Introduction
miRNAs are a class of endogenous small non-coding
RNAs (approximately 21–25 nucleotides in length), which are derived
from longer transcripts termed pri-miRNAs and pre-miRNAs (1–5).
miRNAs combine target mRNAs through partial complementarity to
specific sequences located in the 3′ untranslated region (3′-UTR)
and act as post-transcriptional regulators of gene expression
(6–9). It has been shown that human miRNA
genes regions along with perturbed miRNA expression patterns have
been detected in many human benign and malignant cancers (10). Therefore, it is of potential
importance to elucidate the biological functions of miRNAs.
Additionally, miRNAs have been shown to play
important roles in invasion and metastasis of cancer (11–15).
For example, miR-155 may take part in the TGF-β-induced
epithelial-mesenchymal transition (EMT) and in cell migration and
invasion through targeting of the RhoA transcript (16). MiR-21 has the ability to stimulate
cell invasion and metastasis in several tumor models, including
breast cancer (12), colon cancer
(17), and glioma (18). The pro-metastatic transcription
factor TWIST1 is able to activate miR-10b, which is essential for
TWIST1-induced EMT involved in promotion of cell motility and
invasiveness (19). Tumor invasion
and metastasis are the critical indicators that define the
prognosis of cancer patients. Therefore, it is very important to
understand the specific roles of miRNAs in cancer progression, and
it could lead to the identification of predictive markers and the
development of novel therapeutic strategies for patients with
metastases.
In mammalians, the miR-34 family consists of three
miRNAs which are encoded by two different genes: miR-34a is encoded
by its own transcript, whereas miR-34b/c share the common primary
one. In mice, miR-34a is highly expressed in brain tissues
(20), whereas miR-34b/c is mainly
expressed in lung tissues (21).
These analyses also pointed out that miR-34a is expressed at higher
levels than miR-34b/c, with the exception of the lung. miR-34 genes
may be the important targets for other signaling pathways involved
in normal life progression, however this remains to be determined
by genetic analysis.
The aim of this study is to determine the role of
miR-34b in the non-small cell lung cancer. In our study, the
expression level of miR-34b was significantly decreased in NSCLC
tissues in comparison with pericarcinous tissues of lung cancer.
In vitro gain-of-function experiments indicated that miR-34b
functioned as a tumor suppressor and inhibited cell proliferations
by inducing cell apoptosis. Thus, we analyzed the relations between
miR-34b and Met and relevant proteins using IHC technology. The
results suggest a miR-34b/Met axis with potential therapeutic
implications.
Materials and methods
Samples
Twenty-nine pairs of NSCLC specimens and
corresponding pericarcinous tissues of lung cancer (PTLC) were
collected at the time of surgery and prior to chemotherapy.
Specimens were obtained from patients in Shandong Provincial
Hospital Affiliated to Shandong University from 2010 to 2011 with
informed consent. All tissue samples were from untreated patients
undergoing surgery and all clinicopathological information was
available. The study was approved by the Hospitals’ Ethics Review
Committee. Part of the tissue specimens were snap frozen in liquid
nitrogen and stored at −80°C, and the remaining were fixed with
formalin and embedded in paraffin.
Cell culture
Human NSCLC cell lines (A549 and SPC-A-1) were
provided by Cell Bank for Chinese Academy of Sciences. A549 was
grown in DMEM (Hyclone) and SPC-A-1 in 1640 (Hyclone) supplemented
with 10% fetal bovine serum (Gibco) and 100 units/ml penicillin and
1% streptomycin (Invitrogen) and maintained at 37°C with 5%
CO2.
Quantitative real-time polymerase chain
reaction (qRT-PCR)
Total RNA was obtained from NSCLC frozen tumor
tissues, pericarcinous tissues of lung cancer and cell lines using
TRIzol reagent (Invitrogen), according to the manufacturer’s
instructions. RNA concentration was determined
spectrophotometrically, and integrity was checked by gel
electrophoresis. RNA quality was confirmed in an Agilent 2100
Bioanalyzer (Agilent Technologies). The expression levels of the
mature miRNAs were determined using TaqMan MicroRNA Assays (Applied
Biosystems) and cDNAs were synthesized using the TaqMan miRNA RT
kit based on the specific stem-loop RT primer design. Reverse
transcriptase reactions contained 10 ng RNA samples, 3 μl stem-loop
RT primers, 1.5 μl of 10X RT buffer, 0.15 μl of 100
mM dNTPs, 1 μl MultiScribe reverse transcriptase, 0.19
μl RNase inhibitor and 4.16 μl nuclease free water.
The 15 μl reactions were incubated for 30 min at 16°C, 30
min at 42°C, 5 min at 85°C, and then held at 4°C. The 20 μl
PCR reaction included 1.33 μl RT product, 1X TaqMan
Universal PCR master mix and 1 μl primers and probe mix of
the TaqMan MicroRNA Assay kit. The initial PCR step was a 10 min
hold at 95°C; then 40 cycles consisted of a 15-sec denaturation
step at 95°C followed by 1-min of annealing/extension at 63°C. The
PCR reactions were run on a 7500 real-time PCR machine (Applied
Biosystems) and analyzed using 7500 System SDS software. The miRNA
expression level was normalized to the expression level of U6 small
nuclear RNA (RNU6B). Primers used for hsa-miR-34b and RNU6B were
purchased from Applied Biosystems. All reactions were performed in
triplicate and included a negative control lacking cDNA.
Cell transfection
SPC-A-1 cell line was transfected with double
stranded synthetic miRNA mimics (syn-hsa-miR-34b miScript miRNA)
and scrambled controls (Qiagen) using HiPerFect transfection
reagent (Qiagen) according to the manufacturer’s protocol for
overexpression. Approximately 5×105 cells were seeded
and cultured in 6-well plates. Complexes containing the mimics were
added directly to each well at a final miRNA mimic concentration of
5 nM. Incubate the cells with the transfection complexes under
their normal growth conditions. All groups were performed in
triplicate.
Cell apoptosis
For cell apoptosis assay, SPC-A-1 cells were seeded
in 6-well plates, both adherent and floating cells were harvested
at 72 h after transfection, and stained with Annexin V-FITC
(Clontech) and propidium iodide (PI) (Clontech) for 15 min in the
dark at room temperature followed by flow cytometric analysis. The
cell line experiment was performed at least three times depending
on the reproducibility.
Immunohistochemistry (IHC)
Antibodies against phospho-Met, p53 (phospho S392)
and Mdm2 are rabbit polyclonal antibodies (Abcam). In brief, the
slides were dewaxed, and endogenous peroxidase activity was then
quenched with 3% H2O2. Tissue samples were
heated in 1 mmol/l ethylenediaminetetraacetic acid (EDTA) buffer
for 15 min in a water bath (96–98°C) to retrieve antigens, and
cross-reactivity was blocked with normal goat serum. The slides
were then incubated overnight at 4°C with primary antibodies (1:500
for primary antibody). The subsequent steps were according to the
instructions of Zymed (Streptavidin-Perosidase Method). The primary
antibodies were replaced by normal serum or phosphate-buffered
saline (PBS) as negative controls.
Evaluation of immunostaining
The criterion for a positive reaction was clear
cytoplasm or/and nucleus staining. The samples with more than 10%
of the tumor cells stained were considered to be positive.
Statistical analysis
A statistical analysis was done using the SPSS 18.0
statistical software package. For qRT-PCR data, the statistical
analysis of miR-34b expression level in NSCLC tissues and PTLC
tissues were log2 transformed. Values are expressed as the mean ±
SEM. Differences in miR-34b between NSCLC and PTLC were analyzed
using two-sample t-test. Pearson χ2 and Fisher’s exact
tests were used to determine the correlation between miR-34b
expression and clinical stage and lymph node metastasis status.
Spearman correlation analysis was used to determine the correlation
between miR-34b expression and levels of phospho-Met, pS392p53 and
Mdm2. Other results were analyzed using independent sample t-test.
P<0.05 was defined as being significant.
Results
Downregulation of miR-34b expression in
NSCLC
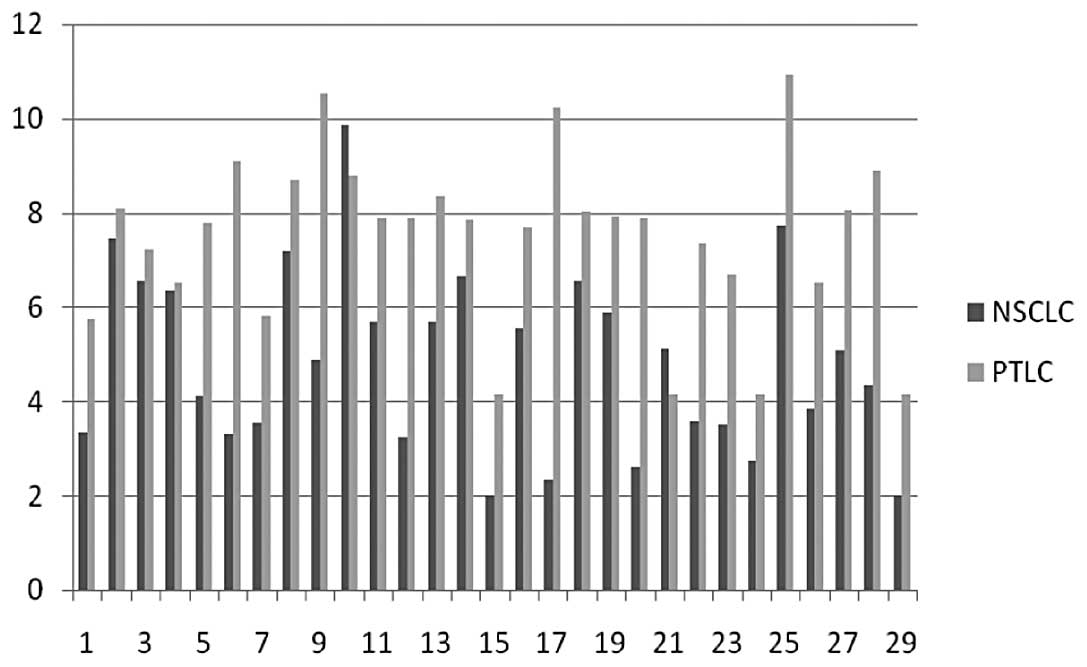
Compared with PTLC, a significant downregulation of
miR-34b expression in NSCLC was noted. Of 29 matched cancer and
normal tissues, the expressions of miR-34b in 27 cancer tissues
were decreased in comparison with the matched PTLC (p<0.001)
(Fig. 1). The mean level of
miR-34b in NSCLC was decreased about 66.11±4.31% compared to that
in PTLC.
Correlations between miR-34b expression
and clinicopathological characteristics of NSCLC
For further analysis, all patients were divided into
2 groups (miR-34b high and low expression) on the basis of the mean
level of miR-34b expression in 29 NSCLC, and the
clinicopathological characteristics of these groups were
summarized. A significant association between the miR-34b
expression level and lymph node metastasis was observed (Table I). In 8 cases without lymph node
metastasis, 1 (12.50%) has high level of miR-34b expression,
whereas in 21 cases with lymph node metastasis, 12 (57.14%) cases
have high expression levels of miR-34b (P=0.031). No correlation
was observed between miR-34b expressions and gender, age,
differentiation or pathologic TNM stage.
 | Table IRelations between miR-34b expression
and clinicopathological characteristics in patients with NSCLC. |
Table I
Relations between miR-34b expression
and clinicopathological characteristics in patients with NSCLC.
| Characteristics | Patients | miR-34b
expression | P-value |
|---|
| High | Low |
|---|
| Gender | | | | 0.525 |
| Male | 23 | 11 | 12 | |
| Female | 6 | 2 | 4 | |
| Age | | | | 0.897 |
| <60 | 13 | 6 | 7 | |
| ≥60 | 16 | 7 | 9 | |
| Differentiation | | | | 0.198 |
| Well/ Moderate | 15 | 5 | 10 | |
| Poor | 14 | 8 | 6 | |
| Invasion depth | | | | 0.957 |
| T1 | 6 | 3 | 3 | |
| T2 | 14 | 6 | 8 | |
| T3, T4 | 9 | 4 | 5 | |
| Lymph node
metastasis | | | | 0.031 |
| Positive | 21 | 12 | 9 | |
| Negative | 8 | 1 | 7 | |
| TNM stage | | | | 0.384 |
| I | 6 | 2 | 4 | |
| II | 7 | 2 | 5 | |
| III, IV | 16 | 9 | 7 | |
miR-34b transfection induces cell
apoptosis in NSCLCs
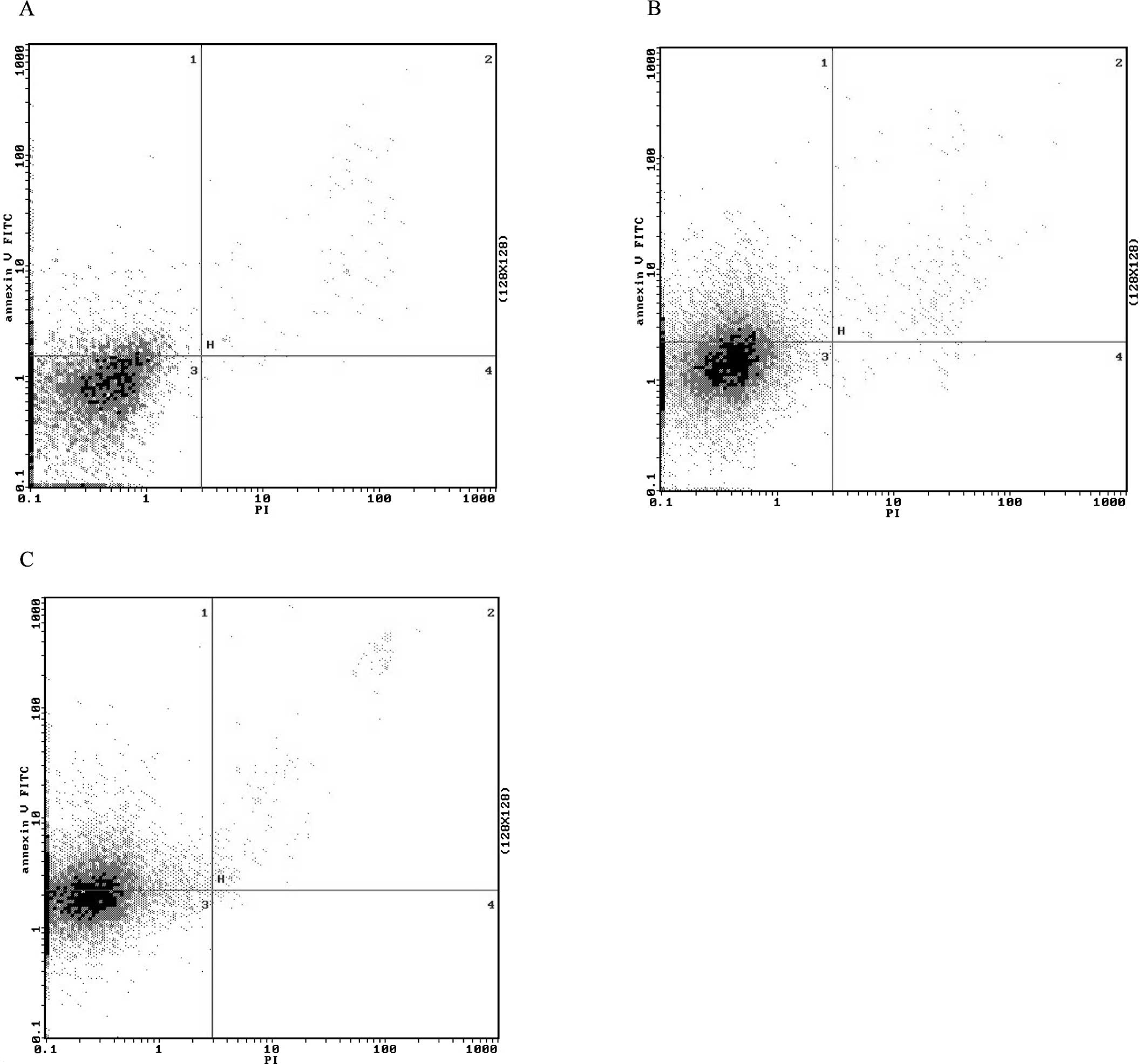
Next, we examined the effects of ectopic miR-34b on
the cell growth arrest by flow cytometry. Overexpression of miR-34b
resulted in a significant increase in the proportion of
FITC-annexin V-positive cells and FITC-annexin V-negative/propidium
iodide (PI)-positive cells, and a decrease in the proportion of
FITC-annexin V-negative/PI-negative cells (Fig. 2), which indicated that
overexpression of miR-34b may lead to apoptosis and necrosis. Taken
together, the results suggest that miR-34b may inhibit
proliferation of NSCLC cells mainly by inducing apoptosis.
phospho-Met, p53 (phospho S392) and Mdm2
are over-expressed corresponding to miR-34b reduction
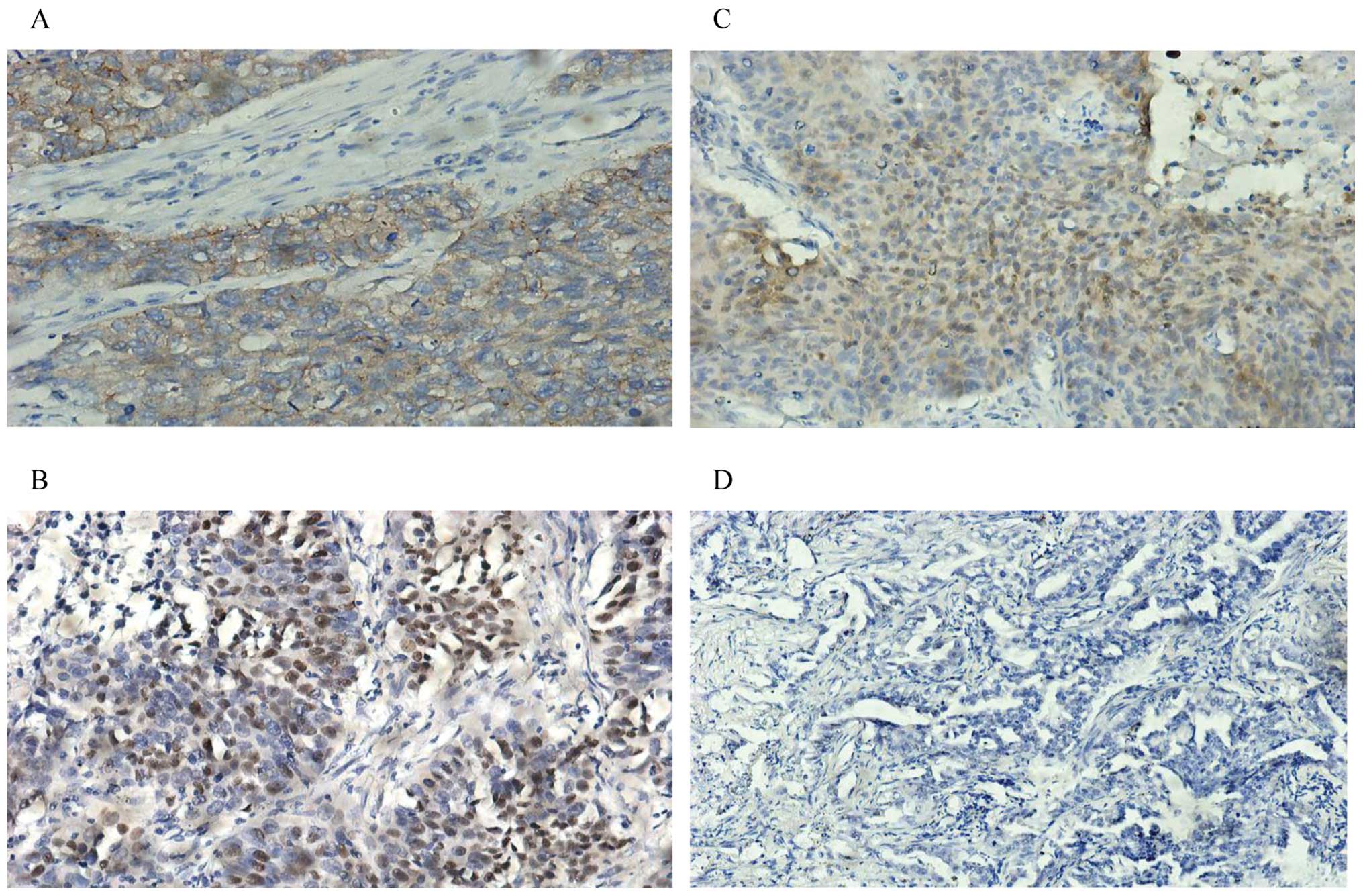
Immunohistochemistry technology was used to examine
the expression of phospho-Met, p53 (phosphoS392) and Mdm2 in
clinical samples with NSCLC and matched PTLC (Fig. 3). There were positive expressions
of phospho-Met in 44.8% (13/29), pS392p53 in 58.62% (17/29) and
Mdm2 in 79.31% (23/29) of samples with NSCLC, respectively. In
addition, Met IHC scoring was correlated with miR-34b level in
samples with NSCLC (P=0.012). Spearman correlation analysis was
used to determine the correlations between miR-34b expression and
levels of phospho-Met, pS392p53 and Mdm2.
Discussion
It has been provn that miRNAs to function as
important regulators of gene post-transcriptional regulation.
Accumulated studies have indicated that miRNAs act as tumor
suppressors or oncogenes depending on whether their specific
targets act as oncogenes or tumor suppressors, suggesting that the
aberrant expressions of several miRNAs might contribute to human
carcinogenesis (12,22). Thus, understanding of the specific
miRNAs involved in the process of tumor development would provide
significant insights for the diagnosis and treatment of patients
with tumors. Our data showed that miR-34b functioned as a tumor
suppressor and was correlated with tumor progression.
Additionally, as expected, NSCLC cells transfected
with miR-34b mimics induced apoptosis, indicating that miR-34b had
drastic effects on cell survival. The ectopic expression of either
miR-34a or miR-34b/c led to substantial apoptosis and growth arrest
(23). Previous studies have
proved that ectopic miR-34b caused a cell cycle arrest in the G1
phase (21,24). As apoptosis and cell cycle arrest
are common end points of p53 signal pathway activation, miR-34
genes could be the potent regulators of tumor suppression by p53
(23). The induction of miR-34
genes permits p53 to regulate the expression of a large number of
proteins. Furthermore, targeting of p53-induced mRNAs by miR-34 may
affect p53 preventing an uncontrolled response of p53 activation
(25).
Receptor tyrosine kinase (RTK) signaling is a core
pathway frequently altered in cancer. Over recent years,
therapeutic approaches based on compounds selectively targeting
oncogenic RTKs have been developed. As RTKs share several effectors
that participate in the oncogenic process and in drug response, an
alternative strategy would rely on the identification of
drug-treatable nodal points required for RTK-triggered
tumorigenesis.
The Met-RTK and its ligand HGF have essential
functions during embryogenesis and regenerative processes by
regulating cell development, scattering, migration, angiogenesis,
survival, proliferation and differentiation (26). Met is a target of miR-34b and is
regulated by miR-34b combining Met mRNAs through partial
complementarity to sequences located in the 3′-UTR of Met. It has
been shown that c-Abl activation by Met triggers p53
phosphorylation on Ser392, which elevates the transcriptional
activities of p53 and drives the transcriptional upregulation of
Mdm2 and protection from cell death (27). Furthermore, p53 enhances the
expression of miR-34 genes, which in turn mediates downregulation
and degradation of Met. However, there is no increased expression
level of miR-34b corresponding to the overexpression of pS392
observed in the samples with NSCLC. We suspect that miR-34b may be
modulated by other effectors in cellular signal pathways, or
phosphrylation on S392 decrease the transcriptional activities of
p53 in lung cancer tissues.
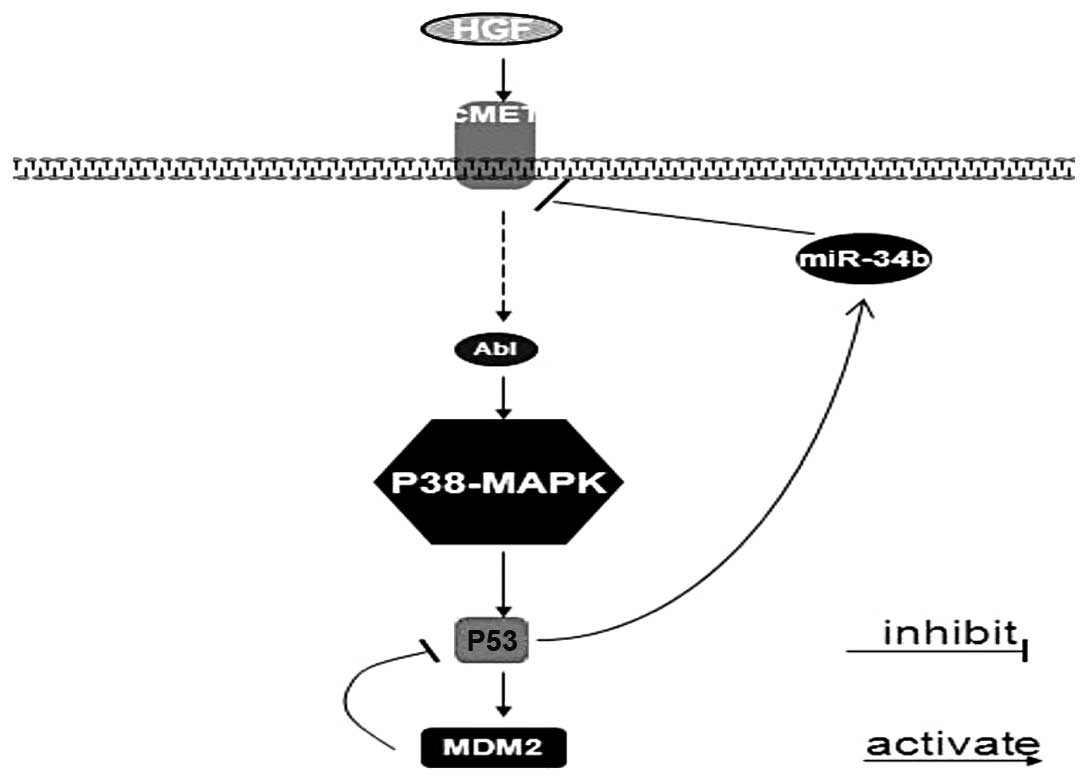
miR-34b regulates the HGF-MET signal pathway, in
which p53 could be modulated through phosphorylation of p53 on
Ser392 by p38-MAPK and inversely p53 possesses anti-survival
potential by upregulating miR-34b, subsequently miR-34b has an
effect on Met in a feedback loop (Fig.
4). Collectively, Met and miR-34b work as key nodal points in
p53 network, which provides new insights into therapeutic
strategies of HGF-MET signal pathway and functional restoration of
miR-34b is an effective novel therapy.
Acknowledgements
This study was supported by grants
from the National Natural Science Foundation of China (81141100),
Natural Science Foundation of Shandong Province of China
(ZR2010HM067 and ZR2011HM077) and Provincial Science and Technology
Foundation of Shandong (2011GGH21819).
References
|
1
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee Y, Ahn C, Han J, et al: The nuclear
RNase III Drosha initiates microRNA processing. Nature.
425:415–419. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kong W, Zhao JJ, He L and Cheng JQ:
Strategies for profiling microRNA expression. J Cell Physiol.
218:22–25. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hutvágner G, McLachlan J, Pasquinelli AE,
Bálint E, Tuschl T and Zamore PD: A cellular function for the
RNA-interference enzyme Dicer in the maturation of the let-7 small
temporal RNA. Science. 293:834–838. 2001.PubMed/NCBI
|
|
5
|
Grishok A, Pasquinelli AE, Conte D, et al:
Genes and mechanisms related to RNA interference regulate
expression of the small temporal RNAs that control C. elegans
developmental timing. Cell. 106:23–34. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schwarz DS, Hutvágner G, Du T, Xu Z,
Aronin N and Zamore PD: Asymmetry in the assembly of the RNAi
enzyme complex. Cell. 115:199–208. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ørom UA, Nielsen FC and Lund AH:
MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and
enhances their translation. Mol Cell. 30:460–471. 2008.PubMed/NCBI
|
|
8
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:E852005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs genomics, biogenesis,
mechanism, and function. Cell. 116:281–297. 2004.PubMed/NCBI
|
|
10
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Crawford M, Brawner E, Batte K, et al:
MicroRNA-126 inhibits invasion in nonsmall cell lung carcinoma cell
lines. Biochem Biophys Res Commun. 373:607–612. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tavazoie SF, Alarcón C, Oskarsson T, et
al: Endogenous human microRNAs that suppress breast cancer
metastasis. Nature. 451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Valastyan S, Reinhardt F, Benaich N, et
al: A pleiotropically acting microRNA miR-31, inhibits breast
cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kong W, Yang H, He L, et al: MicroRNA-155
is regulated by the transforming growth factor beta/ Smad pathway
and contributes to epithelial cell plasticity by targeting RhoA.
Mol Cell Biol. 28:6773–6784. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Asangani IA, Rasheed SA, Nikolova DA, et
al: MicroRNA-21 (miR-21) posttranscriptionally downregulates tumor
suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene. 27:2128–2136. 2008.
View Article : Google Scholar
|
|
18
|
Gabriely G, Wurdinger T, Kesari S, et al:
MicroRNA 21 promotes glioma invasion by targeting matrix
metalloproteinase regulators. Mol Cell Biol. 28:5369–5380. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang J, Mani SA and Weinberg RA: Exploring
a new twist on tumor metastasis. Cancer Res. 66:4549–4552. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lodygin D, Tarasov V, Epanchintsev A, et
al: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell Cycle. 7:2591–2600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bommer GT, Gerin I, Feng Y, et al:
p53-mediated activation of miRNA34 candidate tumor-suppressor
genes. Curr Biol. 17:1298–1307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tarasov V, Jung P, Verdoodt B, et al:
Differential regulation of microRNAs by p53 revealed by massively
parallel sequencing: miR-34a is a p53 target that induces apoptosis
and G1-arrest. Cell Cycle. 6:1586–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cohen SM, Brennecke J and Stark A:
Denoising feedback loops by thresholding-a new role for microRNAs.
Genes Dev. 20:2769–2772. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Naran S, Zhang X and Hughes S J:
Inhibition of HGF/MET as therapy for malignancy. Expert Opin Ther
Targets. 13:569–581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Furlan A, Stagni V, Hussain A, et al: Abl
interconnects oncogenic Met and p53 core pathways in cancer cells.
Cell Death Differ. 18:1608–1616. 2011. View Article : Google Scholar : PubMed/NCBI
|