Introduction
Primary liver cancer, which is frequently
hepatocellular carcinoma (HCC), is the sixth most common cancer and
third most frequent cause of cancer-related death worldwide, and it
is becoming more prevalent not only in East Asia, South-East Asia,
and Africa but also in Western countries (1–3).
Recently, the multikinase inhibitor sorafenib was demonstrated to
prolong overall survival (OS) in patients with advanced HCC, and it
has become the standard drug for first-line systemic treatment
(4–6). However, based on the Response
Evaluation Criteria in Solid Tumors (RECIST), the response rate for
sorafenib is rather low, and the incidence of adverse events is
relatively high, especially in elderly patients (7). Therefore, the generation of a novel
effective therapy for HCC is a priority.
Immunotherapy is an attractive option for treating
HCC. Many of the tumor antigens associated with HCC are potential
candidates for peptide vaccines (8,9). The
carcinoembryonic antigen Glypican-3 (GPC3), which is a 65-kDa
protein of 580 amino acids, belongs to the family of
glycosyl-phosphatidylinositol (GPI)-anchored heparan sulfate
proteoglycans (HSPG) (10,11). GPC3 is specifically overexpressed
in HCC (72–81% of cases) and correlates with poor prognosis
(12–16). This suggests that GPC3 is an ideal
target for anti-HCC immunotherapy.
We have previously demonstrated the antigenicity of
GPC3, and that the HLA-A*24:02-restricted
GPC3298–306 (EYILSLEEL) peptide and the
HLA-A*02:01-restricted GPC3144–152
(FVGEFFTDV) peptide can induce GPC3-reactive CTLs without inducing
autoimmunity (17–21).
HLA-A2 is the most frequent HLA-A type in all ethnic
groups (22). HLA-A2 is also
expressed in about 40% of Japanese persons (23,24)
and in about 50% of Caucasians (25). Among Caucasians, >90% of
HLA-A2-positive individuals carry the HLA-A*02:01 allele
(25), whereas among the Japanese,
there are multiple common and well-documented (CWD) allelic
variants, including HLA-A*02:01, HLA-A*02:06
and HLA-A*02:07 (26).
The frequencies of the HLA-A*02:01,
HLA-A*02:06 and HLA-A*02:07 alleles in the
Japanese population are 19, 14 and 7%, respectively (26). Therefore, we confirmed that the
HLA-A*02:01-restricted GPC3144–152
(FVGEFFTDV) peptide could also bind to HLA-A*02:06 and
HLA-A*02:07 using a binding assay (unpublished
data).
On the basis of these results, we conducted a phase
I clinical trial of a GPC3-derived peptide vaccine in 33 patients
with advanced HCC. The HLA-A*24:02-restricted
GPC3298–306 peptide was used for
HLA-A*24:02-positive patients and the
HLA-A*02:01-restricted GPC3144–152 peptide
was used for HLA-A*02:01, HLA-A*02:06 and
HLA-A*02:07-positive patients. We found that GPC3
vaccination was well-tolerated, and that the GPC3 peptide vaccine
induced a GPC3-specific CTL response in almost all of the patients
(27–30). Moreover, the vaccination-induced
GPC3-specific CTL response correlated with overall survival (OS);
the OS was significantly longer in patients with high GPC3-specific
CTL frequencies than in those with low GPC3-specific CTL
frequencies (27). In terms of
clinical responses, one patient showed a partial response (PR) and
19 patients showed stable disease 2 months after initiation of
treatment. One patient with HCC who showed a PR was
HLA-A*02:07-positive. In addition, several
HLA-A*02:01-restricted GPC3 peptide-specific CTL clones
with cytotoxic activities against GPC3 were established from the
peripheral blood mononuclear cells (PBMCs) of patients vaccinated
in this trial (27).
The aims of the present study were: i) to establish
GPC3-derived, peptide-specific CTL clones from the PBMCs of an
HLA-A*02:07-positive patient with HCC who showed a PR in
the phase I clinical trial; and ii) to analyze the functions of
these CTL clones.
Materials and methods
Ethics information
This study was approved by the Ethics Committee of
the National Cancer Center and conformed to the ethical guidelines
of the 1975 Declaration of Helsinki. All the patients gave written
informed consent before entering the study at the National Cancer
Center Hospital East (Chiba, Japan). The trial has been registered
with the University Hospital Medical Information Network Clinical
Trials Registry (UMIN-CTR no. 000001395).
PBMCs collection
Peripheral blood samples were obtained pre- and
post-vaccination from the patient with HCC who was
HLA-A*02:07-positive. Post-vaccination, blood samples
were collected from the patient every 2 weeks. The GMP-grade
peptide GPC3144–152 (FVGEFFTDV) (American Peptide Co.,
Sunnyvale, CA, USA) was emulsified in IFA (Montanide ISA-51 VG;
SEPPIC, Paris, France) and injected intradermally at 30 mg/body
three times at 14-day intervals (27,28).
PBMCs were isolated by density centrifugation using Ficoll-Hypaque
(Pharmacia, Uppsala, Sweden) and frozen in liquid nitrogen until
use.
Cell lines
The human lung cancer cell line 1–87
(GPC3−,
HLA-A*02:07+/A*11:01+)
and hepatitis B virus (HBV)-integrated human hepatocellular
carcinoma cell line JHH-7 (GPC3+,
HLA-A*24:02+/A*31:01+)
were conserved in our laboratory and cultured in Dulbecco’s
modified Eagle’s medium (DMEM; Sigma Chemical Company, St. Louis,
MO, USA) that was supplemented with 10% heat-inactivated fetal
bovine serum (FBS; Gibco, Carlsbad, CA, USA).
Plasmids and transfection
The expression vectors pcDNA3.1 (Invitrogen,
Carlsbad, CA, USA) and pcDNA3.1 that contained the
HLA-A*02:07 cDNA were used for the transfection
experiments. The pcDNA3.1 construct that contained
HLA-A*02:07 was kindly provided by Dr Ryo Abe (Tokyo
University of Science, Chiba, Japan). The
JHH-7/HLA-A*02:07 cell line was obtained by transfection
of JHH-7 cells with the expression vector using FuGENE HD (Roche
Applied Science, Mannheim, Germany). JHH-7/mock and
JHH-7/HLA-A*02:07 cells were cultured in DMEM that was
supplemented with 10% heat-inactivated FBS and 1 mg/ml G418
(Calbiochem, Darmstadt, Germany).
Induction of GPC3144–152
peptide-specific CTLs from PBMCs
The PBMCs were cultured (2×106
cells/well) with the GPC3144–152 peptide in RPMI-1640
(Sigma Chemical Company) that was supplemented with 10%
heat-inactivated FBS, 100 IU/ml recombinant human IL-2 (Nipro,
Osaka, Japan), and 10 ng/ml recombinant human IL-15 (PeproTech Inc,
Rocky Hill, NJ, USA) for 14 days.
CD107a staining and flow cytometry
analysis
CD8+ T cells were isolated using human
CD8 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) from
PBMCs that were stimulated with the GPC3144–152 peptide
for 14 days. The CD8+ T cells were incubated with
GPC3144–152-pulsed or HIV19–27-pulsed 1–87
cells at a ratio of 2:1 for 3.5 h at 37°C. CD107a-specific
antibodies (BD Biosciences, San Jose, CA, USA) were included in the
mixture during the incubation period.
Generation of CTL clones
CD8+ CD107a+ cells were sorted
using a FACSAria cell sorter (BD Biosciences). Sorted CTLs were
stimulated and the CTL clones were established as previously
described (28).
Cytotoxicity assay
Cytotoxic capacity was analyzed with the Terascan
VPC system (Minerva Tech, Tokyo, Japan). The CTL clone was used as
the effector cell type. Target cells were labeled in calcein-AM
solution for 30 min at 37°C. The labeled cells were then
co-cultured with the effector cells for 4–6 h. Fluorescence
intensity was measured before and after the culture period, and
specific cytotoxic activity was calculated as previously described
(28).
IFN-γ ELISPOT assay
Specific secretion of IFN-γ from human CTLs in
response to stimulator cells was assayed using the IFN-γ ELISPOT
kit (BD Biosciences), according to the manufacturer’s instructions.
Stimulator cells were pulsed with or without peptide for 1.5 h at
room temperature and then washed three times. Responder cells were
incubated with stimulator cells for 20 h. The resulting spots were
counted using an ELIPHOTO counter (Minerva Tech).
Determination of recognition
efficiency
Calcein-AM-labeled target cells were pulsed with
various concentrations of peptide, starting at 10−6 M
and decreasing in log steps to 10−14 M. The CTL clones
were incubated with the target cells at an effector:target (E/T)
ratio of 10:1 for 4 h. The recognition efficiencies of the CTL
clones were defined as previously described (28).
RNA interference
Human GPC3-specific siRNAs were chemically
synthesized as double-strand RNA (Invitrogen). A non-silencing
siRNA, AllStras Neg. Control siRNA, was obtained from Qiagen
(Valencia, CA, USA). The following GPC3-specific siRNA sequences
were used: GPC3-siRNA (#4149), 5′-UUAUCAUUCCAUCACCAGAGCCUCC-3′;
GPC3-siRNA (#4150), 5′-GGAGGCUCUGGUGAUGGAAU GAUAA-3′: and
GPC3-siRNA (#4151), 5′-UAUAGAUGACUG GAAACAGGCUGUC-3′. Synthetic
siRNA duplexes were transfected using Lipofectamine RNAiMAX
(Invitrogen), according to the manufacturer’s protocols.
RT-PCR
Using the TRIzol reagent (Invitrogen), we extracted
total cellular RNA from untreated or siRNA (GPC3-siRNA or
negative-siRNA)-treated JHH-7/HLA-A*02:07. cDNA was
synthesized using the PrimeScript II 1st Strand cDNA Synthesis kit
(Takara, Kyoto, Japan) according to the manufacturer’s
instructions. The cDNA was added to a reaction mix that contained
10X Ex Taq Buffer (Takara), 2.5 mM dNTP mixture (Takara), 5 units
Ex Taq (Takara), and 10 μM of the GPC3- or
β-actin-specific PCR primers. The following primer sequences
(sense and antisense, respectively) were used: for GPC3,
5′-AGCCAAAAGGCAGCAAGGAA-3′ and 5′-AAGA AGAAGCACACCACCGA-3′; and for
β-actin, 5′-CCTCGCCT TTGCCGATCC-3′ and
5′-GGATCTTCATGAGGTAGTC AGTC-3′. PCR was performed using the 96-well
Gene Amp PCR System 9700 (Applied Biosystems, Carlsbad, CA, USA).
PCR was performed for 20 cycles of 98°C for 10 sec, 64°C for 30 sec
and 72°C for 30 sec, followed by a step of 72°C for 10 sec.
Sequence analysis of TCR-β gene
Using the TRIzol reagent (Invitrogen), total
cellular RNA was extracted from established CTL clones. The cDNA of
the TCR-β gene was synthesized using the PrimeScript II 1st
Strand cDNA Synthesis Kit (Takara) according to the manufacturer’s
instructions, with the modification that we used 200 nM of the
primer specific for the TCR-β chain constant region. The cDNA
products were subjected to 2-step PCR, as previously described by
Yukie Tanaka-Harada (35,36), and the PCR products were purified
and sequenced in the Applied Biosystems 3500 Genetic Analyzer
(Applied Biosystems). The TCR-β variable (TRBV) gene,
TCR-β joining (TRBJ) gene, TCR-β diversity
(TRBD) alleles, and complementarity-determining region 3
(CDR3) sequences were identified using the IMGT databases
(http://www.imgt.org/).
Results
GPC3144–152 peptide-specific
CTLs in the peripheral blood of the patient exert a clinical
effect
We analyzed the immune responses of the patient who
showed a PR following GPC3144–152 peptide vaccination.
In this patient, the supraclavicular lymph node metastases markedly
regressed, two liver tumors disappeared, and the thoracic bone
metastasis showed necrosis after the third vaccination (27). The levels of DCP decreased in the
patients over the 2-month period. We evaluated the
GPC3144–152-specific immune responses in the peripheral
blood using the ex vivo IFN-γ ELISPOT assay. For the
HLA-A*02:07-positive patient with advanced HCC, the
number and area of the spots increased after two rounds of
vaccination, as compared with the pre-vaccination values, and the
peak values were noted 10 weeks after the start of the treatment
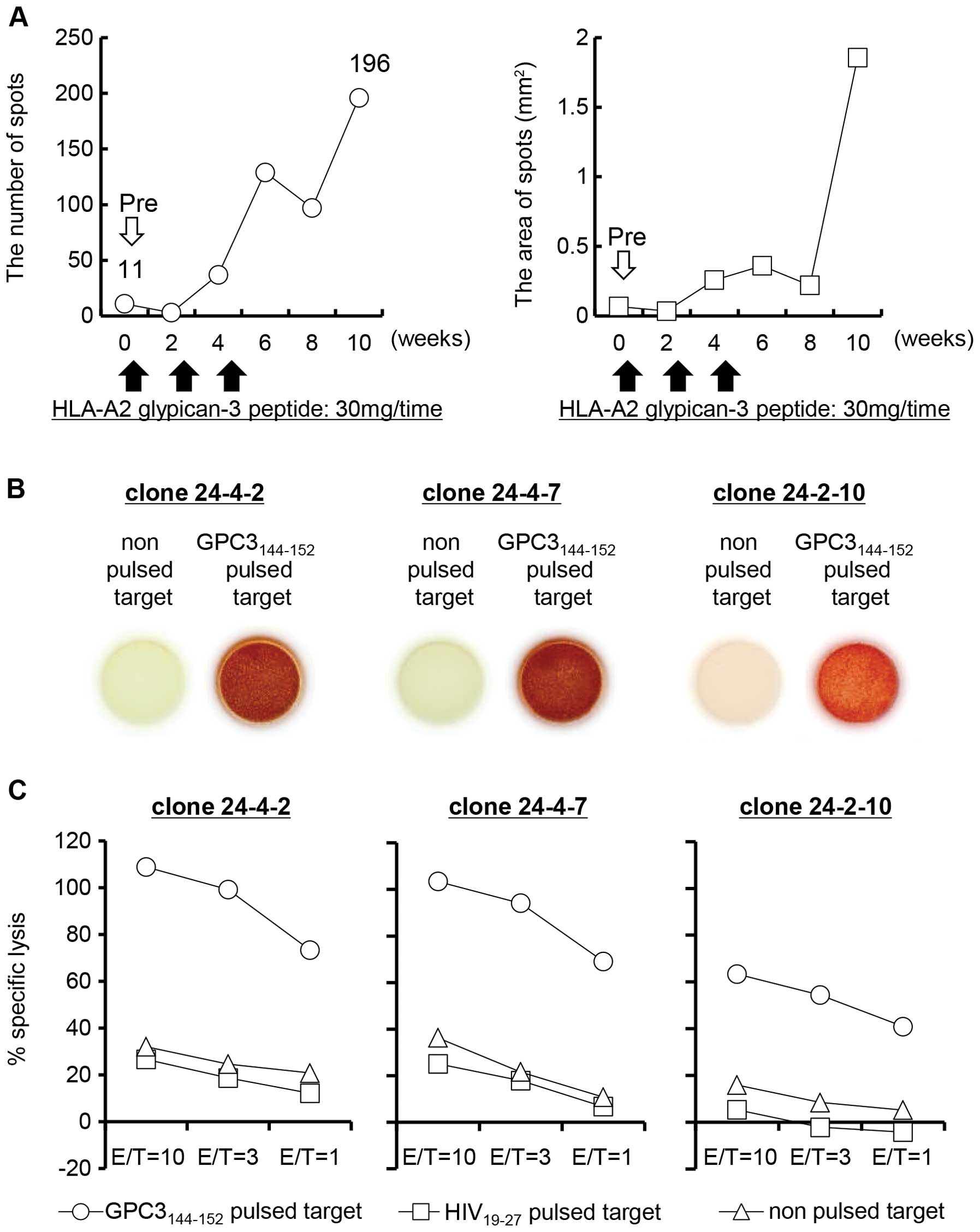
(Fig. 1A).
Establishment of
GPC3144–152-specific CTL clones from the PBMCs of the
patient
To investigate the ability of the
GPC3144–152-specific CTLs induced by peptide vaccination
to recognize antigen, we established CTL clones from the PBMCs of
this patient 10 weeks after the start of treatment. The PBMCs were
stimulated with the GPC3144–152 peptide in vitro
for 14 days. CD8+ T cells were isolated from the
stimulated PBMCs, and then incubated with peptide-pulsed 1–87
cells. CD8+ CD107a+ cells that reacted with
the GPC3144–152-pulsed 1–87 cells were sorted to the
single-cell level. Thus, we established GPC3144–152
peptide-specific CTL clones.
Three established CTL clones were analyzed for
function using the IFN-γ ELISPOT assay and cytotoxicity assay. All
of the CTL clones released IFN-γ in response to the
GPC3144–152-pulsed 1–87 cells, but not in response to
non-pulsed 1–87 cells (Fig. 1B).
Moreover, these CTL clones showed cytotoxicity against
GPC3144–152-pulsed 1–87 cells, but not against
non-pulsed or HIV19-27-pulsed 1–87 cells (Fig. 1C). These results indicate that the
CTL clones 24-4-2, 24-4-7 and 24-2-10 have specificity for the
GPC3144–152 peptide.
Functional avidity of the
GPC3144–152-specific CTL clones
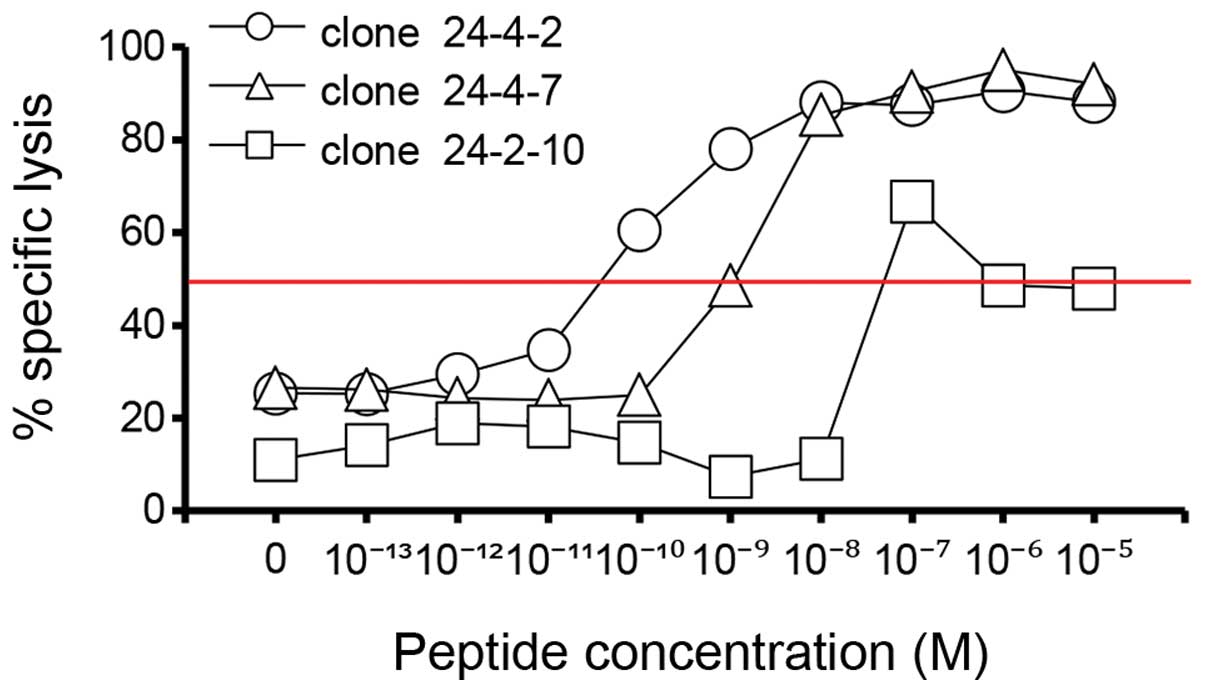
We evaluated the cytotoxicity profiles of the CTL
clones for 1–87 cells pulsed with a decreasing concentration series
(from 10−6 to 10−14 M) of the
GPC3144–152 peptide. The peptide concentration at which
the curve reached 50% cytotoxicity was defined as the recognition
efficiency of the clone. The recognition efficiencies of CTL clones
24-4-2, 24-4-7 and 24-2-10 were 10−11, 10−9
and 10−8 M, respectively (Fig. 2). This result suggests that CTL
clone 24-4-2 has a higher avidity than the other two clones and,
conversely, that CTL clone 24-2-10 has a lower avidity than the
other two clones.
A GPC3144–152-specific CTL
clone recognizes cancer cells that endogenously express GPC3
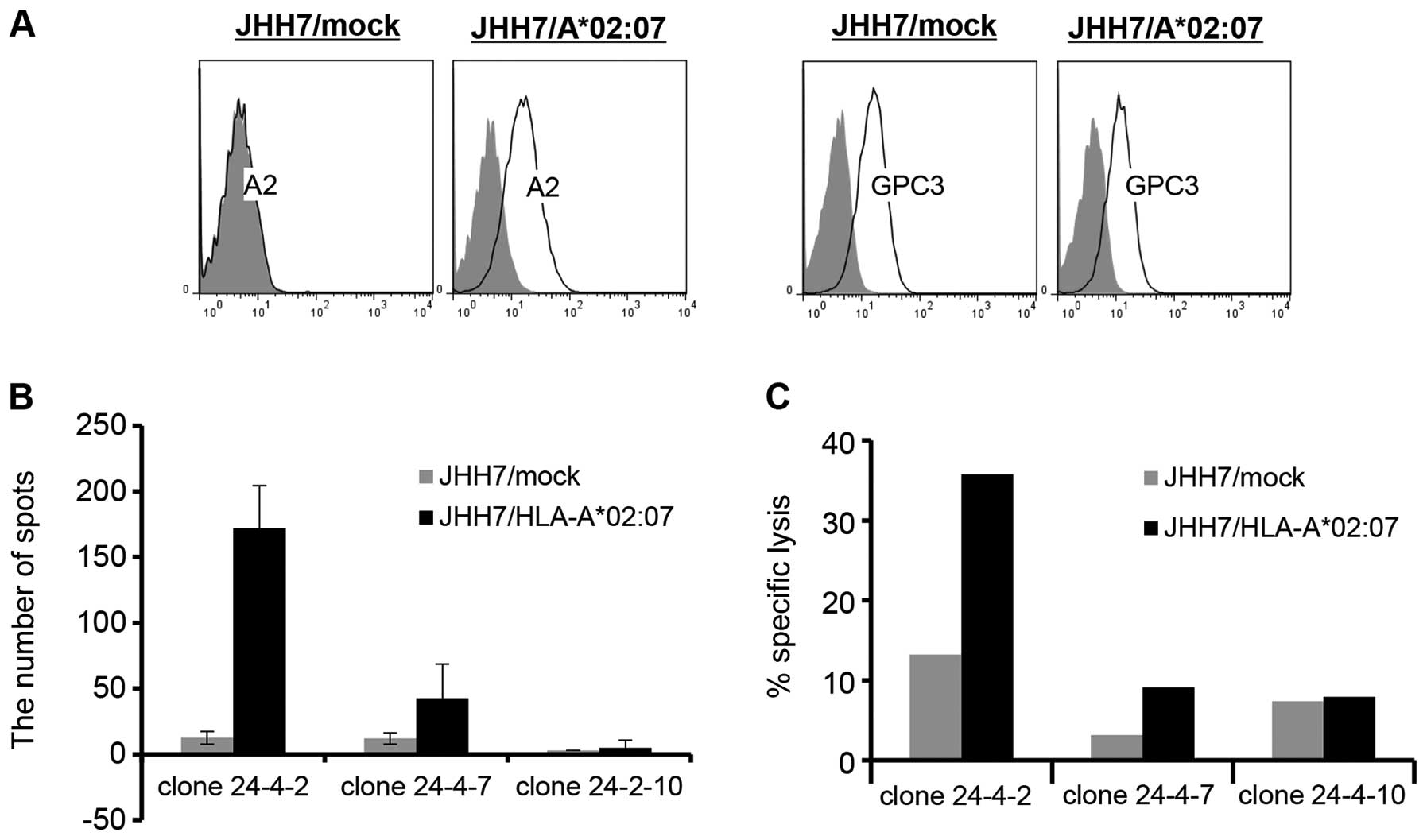
Next, we tested the reactivities of these CTL clones
against cancer cell lines that expressed GPC3 and
HLA-A*02:07. We used the JHH-7/mock (GPC3+,
HLA-A*02:07-) and JHH-7/HLA-A*02:07
(GPC3+, HLA-A*02:07+)
transfectants as the target cells (Fig. 3A). The CTL clone 24-4-2 (with high
avidity) produced IFN-γ and was cytotoxic for
JHH-7/HLA-A*02:07 cells but not for JHH-7/mock cells
(Fig. 3B and C). The other clones
did not produce IFN-γ and did not exhibit cytotoxicity for the two
target cell lines. These results suggest that only high-avidity
CTLs recognize cancer cells that express GPC3 peptide
endogenously.
CTL clone 24-4-2 shows specificity for
GPC3
To ascertain the GPC3 antigen-specific response of
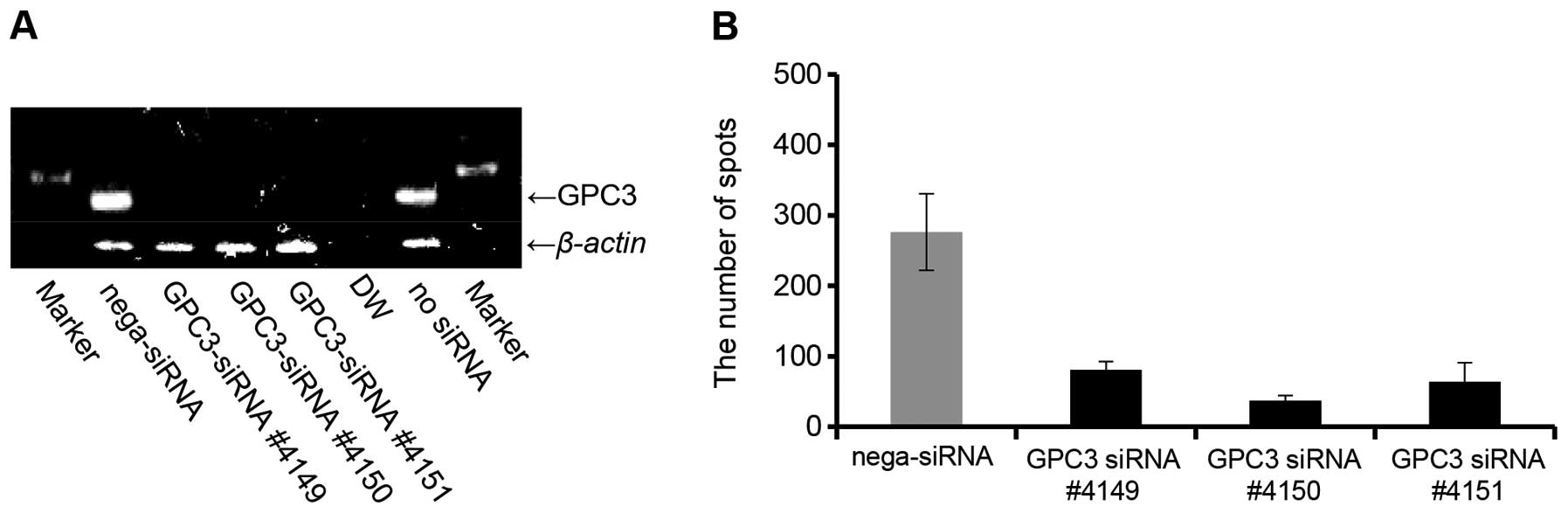
CTL clone 24-4-2, we created a GPC3 knockdown via siRNA treatment
of the JHH-7/HLA-A*02:07 cells. GPC3 expression by the
JHH-7/HLA-A*02:07 cells was clearly decreased by the
GPC3-siRNA, as assessed by RT-PCR (Fig. 4A). We examined the IFN-γ production
levels of CTL clone 24-4-2 against JHH-7/HLA-A*02:07
cells treated with GPC3-siRNA. IFN-γ production by CTL clone 24-4-2
was significantly decreased by the GPC3-siRNA (Fig. 4B). These results indicate that the
HLA-A2-restricted GPC3144–152 peptide is processed
naturally by cancer cells, and that both HLA-A*02:07 and
HLA-A*02:01 can present the GPC3144–152
peptide on the surfaces of cancer cells.
Established CTL clones have different
sets of TCR-β alleles
We analyzed the TCR-β gene sequences of the
established CTL clones. The TRBV, TRBJ and
TRBD alleles were identified using the IMGT databases. Thus,
we identified the TRBV, TRBD and TRBJ alleles
of the CTL clones (Table I). Each
of the established CTL clones had different allele sets.
 | Table I.TCR-β chain sequencing for
established CTL clones. |
Table I.
TCR-β chain sequencing for
established CTL clones.
| No. | TRBV | TRBJ | TRBD |
|---|
| Clone 24-4-2 |
18*01 |
1–2*01 |
1*01 |
| Clone 24-4-7 |
7-3*01 |
2–7*01 |
1*01 |
| Clone 24-2-10 |
7-6*01 |
2-1*01 |
2*01 |
CTL clone 24-4-2 is subject to
HLA-A*02:07 restriction
We investigated whether CTL clone 24-4-2 recognized
the GPC3144–152 peptide-HLA-A*02:01 complex
and the GPC3144–152 peptide-HLA-A*02:06
complex, as well as the GPC3144–152
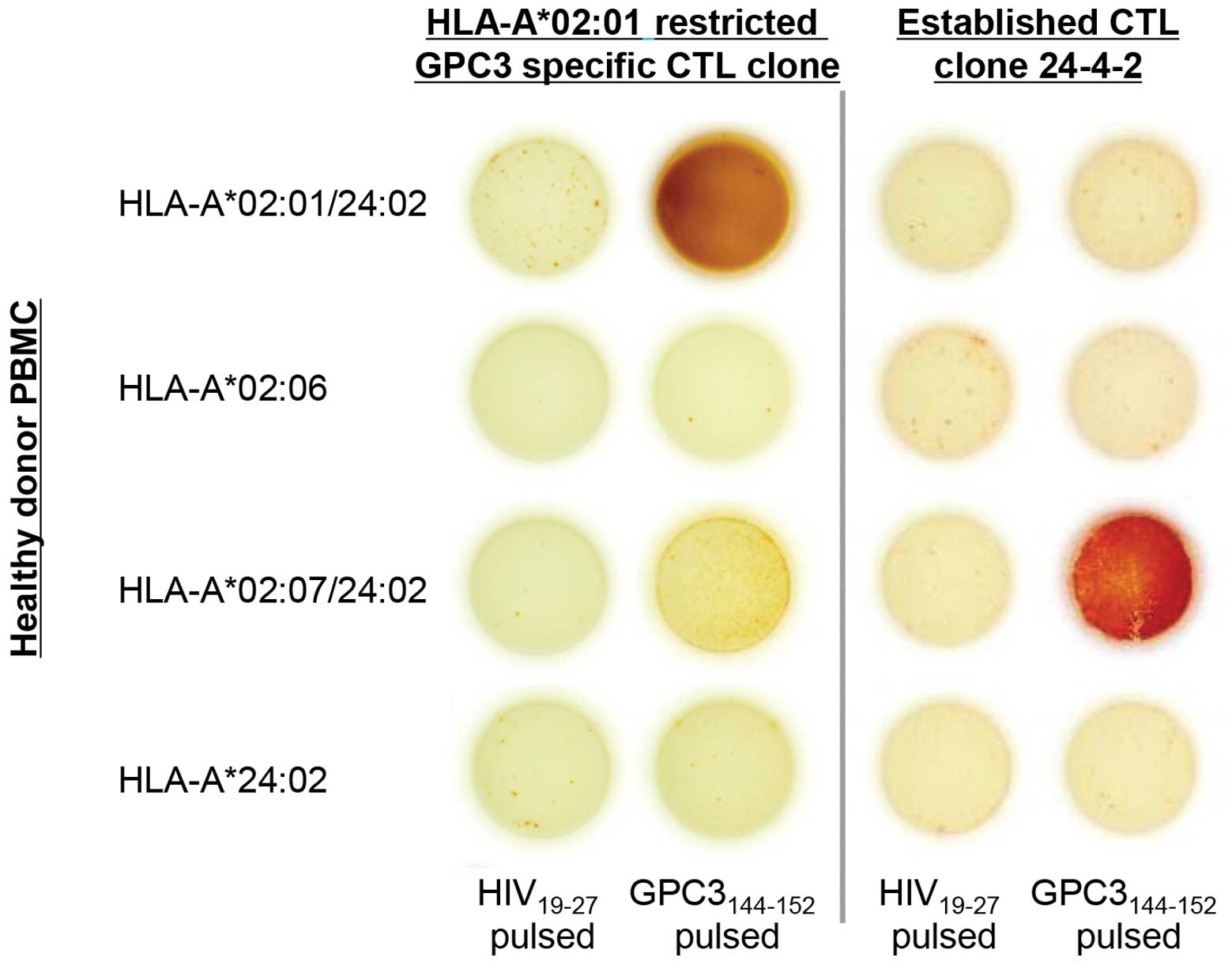
peptide-HLA-A*02:07 complex. Healthy donor PBMCs with
HLA-A*02:01, HLA-A*02:06,
HLA-A*02:07 and HLA-A*24:02 were used as the
targets, and an HLA-A*02:01-restricted, GPC3-specific
CTL clone, which is a previously established CTL clone (26), was used as the control. The
HLA-A*02:01-restricted CTL clone recognized only the
GPC3144–152 peptide-HLA-A*02:01 complex, and
CTL clone 24-4-2 recognized only the GPC3144–152
peptide-HLA-A*02:07 complex (Fig. 5). These outcomes indicate that CTL
clone 24-4-2 has HLA-A*02:07 restriction.
Discussion
Clinical trials of peptide-based vaccines are
underway in several parts of the world. However, the monitoring of
individual CTL post-vaccination has scarcely been reported in
immunotherapy trials. In the present study, we established
HLA-A*02:07+ GPC3144–152-specific
CTL clones from the PBMCs of a patient who showed a PR following
GPC3-derived peptide vaccination and we performed functional
analyses against established CTL clones.
This patient showed an increase in the number of
CTLs specific for the GPC3-derived peptide in the peripheral blood
after vaccination (Fig. 1A)
(27,28). Ten weeks after the start of
treatment, the GPC3144–152-specific CTL counts had
increased approximately 18-fold, as compared with the
pre-vaccination counts. In this case, analysis of the established
CTL clones after vaccination could lend support to the notion that
the vaccine-induced CTLs exert an antitumor effect, since few
GPC3144–152-specific CTLs were detected before
vaccination.
In the present study, we confirmed that
GPC3144–152-specific CTL clones are cytotoxic for both
GPC3144–152-pulsed 1–87 cells and
JHH-7/HLA-A*02:07 cells that express GPC3 peptide
endogenously. Confirming that the GPC3 peptide-specific CTL clones
kill cancer cells that express endogenously the antigen peptide is
important because antigen-derived and CTL-inducible peptides are
not necessarily presented by cancer cells that endogenously express
the antigen (31–33). Three established CTL clones showed
cytotoxic activities related to their avidity for
GPC3144–152-pulsed 1–87 cells and
JHH-7/HLA-A*02:07 cells that expressed the GPC3 peptide
endogenously. These results show that although CTLs with different
avidity can be isolated, only those CTLs with high avidity can kill
cancer cells that express the antigen peptide endogenously. Several
investigators have demonstrated a correlation between T-cell
avidity and target recognition by T-cell populations that recognize
murine tumor models and human cancers (34). Our results strongly support this
observation.
The TCR usage of antigen-specific T cells is thought
to be influenced by the affinity of the TCR for the antigen
peptide-HLA class I complex. Several studies on the TCR usage of
tumor-associated antigen (TAA)-specific T cells have used the
TRBV gene family (35–41).
These studies mainly analyzed the frequencies of TAA tetramer
positive CD8+ T cells. Although it is important to
examine quantitative aspects, such as the frequencies of TAA
tetramer positive CD8+ T cells, the cytotoxicity of
these T cells against cancer cells that express the TAA peptide
endogenously cannot be confirmed. Moreover, GPC3 dextramer positive
CD8+ T cells were not detected in the PBMCs of the
patients with HCC before GPC3 peptide vaccination (27,28).
To analyze biased usage of the TCR gene of GPC3 dextramer
positive CD8+ T cells in the patients with HCC before
and after GPC3 peptide vaccination, a new detection system with
greater sensitivity ex vivo will be required. In the present
study, we analyzed the TCR-β genes of the established
GPC3144–152-specific CTL clones, to confirm that these
CTL clones have different TCRs. Our experiments show that the
established CTL clones have different TCR-β-chain allele sets,
i.e., TRBV, TRBD and TRBJ alleles (Table I), and different CDR3 sequences
(data not shown). These results suggest that various GPC3-specific
CTLs are induced by GPC3144–152 peptide vaccination.
A*HLA-A*02:07 differs from
HLA-A*02:01 by a single non-conservative change (Y to C)
at residue 99. X-ray crystallographic data have identified position
99 as one of the residues forming the D secondary pocket, which
engages the residue at position 3 on peptide ligands (42–44).
Although hHLA-A*02:07 was originally not included in the
HLA-A2 supertype, cross-reactivity between HLA-A*02:07
and other A2 subtypes was detected at the functional level
(44,45). Moreover, this HLA molecule indeed
binds a subset of the peptide repertoire bound by other A2 subtypes
(44). For these reasons,
HLA-A*02:07 should also be included in the A2 supertype
(46). Ito et al (47) and Nonaka et al (48) reported that an HLA-A2-restricted
CTL line established from the tumor-infiltrating lymphocytes (TIL)
of an HLA-A*02:07-positive patient showed significant
cytotoxicities for HLA-A*02:01-, HLA-A*02:06-
and HLA-A*02:07-positive cancer cells. Therefore, we
examined whether the GPC3144–152-specific CTL clone
24-4-2, which was established from the PBMCs of an
HLA-A*02:07- positive patient with HCC, could recognize
HLA A-A*02:01 or HLA-A*02:06. However, this
CTL clone failed to recognize HLA-A*02:01 or
HLA-A*02:06.
We have reported previously on the detection via
immunohistochemical staining of massive infiltration of
CD8-positive T cells into the remaining liver tumor of this patient
(27). It was difficult to confirm
that these tumor-infiltrating CD8+ T cells have
specificity for GPC3. Currently, we are conducting clinical testing
of liver biopsies taken before and after GPC3 peptide vaccination
of patients with advanced HCC. Our aim is to reveal the GPC3
peptide-specific immune responses induced by the GPC3-derived
peptide vaccine in both the peripheral blood and the tumor. We are
analyzing the TCR gene sequences of CD8 or GPC3 dextramer
positive T cells in both the peripheral blood and tumor. Already in
this trial, a remarkable clinical effect has been observed for an
HLA-A*02:07-positive patient with HCC who received
GPC3144–152 peptide vaccination (49).
HLA-A*02:07 is present in the
populations of East Asia, South-East Asia (7%), and northern India
(11.5%) (26,50–52).
In southern China, the frequency of the HLA-A*02:07
allele is reported to be even higher than the frequency of the
HLA-A*02:01 allele (53,54).
In addition, about 75% of liver cancer cases occur in South-East
Asia, including China, Hong Kong, Taiwan, Korea, India and Japan
(55). Taking together these
previous reports and our results, it appears that
HLA-A*02:07-positive patients with HCC are good
candidates for GPC3144–152 peptide vaccination. Further
studies will be necessary to prove the clinical efficacy of GPC3
peptide vaccination for advanced HCC.
In conclusion, we present substantial evidence that
GPC3144–152-specific CTLs with different TCR allele sets
that are induced in patients with HCC who show a PR following
GPC3144–152 peptide vaccination indicate not only high
avidity but also natural antigen-specific killing activity against
tumor cells.
Acknowledgements
We thank Dr Ryo Abe and Dr Toshihiro
Suzuki for providing the pcDNA3.1 construct that expresses
HLA-A*02:07. This study was supported in part by Health
and Labor Science Research Grants for Clinical Research and Third
Term Comprehensive Control Research for Cancer from the Ministry of
Health, Labor and Welfare, Japan and the National Cancer Center
Research and Development Fund.
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Zidan A, Scheuerlein H, Schüle S,
Settmacher U and Rauchfuss F: Epidemiological pattern of hepatitis
B and hepatitis C as etiological agents for hepatocellular
carcinoma in iran and worldwide. Hepat Mon. 12:e68942012.PubMed/NCBI
|
|
4.
|
Llovet JM, Ricci S, Mazzaferro V, et al:
Sorafenib in advanced hepatocellular carcinoma. N Engl J Med.
359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Cheng AL, Kang YK, Chen Z, et al: Efficacy
and safety of sorafenib in patients in the Asia-Pacific region with
advanced hepatocellular carcinoma: a phase III randomised,
double-blind, placebo-controlled trial. Lancet Oncol. 10:25–34.
2009. View Article : Google Scholar
|
|
6.
|
Kim HY and Park JW: Molecularly targeted
therapies for hepatocellular carcinoma: Sorafenib as a stepping
stone. Dig Dis. 29:303–309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Morimoto M, Numata K, Kondo M, et al:
Higher discontinuation and lower survival rates are likely in
elderly Japanese patients with advanced hepatocellular carcinoma
receiving sorafenib. Hepatol Res. 41:296–302. 2011. View Article : Google Scholar
|
|
8.
|
Greten TF, Manns MP and Korangy F:
Immunotherapy of hepatocellular carcinoma. J Hepatol. 45:868–878.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Mizukoshi E, Nakamoto Y, Arai K, et al:
Comparative analysis of various tumor-associated antigen-specific
t-cell responses in patients with hepatocellular carcinoma.
Hepatology. 53:1206–1216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Filmus J, Shi W, Wong ZM and Wong MJ:
Identification of a new membrane-bound heparan sulphate
proteoglycan. Biochem J. 311:561–565. 1995.
|
|
11.
|
Filmus J and Selleck SB: Glypicans:
proteoglycans with a surprise. J Clin Invest. 108:497–501. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Nakatsura T, Yoshitake Y, Senju S, et al:
Glypican-3, over-expressed specifically in human hepatocellular
carcinoma, is a novel tumor marker. Biochem Biophys Res Commun.
306:16–25. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Capurro M, Wanless IR, Sherman M, Deboer
G, Shi W, Miyoshi E and Filmus J: Glypican-3: a novel serum and
histochemical marker for hepatocellular carcinoma.
Gastroenterology. 125:89–97. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Nakatsura T and Nishimura Y: Usefulness of
the novel oncofetal antigen glypican-3 for diagnosis of
hepatocellular carcinoma and melanoma. BioDrugs. 19:71–77. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Shirakawa H, Kuronuma T, Nishimura Y, et
al: Glypican-3 is a useful diagnostic marker for a component of
hepatocellular carcinoma in human liver cancer. Int J Oncol.
34:649–656. 2009.PubMed/NCBI
|
|
16.
|
Shirakawa H, Suzuki H, Shimomura M, et al:
Glypican-3 expression is correlated with poor prognosis in
hepatocellular carcinoma. Cancer Sci. 100:1403–1407. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Motomura Y, Senju S, Nakatsura T, et al:
Embryonic stem cell-derived dendritic cells expressing glypican-3,
a recently identified oncofetal antigen, induce protective immunity
against highly metastatic mouse melanoma, B16-F10. Cancer Res.
66:2414–2422. 2006. View Article : Google Scholar
|
|
18.
|
Motomura Y, Ikuta Y, Kuronuma T, et al:
HLA-A2 and -A24-restricted glypican-3-derived peptide vaccine
induces specific CTLs: preclinical study using mice. Int J Oncol.
32:985–990. 2008.PubMed/NCBI
|
|
19.
|
Iwama T, Horie K, Yoshikawa T, et al:
Identification of an H2-Kb or H2-Db restricted and
glypican-3-derived cytotoxic T-lymphocyte epitope peptide. Int J
Oncol. 42:831–838. 2013.PubMed/NCBI
|
|
20.
|
Komori H, Nakatsura T, Senju S, et al:
Identification of HLA-A2- or HLA-A24-restricted CTL epitopes
possibly useful for glypican-3-specific immunotherapy of
hepatocellular carcinoma. Clin Cancer Res. 12:2689–2697. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Nakatsura T, Komori H, Kubo T, et al:
Mouse homologue of a novel human oncofetal antigen, glypican-3,
evokes T-cell-mediated tumor rejection without autoimmune reactions
in mice. Clin Cancer Res. 10:8630–8640. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Imanish T, Akaza T, Kimura A, Tokunaga K
and Gojobori T: Allele and haplotype frequencies for HLA and
complement loci in various ethnic groups. HLA. 1991, 1. Tsuji K,
Aizawa M and Sasazuki T: Oxford University Press; Oxford: pp.
1065–1220. 1992
|
|
23.
|
Sidney J, Grey HM, Kubo RT and Sette A:
Practical, biochemical and evolutionary implications of the
discovery of HLA class I supermotifs. Immunol Today. 17:261–266.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Yasuda N, Tsuji K, Aizawa M, et al: HLA
antigens in Japanese populations. Am J Hum Genet. 28:390–399.
1976.
|
|
25.
|
Ellis JM, Henson V, Slack R, Ng J,
Hartzman RJ and Katovich Hurley C: Frequencies of HLA-A2 alleles in
five U.S. population groups. Predominance of A*02011 and
identification of HLA-A*0231. Hum Immunol. 61:334–340.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Mehra NK, Jaini R, Rajalingam R,
Balamurugan A and Kaur G: Molecular diversity of
HLA-A*02 in Asian Indians: predominance of
A*0211. Tissue Antigens. 57:502–507. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Sawada Y, Yoshikawa T, Nobuoka D, et al:
Phase I trial of a glypican-3-derived peptide vaccine for advanced
hepatocellular carcinoma: immunologic evidence and potential for
improving overall survival. Clin Cancer Res. 18:3686–3696. 2012.
View Article : Google Scholar
|
|
28.
|
Yoshikawa T, Nakatsugawa M, Suzuki S, et
al: HLA-A2-restricted glypican-3 peptide-specific CTL clones
induced by peptide vaccine show high avidity and antigen-specific
killing activity against tumor cells. Cancer Sci. 102:918–925.
2011. View Article : Google Scholar
|
|
29.
|
Nobuoka D, Yoshikawa T, Sawada Y, Fujiwara
T and Nakatsura T: Peptide vaccines for hepatocellular carcinoma.
Hum Vaccin Immunother. 9:210–212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Sawada Y, Sakai M, Yoshikawa T, Ofuji K
and Nakatsura T: A glypican-3-derived peptide vaccine against
hepatocellular carcinoma. Oncoimmunology. 1:1448–1450. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Purbhoo MA, Li Y, Sutton DH, et al: The
HLA A*0201-restricted hTERT(540–548) peptide is not
detected on tumor cells by a CTL clone or a high-affinity T-cell
receptor. Mol Cancer Ther. 6:2081–2091. 2007.PubMed/NCBI
|
|
32.
|
Nakatsugawa M, Horie K, Yoshikawa T, et
al: Identification of an HLA-A*0201-restricted cytotoxic
T lymphocyte epitope from the lung carcinoma antigen, Lengsin. Int
J Oncol. 39:1041–1049. 2011.
|
|
33.
|
Guo Y, Zhu Y and Sun S: Identification and
functional studies of HLA-A0201 restricted CTL epitopes in the X
protein of hepatitis B virus. Acta Virol. 55:107–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
McKee MD, Roszkowski JJ and Nishimura MI:
T cell avidity and tumor recognition: implications and therapeutic
strategies. J Transl Med. 3:352005. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Harada Y and Kawase I: Single cell-based T
cell receptor gene analysis reveals existence of expanded WT1
(Wilms’ tumor gene) product-specific T cell clones in leukemia
patients but not healthy volunteers. Med J Osaka Univ. 50:1–12.
2007.
|
|
36.
|
Tanaka-Harada Y, Kawakami M, Oka Y, et al:
Biased usage of BV gene families of T-cell receptors of WT1 (Wilms’
tumor gene)-specific CD8+ T cells in patients with
myeloid malignancies. Cancer Sci. 101:594–600. 2010.
|
|
37.
|
Morimoto S, Oka Y, Tsuboi A, et al: Biased
usage of T cell receptor β-chain variable region genes of Wilms’
tumor gene (WT1)-specific CD8+ T cells in patients with
solid tumors and healthy donors. Cancer Sci. 103:408–414. 2012.
|
|
38.
|
Valmori D, Dutoil V, Lienard D, et al:
Tetramer-guided analysis of TCR beta-chain usage reveals a large
repertoire of melan-A-specific CD8+ T cells in melanoma
patients. J Immunol. 165:533–538. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Mandruzzato S, Rossi E, Bernardi F, et al:
Large and dissimilar repertoire of Melan-A/MART-1-specific CTL in
metastatic lesions and blood of a melanoma patient. J Immunol.
169:4017–4024. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Zhou J, Dudley ME, Rosenberg SA and
Robbins PF: Selective growth, in vitro and in vivo, of individual T
cell clones from tumor-infiltrating lymphocytes obtained from
patients with melanoma. J Immunol. 173:7622–7629. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Akiyama Y, Maruyama K, Tai S, et al:
Characterization of a MAGE-1-derived HLA-A24 epitope-specific CTL
line from a Japanese metastatic melanoma patient. Anticancer Res.
29:647–655. 2009.PubMed/NCBI
|
|
42.
|
Saper MA, Bjorkman PJ and Wiley DC:
Refined structure of the human histocompatibility antigen HLA-A2 at
2.6 A resolution. J Mol Biol. 219:277–319. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Madden DR, Garboczi DN and Wiley DC: The
antigenic identity of peptide-MHC complexes: a comparison of the
conformations of five viral peptides presented by HLA-A2. Cell.
75:693–708. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Sidney J, del Guercio MF, Southwood S,
Hermanson G, Maewal A, Appella E and Sette A: The HLA-A0207 peptide
binding repertoire is limited to a subset of the A0201 repertoire.
Hum Immunol. 58:12–20. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Rivoltini L, Loftus DJ, Barracchini K, et
al: Binding and presentation of peptides derived from melanoma
antigens MART-1 and glycoprotein-100 by HLA-A2 subtypes:
implications for peptide-based immunotherapy. J Immunol.
156:3882–3891. 1996.PubMed/NCBI
|
|
46.
|
Sette A and Sidney J: HLA supertypes and
supermotifs: a functional perspective on HLA polymorphism. Curr
Opin Immunol. 10:478–482. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Ito M, Shichijo S, Tsuda N, Ochi M,
Harashima N, Saito N and Itoh K: Molecular basis of T cell-mediated
recognition of pancreatic cancer cells. Cancer Res. 61:2038–2046.
2001.PubMed/NCBI
|
|
48.
|
Nonaka Y, Tsuda N, Shichijo S, et al:
Recognition of ADP-ribosylation factor 4-like by HLA-A2-restricted
and tumor-reactive cytotoxic T lymphocytes from patients with brain
tumors. Tissue Antigens. 60:319–327. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Sawada Y, Yoshikawa T, Fujii S, et al:
Remarkable tumor lysis in a hepatocellular carcinoma patient
immediately following glypican-3-derived peptide vaccination: an
autopsy case. Hum Vaccin Immunother. 9:Mar 6–2013.(Epub ahead of
print).
|
|
50.
|
Krausa P, Brywka M III, Savage D, et al:
Genetic polymorphism within HLA-A*02: significant
allelic variation revealed in different populations. Tissue
Antigens. 45:223–231. 1995.PubMed/NCBI
|
|
51.
|
Chang CX, Tan AT, Or MY, et al:
Conditional ligands for Asian HLA variants facilitate the
definition of CD8(+) T-cell responses in acute and chronic viral
diseases. Eur J Immunol. 43:1109–1120. 2013.PubMed/NCBI
|
|
52.
|
Chen KY, Liu J and Ren EC: Structural and
functional distinctiveness of HLA-A2 allelic variants. Immunol Res.
53:182–190. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Shieh DC, Lin DT, Yang BS, Kuan HL and Kao
KJ: High frequency of HLA-A*0207 subtype in Chinese
population. Transfusion. 36:818–821. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Cheng LH, Jin SZ, Gao SQ, Li Z, Zou HY,
Wang DM and Wu GG: Difference in HLAA*02 allele
distribution between Han populations in south and north China. Di
Yi Jun Yi Da Xue Xue Bao. 25:321–324. 2005.(In Chinese).
|
|
55.
|
Mohana Devi S, Balachandar V, Arun M,
Suresh Kumar S, Balamurali Krishnan B and Sasikala K: Analysis of
genetic damage and gene polymorphism in hepatocellular carcinoma
(HCC) patients in a South Indian population. Dig Dis Sci.
58:759–767. 2013.PubMed/NCBI
|