Introduction
Colorectal cancer (CRC) is a common malignant
disease. Five-year survival rate and cancer-specific 5-year
survival rate of patients with CRC (independent of stage and cause
of death) are 56 and 64%, respectively. Overall survival is 96% in
stage 1, 92% in stage 2, 58% in stage 3, and 0% for patients with
metastatic disease at the time of diagnosis. Cancer-specific
survival rates range from 100% for patients in stage 1 to 0% for
patients with metastatic disease. Local and distant metastasis are
the main risk factors reducing survival of patients with CRC
(1).
The process of cancer metastasis includes cancer
cell proliferation, local invasion, intravasation and cancer cell
survival, extravasation and attachment to secondary organs, and
metastatic growth in a new environment (2). CXC chemokine receptor 4 (CXCR4) and
its ligand stromal cell-derived factor-1α (SDF-1α, also known as
CXCL12) play an important role in cancer growth and dissemination,
and CXCR4/SDF-1α is positively correlated with metastasis in many
types of cancer including thyroid, lung, ovarian, renal, prostate,
breast, pancreatic, gastric and colorectal cancer (3–12).
CXCR4 and SDF-1 have been investigated as growth and
metastasis inhibitors of many different cancers. Treatment of
anaplastic thyroid carcinoma cells with SDF-1 induces
proliferation, which is blocked by the specific CXCR4 antagonist
AMD3100 and by CXCR4 RNA interference, and AMD3100 effectively
reduces tumor growth in nude mice inoculated with different
anaplastic thyroid carcinoma cells (13). Blocking of the SDF-1/CXCR4
interaction with AMD3100 inhibits meta-static tumor growth in a
mouse hepatic metastasis model of colon cancer (11). AMD3100 significantly inhibits the
invasion ability of SW480 cells and markedly reduces the expression
of VEGF and MMP-9 but not MMP-2 (14). Systemic administration of
d-Arg3FC131, a CXCR4 antagonist, inhibits the growth of GH3
somatotrope tumor cell xenografts in immunodeficient nude mice by
inducing apoptosis and suppressing the proliferation of tumor cells
(15). SW620 cell lines with
reduced expression of CXCR3 and/or CXCR4 created using microRNA,
CXCR3-, CXCR4-, and CXCR3/ CXCR4 double-knockdowns significantly
reduces metastasis to lymph nodes, liver and lungs, and
significantly decreases the dissemination of cancer cells to liver
and lungs (16). Functional CXCR4
knockdown using lentiviral short hairpin RNA (shRNA) vectors
significantly decreases the migration behavior in CRC SW480 and
SW620 cell lines, and pharmacologic inhibition of the SDF-1α/CXCR4
interaction by the bicyclam plerixafor at 100 μM
significantly abrogates CXCR4-dependent migration and invasion
(17). Blocking CXCR4 expression
at the mRNA level by a combination of two siRNAs impairs invasion
of breast cancer cells in the Matrigel invasion assay and inhibits
breast cancer metastasis in an animal model (18). Knockdown of CXCR4 significantly
limits the growth of orthotopically transplanted breast cancer
cells and prevents primary tumor formation in some mice, and all
mice transplanted with CXCR RNA interference (RNAi) cells survived
without developing macroscopic metastases (19). CXCR4-miRNA-transfected breast tumor
cells show reduced migration and invasion ability in vitro
and formed fewer lung metastases in vivo compared to
ctrl-miRNA-transfected cells (20).
To date, the relationship between CXCR4 RNAi and the
inhibition of metastasis of CRC to the liver has not been well
documented. In this study, a lentivirus vector was successfully
constructed to introduce CXCR4 RNAi into CRC xenografts. The
results demonstrate the efficiency of the lentivirus system to
silence CXCR4 and inhibit the growth of hepatic metastasis from
grafted CRC. This approach may have therapeutic potential in
CRC.
Materials and methods
Main reagents and instruments
Mouse monoclonal anti-human CXCR4 antibody, GAPDH,
RPMI-1640, fetal bovine serum (FBS), penicillin, streptomycin,
Polybrene, phosphate-buffered saline (PBS; Hyclone, Logan, UT,
USA), Matrigel (BD Biosciences, San Jose, CA, USA), RNA extraction
kit, reagents for reverse transcription, western blotting kit
(Boster Biological Technology Co., Wuhan, China), human colon
cancer cell line (SW480 cells), nude (nu/nu) BALB/c mice
(Shanghai Cancer Institute, Shanghai, China), Lenti-X Bicistronic
Expression System (Clontech, Palo Alto, CA, USA), Lipofectamine
2000, Lipofectamine™ RNAiMAX transfection reagents (Invitrogen
Corp., Carlsbad, CA, USA), Opti-MEM medium (Gibco-BRL,
Gaithersburg, MD, USA), CellTiter96AQ cell proliferation detection
kit (Promega, Madison, WI, USA), Transwell plates (BD), SYBR Green
PCR Master Mix (Toyobo Biologics, Osaka, Japan), and thermal cycler
(ABI PRISM® 7700 Sequence Detection System, Applied
Biosystems, Foster City, CA, USA) were used in the present study.
This study was approved by the ethics board at the First Affiliated
Hospital of Sun Yat-sen University.
Synthesis of siRNA
Three pairs of siRNAs targeting CXCR4 [the human
CXCR4 gene (NM-004363.2)] were synthesized by Qiagen:
sequence A (siCXCR4-A): 5’-UAAAAUCUUCCUGC CCAC CdTdT-3’ (sense) and
3’-dTdTAUUUUAGAAGGACGG GUGG-5’ (antisense); sequence B (siCXCR4-B):
5’-CAAGGAA GCUGUUGGCUGAdTdT-3’ (sense) and 3’-dTdTGUUCCUU
CGACAACCGACU-5’ (antisense); sequence C (siCXCR4-C):
5’-CUGUCCUGCUAUUGCAUUAdTdT-3’ (sense) and 3’-dTd
TGACAGGACGACGAUAACGUAAU-5’ (antisense). In addition, negative
control siCXCR4 was synthesized by Guangzhou RiboBio Co.
(Guangzhou, China) for monitoring the influence of exogenous
genes.
Culture of SW480 cells and siRNA
transfection
SW480 cells were routinely cultured and seeded on
6-well plates at a density of 5×104 cells/well. Then,
nonsense control siCXCR4(NCsiCXCR4), siCXCR4-A, siCXCR4-B and
siCXCR4-C at different concentrations (25, 50 and 100 nM) were
separately added to each well. When 40% confluent, SW480 cells were
transfected. The complete medium was removed and cells were washed
in PBS twice and maintained in 1 ml of high glucose DMEM containing
20% FBS. The solutions in tube A [i.e., siRNA solution (20
μM) prepared with RNAase-free deionized water, diluted in
500 μl of Opti-MEM, and kept at room temperature] and tube B
(5 μl of Lipofectamine 2000 mixed in 500 μl of
Opti-MEM and kept at room temperature for ≤5 min) were mixed, kept
at room temperature for 20 min, added to each well, and incubated
4-6 h. Then, the medium was removed, the cells were washed in PBS
twice, and 2 ml of complete medium was added. At 24 h after
transfection, the medium was removed, 1 ml of TRIzol reagent was
added, and quantitative PCR was performed to detect mRNA
levels.
RT-PCR
Total RNA was extracted using a kit according to the
manufacturer’s instructions and the quality of RNA determined. In
brief, a solution of 1.0 μg of RNA in distilled water
(dH2O) was diluted to a final volume of 12 μl
with dH2O, was heated to and kept at 65°C for 5 min for
RNA denaturation, kept on ice to avoid RNA renaturation, and
incubated with 0.5 μl of Oligo(dT) (Promega) random primer
(Promega), 2.0 μl of 10 mM dNTP, 0.5 μl of RNase
inhibitor, 4.0 μl of 5X buffer and 0.5 μl of M-MLV at
30°C for 10 min, 42°C for 60 min and 80°C for 10 min. The primers
were as follows: 18S rRNA-112 bp (F: CCTGGATACCGCAGCTAGGA; R: GCG
GCGCAATACGAATGCCCC). CXCR4-159 bp: (F: ATCAG TCTGGACCGCTACCT; R:
GTCATCTGCCTCACTGA CGT). The reaction mixture (including 0.5
μl of cDNA, 0.5 μl of forward primer, 0.5 μl
of reverse primer, 10.0 μl of 2X SYBR Green PCR Master Mix,
and 4.0 μl of dH2O) was kept at 95°C for 5 min,
and PCR conditions were as follows: 40 cycles of 95°C for 15 sec,
60°C for 15 sec and 72°C for 32 sec. The melting temperature was
60–95°C. All experiments were performed in triplicate. Cells
transfected with 25 nM siCXCR4-C had the lowest expression of CXCR4
and therefore it was used in the following experiment.
Western blot assays
Transfection with siCXCR4-C and
NCsiCXCR4
SW480 cells were passaged 24 h prior to
transfection. When 30–50% confluent, cells were transfected with 25
nM siCXCR4-C or NCsiCXCR4 in the presence of Lipofectamine™ RNAiMAX
in Opti-MEM for 4 h, and then the medium was replaced with
RPMI-1640 containing 10% FBS. Protein extraction, SDS-PAGE, and
protein transfer were done according to the manufacturer’s
instructions. The membrane was washed with Tris-buffered saline
with Tween-20 (TBST; 3X, 5 min each time), incubated with 5%
non-fat milk at room temperature overnight to block nonspecific
binding, washed with TBST (3X, 5 min each time), treated with
primary antibody (anti-CXCR4 or anti-GAPDH) at 37°C for 2 h, washed
with TBST (3X, 5 min each time), treated with secondary antibody at
37°C for 1 h, washed with TBST (3X) and then with distilled water
(3X, a total of 2 min), placed on a plastic board, incubated with
chemiluminescence substrate for 5 min, and visualized by an imaging
system in the dark.
Construction and identification of
pLenti-CXCR4-siRNA Splicing of vector GV115 with restriction
enzymes
Vector GV115 was digested with
AgeI/EcoRI by incubating 3 μl of vector GV115
with 0.5 μl of EcoRI, 0.5 μl of AgeI, 1
μl of 10X D buffer, and 5 μl of deionized water or
distilled water at 37°C overnight, and the products were separated
by 1% gel electrophoresis.
Insertion of siRNA into vector
GV115
The following reagents were added to a 0.2-ml EP
tube: PCR products (3 μl), recycled vector GV115 (2
μl), 10X ligase buffer (1 μl), T4 DNA ligase (1
μl), and deionized water (3 μl). Ligation was done at
16°C for 2 h. The lentiviral plasmid product was
pLenti-CXCR4-siRNA. In the control group, water was added instead
of PCR products.
Transduction of
pLenti-CXCR4-siRNA
On ice, 5 μl of ligation products was added
to 50 μl of DH5α competent cells and incubated on ice for 30
min and at 42°C for 90 sec. This solution was rapidly transferred
to ice, incubated on ice for 2 min, incubated with 200 μl of
LB medium at 37°C for 1 h with mixing at 200 rpm, and transferred
to an LB plate containing 100 μg/ml ampicillin (Amp). The
plate was incubated at room temperature until the solution was
absorbed, and inverted and placed in a 37°C incubator
overnight.
Identification of
pLenti-CXCR4-siRNA
One colony was collected from the plate, transferred
to a 3-ml LB tube, and incubated overnight with continuous shaking
for plasmid extraction. The sequencing of pLenti-CXCR4-siRNA was
performed by the BGI Gene Tech Co., Ltd. (Beijing, China). There
was a XhoI restriction site in the pLenti-CXCR4-siRNA
sequence, and thus XhoI was used for identification. Once
the sequence was cleaved, these plasmids were either positive or
negative for pLenti-CXCR4-siRNA. The plasmid was cut with
restriction enzyme XhoI at 37°C overnight in the following
solution: 3.6 μl of ddH2O, 1 μl of 10X H
buffer, 1 μl of 10X BSA, 4 μl of plasmid and 0.4
μl of XhoI.
Packaging of Lenti-CXCR4-siRNA
293T cells in the logarithmic phase (cell
confluence: 70–80%) were washed with 5 ml of PBS, treated with 2–3
ml of trypsin until these cells became round, switched to complete
medium [90% DMEM (high glucose: 4.5g/l) + Glu Max + 10% FBS +
penicillin + streptomycin (100X)] to stop the cutting, and
dispersed into a single cell suspension, counted to determine cell
density, and seeded at 3×105 cells/ml into flasks (10
ml/flask). At 30 min before transfection, the medium was replaced
with complete medium (5 ml/flask). The packaging of lentiviral
vector was done according to the instructions provided with the
Lenti-X Bicistronic Expression System. In brief, 1.5 μg of
packaging plasmid, 0.5 μg of expression plasmid, and 250
μl of serum-free medium in a 1.5-ml sterilized EP tube were
incubated at room temperature for 5 min, while 9 μl of
Lipofectamine 2000 and 250 μl of serum-free medium in a
separate 1.5-ml sterilized EP tube were incubated at room
temperature for 5 min. The DNA solution and Lipofectamine were
mixed and incubated at room temperature. To each well of a 6-well
plate, the following was added with overnight incubation at 37°C: 1
ml of growth medium containing serum and DNA-Lipofectamine mixture
and 1 ml of re-suspended 293T cells (1×106 cells/ml).
The medium containing DNA-Lipofectamine was removed and replaced by
DMEM. At 48–72 h after transfection, the supernatant was
collected.
Condensation and determination of
titer of Lenti-CXCR4-siRNA
NaCl (5 mol/l) was added to the above supernatant to
a final concentration of 0.15 mol/l. A solution of 20% (w/v)
PEG8000 was added and the final mixture was incubated at 4°C
overnight with continuous shaking, then centrifuged at 4°C for 30
min at 10,000 g. The supernatant was collected with a vacuum pump
in a biological safety cabinet and the sediment was diluted in an
appropriate volume of OptiMEM, covered with Parafilm membrane, and
stored at 4°C. The Lenti-CXCR4-siRNA was titered using a kit
according to the manufacturer’s instructions.
Effects of Lenti-CXCR4-siRNA on growth,
migration, invasiveness, and liver metastasis of SW480 cells
Culture of SW480 cells
SW480 cells were seeded into 20 wells of a 96-well
plate at a density of 3–5×103 cells/ml (90
μl/well).
Transfection of SW480 cells with
Lenti-CXCR4-siRNA
Conventional medium was added to 2 sterilized 1.5-ml
EP tubes (45 μl per tube). Then, 5 μl of
1×108 TU/ml Lenti-CXCR4-siRNA was serially diluted to
1×107 TU/ml and 1×106 TU/ml by addition to
the first tube followed by gentle vortexing to avoid foaming and
subsequent transfer of 5 μl of solution in the first tube to
the second tube followed by vortexing. In another tube, 2 μl
of Polybrene was diluted to 400 and 10 μl added to each
well. The volume of Polybrene per well of 100 μl was diluted
2,000 times to a final concentration of 5 μg/ml. Then, 10
μl of lentivirus solution at three different concentrations
(1×106, 1×105 and 1×104 TU,
respectively), was added to the corresponding wells. The cell count
per well was ∼1×104. Thus, the multiplicity of infection
(MOI) per well was 100, 10 and 1, respectively. In the Polybrene
well, 10 μl of diluted Polybrene was added. The plates were
gently vortexed and placed in an incubator for 8–12 h. The cells
were observed and the supernatant was removed. The medium was
refreshed. At 3–4 days after transfection, fluorescence was
detectable under a microscope.
Screening of colonies
SW480 cells were seeded into a 6-well plate and
maintained in medium containing penicillin and streptomycin on the
first day, transfected when 50–60% confluent on the second day,
visualized (i.e., counted and marked) as fluorescent colonies on
the plate under a fluorescence microscope on the third day. The
marked cell colonies were digested with 0.25% trypsin, transferred
to a 24-well plate, and observed again under a fluorescence
microscope. Those with fluorescence were selected for
passaging.
Detection of CXCR4 expression by
qRT-PCR and western blot assays
The expression of CXCR4 mRNA and protein was assayed
by real-time reverse transcription (qRT)-PCR and western blotting
as described above.
Detection of cell viability (MTS)
Cells were divided into the SW480 group, negative
control (NC) group, and Lenti-CXCR4-siRNA group. Cells were
digested with trypsin, adjusted to a density of 1×105
cells/ml, seeded into a 96-well plate (100 μl/well;
1×104 cells/well), allowed to adhere, collected at 24,
48, 72 and 96 h, incubated 4 h with MTS solution (ratio: 1:10), and
assessed for cell proliferation by optical density (OD) measurement
at 490 nm with a micro-plate reader.
Detection of cell migration after
Lenti-CXCR4-siRNA transfection by Transwell assay
Cells were grouped as described above. Two days
after transfection of 1×105 cells with
Lenti-CXCR4-siRNA, the cells were re-suspended in 100 μl of
serum-free RPMI-1640, transferred to the upper chamber of a
Transwell system with 600 μl of complete medium added to the
lower chamber, and incubated at 37°C in an atmosphere containing 5%
CO2 for 24 and 48 h. The cells in the upper chamber were
removed with a swab, fixed in 4% paraformaldehyde for 15 min, and
washed in PBS once. The upper chamber was stained with crystal
violet for 10 min and washed in PBS once. The cells migrating from
the upper chamber were counted under a microscope.
Detection of invasiveness of cells
transfected with Lenti-CXCR4-siRNA by Transwell assays
Cells were divided into 3 groups as described above.
Approximately 40 μl of Matrigel solution (Matrigel kept at
4°C overnight and then mixed with serum-free medium at a ratio of
1:3) was added to a pre-cooled Transwell system and incubated at
37°C for 2 h. After excess solution was removed from the chamber,
serum-free medium (100 and 600 μl, respectively) was added
to the upper and lower chambers, and the Transwell plate was
incubated at 37°C overnight. The procedures described above were
performed.
Liver metastasis of SW480 cells after
Lenti-CXCR4-siRNA transfection in BALB/c nu/nu mice
A total of 30 BALB/c nu/nu mice aged 4–6
weeks and weighing 14–23 g were housed in a specific pathogen-free
(SPF) environment. These animals were divided into three groups as
described above (n=10). Cells in the logarithmic growth phase were
digested in 0.25% trypsin to prepare a single cell suspension. The
cell viability was confirmed to be ≥95% by trypan blue exclusion
staining. The cell density was adjusted to 1×107/ml. The
procedures were done under aseptic conditions. Mice were
intraperitoneally anesthetized with 1% pentobarbital sodium (35
mg/kg). A midline incision was made in the upper abdomen, and the
spleen was exposed and slowly injected with cell suspension (0.2
ml; 2×106 cells). The injection site was compressed with
a tampon soaked in 95% alcohol and the wound was closed. These mice
were returned to SPF housing conditions and monitored for activity,
food intake, and change in body weight. At 4 weeks after surgery,
laparotomy and subsequent exploration were done to observe liver
metastasis. The number of metastatic sites and hepatic metastasis
mean weight (in g) were determined.
Statistical analysis
Statistical analysis was done using SPSS version
11.0 for Windows. Comparisons were made by t-test. A value of
P<0.05 was considered statistically significant.
Results
RT-PCR
The mRNA expression was the lowest in the sequence C
group and ∼5.4% of that in the NCsiCXCR4 group. Thus, cells with
sequence C were used in the following experiments and designated
the siCXCR4 group.
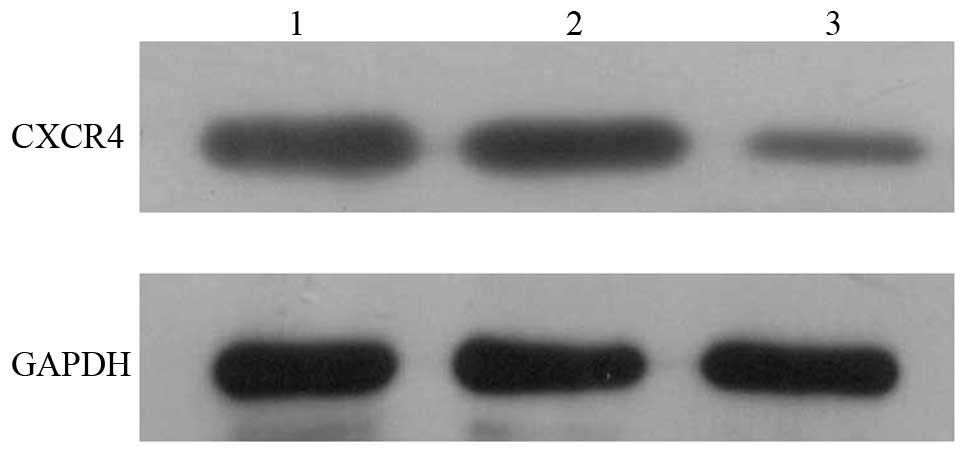
Western blot assays
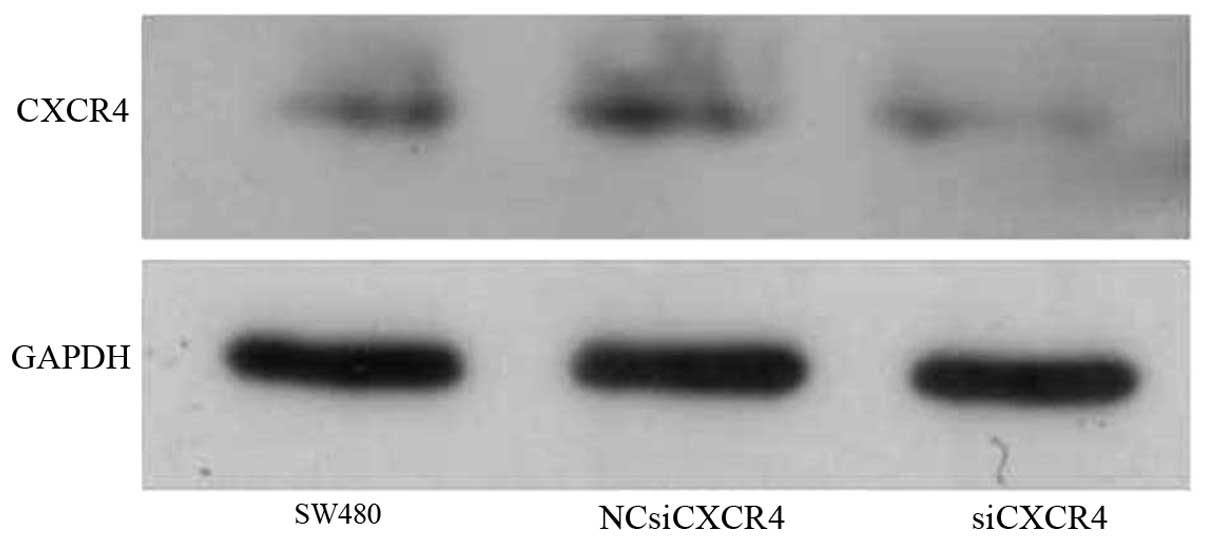
The SW480, NCsiCXCR4, and siCXCR4 groups had a GAPDH
optical density (OD) of 367.67, 3593.84 and 3190.51, respectively
and a CXCR4 OD of 820.54, 1106.66 and 604.48, respectively. Thus,
the relative expression of CXCR4 was 22.34, 30.80 and 18.95%,
respectively. These findings demonstrated that siCXCR4 could
effectively inhibit expression of CXCR4 protein (Fig. 1).
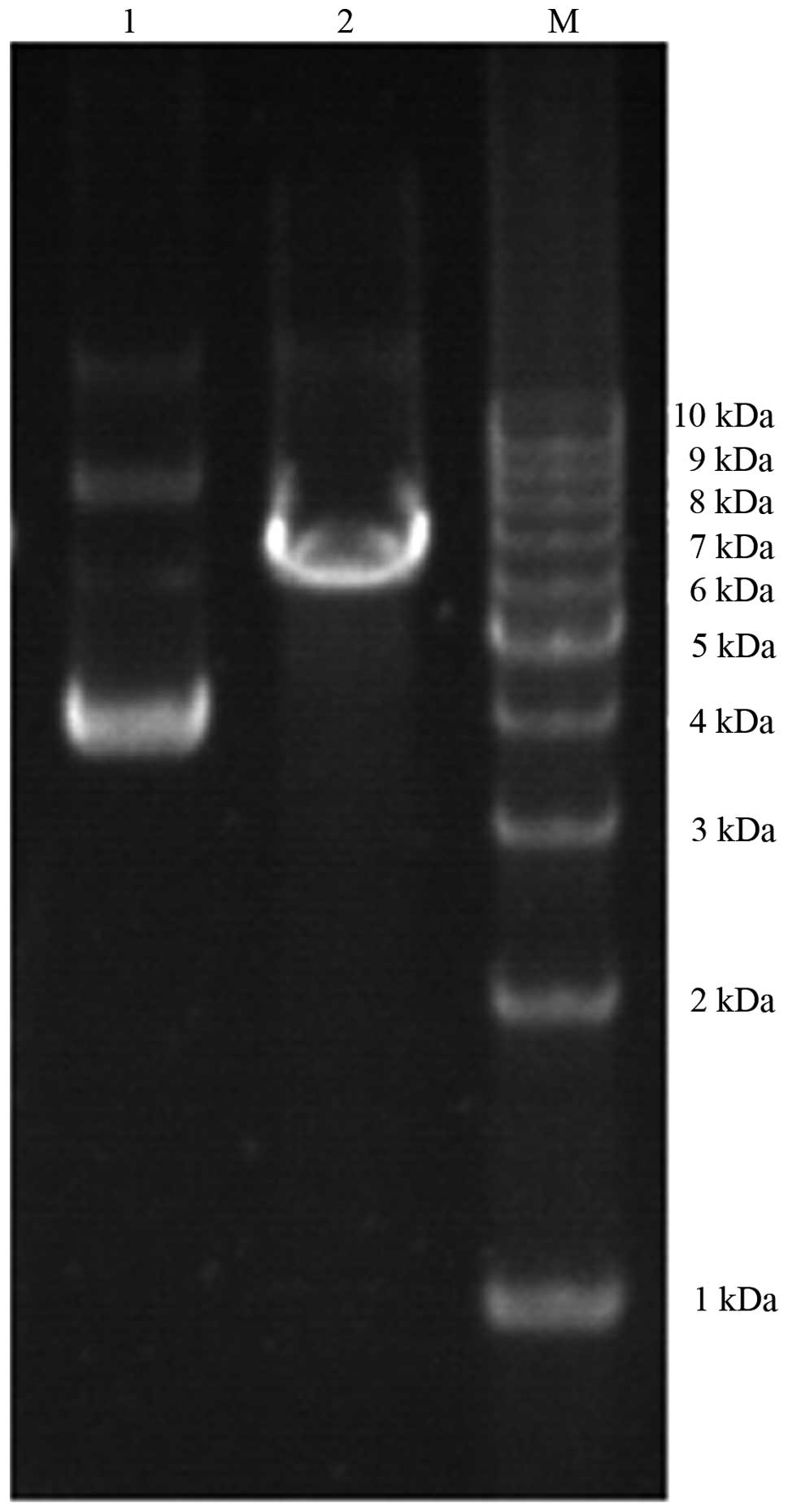
Cutting of pLenti-CXCR4-siRNA
The vector GV115 (7.5 kb in size) had no XhoI
restriction site. However, the siRNA sequence had a XhoI
site that could be cut by XhoI. Blank GV115 was circular and
therefore migrated more rapidly. Thus, blank GV115 migrated ahead
of the cut GV115 (linear sequence), suggesting successful cloning
(Fig. 2).
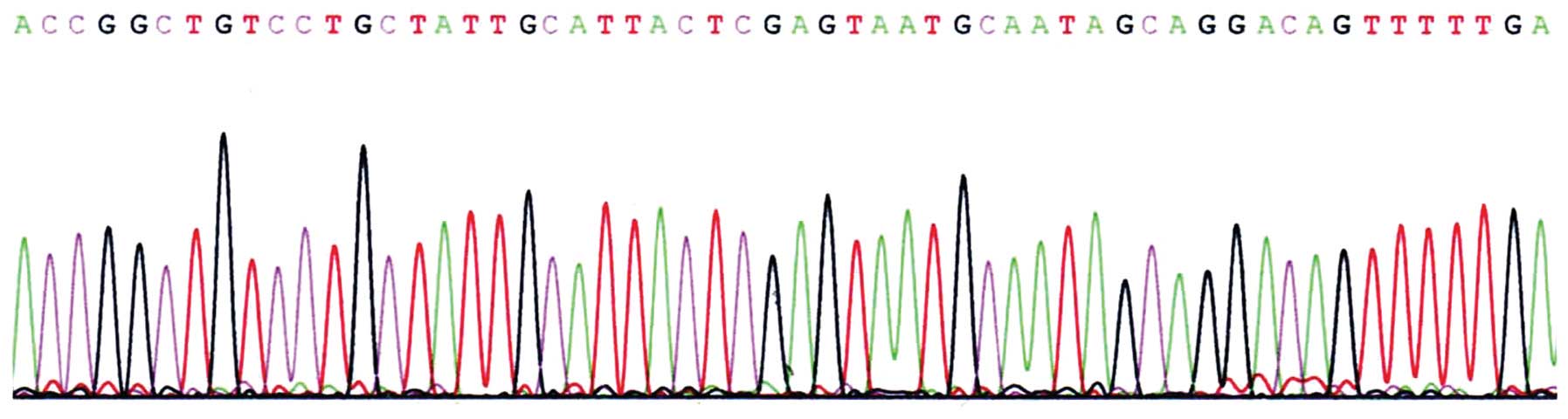
Sequencing of pLenti-CXCR4-siRNA
All the bases in siRNA were correct. BLAST analysis
showed successful cloning and that there was no mutation. Thus,
pLenti-CXCR4-siRNA could be used in the experiments that followed
(Fig. 3).
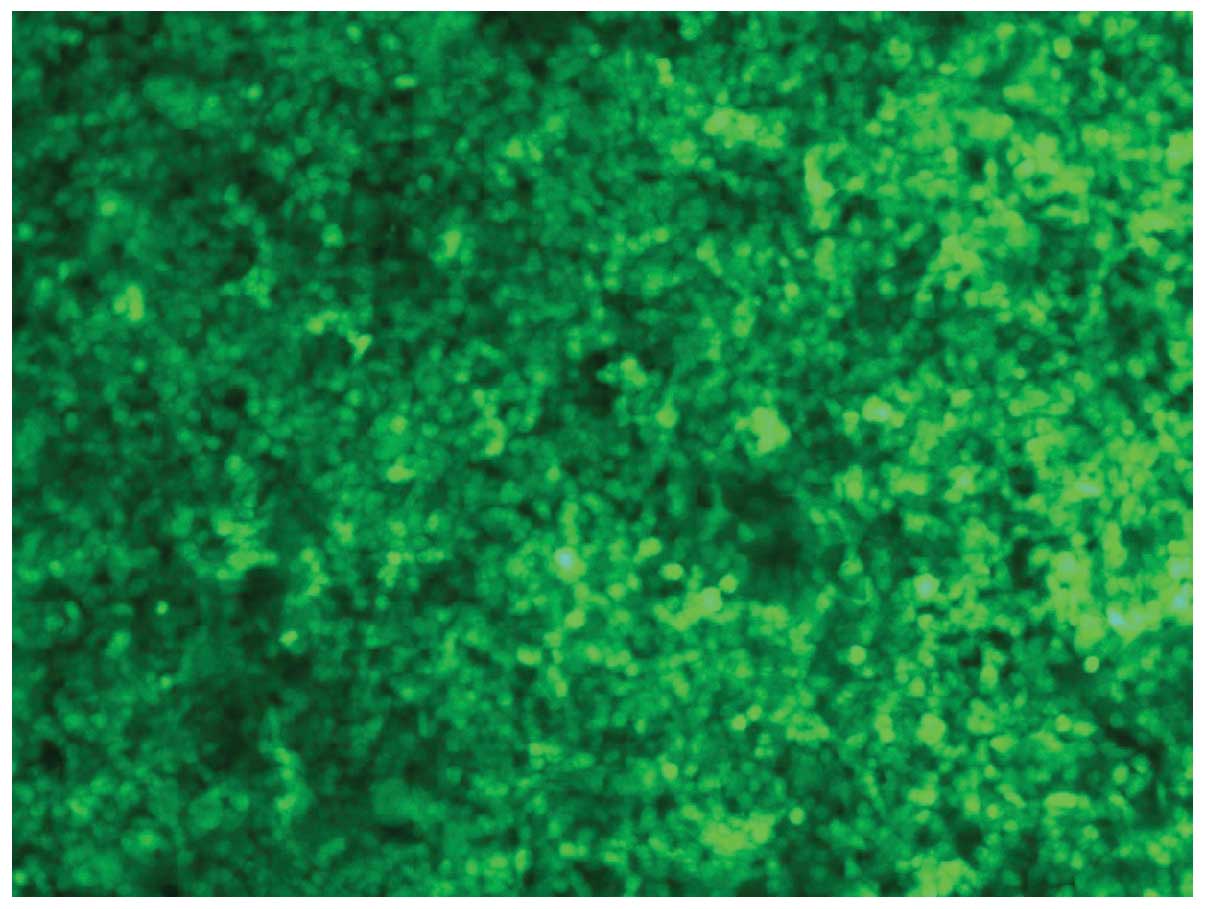
Transfection of 293T cells with
pLenti-CXCR4-siRNA
All 293T cells transfected with pLenti-CXCR4-siRNA
were shown to exhibit green fluorescence under a fluorescence
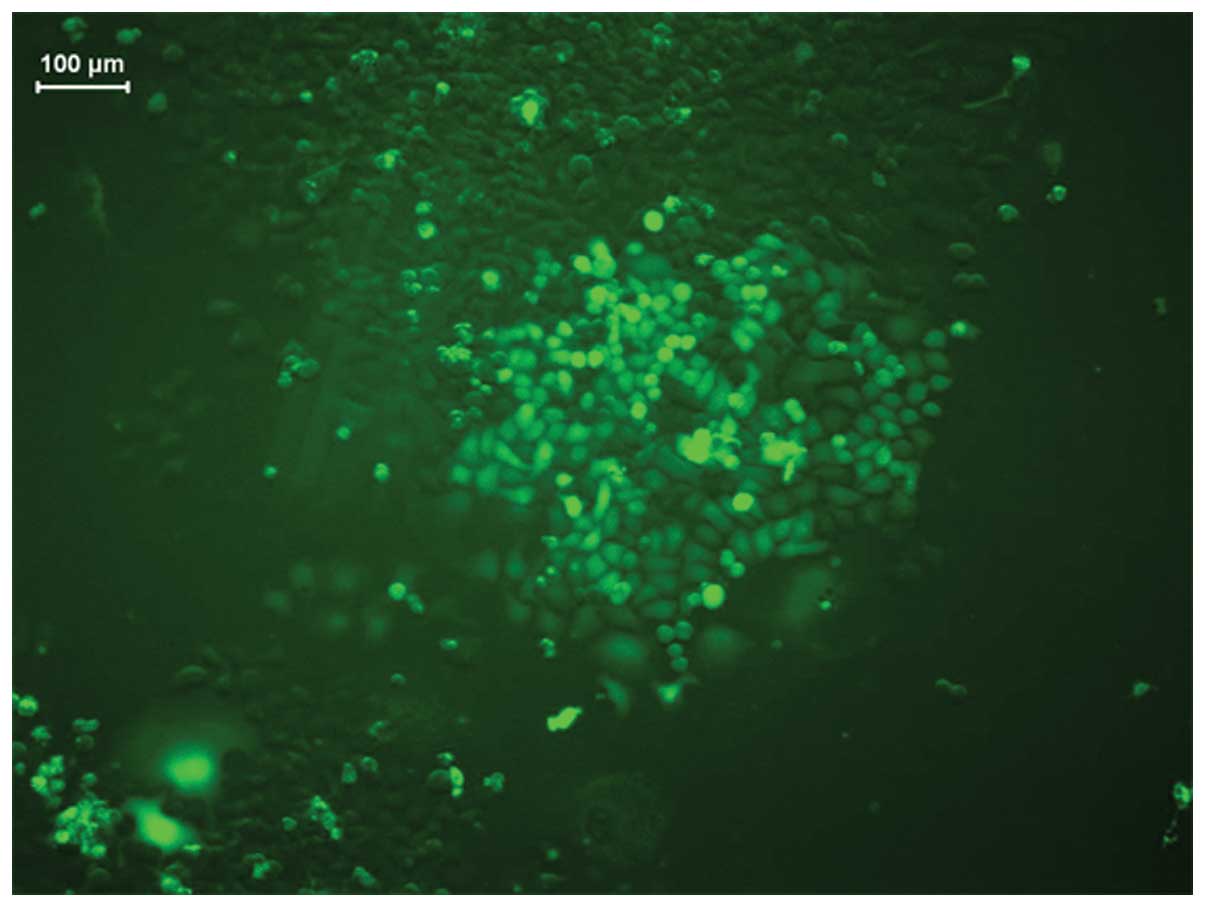
microscope (Fig. 4).
Purified Lenti-CXCR4-siRNA (titer
2×109 TU/ml) was used for transfection of SW480
cells
All infected cells showed green fluorescence under a
fluorescence microscope, suggesting successful transfection
(Fig. 5).
Detection of CXCR4 mRNA
In the SW480, NC, and Lenti-CXCR4-siRNA groups, the
expression of CXCR4 mRNA was 0.54±0.06, 1.00±0.03 and 0.11±0.04,
respectively. The expression of CXCR4 mRNA was the lowest in the
CXCR4 RNA group and differed markedly from the SW480 and NC groups
(P=0.0001).
Detection of CXCR4 protein
In the SW480, NC, and Lenti-CXCR4-siRNA groups, the
expression of CXCR4 protein (in μg/μl) was 4.61, 3.58
and 3.08, respectively. Western blot assays showed that the
expression of CXCR4 protein was the lowest in the Lenti-CXCR4-siRNA
group. The relative expression of CXCR4 protein was 0.18±0.02,
0.6±0.03 and 0.72±0.03, respectively, in the Lenti-CXCR4-siRNA,
SW480, and NC groups (Fig. 6,
P=0.0001).
Detection of MTS
At 4 days after infection, proliferation (OD of MTS)
was markedly lower in the Lenti-CXCR4-siRNA group than in the SW480
group (0.92±0.06 vs 1.38±0.04, P=0.0050) and NC group (0.92±0.06 vs
1.28±0.05, P=0.0256). This suggests that RNA interference (RNAi)
significantly inhibits the growth of SW480 cells.
Detection of cell migration
The number of migrated cells was significantly lower
in the Lenti-CXCR4-siRNA group than in the SW480 group (0.75±0.71
vs 32±6.85, P=0.0000) and NC group (0.75±0.71 vs 32.63±1.69,
P=0.0000). This suggests that RNAi significantly suppresses the
migration of SW480 cells (Fig.
7).
Detection of cell invasiveness
The number of migrated cells was dramatically lower
in the Lenti-CXCR4-siRNA group than in the SW480 group (0.63±0.74
vs 29.13±10.30) and NC group (0.63±0.74 vs 30.38±6.09; P=0.0000).
This suggests that CXCR4 RNAi markedly inhibits the invasiveness of
SW480 cells.
Liver metastasis of SW480 cells after
Lenti-CXCR4-siRNA transfection
The number of metastatic lesions was significantly
lower in the Lenti-CXCR4-siRNA group than in the SW480 group
(3.50±2.51 vs 7.10±3.98, P=0.034) and NC group (3.50±2.51 vs
7.50±4.09, P=0.019). The mean weight of metastatic tumors (in g)
was dramatically lower in the Lenti-CXCR4-siRNA group than in the
SW480 group (1.45±2.07 vs 2.25±2.51, P=0.000) and NC group
(1.45±2.07 vs 2.11±2.38, P=0.000). These findings demonstrate that
Lenti-CXCR4-siRNA transfection markedly inhibits the liver
metastasis of SW480 cells.
Discussion
As chemoattractant cytokines, chemokines can
facilitate cell activation, differentiation, and trafficking.
Several organs including lung, lymph nodes, liver, skeletal muscle,
brain, kidney, heart, skin, and bone marrow, express CXCL12.
CXCR4 is expressed in most cancer cells, and hypoxia
and injury stimulate its production. The binding of CXCL12 to CXCR4
initiates several signaling pathways and promotes chemotaxis, cell
proliferation, increase in intracellular calcium concentration and
gene transcription (21).
CXCR4-positive cancers metastasize to the lymph nodes, liver, and
bones in a CXCL12-dependent manner (22). The CXCL12/CXCR4 pathway plays a
pivotal role in several aspects of tumor progression including
vascularization, metastasis and survival (23,24).
In CRC tissues, CXCL12 is significantly downregulated and CXCR4 is
significantly upregulated compared to the corresponding normal
tissues (25). The frequency of
cytoplasmic and nuclear expression of CXCR4 in CRC was shown to be
35.6 and 36.9%, respectively, and nuclear but not cytoplasmic
expression of CXCR4 has been associated with advanced CRC and
lymphovascular invasion (26). The
incidence of nodal metastasis was significantly higher in CRC
patients with CXCR4-positive tumors than in those with
CXCR4-negative tumors, and a significant correlation was observed
between CXCR4 and vascular endothelial growth factor C expression
and lymphatic invasion (27). It
was reported that there are more CXCR4-positive cells at metastatic
sites in the liver than at primary sites of CRC (28). All brain metastases from CRC were
reported to strongly express CXCR4 (29). A high expression of CXCR4 in the
primary CRC tumor is considered an independent prognostic factor
for poor disease-free survival, and nuclear distribution of CXCR4
is reported to have an inverse relationship with disease-free and
overall survival (30). Therefore,
CXCR4 is an important mediator of invasion and metastasis of
CXCR4-expressing CRC, and it is possible to prevent the development
of CRC metastasis through inhibition of CXCR4. The mechanism of
growth, invasion, migration and metastasis of CXCL12/CXCR4 pathway
includes regulating the expression of MMPs and integrins. CXCR4 and
its ligand SDF-1α are inversely expressed in CRC cell lines. SDF-1α
activates matrix metalloproteinase-9 and increases vascular
endothelial growth factor (VEGF) expression and cell proliferation
(31). The concomitant and high
expression of CXCR4 and VEGF is a strong and independent predictor
of early distant relapse in CRC. CXCR4 triggers a plethora of
phenomena, including stimulation of clonogenic growth, induction of
VEGF release and ICAM-1 upregulation (32). CXCR4 silencing could upregulate the
mRNA and protein expression of E-cadherin, and downregulate the
mRNA and protein expression of MMP-2/-9 (33). MMP-2 and MMP-9 expression
significantly increased when cells were cultured in the presence of
SDF-1α, suggesting that CXCL12-CXCR4 axis triggers increased
expression of these genes to promote invasion (34). The CXCR4 ligand SDF-1α doubled
secreted VEGFA under hypoxic conditions in human chondrosarcoma
cell line and these effects were inhibited by CXCR4 siRNA (35). SDF-1/CXCR4 induces directional
migration and liver metastasis of CRC cells by upregulating
integrin αvβ6 through ERK/Ets-1 pathway (36). SDF-1 enhanced ovarian cancer cell
invasion through αvβ6 integrin-mediated uPA expression via the p38
MAPK and PI3 K/Akt pathway (37).
Integrin can modulate tumor cell morphology, and regulate the
expression of CXCR4 which is associated with the invasive phenotype
and progression of prostate cancer (38).
RNAi is a process in which double-stranded RNA is
used to enhance the degradation of cognate mRNA. Synthetic 21–23
nucleotide (siRNA) has been demonstrated to induce transient and
efficient RNAi (39).
SiRNA-producing plasmid vectors are associated with transient siRNA
expression and low transfection efficiency and are widely used in
gene interventions, including RNAi (17). In the present study, DNA sequencing
and agarose gel electrophoresis provided strong evidence that CXCR4
siRNA was correctly inserted into the multiple cloning site of the
GV115 expression plasmid. The physical particle titer of the
recombinant virus Lenti-CXCR4-siRNA was 2×109 TU/ml. In
the present study, Lenti-CXCR4-siRNA effectively inhibited the
expression of CXCR4 mRNA and reduced the level of CXCR4 protein.
Lentiviral vector-mediated Met oncogene-specific, stable RNA
interference impaired spontaneous motility and invasiveness of
canine osteosarcoma cells (40).
Lentiviral transgenic systems effectively transferred shRNA
targeting human telomerase reverse transcriptase (hTERT) to oral
squamous cell carcinoma KB cell lines with >80% gene transfer
efficiency, significantly and specifically inhibited hTERT
expression both at the mRNA and protein levels, and increased rates
of KB cell apoptosis by 206.33% (41). Thus, it is reasonable to conclude
that lentivirus can effectively induce gene RNAi.
The increase of tumor cell proliferation and the
decrease of apoptosis are essential characteristics of malignant
lesions. Our data show that Lenti-CXCR4-siRNA reduces SW480 cell
proliferation, which is evidence that the CXCR4/SDF-1 axis
stimulates cell multiplication and that inhibition of CXCR4 was
able to reduce SW480 cell growth. One study showed that CXCR4
protein expression on the CD34(+) cell surface is lower in the
low-grade myelodysplastic syndrome than in high-grade
myelodysplastic syndrome; CD34(+) cell apoptosis is significantly
higher in low-grade myelodysplastic syndrome, and apoptosis is
negatively correlated with CXCR4 expression (42). In another study, CXCR4 RNAi blocked
SDF-1-induced proliferation of anaplastic thyroid carcinoma cells
and increase in phosphorylation of extracellular signal-regulated
kinases (13). A CXCR4 antagonist,
d-Arg3FC131, was shown to induce apoptosis and suppress the
proliferation of somatotrope tumor cells (15). Thus, CXCR4 RNAi and its antagonist
can be used to inhibit tumor cell proliferation and growth.
In the present study, Lenti-CXCR4-siRNA dramatically
suppressed the migration and invasion ability of SW480 cells in
vitro, and reduced the number and mean weight of hepatic
metastasis in the liver metastasis model in Balb/c mice.
Previously, recombinant CXCR4-RNAi plasmids were found to reduce
CXCR4, inhibit cell growth, invasiveness, and migration, and induce
cell apoptosis in renal cell carcinoma in vitro (7).
Using a recombinant lentiviral RNA interference
vector of the CXCR4 gene in highly aggressive (Tca8113 and SCC-9)
tumor cells, significant inhibition of the proliferation of both
cell lines in vitro and in vivo was shown (43). Lentivirus-mediated CXCR4 RNAi
reduced the expression of CXCR4 and tumor growth, and inhibited
metastasis, particularly of bone metastasis of a prostate cancer
cell line (44). In a lentiviral
CXCR4 overexpression and knockdown model established in SW480,
SW620, and RKO cells, CXCR4 overexpression favored chemotaxis and
SDF-1α gradient-dependent invasion of cells, while
lentiviral-mediated CXCR4 RNAi decreased cell migration (17). Knockdown of CXCR4 with RNAi
impaired invasion of breast cancer cells and significantly limited
the growth and metastasis to the liver and lung in vivo
(18,19). CXCL12 stimulation had no impact on
Caco-2 cells but significantly increased migration of CXCR4-bearing
SW480 and HT-29 cells (CXCR4 expression being less pronounced in
HT-29 cells), and this effect was significantly abrogated by
neutralizing anti-CXCR4 antibody as well as by CXCR4 siRNAs
(25).
In conclusion, hepatic metastasis of CRC is a
prevalent and serious problem, and efficacious therapy is required
to deal with it. The recombinant lentivirus Lenti-CXCR4-siRNA was
correctly constructed in the present study and potently inhibited
CXCR4 expression and growth, migration, invasion, and liver
metastasis of CRC. Lentiviral mediated CXCR4 RNAi is a potential
treatment for CRC.
Abbreviations:
|
CXCR4
|
CXC chemokine receptor 4;
|
|
CRC
|
colorectal cancer;
|
|
siRNA
|
small interfering RNA;
|
|
siCXCR4
|
small interfering CXCR4 RNA
|
Acknowledgements
This study was supported by grants
from the Science and Technology of Guangdong Province Funds (no.
2010B060900100 and no. 2012B010300011).
References
|
1.
|
Wichmann MW, Beukes E, Esufali ST,
Plaumann L and Maddern G: Five-year results of surgical colorectal
cancer treatment in rural Australia. ANZ J Surg. 83:112–117.
2013.
|
|
2.
|
Shin HN, Moon HH and Ku JL: Stromal
cell-derived factor-1α and macrophage migration-inhibitory factor
induce metastatic behavior in CXCR4-expressing colon cancer cells.
Int J Mol Med. 30:1537–1543. 2012.
|
|
3.
|
Yasuoka H, Kodama R, Hirokawa M, Takamura
Y, Miyauchi A, Sanke T and Nakamura Y: CXCR4 expression in
papillary thyroid carcinoma: induction by nitric oxide and
correlation with lymph node metastasis. BMC Cancer. 8:2742008.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Cavallaro S: CXCR4/CXCL12 in
non-small-cell lung cancer metastasis to the brain. Int J Mol Sci.
14:1713–1727. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Sekiya R, Kajiyama H, Sakai K, Umezu T,
Mizuno M, Shibata K, Yamamoto E, Fujiwara S, Nagasaka T and Kikkawa
F: Expression of CXCR4 indicates poor prognosis in patients with
clear cell carcinoma of the ovary. Hum Pathol. 43:904–910. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Jung SJ, Kim CI, Park CH, Chang HS, Kim
BH, Choi MS and Jung HR: Correlation between chemokine receptor
CXCR4 expression and prognostic factors in patients with prostate
cancer. Korean J Urol. 52:607–611. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Wang L, Huang T, Chen W, Gao X, Zhou T, Wu
Z and Sun Y: Silencing of CXCR4 by RNA interference inhibits cell
growth and metastasis in human renal cancer cells. Oncol Rep.
28:2043–2048. 2012.PubMed/NCBI
|
|
8.
|
Gros SJ, Kurschat N, Drenckhan A, Dohrmann
T, Forberich E, Effenberger K, Reichelt U, Hoffman RM, Pantel K,
Kaifi JT and Izbicki JR: Involvement of CXCR4 chemokine receptor in
metastastic HER2-positive esophagealcancer. PLoS One. 7:e472872012.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Zhong W, Chen W, Zhang D, Sun J, Li Y,
Zhang J, Gao Y, Zhou W and Li S: CXCL12/CXCR4 axis plays pivotal
roles in the organ-specific metastasis of pancreatic
adenocarcinoma: a clinical study. Exp Ther Med. 4:363–369.
2012.
|
|
10.
|
Lee HJ and Jo DY: The role of the
CXCR4/CXCL12 axis and its clinical implications in gastric cancer.
Histol Histopathol. 27:1155–1161. 2012.PubMed/NCBI
|
|
11.
|
Zhang SS, Han ZP, Jing YY, Tao SF, Li TJ,
Wang H, Wang Y, Li R, Yang Y, Zhao X, Xu XD, Yu ED, Rui YC, Liu HJ,
Zhang L and Wei LX: CD133(+)CXCR4(+) colon cancer cells exhibit
metastatic potential and predict poor prognosis of patients. BMC
Med. 10:852012.
|
|
12.
|
Wang TB, Chen ZG, Wei XQ, Wei B and Dong
WG: Serum vascular endothelial growth factor-C and
lymphangiogenesis are associated with the lymph node metastasis and
prognosis of patients with colorectal cancer. ANZ J Surg.
81:894–899. 2011.PubMed/NCBI
|
|
13.
|
De Falco V, Guarino V, Avilla E,
Castellone MD, Salerno P, Salvatore G, Faviana P, Basolo F, Santoro
M and Melillo RM: Biological role and potential therapeutic
targeting of the chemokine receptor CXCR4 in undifferentiated
thyroid cancer. Cancer Res. 67:11821–11829. 2007.PubMed/NCBI
|
|
14.
|
Li JK, Yu L, Shen Y, Zhou LS, Wang YC and
Zhang JH: Inhibition of CXCR4 activity with AMD3100 decreases
invasion of human colorectal cancer cells in vitro. World J
Gastroenterol. 14:2308–2313. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Kim JM, Lee YH, Ku CR and Lee EJ: The
cyclic pentapep-tide d-Arg3FC131, a CXCR4 antagonist, induces
apoptosis of somatotrope tumor and inhibits tumor growth in nude
mice. Endocrinology. 152:536–544. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Murakami T, Kawada K, Iwamoto M, Akagami
M, Hida K, Nakanishi Y, Kanda K, Kawada M, Seno H, Taketo MM and
Sakai Y: The role of CXCR3 and CXCR4 in colorectal cancer
metastasis. Int J Cancer. 132:276–287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Heckmann D, Laufs S, Maier P, Zucknick M,
Giordano FA, Veldwijk MR, Eckstein V, Wenz F, Zeller WJ, Fruehauf S
and Allgayer H: A lentiviral CXCR4 overexpression and knockdown
model in colorectal cancer cell lines reveals plerixafor-dependent
suppression of SDF-1α-induced migration and invasion. Onkologie.
34:502–508. 2011.PubMed/NCBI
|
|
18.
|
Liang Z, Yoon Y, Votaw J, Goodman MM,
Williams L and Shim H: Silencing of CXCR4 blocks breast cancer
metastasis. Cancer Res. 65:967–971. 2005.PubMed/NCBI
|
|
19.
|
Smith MC, Luker KE, Garbow JR, Prior JL,
Jackson E, Piwnica-Worms D and Luker GD: CXCR4 regulates growth of
both primary and metastatic breast cancer. Cancer Res.
64:8604–8612. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Liang Z, Wu H, Reddy S, Zhu A, Wang S,
Blevins D, Yoon Y, Zhang Y and Shim H: Blockade of invasion and
metastasis of breast cancer cells via targeting CXCR4 with an
artificial microRNA. Biochem Biophys Res Commun. 363:542–546. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Beverly AT and Simon PF: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Meads MB, Hazlehurst LA and Dalton WS: The
bone marrow microenvironment as a tumor sanctuary and contributor
to drug resistance. Clin Cancer Res. 14:2519–2526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Balkwill F: Cancer and the chemokine
network. Nat Rev Cancer. 4:540–550. 2004. View Article : Google Scholar
|
|
24.
|
Petit I, Jin D and Rafii S: The
SDF-1-CXCR4 signaling pathway: a molecular hub modulating
neo-angiogenesis. Trends Immunol. 28:299–307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Rubie C, Frick VO, Ghadjar P, Wagner M,
Justinger C, Faust SK, Vicinus B, Gräber S, Kollmar O and Schilling
MK: CXC receptor-4 mRNA silencing abrogates CXCL12-induced
migration of colorectal cancer cells. J Transl Med. 9:222011.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Wang SC, Lin JK, Wang HS, Yang SH, Li AF
and Chang SC: Nuclear expression of CXCR4 is associated with
advanced colorectal cancer. Int J Colorectal Dis. 25:1185–1191.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Fukunaga S, Maeda K, Noda E, Inoue T, Wada
K and Hirakawa K: Association between expression of vascular
endothelial growth factor C, chemokine receptor CXCR4 and lymph
node metastasis in colorectal cancer. Oncology. 71:204–211. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Matsusue R, Kubo H, Hisamori S, Okoshi K,
Takagi H, Hida K, Nakano K, Itami A, Kawada K, Nagayama S and Sakai
Y: Hepatic stellate cells promote liver metastasis of colon cancer
cells by the action of SDF-1/CXCR4 axis. Ann Surg Oncol.
16:2645–2653. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Mongan JP, Fadul CE, Cole BF, Zaki BI,
Suriawinata AA, Ripple GH, Tosteson TD and Pipas JM: Brain
metastases from colorectal cancer: risk factors, incidence, and the
possible role of chemokines. Clin Colorectal Cancer. 8:100–105.
2009. View Article : Google Scholar
|
|
30.
|
Speetjens FM, Liefers GJ, Korbee CJ,
Mesker WE, van de Velde CJ, van Vlierberghe RL, Morreau H,
Tollenaar RA and Kuppen PJ: Nuclear localization of CXCR4
determines prognosis for colorectal cancer patients. Cancer
Microenviron. 2:1–7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Brand S, Dambacher J, Beigel F, Olszak T,
Diebold J, Otte JM, Göke B and Eichhorst ST: CXCR4 and CXCL12 are
inversely expressed in colorectal cancer cells and modulate cancer
cell migration, invasion and MMP-9 activation. Exp Cell Res.
310:117–130. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Ottaiano A, Franco R, Aiello Talamanca A,
Liguori G, Tatangelo F, Delrio P, Nasti G, Barletta E, Facchini G,
Daniele B, Di Blasi A, Napolitano M, Ieranò C, Calemma R, Leonardi
E, Albino V, De Angelis V, Falanga M, Boccia V, Capuozzo M, Parisi
V, Botti G, Castello G, Vincenzo Iaffaioli R and Scala S:
Overexpression of both CXC chemokine receptor 4 and vascular
endothelial growth factor proteins predicts early distant relapse
in stage II-III colorectal cancer patients. Clin Cancer Res.
12:2795–2803. 2006. View Article : Google Scholar
|
|
33.
|
Zhu Y, Yang P, Zhang X, Zhang L, Cui G,
Wang Q, Lv L, Zhang Y, Xin X, Yan T, Zhao M and Zhang N: The effect
and mechanism of CXCR4 silencing on metastasis suppression of human
glioma u87 cell line. Anat Rec (Hoboken). 296:1857–1864. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Shen B, Zheng MQ, Lu JW, Jiang Q, Wang TH
and Huang XE: CXCL12-CXCR4 promotes proliferation and invasion of
pancreatic cancer cells. Asian Pac J Cancer Prev. 14:5403–5408.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Sun X, Charbonneau C, Wei L, Yang W, Chen
Q and Terek RM: CXCR4-targeted therapy inhibits VEGF expression and
chondrosarcoma angiogenesis and metastasis. Mol Cancer Ther.
12:1163–1170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Wang B, Wang W, Niu W, Liu E, Liu X, Wang
J, Peng C, Liu S, Xu L, Wang L and Niu J: SDF-1/CXCR4 axis promotes
directional migration of colorectal cancer cells through
upregulation of integrin αvβ6. Carcinogenesis. 35:282–291.
2014.PubMed/NCBI
|
|
37.
|
Xue B, Wu W, Huang K, Xie T, Xu X, Zhang
H, Qi C, Ge J and Yu Y: Stromal cell-derived factor-1 (SDF-1)
enhances cells invasion by αvβ6 integrin-mediated signaling in
ovarian cancer. Mol Cell Biochem. 380:177–184. 2013.
|
|
38.
|
Kiss DL, Windus LC and Avery VM: Chemokine
receptor expression on integrin-mediated stellate projections of
prostate cancer cells in 3D culture. Cytokine. 64:122–130. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Paddison PJ, Caudy AA, Bernstein E, Hannon
GJ and Conklin DS: Short hairpin RNAs (shRNAs) induce
sequence-specific silencing in mammalian cells. Genes Dev.
16:948–958. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
De Maria R, Miretti S, Iussich S, Olivero
M, Morello E, Bertotti A, Christensen JG, Biolatti B, Levine RA,
Buracco P and Di Renzo MF: Met oncogene activation qualifies
spontaneous canine steosarcoma as a suitable pre-clinical model of
human steosarcoma. J Pathol. 218:399–408. 2009.PubMed/NCBI
|
|
41.
|
Chen D, Huang H, Pan C, Wang J, Zhang B
and Wang A: Antitumor effects of targeting hTERT
lentivirus-mediated RNA interference against KB cell lines. Oncol
Res. 17:621–630. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Chunkang C, Rui Y, Feng X, Juan G, Xi Z,
Lingyun W, Xiao L and Jianmin W: The roles of SDF-1/CXCR4 axis and
its relationship with apoptosis in the myelodysplastic syndromes.
Med Oncol. 28(Suppl 1): 494–500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Yu T, Wu Y, Huang Y, Yan C, Liu Y, Wang Z,
Wang X, Wen Y, Wang C and Li L: RNAi targeting CXCR4 inhibits tumor
growth through inducing cell cycle arrest and apoptosis. Mol Ther.
20:398–407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Wang Q, Diao X, Sun J and Chen Z:
Regulation of VEGF, MMP-9 and metastasis by CXCR4 in a prostate
cancer cell line. Cell Biol Int. 35:897–904. 2011. View Article : Google Scholar : PubMed/NCBI
|