Introduction
Breast cancer is a heterogeneous disease, and it has
long been appreciated that tumors with different biological
features have different clinical outcomes and responses to therapy.
At present, prognosis and treatment selection in breast cancer are
based on characterization of tumor growth factor receptor status
involving estrogen receptor (ER), progesterone receptor (PR) and
C-erbB2. These markers can be used to define four functional groups
of tumors: i) hormone receptor-positive; ii) C-erbB2-negative; iii)
hormone receptor-negative, C-erbB2-negative (triple-negative
tumors); and iv) C-erbB2 overexpressing tumors with or without
hormone-receptor expression (1).
Triple-negative breast cancer, which is defined as
being negative for ER, PR and C-erbB2, is associated with
aggressive clinical behavior and poor prognosis. These cancers have
become the subject of research interest because they do not benefit
from hormonal therapies or treatments targeted against C-erbB2
receptors, and because they appear to be prevalent in breast
cancer; one study reported 25 triple-negative cell lines out of 51
breast cancer cell lines that were examined (2). These triple-negative cell lines will
be useful in research on tumor biology that relates to aggressive
clinical behavior and poor prognosis of the tumors, as well as
prediction of response to therapy and discovery of new therapeutic
targets.
Breast tumor cells frequently co-exist with
surrounding stroma such as normal epithelial, fibroblast and
mesothelial cells (3,4). Many breast tumor derived cell lines
have been established from metastatic tumors, raising questions as
to their relationship to primary tumors (3). This is clearly unrepresentative of
the diverse types of tumor reflected by the specific types, various
grades or stages and indications for tumor progression that are
observed in primary breast cancer. For these reasons it would be
more clinically relevant to use cells that are derived directly
from a primary tumor, that is the target of most drug therapies
(5).
We report the characterization of seven human breast
cancer cell lines designated SNU-306, SNU-334, SNU-1528, SNU-1553,
SNU-1581, SNU-1958 and SNU-2372 including two triple-negative cell
lines (SNU-1958 and SNU-2372), which were derived from three
primary breast carcinomas, two pleural effusions, one pericardial
effusion, and one ascitic fluid obtained from Korean breast
carcinoma patients.
We describe the cell phenotypes including the
histopathology of the primary tumors and their in vitro
growth characteristics; DNA fingerprinting analysis to verify the
authenticity of each of the seven breast cancer cell lines;
expressions levels of ER-α, PR, C-erbB2,
E-cadherin, COX-2, MDR and MXR(BCRP)
genes; and alteration of p53 and epidermal growth factor
receptor (EGFR) genes.
Materials and methods
Cell line establishment and
maintenance
Cell lines were established from three primary
breast carcinomas, one pleural effusion and one pericardial
effusion of breast carcinomas. Solid tumors were finely minced with
scissors and dissociated into small aggregates by pipetting.
Appropriate amounts of finely minced neoplastic-tissue fragments
were seeded into 25-cm2 flasks. Pleural effusions were
collected, pelleted, washed and resuspended in growth medium. Tumor
cells were initially cultured in ACL-4 medium supplemented with 5%
heat-inactivated fetal bovine serum (6–8).
After establishment, these cell lines were maintained in RPMI-1640
containing 10% heat-inactivated fetal bovine serum. Initial cell
passages were performed when heavy tumor cell growth was observed
and subsequent passages were performed every one or two weeks.
Adherent cultures were passaged at subconfluence after
trypsinization. Cultures were maintained in humidified incubators
at 37°C in an atmosphere of 5% CO2 and 95% air. Breast
cancer cell lines MCF-7, MDA-MD231 and SK-BR3 obtained from the
Korean Cell Line Bank were used as polymerase chain reaction (PCR)
controls.
Growth properties and morphology in
vitro
Population doubling times were determined by seeding
0.5–3×105 viable cells into 25-cm2 flasks and
counting daily for at least 14 days. Cultures were fed every three
or four days and 24 h prior to counting. Cell viability was
determined by a dye-exclusion method using 0.4% trypan blue. PCR
and microscopic examination were used to test for mycoplasma
(e-Myco Mycoplasma Detection kit; Intron Biotechnology, Gyonggi,
Korea) or bacterial contamination, respectively. For morphological
studies, cells were grown on 75-cm2 culture flasks and
observed daily by phase-contrast microscopy.
Nucleic acid isolation and cDNA
synthesis
Genomic DNA and total RNA were isolated from washed
cell pellets. Total genomic DNA was extracted according to a
standard sodium dodecyl sulfate-proteinase K procedure, and total
cellular RNA was extracted according to the manufacturer’s
instructions (Intron Biotechnology). For cDNA synthesis, 2
μg of total RNA was reverse transcribed using random oligo
(dT) primer, dNTPs, and 1 μl (200 units) of Superscript™ II
reverse transcriptase (Life Technologies, Frederick, MD, USA) in a
final volume of 20 μl for 75 min at 42°C after a 10-min
denaturation at 70°C. A total of 80 μl of distilled water
was then added to the reverse-transcription reaction mixture.
DNA profiles
DNA was PCR amplified at loci containing the highly
polymorphic microsatellite markers D1S1586 and D3S1765. PCR
products were denatured using 95% formamide and electrophoresed on
a sequencing gel for 2 h at a constant 60 W. Gels were dried and
visualized autoradiographically. DNA was also amplified using
AmpFlSTR identifiler PCR amplification kit (Applied Biosystems,
Foster City, CA, USA). PCR amplified 15 tetranucleotide repeat loci
and gender determining marker at loci containing highly polymorphic
microsatellite markers. Amplified products were analyzed using an
ABI 3730 Genetic analyzer (Applied Biosystems).
Expression of ER-α, PR, C-erbB2, COX-2
and E-cadherin genes
For the mRNA expression analysis of ER-α,
ER-β (9), PR
(10), C-erbB2 (11), E-cadherin (12), COX-2 (13), MDR1 (14) and MXR (15) genes in the seven cell lines, cDNA
was amplified in 25 μl of a PCR reaction mix using 1
μl of reverse-transcription reaction, primers and 0.5 units
of Taq DNA polymerase. PCR amplification was carried out in a
programmable thermal cycler. Primers for β-actin were used
to confirm RNA integrity. Both genes and β-actin RT-PCR
reactions used the same cDNA synthesis. Amplified DNA fragments
were fractionated in a 2% agarose gel and stained with ethidium
bromide.
Western blot analysis
Western blot analysis was performed as described
previously (16). Briefly, cell
homogenates containing equivalent amounts of protein were
centrifuged at 4,000 x g, and the supernatant fractions subjected
to SDS-PAGE. Following electrophoresis, proteins were transferred
to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica,
MA, USA) blocked by incubation for 2 h at 48°C in 1% Tween-20-TBS
buffer containing 1.5% non-fat dry milk (Bio-Rad, Hercules, CA,
USA) and 1 mM MgCl2. Membranes were incubated for 2 h at
room temperature with primary antibodies against progesterone
receptor (Ventana, Tucson, AZ, USA), estrogen receptor α, C-erbB2
(both from Dakocytomation, Carpinteria, CA, USA), or actin
(Sigma-Aldrich, St. Louis, MO, USA). Next, membranes were washed
for 3×15 min with blocking solution, and incubated with diluted
HRP-conjugated secondary antibody (Southern Biotech, Birmingham,
UK) for 1 h at room temperature. This was followed by washing with
blocking solution (3×15 min), incubation with WEST-ZOL plus
chemiluminescence reagent (Intron Biotechnology) for 1 min, and
exposure to film (Kodak Blue XB-1).
Detection of alterations in the p53 and
EGFR genes
Mutational screening of exons 4–8 of p53 was
performed by direct sequencing analysis. Oligonucleotide primers
for the genomic PCR and PCR procedures were as described previously
(17). Mutations of EGFR were also
screened through exons 18–24 by direct sequencing analysis
(18). PCR reactions were carried
out in 25 μl containing 100 ng genomic DNA, 2.5 pmoles of
each primer, four dNTPs at 250 μM each, 0.5 units of Taq
polymerase and PCR reaction buffer. Reactions were initiated by
denaturation for 5 min at 94°C and amplification was conducted over
35 cycles in a programmable thermal cycler. Fresh PCR products were
sequenced using a Taq dideoxy terminator cycle sequencing kit on an
ABI 3730 DNA sequencer (Applied Biosystems).
Taxol cytotoxicity assay
A colorimetric assay using the tetrazolium salt
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
(Sigma-Aldrich) was used to assess the cytotoxicity of taxol
(Sigma-Aldrich).
Results
A total of seven breast cancer cell lines derived
from Korean patients were established in AR5 medium. Population
doubling times ranged from 47–152 h and cell viability after
thawing was about 85% (Table I).
All cell lines were free of contamination from either bacteria or
mycoplasma.
 | Table I.Origin and in vivo
characteristics of seven SNU breast cancer cell lines. |
Table I.
Origin and in vivo
characteristics of seven SNU breast cancer cell lines.
| Cell line | Gender/age | Tumor origin | Date of
initiation | Histology | Size | TNM stage | Survival
(months) | Remark |
|---|
| SNU-306 | F/28 | Primary | 1989.12.18 | Infiltrating ductal
carcinoma | 9 cm | pT3N3(16/25)M0 | 24 | |
| SNU-334 | F/40 | Primary | 1990.02.01 | Infiltrating ductal
carcinoma | 12 cm | pT3N2(9/13)M0 | 12 | |
| SNU-1528 | F/46 | Primary | 1998.02.13 | Infiltrating ductal
carcinoma | 3.5 cm | pT2N3(35/35)M0 | 7 | |
| SNU-1553a | F/43 | Pleural
effusion | 1998.11.05 | Metastatic
carcinoma | | rpM1 | | Resection 1 year
previously |
| | | Infiltrating ductal
carcinoma | 11 cm | pT2N2(7/12)M0 | 14 | |
| SNU-1581b | F/50 | Pericardial
effusion | 1999.03.18 | Metastatic
carcinoma | | rpM1 | | Resection 3 years
previously |
| | | Infiltrating ductal
carcinoma | 4 cm | T2N0(0/9)M0 | 27 | |
| SNU-1598c | F/55 | Ascitic fluid | 2002.03.08 | Poorly
differentiated metastatic carcinoma | | rpM1 | | Resection 12 years
previously |
| | | Infiltrating ductal
carcinoma | 3 cm | T2N1(5/8)M0 | 148 | |
| SNU-2372d | F/55 | Pleural
effusion | 2007.11.07 | Metastatic
carcinoma | | rpM1 | | |
| | | Infiltrating ductal
carcinoma | 4.2 cm | T2N1(2/15) | 28 | Resection 2 years
previously |
Three of the tumors were obtained from primary
breast carcinomas, while SNU-1553 and SNU-2372 were obtained from
pleural effusion, SNU-1581 from a pericardial effusion and SNU-1958
from ascitic fluid (Fig. 1F). The
three tumors from primary breast cancer were infiltrating ductal
carcinoma. All showed marked nuclear and histologic atypism. Ductal
carcinoma in situ component was present in the cell lines
derived from all patients except SNU-334. In the patient from whom
the SNU-1581 cell line was derived, the stage IIA infiltrating
ductal carcinoma had been removed 3 years prior to the occurrence
of malignant pericardial effusion. In the patient from whom the
SNU-1958 cell line was derived, stage IIA infiltrating ductal
carcinoma was removed 10 years prior to the recurrence in the
peritoneal cavity with ascites. In the patient from whom the
SNU-2372 cell line was derived, multiple cervical, axillary lymph
node, and chest wall recurrence was detected 1 month after
resection of stage IIA breast cancer, and the cell line was
established from the pleural effusion. Characteristics of the cell
lines are summarized in Table
II.
 | Table II.In vitro characteristics of
seven SNU breast cancer cell lines. |
Table II.
In vitro characteristics of
seven SNU breast cancer cell lines.
| Cell line | Growth pattern | Viability | Doubling time | Cell
morphology |
|---|
| SNU-306 | Adherent | 85 | 152 | Pleomorphic |
| SNU-334 | Floating
aggregates | 88 | 80 | Round to oval |
| SNU-1528 | Adherent | 83 | 110 | Polygonal |
| SNU-1553 | Adherent | 91 | 89 | Pleomorphic |
| SNU-1581 | Adherent | 89 | 47 | Spindle to
pleomorphic |
| SNU-1958 |
Adherent/floating | 87 | 53 | Polygonal, round to
oval |
| SNU-2372 | Adherent | 82 | 78 | Polygonal |
Table II and
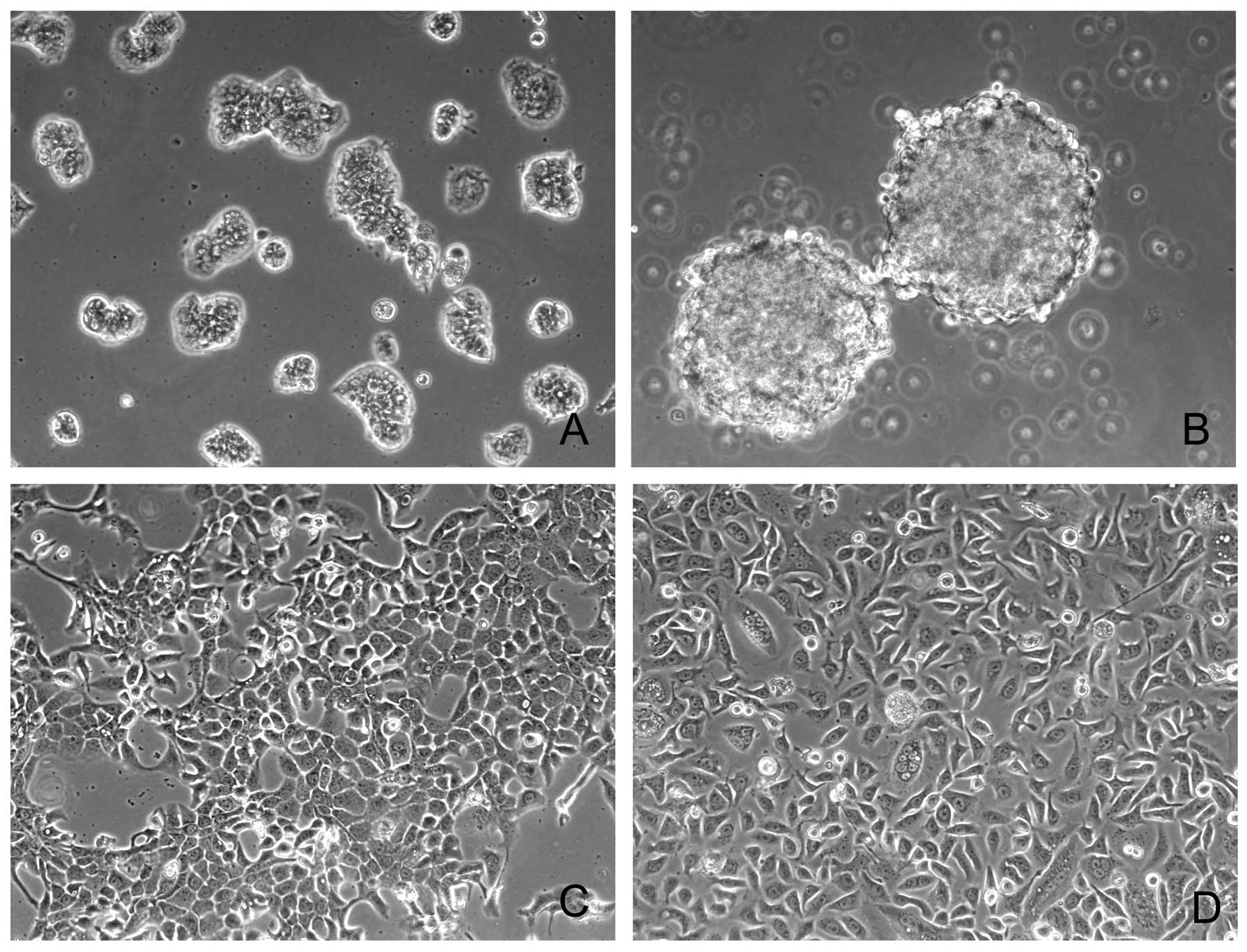
Fig. 1 summarize the morphologic
observations. Briefly, SNU-306, SNU-1528, SNU-1553 and SNU-2372
grew in vitro as adherent monolayers, the SNU-334 grew as
floating aggregates, and SNU-1581 and SNU-1958 cell lines grew as
both floating aggregates and monolayers (Fig. 1A–E, G and H). SNU-306 cell line
grew as various sized colonies consisting of tightly packed small
cells (Fig. 1A). SNU-334 cells
were round or oval (Fig. 1B).
SNU-1528 epithelial cells were spindle- or polygonal-shaped
(Fig. 1C). SNU-1553 cells were
polygonal in shape and displayed prominent nucleoli; also some
giant cells containing several nuclei were evident (Fig. 1D). SNU-1581 epithelial cells had a
spindle or polygonal shape (Fig.
1E). SNU-1958 cells were pleomorphically shaped (Fig. 1G) and SNU-2372 cells were polygonal
in shape and displayed prominent nucleoli (Fig. 1H).
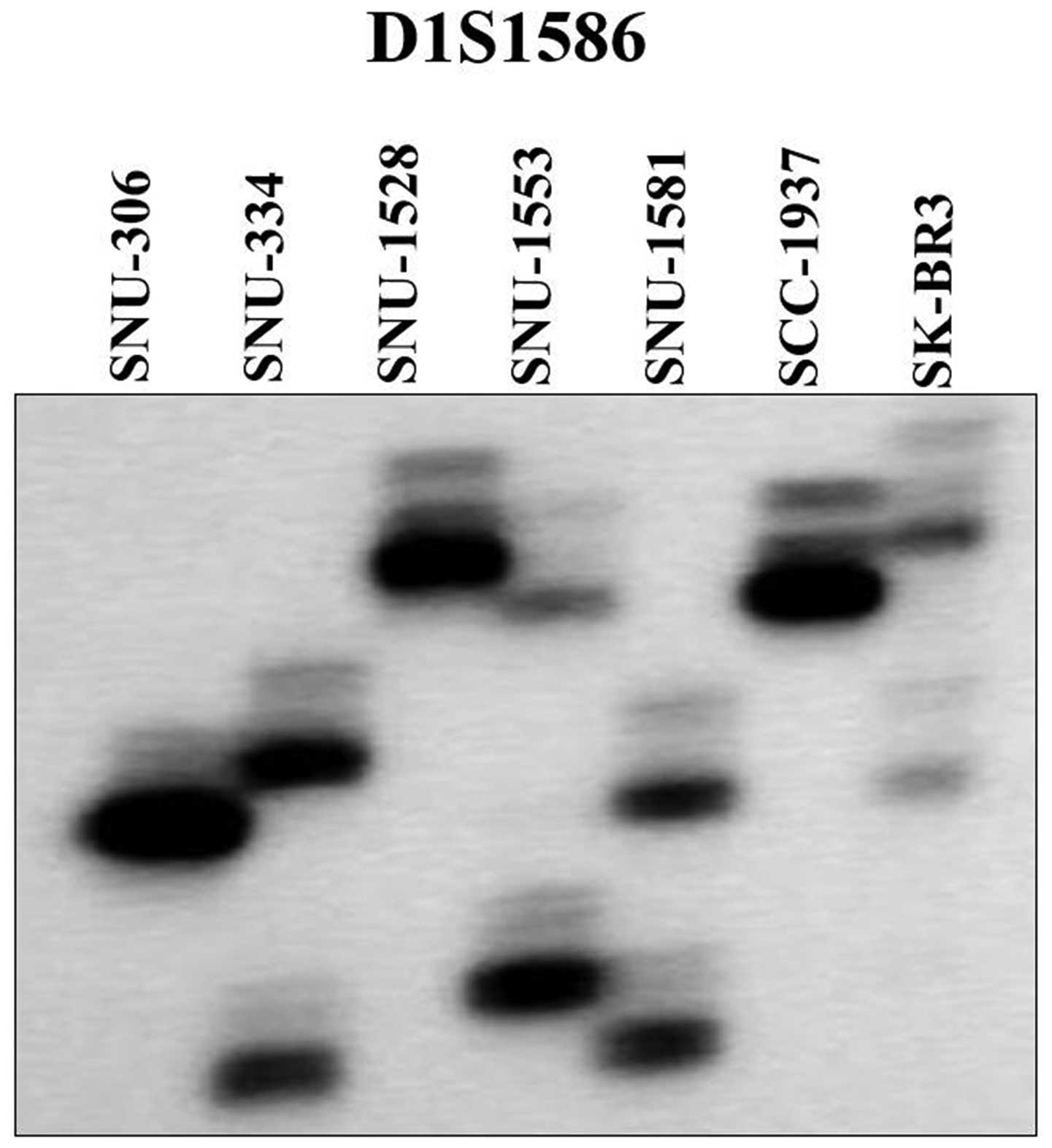
Use of two highly polymorphic microsatellite markers
showed that the seven breast cancer cell lines were unique and
unrelated (Fig. 2), and helped
exclude the possibility of cross-contamination among the cell
lines. DNA fingerprinting using the AmpFlSTR identifiler PCR
amplification kit revealed the heterogeneous distribution of 15
tetranucleotide repeat loci and Amelogen gender determining marker
in each cell line, and confirmed the lack of cross-contamination
(Table III).
 | Table III.DNA fingerprinting analysis using 16
STR loci for the seven newly established breast cancer cell
lines. |
Table III.
DNA fingerprinting analysis using 16
STR loci for the seven newly established breast cancer cell
lines.
| Loci | SNU-306 | SNU-334 | SNU-1528 | SNU-1553 | SNU-1581 | SNU-1958 | SNU-2372 |
|---|
| D8S1179 | 13, 14 | 13, 15 | 13, 14 | 12 | 16 | | |
| D21S11 | 30 | 30, 32 | 30, 30.2 | 30 | 30, 32.2 | | |
| D7S820 | 11, 12 | 10, 11 | 11 | 12 | 8, 10 | 8, 11 | 11 |
| CSF1P0 | 11 | 12 | 9, 10 | 11 | 10, 11 | 10, 13 | 12 |
| D3S1358 | 15, 17 | 15 | 16 | 17 | 16 | 15 | 15.2, 18.2 |
| TH01 | 9 | 9 | 5.3 | 7, 9 | 6, 8 | | |
| D13S317 | 8, 10 | 12 | 9 | 11 | 8 | 9, 10 | 9, 11 |
| D16S539 | 13 | 9 | 9, 11 | 13 | 10 | | |
| D2S1338 | 25 | 17 | 25 | 19 | 18 | | |
| D19S433 | 13 | 14, 14.2 | 14 | 13, 14 | 13 | | |
| vWA | 16, 17 | 18 | 17 | 16, 17 | 17 | 16, 17 | 17 |
| TPOX | 8 | 11 | 11 | 11, 12 | 8, 9 | 8 | 8 |
| D18S51 | 13 | 14, 15 | 18 | 13 | 13 | | |
| Amelogenin | X, X | X, X | X, X | X, X | X, X | X, X | X, X |
| D5S818 | 11 | 11 | 11, 12 | 10 | 12, 13 | 9, 11 | 12 |
| FGA | 23 | 20 | 24 | 22, 23 | 19, 24 | 19, 21 | 25 |
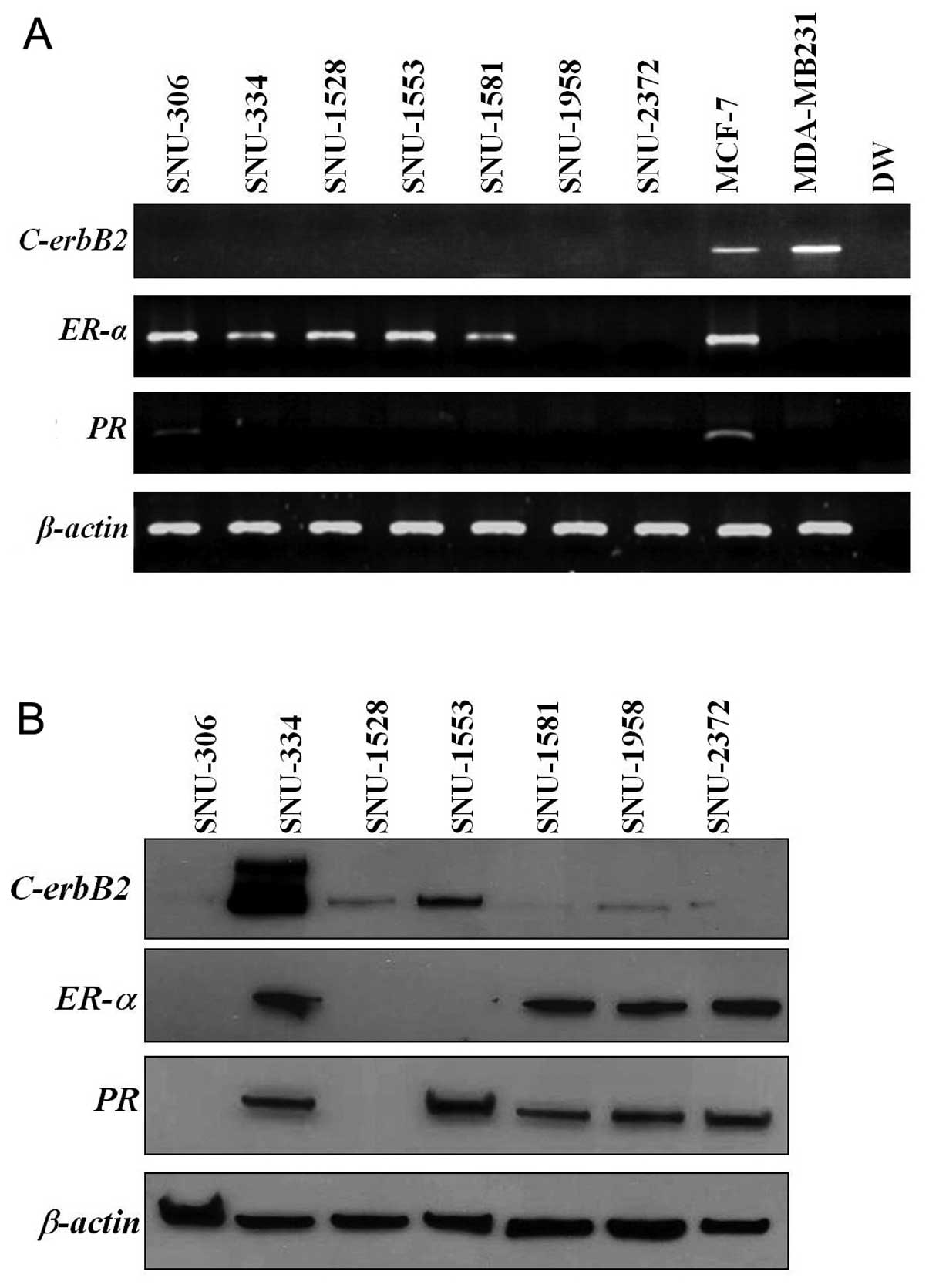
In RT-PCR analysis, ER-α was expressed in
SNU-306, SNU-334, SNU-1528, SNU-1553 and SNU-1581. PR was
expressed only in the SNU-306 and C-erbB2 was not expressed
in any of the cell lines (Fig.
3A). These combinations revealed three cell line groups:
ER-α and PR expression without C-erbB2
expression (SNU-306), ER-α expression without PR and
C-erbB2 expression (SNU-334, SNU-1528, and SNU-1553), and no
expression of ER-α, PR and C-erbB2
(triple-negative; SNU-1958 and SNU-2372) (Table IV). In western blot analysis,
C-erbB2 was highly expressed in SNU-334 and weakly expressed
in SNU-1528, SNU-1553 and SNU-1958 cell lines. ER-α was
expressed in SNU-334, SNU-1581, SNU-1958 and SNU-2372 cell lines.
PR was expressed in the SNU-334, SNU-1553, SNU-1581,
SNU-1958 and SNU-2372 cell lines (Fig.
3B).
 | Table IV.Expressions of genes in breast cancer
cell lines. |
Table IV.
Expressions of genes in breast cancer
cell lines.
| Cell line | C-erbB2a | ER-αa | PRa | COX-2 | MDR1 | MXR | E-cadherin |
|---|
| SNU-306 | −/− | +/− | +/− | + | − | − | + |
| SNU-334 | −/+ | +/+ | −/+ | − | − | − | + |
| SNU-1528 | −/+ | +/− | −/− | + | − | − | + |
| SNU-1553 | −/+ | +/− | −/+ | − | − | + | + |
| SNU-1581 | −/− | +/+ | −/+ | − | − | − | − |
| SNU-1958 | −/+ | −/+ | −/+ | − | + | − | + |
| SNU-2372 | −/− | −/+ | −/+ | + | − | − | + |
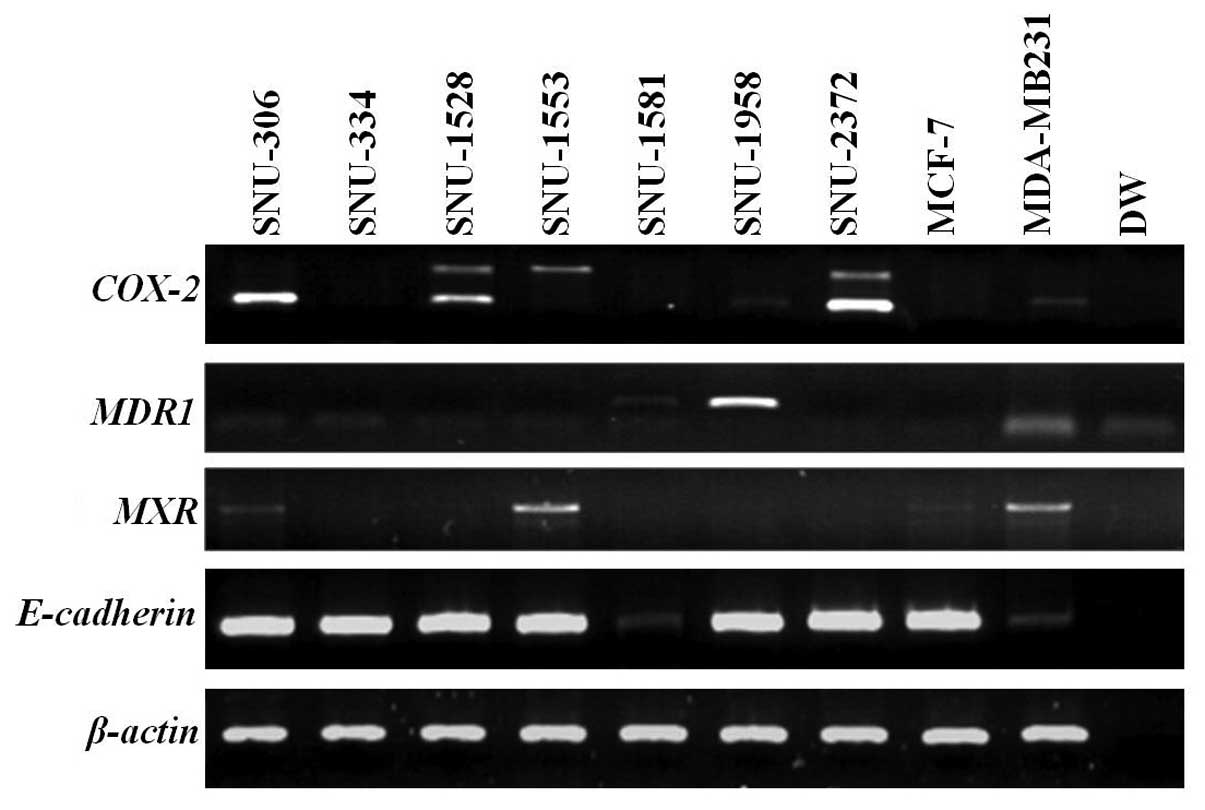
COX-2 was expressed in SNU-306, SNU-1528,
SNU-1958 and SNU-2372. MDR1 was highly overexpressed in the
SNU-1958 and weakly expressed in the SNU-1581. MXR was
expressed in SNU-306 and SNU-1553. E-cadherin was not
expressed in the SNU-1581 (Fig.
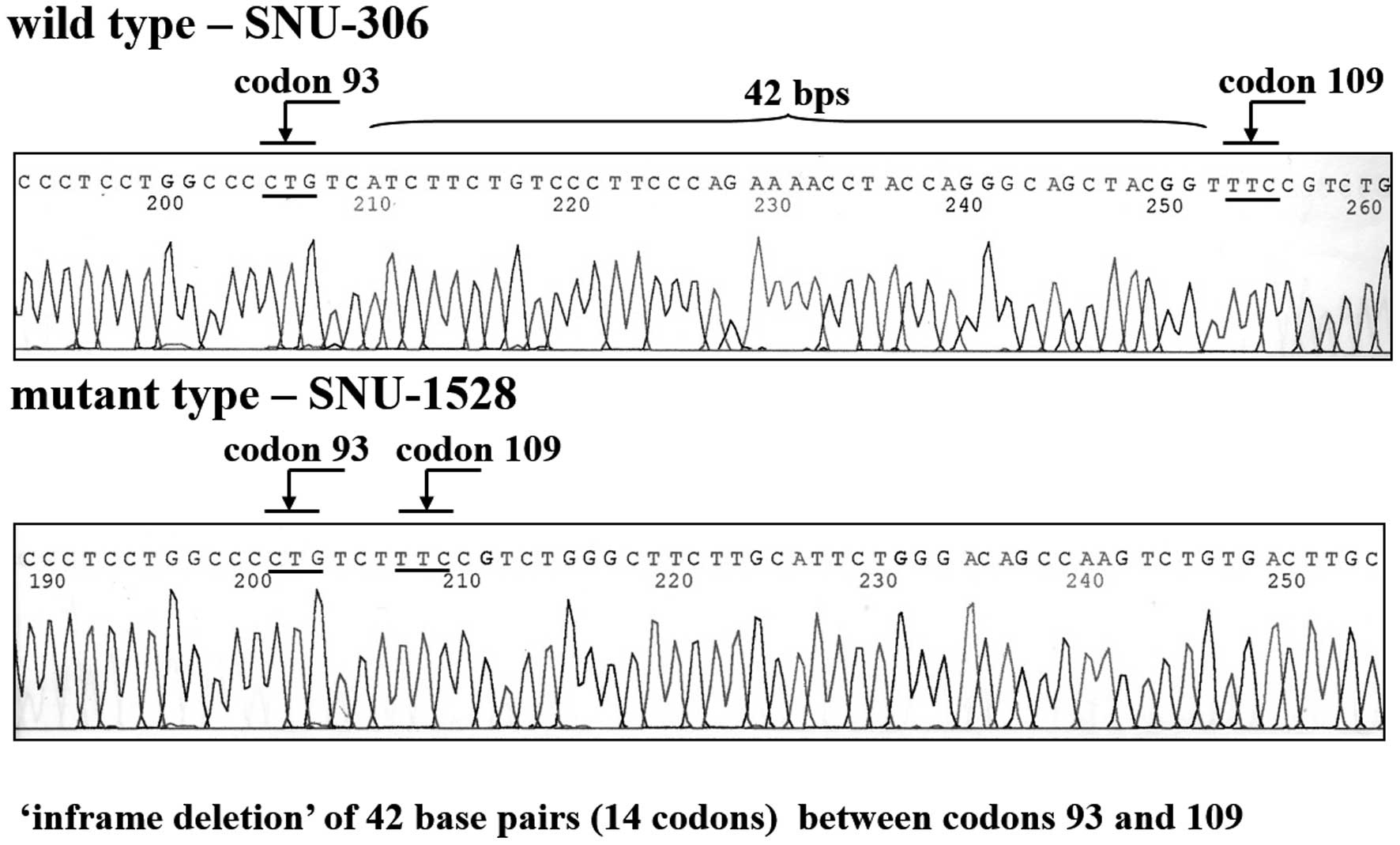
4). SNU-1528 had a mutation in exon 4. Specifically, cells
displayed an inframe deletion of 42 base pairs from codons 93–109
in exon 4 (Fig. 5). SNU-306,
SNU-334 and SNU-1581 possessed arginine at codon 72 and the
SNU-1553 cell line harbored proline at codon 72. There were no
mutations in the EGFR gene in these cell lines (data not
shown). SNU-1528 displayed more cross resistance for paclitaxel
than SNU-334, SNU-1533, and SNU-1581 cell lines (data not shown).
Taxol IC50 (nM/ml) values were >1161.298 for
SNU-1528, 41.905±9.264 for SNU-1553, 41.063±4.681 for SNU-334, and
26.432±11.397 for SNU-1581.
Discussion
Much of the current knowledge on biology of breast
carcinomas is based on in vivo and in vitro studies
performed with breast cancer cell lines (4). The present study reports on seven
cell lines obtained from three primary carcinomas, two pleural
effusions, one pericardial effusion and one ascitic fluid. Each
cell line was shown to be unique at the DNA level using
fingerprinting analysis, two highly polymorphic markers, and 15
short tandem repeat markers. None of the cell lines was
contaminated by mycoplasma or bacteria.
The presence or absence of tumor growth factor
receptors (specifically, ER, PR and C-erbB2)
is important for prediction of prognosis and treatment selection in
breast cancer patients. ER-α remains a very effective
biologic target for breast cancer treatment and prevention, and
anti-estrogens are incorporated into the recommended treatment of
all ER-α-expressing tumors. Estrogen is a steroid hormone
that has a profound proliferative effect on normal human mammary
epithelium through its activation of ER-α, a classic nuclear
hormone receptor. ER-α is overexpressed in as many as 70% of
breast cancers; amplification of the ER-α gene appears to be
a prominent mechanism, although it does not account for all cases
of ER-α overexpression (1).
The significance of PR expression in breast
cancer has been less recognized. PR is an estrogen-dependent
protein synthesized after the stimulation of target cells with
estrogen. ER-α-negative and PR-positive breast cancer
cases carry the worst prognosis. Detection of overexpressed
PR in tumors serves as a functional indicator of an intact
ER pathway, even if the tumor is reported as
ER-α-negative.
Cumulative data from a number of studies have
revealed that steroid receptors are distributed in breast tumors as
follows: 50–60% ER+/PR+; 10–20%
ER+/PR−; 5–15% ER−/PR+;
and 15–25% ER−/PR−. In the present study, the
steroid receptor combinations were: ER+/PR+
(SNU-306), ER+/PR− (SNU-334, SNU-1528,
SNU-1553 and SNU-1581), and ER−/PR− (SNU-1958
and SNU-2372) in RT-PCR analysis (Fig.
3A); ER+/PR+ (SNU-334, SNU-1581, SNU-1958
and SNU-2372), ER+/PR− (none),
ER−/PR+ (SNU-1553) and
ER−/PR− (SNU-306 and SNU-1528) in western
blot analysis (Fig. 3B). In this
study, mRNA levels and their corresponding protein levels was not
significantly correlated. Discordant protein and mRNA expression
has been reported in literature (19,20).
This discrepancy might reflect differences in the regulation of
gene products by transcriptional, translational and
post-translational mechanism among different cells.
C-erbB2, which is localized on chromosome
17q12-21 and encodes for a transmembrane tyrosine kinase receptor
protein, is a useful target for the monoclonal anti-C-erbB2
antibody trastuzumab (Herceptin). In vitro, overexpression
of C-erbB2 in epithelial cells affects the regulation of
cell proliferation, apoptotic pathway, motility and adhesion
(21). The absence of
C-erbB2 results in impaired ductal growth accompanying
puberty in mouse mammary glands (22). C-erbB2 amplification and/or
protein overexpression, which is apparent in 20–30% of invasive
breast cancers, is clearly associated with accelerated cell growth
and proliferation, as well as an increased risk of disease
recurrence with shortened overall patient survival. At a molecular
level, amplification is associated with deregulation of G1/S phase
cell cycle control via upregulation of cyclins D1, E and cdk6, as
well as p27 degradation. C-erbB2 also interacts with
important second messengers including SH2 domain-containing
proteins (e.g., Src kinases) that provide potential additional
targets for breast cancer therapy (1). In several studies, C-erbB2
amplification/overexpression in metastatic breast cancer has been
shown to be an independent marker of response to the monoclonal
anti-C-erbB2 antibody for trastuzumab. C-erbB2 was not
expressed in any of the seven cell lines by RT-PCR analysis in this
study. However, this gene was highly expressed in SNU-334 and
weakly expressed in SNU-1528, SNU-1553 and SNU-1958 cell lines by
western blot analysis. C-erbB2 was detected in a primary
tumor of SNU-1553 by immunohistochemistry (data not shown). This
discrepancy between mRNA and protein expression might also reflect
transcriptional or post-translational modulation of c-erbB2
expression.
COX-2 expression is induced during
inflammation by pro-inflammatory cytokines and growth factors, and
is detectable in most tissues. COX-2 overexpression is
common to a variety of human malignancies including cancer of the
colon, and promotes tumor cell growth, angiogenesis, tumor invasion
and metastasis (reviewed in ref. 23). Overexpression of COX-2 is
significantly associated with reduced disease-free survival but not
with overall disease-specific survival. In mouse models,
COX-2 expression is associated with lymph node metastasis.
COX-2 has also been implicated in vascular endothelial
growth factor production that stimulates angiogenesis, with
COX-2 antagonists possessing anti-angiogenic activity.
Inhibition of COX-2 can reverse resistance to apoptosis.
Reduced breast cancer incidence with the use of non-steroidal
anti-inflammatory drugs has been reported (24). In our study, COX-2 was
overexpressed in four of the seven cell lines (SNU-306, SNU-1528,
SNU-1958 and SNU-2372) (Fig.
4).
The most frequently reported alteration associated
with multidrug resistance is the increased expression of a 170-kDa
membrane P-glycoprotein encoded by the MDR1 gene.
P-glycoprotein functions as an energy-dependent drug efflux pump
that reduces intracellular drug accumulation, thereby causing
resistance to many structurally different drugs (14), and was shown by us to be highly
overexpressed in SNU-1958 cells and weakly expressed in SNU-1581
cells.
MXR, also called ABCG2, ABCP or
BCRP is an ABC transporter that has an N-terminal ATP
binding domain and a C-terminal transmembrane domain (15). BCRP/MXR overexpression has
been reported in various drug-resistant cells selected with
mitoxantrone, doxorubicin and topotecan. BCRP/MXR presumably
acts as an efflux pump, resulting in decreased intracellular
concentrations. BCRP/MXR was overexpressed in SNU-306 and
SNU-1553 cell lines (Fig. 4).
E-cadherin gene located on chromosome 16q22.1 encodes a
protein that is important in the maintenance of the epithelial
phenotype mediated by a Ca2+-dependent, homotypic
cell-cell adhesion. The gene has been termed a ‘metastasis
suppressor’ gene, because the E-cadherin protein can suppress tumor
cell invasion and metastasis. E-cadherin gene expression is
reduced or silenced in carcinomas of the breast and liver, and many
cell lines including those from colon, stomach and prostate
(12). Of the seven presently
studied breast cancer cell lines, E-cadherin was not
expressed in SNU-1581 cells (Fig.
4).
p53 tumor suppressor protein is the most
commonly mutated protein in diverse cancers and has been implicated
in the late stage of malignant transformation (25). In this study, a p53 mutation
comprising an inframe deletion of 42 nucleotides from codons 93–109
in exon 4 was evident in the SNU-1528 cell line. In human
populations, the p53 gene is polymorphic at amino acid 72 of
the encoded protein. Arg72 variant was found in the SNU-306,
SNU-334, and SNU-1581 cell lines, and a Pro72 variant was found in
the SNU-1553 cell line. p53 with Pro72 is structurally
different from p53 with Arg72, as this is reflected by its
altered electrophoretic mobility; p53 with Arg72 migrates
more rapidly than p53 with Pro72 (26). The Arg72 variant also induces
apoptosis markedly better than the Pro72 variant, and the two
polymorphic variants of p53 are functionally distinct. These
differences may influence cancer risk or treatment, but most
studies on p53 have involved Pro72 variants because it was
the first form of human p53 to be cloned, whereas few
functional studies have included the Arg72 form (27). In breast cancer patients, Arg72
homozygosity is associated with breast cancers and could be a
potential risk factor for tumorigenesis of the breast (26). Characterization of polymorphic
variation of p53 in the seven cell lines will be helpful for
discerning functional differences of breast cancer by variation of
p53.
Many of the currently used breast cancer cell lines
were established in the late 1970s, and MCF-7, T-47D and
MDA-MB-231, account for more than two-thirds of all abstracts
reporting studies on breast cancer cell lines. These cell lines
were not derived from primary breast tumors, but from tumor
metastases, especially aspirates of pleural effusions. This means
that the majority of commonly used cell lines are derived from more
aggressive and often metastatic tumors, rather than the primary
lesion, hence there is legitimate reason to question the
representativeness of these cell lines. Well-characterized cell
lines derived from primary breast tumors will help alleviate this
situation.
The present study report the cellular and molecular
characteristics of the seven newly established cell lines
designated, SNU-306, SNU-334, SNU-1528, SNU-1553, SNU-1581,
SNU-1958 and SNU-2372, which were derived from breast carcinoma
patients. These well-characterized breast cancer cell lines, which
include two triple-negative cell lines, will be useful for the
study of breast cancer biology.
Acknowledgements
This study was supported by a
research grant from the Korean Cell Line Research Foundation (2009)
and the Cancer Research Institute, Seoul National University (2002)
and Priority Research Centers Program through the NRF grant funded
by the MEST (no. 2009-0093820).
References
|
1.
|
DeVita VT, Lawrence TS and Rosenberg SA:
DeVita, Hellman, and Rosenberg’s Cancer: Principles & Practice
of Oncology. Wolters Kluwer/Lippincott Williams & Wilkins;
Philadelphia, PA: 2008
|
|
2.
|
Neve RM, Chin K, Fridlyand J, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Gazdar AF, Kurvari V, Virmani A, et al:
Characterization of paired tumor and non-tumor cell lines
established from patients with breast cancer. Int J Cancer.
78:766–774. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Lacroix M and Leclercq G: Relevance of
breast cancer cell lines as models for breast tumours: an update.
Breast Cancer Res Treat. 83:249–289. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Burdall SE, Hanby AM, Lansdown MR and
Speirs V: Breast cancer cell lines: friend or foe? Breast Cancer
Res. 5:89–95. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Park JG, Lee JH, Kang MS, et al:
Characterization of cell lines established from human
hepatocellular carcinoma. Int J Cancer. 62:276–282. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Ku JL, Yoon KA, Kim IJ, et al:
Establishment and characterisation of six human biliary tract
cancer cell lines. Br J Cancer. 87:187–193. 2002.PubMed/NCBI
|
|
8.
|
Koh CS, Ku JL, Park SY, et al:
Establishment and characterization of cell lines from three human
thyroid carcinomas: responses to all-trans-retinoic acid and
mutations in the BRAF gene. Mol Cell Endocrinol. 264:118–127. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Takano N, Iizuka N, Hazama S, Yoshino S,
Tangoku A and Oka M: Expression of estrogen receptor-alpha and
-beta mRNAs in human gastric cancer. Cancer Lett. 176:129–135.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Brys M, Wojcik M, Romanowicz-Makowska H
and Krajewska WM: Androgen receptor status in female breast cancer:
RT-PCR and Western blot studies. J Cancer Res Clin Oncol.
128:85–90. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
O-charoenrat P, Rhys-Evans PH, Archer DJ
and Eccles SA: C-erbB receptors in squamous cell carcinomas of the
head and neck: clinical significance and correlation with matrix
metalloproteinases and vascular endothelial growth factors. Oral
Oncol. 38:73–80. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Melki JR, Vincent PC, Brown RD and Clark
SJ: Hypermethylation of E-cadherin in leukemia. Blood.
95:3208–3213. 2000.PubMed/NCBI
|
|
13.
|
Hase T, Yoshimura R, Matsuyama M, et al:
Cyclooxygenase-1 and -2 in human testicular tumours. Eur J Cancer.
39:2043–2049. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Yang CH, Schneider E, Kuo ML, Volk EL,
Rocchi E and Chen YC: BCRP/MXR/ABCP expression in
topotecan-resistant human breast carcinoma cells. Biochem
Pharmacol. 60:831–837. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Rajendra R, Gounder MK, Saleem A, et al:
Differential effects of the breast cancer resistance protein on the
cellular accumulation and cytotoxicity of 9-aminocamptothecin and
9-nitrocamptothecin. Cancer Res. 63:3228–3233. 2003.PubMed/NCBI
|
|
16.
|
Yoo BC, Hong SH, Ku JL, et al: Galectin-3
stabilizes heterogeneous nuclear ribonucleoprotein Q to maintain
proliferation of human colon cancer cells. Cell Mol Life Sci.
66:350–364. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Kang MS, Lee HJ, Lee JH, et al: Mutation
of p53 gene in hepatocellular carcinoma cell lines with HBX DNA.
Int J Cancer. 67:898–902. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Tokumo M, Toyooka S, Kiura K, et al: The
relationship between epidermal growth factor receptor mutations and
clinicopatho-logic features in non-small cell lung cancers. Clin
Cancer Res. 11:1167–1173. 2005.PubMed/NCBI
|
|
19.
|
Chen G, Gharib TG, Huang CC, et al:
Discordant protein and mRNA expression in lung adenocarcinomas. Mol
Cell Proteomics. 1:304–313. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wang G, Lai FM, Lai KB, et al: Discrepancy
between intra-renal messenger RNA and protein expression of ACE and
ACE2 in human diabetic nephropathy. Am J Nephrol. 29:524–531. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Fritz P, Cabrera CM, Dippon J, et al:
c-erbB2 and topoisomerase IIalpha protein expression independently
predict poor survival in primary human breast cancer: a
retrospective study. Breast Cancer Res. 7:R374–R384. 2005.
View Article : Google Scholar
|
|
22.
|
Shyamala G, Chou YC, Cardiff RD and Vargis
E: Effect of c-neu/ErbB2 expression levels on estrogen receptor
alpha-dependent proliferation in mammary epithelial cells:
implications for breast cancer biology. Cancer Res. 66:10391–10398.
2006. View Article : Google Scholar
|
|
23.
|
Swamy MV, Herzog CR and Rao CV: Inhibition
of COX-2 in colon cancer cell lines by celecoxib increases the
nuclear localization of active p53. Cancer Res. 63:5239–5242.
2003.
|
|
24.
|
Nassar A, Radhakrishnan A, Cabrero IA,
Cotsonis G and Cohen C: COX-2 expression in invasive breast cancer:
correlation with prognostic parameters and outcome. Appl
Immunohistochem Mol Morphol. 15:255–259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutations in human cancers. Science. 253:49–53.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Papadakis EN, Dokianakis DN and Spandidos
DA: p53 codon 72 polymorphism as a risk factor in the development
of breast cancer. Mol Cell Biol Res Commun. 3:389–392. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Dumont P, Leu JI, Della Pietra AC III,
George DL and Murphy M: The codon 72 polymorphic variants of p53
have markedly different apoptotic potential. Nat Genet. 33:357–365.
2003. View
Article : Google Scholar : PubMed/NCBI
|