryonic stem cells. Cell. 134:521–533. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Cho WC: MicroRNAs in cancer -from research
to therapy. Biochim Biophys Acta. 1805:209–217. 2010.PubMed/NCBI
|
|
15.
|
Cho WC: MicroRNAs: potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–8121. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
MiR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, et al:
Expression profiling identifies microRNA signature in pancreatic
cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Roldo C, Missiaglia E, Hagan JP, et al:
Micro-RNA expression abnormalities in pancreatic endocrine and
acinar tumors are associated with distinctive pathologic features
and clinical behavior. J Clin Oncol. 24:4677–4684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Hiyoshi Y, Kamohara H, Karashima R, et al:
MicroRNA-21 regulates the proliferation and invasion in esophageal
squamous cell carcinoma. Clin Cancer Res. 15:1915–1922. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Schetter AJ, Leung SY, Sohn JJ, et al:
Micro-RNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Lui WO, Pourmand N, Patterson BK and Fire
A: Patterns of known and novel small RNAs in human cervical cancer.
Cancer Res. 67:6031–6043. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Iorio MV, Visone R, Di Leva G, et al:
MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Fulci V, Chiaretti S, Goldoni M, et al:
Quantitative technologies establish a novel microRNA profile of
chronic lymphocytic leukemia. Blood. 109:4944–4951. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Löffler D, Brocke-Heidrich K, Pfeifer G,
et al: Interleukin-6 dependent survival of multiple myeloma cells
involves the Stat3-mediated induction of microRNA-21 through a
highly conserved enhancer. Blood. 110:1330–1333. 2007.PubMed/NCBI
|
|
30.
|
Ziyan W, Shuhua Y, Xiufang W and Xiaoyun
L: MicroRNA-21 is involved in osteosarcoma cell invasion and
migration. Med Oncol. 28:1469–1474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K
and Yang GH: MicroRNA-21 (miR-21) represses tumor suppressor PTEN
and promotes growth and invasion in non-small cell lung cancer
(NSCLC). Clin Chim Acta. 411:846–852. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY and
Yan M: MicroRNA-21 promotes tumor proliferation and invasion in
gastric cancer by targeting PTEN. Oncol Rep. 278:1019–1026.
2012.PubMed/NCBI
|
|
33.
|
Asangani IA, Rasheed SA, Nikolova DA, et
al: MicroRNA-21 (miR-21) posttranscriptionally down-regulates tumor
suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene. 27:2128–2136. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Nagao Y, Hisaoka M, Matsuyama A, et al:
Association of microRNA-21 expression with its targets, PDCD4 and
TIMP3, in pancreatic ductal adenocarcinoma. Mod Pathol. 25:112–121.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Gabriely G, Wurdinger T, Kesari S, et al:
MicroRNA 21 promotes glioma invasion by targeting matrix
metalloproteinase regulators. Mol Cell Biol. 28:5369–5380. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Zhang Z, Li Z, Gao C, et al: MiR-21 plays
a pivotal role in gastric cancer pathogenesis and progression. Lab
Invest. 88:1358–1366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Papagiannakopoulos T, Shapiro A and Kosik
KS: MicroRNA-21 targets a network of key tumor-suppressive pathways
in glioblastoma cells. Cancer Res. 68:8164–8172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Thum T, Gross C, Fiedler J, et al:
MicroRNA-21 contributes to myocardial disease by stimulating MAP
kinase signaling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Sayed D, Rane S, Lypowy J, et al:
MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol
Biol Cell. 19:3272–3282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Liu M, Tang Q, Qiu M, et al: MiR-21
targets the tumor suppressor RhoB and regulates proliferation,
invasion and apoptosis in colorectal cancer cells. FEBS Lett.
585:2998–3005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Song B, Wang C, Liu J, et al: MicroRNA-21
regulates breast cancer invasion partly by targeting tissue
inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res.
29:292010. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Selaru FM, Olaru AV, Kan T, et al:
MicroRNA-21 is over-expressed in human cholangiocarcinoma and
regulates programmed cell death 4 and tissue inhibitor of
metalloproteinase 3. Hepatology. 49:1595–1601. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Slaby O, Svoboda M, Fabian P, et al:
Altered expression of miR-21, miR-31, miR-143 and miR-145 is
related to clinicopathologic features of colorectal cancer.
Oncology. 72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Kulda V, Pesta M, Topolcan O, et al:
Relevance of miR-21 and miR-143 expression in tissue samples of
colorectal carcinoma and its liver metastases. Cancer Genet
Cytogenet. 200:154–160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Fassan M, Pizzi M, Giacomelli L, et al:
PDCD4 nuclear loss inversely correlates with miR-21 levels in colon
carcinogenesis. Virchows Arch. 458:413–419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Liu K, Li G, Fan C, Zhou X, Wu B and Li J:
Increased expression of microRNA-21 and its association with
chemotherapeutic response in human colorectal cancer. J Int Med
Res. 39:2288–2295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Drebber U, Lay M, Wedemeyer I, et al:
Altered levels of the onco-microRNA 21 and the tumor-supressor
microRNAs 143 and 145 in advanced rectal cancer indicate successful
neoadjuvant chemoradiotherapy. Int J Oncol. 39:409–415. 2011.
|
|
51.
|
Chang KH, Miller N, Kheirelseid EA, et al:
MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg
Oncol. 37:597–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Shibuya H, Iinuma H, Shimada R, Horiuchi A
and Watanabe T: Clinicopathological and prognostic value of
microRNA-21 and microRNA-155 in colorectal cancer. Oncology.
79:313–320. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Yamamichi N, Shimomura R, Inada K, et al:
Locked nucleic acid in situ hybridization analysis of miR-21
expression during colorectal cancer development. Clin Cancer Res.
15:4009–4016. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Wang P, Zou F, Zhang X, et al: MicroRNA-21
negatively regulates Cdc25A and cell cycle progression in colon
cancer cells. Cancer Res. 69:8157–8165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Folini M, Gandellini P, Longoni N, et al:
miR-21: an oncomir on strike in prostate cancer. Mol Cancer.
9:122010. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Jovanovic M and Hengartner MO: MiRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Li L, Ernsting BR, Wishart MJ, Lohse Dl
and Dixon JE: A family of putative tumor suppressors is
structurally and functionally conservedin humans and yeast. J Biol
Chem. 272:29403–29406. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Steck PA, Pershouse MA, Jasser SA, et al:
Identification of a candidate tumour suppressor gene, MMAC1, at
chromosome 10q23.3 that is mutated in multiple advanced cancers.
Nat Genet. 15:356–362. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Li DM and Sun H: TEP1, encoded by a
candidate tumor suppressor locus, is a novel protein tyrosine
phosphatase regulated by transforming growth factor beta. Cancer
Res. 57:2124–2129. 1997.
|
|
61.
|
Leslie NR and Downes CP: PTEN: the down
side of PI 3-kinase signalling. Cell Signal. 14:285–295. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
62.
|
Bader AG, Kang S, Zhao L and Vogt PK:
Oncogenic PI3K deregulates transcription and translation. Nat Rev
Cancer. 5:921–929. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Hsu CP, Kao TY, Chang WL, Nieh S, Wang HL
and Chung YC: Clinical significance of tumor suppressor PTEN in
colorectal carcinoma. Eur J Surg Oncol. 37:140–147. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Naguib A, Cooke JC, Happerfield L, et al:
Alterations in PTEN and PIK3CA in colorectal cancers in the EPIC
Norfolk study: associations with clinicopathological and dietary
factors. BMC Cancer. 11:1232011. View Article : Google Scholar : PubMed/NCBI
|
|
65.
|
Wang ZX, Lu BB, Wang H, Cheng ZX and Yin
YM: MicroRNA-21 modulates chemosensitivity of breast cancer cells
to doxorubicin by targeting PTEN. Arch Med Res. 42:281–290. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
66.
|
Hatley ME, Patrick DM, Garcia MR, et al:
Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21.
Cancer Cell. 18:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|