Introduction
Metastasis is the leading cause of death in patients
diagnosed with cancer (1). Cancer
metastasis occurs when tumor cells disassociate from the primary
tumor, enter the circulation, and migrate to distant organs through
the peripheral blood stream or lymphatic drainage. The development
of metastases in patients is believed to result from tumor cells
entering the circulation and migrating to distant organs (2,3).
Circulating cells with the characteristics of tumor cells of
epithelial origin or circulating tumor cells (CTCs) have been
demonstrated to be present in breast, prostate and colon cancer
patients’ blood and bone marrow (4–11).
These cells have been shown not only in patients with metastatic
diseases, but also in those whose tumors are apparently localized.
Although CTCs are rare in patients, as a few as one cell per 100
million or 1 billion blood cells, molecular characterization of
CTCs may provide a greater understanding of the disease metastases,
help identify aggressive tumors, and enable therapeutic selection
and monitoring of disease for patients undergoing treatment
(4,5,7,9).
To develop technologies that identify and
characterize CTCs and to establish the association of their
presence with potential clinical significance have attracted
tremendous interest in cancer research (6). A variety of technologies have been
developed to improve detection and capture of CTCs from peripheral
blood, which include immunomagnetic bead separation using
monoclonal antibodies targeting epithelial cell-surface antigens,
cell sorting using flow cytometry, filtration based size
separation, density gradient centrifugation, microfluidic devices
and fast-scan imaging (12–18).
For example, CellSearch™ was the first rare cell technology that
demonstrated its clinical validity in predicting progression-free
survival and overall survival of metastatic cancer patients based
on CTC enumeration (4,5,7,9).
Despite advances in CTC capturing technologies, the low frequency
of CTCs in cancer patients and the heterogeneity of the tumor and
the CTCs have limited applications of the CTC technology in clinic.
Current technologies for CTC detection suffer from extensive
leukocyte contamination and dependency on either tumor specific or
epithelial specific immune markers for the capture of the target
cells, making it highly unlikely that one single perfect marker
exists that will identify all the CTCs present in the same tumor
and within the same patient (6,19).
For example, the epithelial cell adhesion molecule (EpCAM)
represents the current capturing antibody of choice for the
majority of microfluidic devices that have been developed to
capture CTCs. However, the use of EpCAM as the capturing antibody
has been criticized. There is strong evidence from preclinical and
clinical studies that a small population of the CTCs undergoes an
epithelial to mesenchymal transition (EMT) and spread the tumor to
distant organs (20). Relying on
one capturing antibody is not the best strategy for CTC
identification and the solution lies in multiplexing cancer
biomarkers to identify as many heterogeneous cancer cells as
possible to study their roles in cancer progression.
Furthermore, it is of great interest to go beyond
cell enumeration and characterize the CTCs by assessing gene and
protein markers on CTCs to gain insight into mechanisms of
metastasis and best treatment modalities for patients (21,22).
For example, breast cancer encompasses a group of highly
heterogeneous diseases, which can be demonstrated at molecular,
histopathologic and clinical levels. In no other cancer has there
been so much research linking the role of various biomarkers to
disease progression and patient outcome. Significant progress has
been made over years in breast cancer detection and management,
including annual mammographic screening, effective hormonal and
chemotherapy therapies, and targeted therapies against estrogen
receptor (ER) and HER2. With such progress, it becomes critically
important to determine which patients are most likely to benefit
from which therapies and in identifying subgroups of patients who
have a more aggressive disease thus are at the highest risk for
recurrence. For example, decision regarding the use of adjuvant
therapies requires weighing the risk of recurrence against the
potential benefit and side-effect of a treatment. Established
clinical, pathologic features and biomarkers such as patient age,
tumor size, nodal status, tumor grade, ER, progesterone receptor
(PR), and HER2 status are used to estimate a patient’s risk for
recurrence and to guide treatment options. However, these types of
risk estimates remain imprecise for many patients, which lead to
either over-treatment of some with unnecessarily toxic therapies,
or to under-treatment of others who receive false assurances of a
favorable prognosis. Attempts have been made to identify additional
molecular markers that could predict disease progression and
patient outcome more precisely (1,23–25).
Studies on gene expression microarray have led to the discovery of
distinct subtypes of breast carcinomas, each with unique phenotypes
and clinical outcomes. Similar studies have shown that breast
cancer can also be divided into 5 similar subgroups using
immunohisto-chemical (IHC) analysis with a panel of protein markers
(such as ER, PR, HER2, Ki67, PI3K and others) (23–25).
To detect such molecular markers using a minimally-invasive test
such as CTCs has a great potential for use in routine clinical
practice to guide therapy choice for breast cancer patients.
In this study, we have developed a novel
microfluidic technology that uses a size and deformability based
capture system to characterize CTCs. The JETTA™ microfluidic chip
contains a parallel network of fluidic channels which have about
56,320 capture chambers (26).
Each chamber ensures that smaller blood cells such as red blood
cells and most of the leukocytes escape while larger cancer cells
get trapped and isolated in the chamber. Because the device
captures cells using label free detection, it is wide open to using
a variety of antibodies. In addition, since target cancer cells are
segregated in their own chambers separate from leukocytes, it
alleviates the problem of leukocyte contamination that is
associated to most of current CTC technologies. Most importantly,
the single cell capturing chamber has the potential to allow
downstream molecular analysis such as polymerase chain reaction
(PCR), fluorescence in situ hybridization (FISH) and IHC
assays to be performed on the microfluidic device at the single
cell level. This capability distinguishes the technology from all
other available CTC technologies and provides tremendous hope for
the field to go to the next stage of clinical validation of
CTCs.
To validate this microfluidic technology, different
breast cancer cells including MCF7, MDA-MB-231 and SKBR3, as well
as a panel of breast cancer biomarkers were used to test the device
(27). We found that the device
captured cells in a range of 20–2,000 with high reproducibility.
The capturing efficiency of the cells was greater than 80%. In
addition, background leukocyte in the captured cell population is
minimized. Furthermore, it captured both epithelial cancer cells
such as MCF7 and SKBR3 and mesenchymal cells such as MDA-MB-231.
Immunostaining of the captured cells on the microchannel device
suggested that a panel of breast cancer biomarkers can be used to
characterize differential expression of the captured cells.
Materials and methods
Microfluidic chip fabrication
process
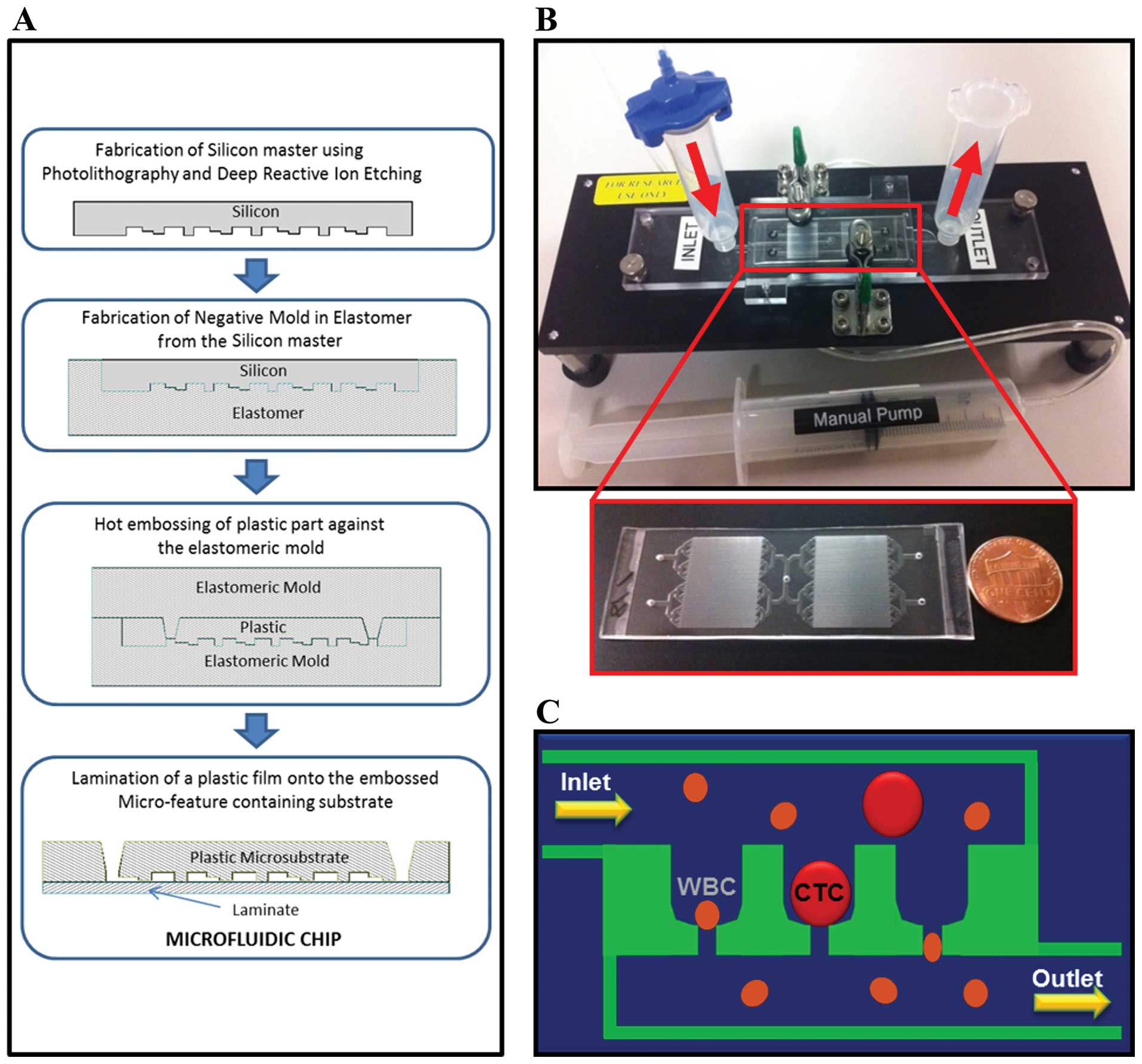
The microfluidic chip fabrication begins with a
silicon master device containing micro-features (Fig. 1A). The micro-features consist of a
fluidic network (∼75 μm deep) leading to multiple cell
trapping chambers (20 × 25 × 30 μm) with individual pore
channels (∼10 × 8 μm). This process uses standard
micro-fabrication techniques (photo-lithography and deep reactive
ion etching). From the master device, a soft elastomeric negative
mold is created by pouring and curing against the silicon master.
The final micro-substrate is created by hot embossing a plastic
plate made of cyclic olefin polymer (COP) against the elastomeric
negative mold. A thin plastic laminate containing
pressure-sensitive adhesive is then laminated against the COP
micro-substrate to create the final microfluidic chip. The
microchannel device is illustrated in Fig. 1B and the size-based filtration for
CTC capturing is described in Fig.
1C.
Cell line and cell culture
Several breast cancer cell lines were used for
microchannel device testing and in spiked-in experiments. Human
mammary carcinoma cell lines MCF7 (ATCC HTB-22), MDA-MB-231 (ATCC
HTB-26), and SKBR3 (ATCC HTB-30) were obtained from American Type
Culture Collection (ATCC, Manassas, VA). MCF7 and MDA-MB-231 cells
were cultured in DMEM medium with 10% deactivated fetal bovine
serum (FBS) (Life Technologies, Carlsbad, CA) and 1% Pen Strep
(Life Technologies). SKBR3 cells were cultured in McCoy’s 5A medium
with 10% deactivated FBS (Life Technologies) and 1% Pen Strep (Life
Technologies). The cultures were maintained at 37°C in a humidified
atmosphere containing 5% CO2 (v/v). The cells were
sub-cultivated every 4 days and the media was replaced every 48 h.
Sub-confluent monolayers were dissociated using 0.25% trypsin
solution (Thermo Scientific, Waltham, MA).
Sample preparation and cell capture
Peripheral blood samples were obtained from healthy
donors using CellSave tubes (Veridex, Raritan, NJ) with written
informed consent (Boca Biolistics, Coconut Creek, FL). A known
amount of cells diluted in cell culture medium were introduced to 2
ml of 1X phosphate-buffered saline (PBS) or 2 ml of normal blood
sample and prefixed in 2 ml 0.8% paraformaldehyde (PFA) using a
tube rocker for 10 min incubation. Prior to sample loading, the
microfluidic device was coated with priming buffer consisting of 1X
PBS, ethylene-diamine-tetraacetic acid (EDTA), and 1.0% bovine
serum albumin (BSA) to coat microchannels and remove bubbles. The
prepared sample was then added into the inlet reservoir, followed
with loading into the microfluidic device at approximately 1 ml/min
volumetric flow rate. Cancer cells owing to their bigger size
compared to blood cells were captured by micro-chambers and the
remaining solution containing red blood cells and most of the
leukocytes is collected by the outlet reservoir after passing
through pore chambers. A background level of larger leukocytes such
as monocytes are also trapped by the micro-chambers but are
distinguished by their surface markers in the subsequent
analysis.
Immunofluorescence staining of CTCs
After being captured in the microchannel device,
prefixed cells were fixed using 4.0% PFA for 10 min at room
temperature. Permeabilization was then achieved by 0.1% Triton
X-100 (Sigma-Aldrich, St. Louis, MO) and 1.0% BSA for 10 min at
room temperature. After blocking with 5% Goat Serum (Life
Technologies) for 25 min, the cells were incubated for 50 min with
mouse monoclonal antibodies. AlexaFlour 488 conjugated antibodies
against either vimentin (Santa Cruz Biotechnology, Santa Cruz, CA)
or E-cadherin (BD Biosciences, San Diego, CA) were diluted 1:100
for staining. Monoclonal IgG1 primary antibodies against HER-2
(BioLegend, San Diego, CA), ER (BD Biosciences), PI3K (Abcam,
Cambridge, MA), and PanCK (Sigma-Aldrich) were diluted 1:200. All
primary antibodies were then detected by anti-mouse AlexaFluor 488
secondary IgG1 antibody with 30 min incubation. The antibody
against leukocyte common antigen, CD45 (mouse IgG2a) (AbD Serotec,
Oxford, UK) was diluted 1:200 and used as a marker for background
leukocytes. CD45 was then detected by anti-mouse AlexaFluor 594
secondary IgG2 antibody (1:500 dilution) (Life Technologies).
Nuclei were counterstained with 1.0 μg/ml Hoechst-33342
(Life Technologies) for 5 min after secondary antibody
incubation.
Microscope imaging, enumeration and
analysis of CTCs
Cells were monitored using an inverted fluorescence
microscope TE2000-U (Nikon, Tokyo, Japan). Bright-field and
fluorescence images and time lapse videos were captured using a HQ2
CCD camera (Photometrics, Tucson, AZ). All images were taken with
the same exposure time and conditions in order to compare the
relative fluorescence intensity. Data collection and imaging
analysis were performed using the NIH ImageJ software. CTC
enumeration following antibody labeling was performed manually.
PanCK+/CD45− nucleated cells were identified
as CTCs. Positive and negative controls for antibody performance
and staining were included in each experiment. Each experiment was
performed in triplicates and results are expressed as means ± SE
for each set of experiments.
Results
Enumeration and capture efficiency of
cells
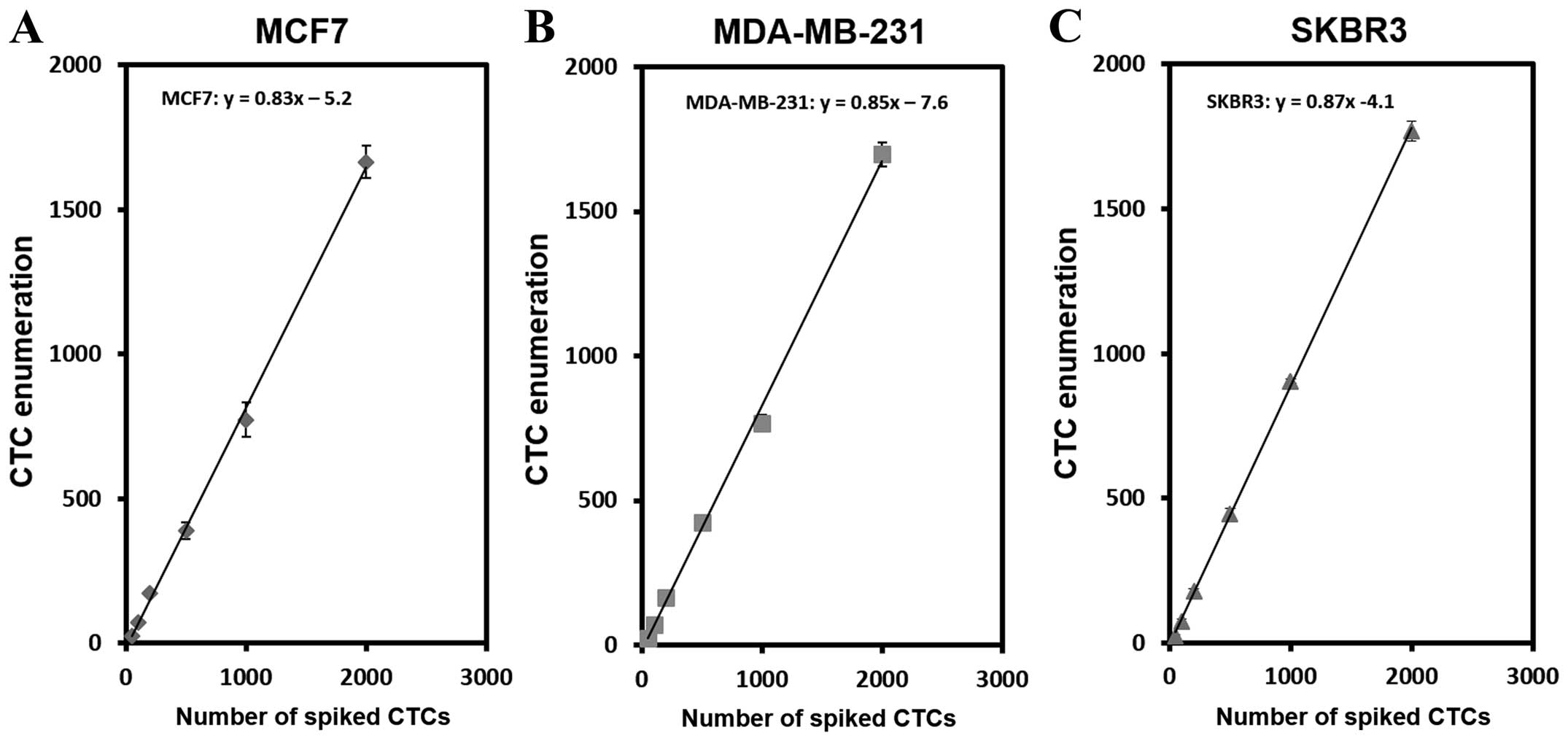
To test the performance of the microchannel device,
we first determined the capture efficiency of cells using cell
lines in 1X PBS (Fig. 2). As shown
in Fig. 2A, different number of
MCF7 cells, ranging from 20 to 2,000 cells per 2 ml 1X PBS, were
analyzed. The average capture efficiency is 83%. Similar
experiments have been also performed with MDA-MB-231 and SKBR3 cell
lines yielding averaged capture efficiency of 85% and 87%,
respectively (Fig. 2B and C).
Table I shows the capture
efficiency of the device at each number of spiked cells. The
efficiency of cell capturing ranged between 75–83% for MCF7, 77–85%
for MDA-MB-231 and 78-89% for SKBR3. Coefficient of variance
obtained by three independent experiments (n=3) varied between 2.5
to 6.7 suggesting high reproducibility of cell capturing with this
device.
 | Table I.Capturing efficiency of breast cancer
cells using cell lines. |
Table I.
Capturing efficiency of breast cancer
cells using cell lines.
| Cell line | No. of spiked
cells | Capturing efficiency,
% | Coefficient of
variance |
|---|
| MCF7 | 50 | 75.0 | 6.7 |
| 100 | 78.7 | 5.6 |
| 200 | 82.3 | 5.8 |
| 500 | 83.2 | 4.4 |
| 1,000 | 80.5 | 3.5 |
| 2,000 | 83.2 | 3.4 |
| MDA-MB-231 | 50 | 77.0 | 6.5 |
| 100 | 79.2 | 6.3 |
| 200 | 82.8 | 5.2 |
| 500 | 85.0 | 4.4 |
| 1,000 | 80.5 | 5.6 |
| 2,000 | 84.9 | 2.5 |
| SKBR3 | 50 | 78.0 | 5.1 |
| 100 | 81.2 | 4.2 |
| 200 | 85.1 | 2.7 |
| 500 | 89.0 | 4.5 |
| 1,000 | 84.1 | 5.3 |
| 2,000 | 86.9 | 2.6 |
Enumeration and capture efficiency of
spike-in cells
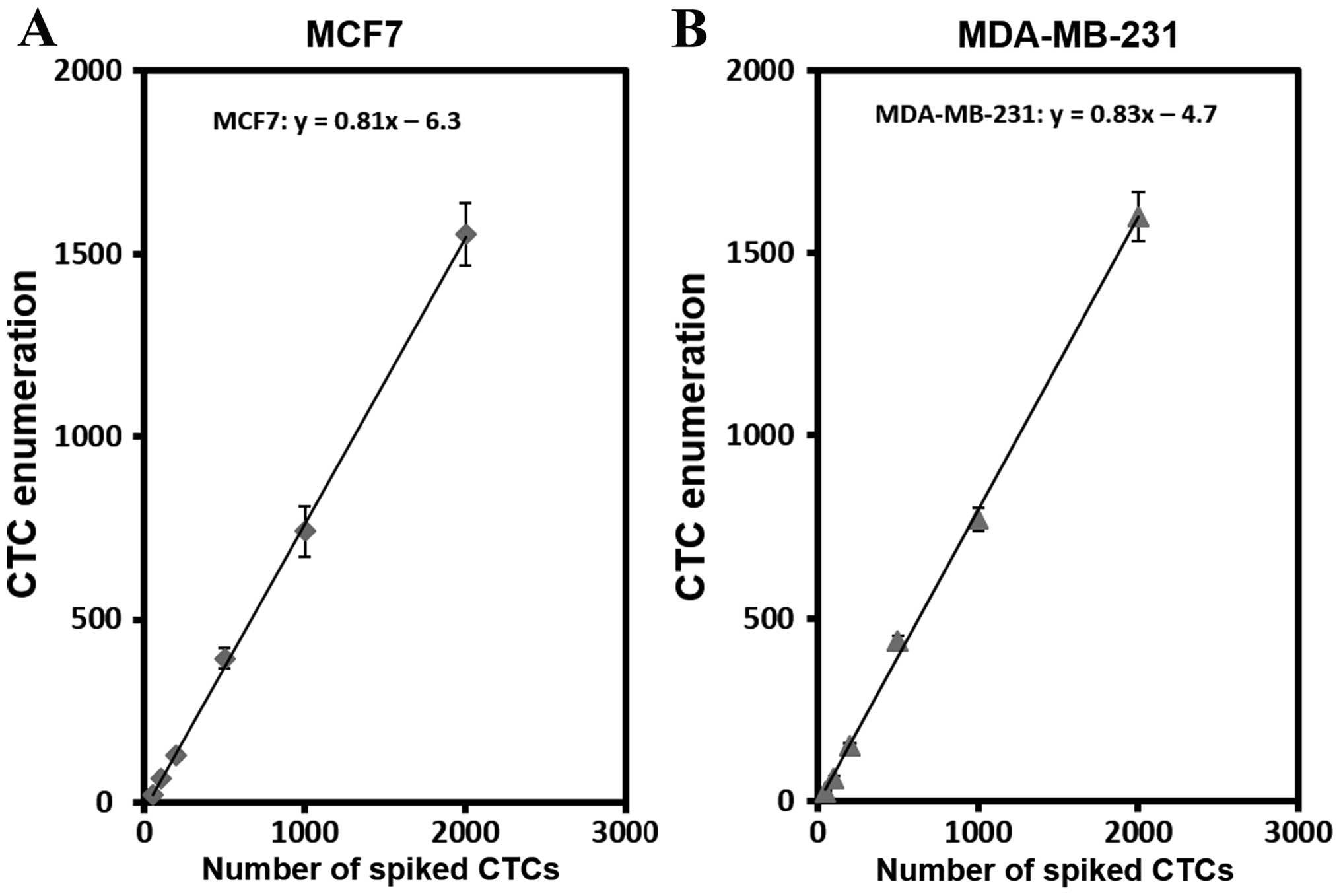
To assess cell capture efficiency under
physiological conditions, we performed a series of spike-in
experiments in which certain number of breast cancer cells
including MCF7 (epithelial) and MDA-MB-231 (mesenchymal) were
spiked into peripheral blood samples from healthy donors. As shown
in Fig. 3, the average cell
capturing efficiency in the spike-in samples was 81% and 83% for
MCF7 and MDA-MB-231 cells, respectively. The results showed that
the capture efficiency of two cell lines was quite comparable
ranging from 74–82% for MCF7 and 75–82% for MDA-MB-231 depending
upon number of spiked cells (Table
II). Low coefficient of variance (1.0–6.5) indicated high
reproducibility of the results (n=3). This data further
demonstrated that the capture efficiency and experimental
reproducibility for each cell spiking number are consistent with
the results we observed for the cells in PBS buffer. We have tested
20 MCF7 cells spiked into 2 ml blood samples, the average cell
capturing efficiency was 84% with the standard deviation of 11.9%
(n=5). In addition, spiked-in samples with 5 MCF7 cells yielded 4
or 5 cells in multiple tests although the accuracy of cell counts
is difficult to achieve at this level (data not shown).
Collectively, high capture efficiency and reproducibility were
evident with the device in both the cell lines and the spike-in
samples.
 | Table II.Capturing efficiency of spike-in
breast cancer cells in normal donor blood. |
Table II.
Capturing efficiency of spike-in
breast cancer cells in normal donor blood.
| Cell line | No. of spiked
cells | Capturing efficiency,
% | Coefficient of
variance |
|---|
| MCF7 | 50 | 74.0 | 5.6 |
| 100 | 77.7 | 3.7 |
| 200 | 79.2 | 3.8 |
| 500 | 78.9 | 4.3 |
| 1,000 | 79.0 | 4.0 |
| 2,000 | 80.9 | 2.7 |
| MDA-MB-231 | 50 | 75.0 | 6.5 |
| 100 | 81.0 | 3.0 |
| 200 | 79.0 | 1.0 |
| 500 | 80.5 | 3.7 |
| 1,000 | 82.6 | 1.5 |
| 2,000 | 82.3 | 4.5 |
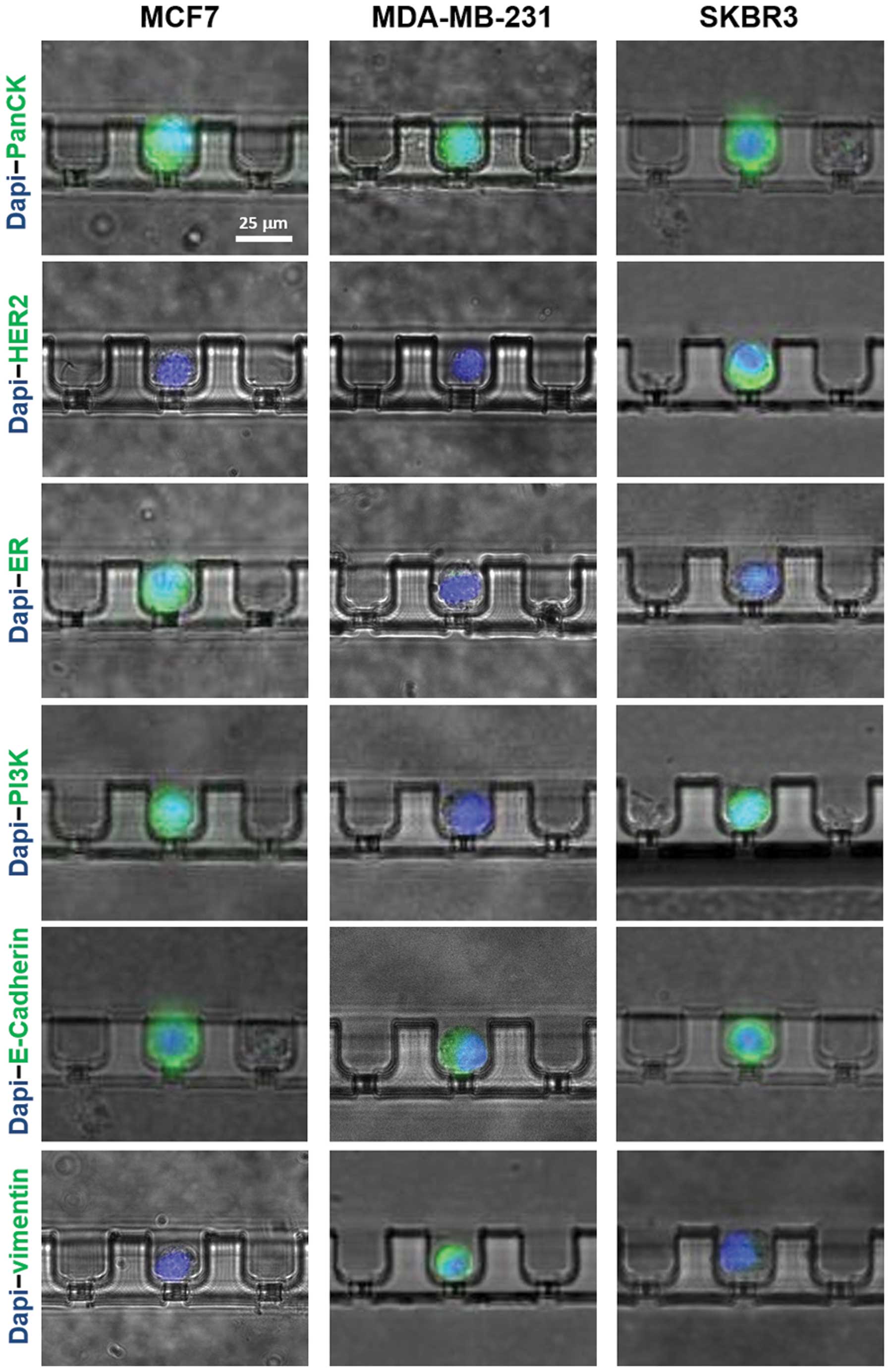
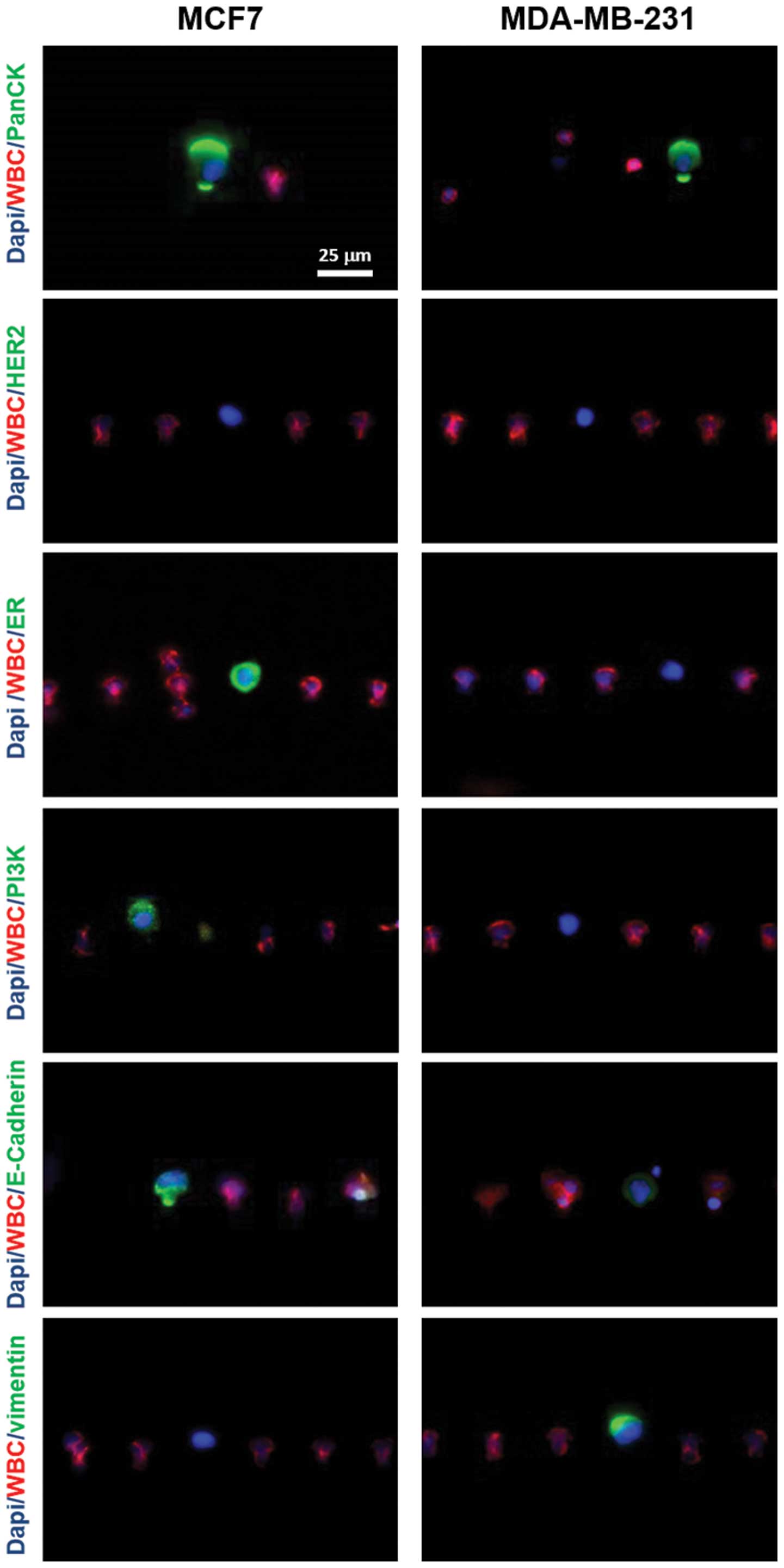
Molecular characteristics of cells
To examine the ability of the microchannel device to
characterize the captured cells with molecular markers, we
performed a series of immunostaining experiments to analyze the
expression of several breast cancer epithelial or
mesenchymal-specific biomarkers. MCF7, MDA-MB-231 and SKBR3 cells
were used in the experiments.
For each cell line, 100 cells were first spiked into
2 ml 1X PBS and then stained with either PanCK, HER-2, ER, PI3K,
E-cadherin or vimentin after being captured by microchambers
(Fig. 4) The cell nuclei were also
stained by 1.0 μm/g Hoechst-33342 in all cases. Our
observation revealed positive staining of PanCK in all three cell
lines. HER-2 was only expressed in SKBR3 cells, but not MCF7 cells.
ER was only expressed in MCF7 cells, but not SKBR3 cells. In
addition, both epithelial cells, MCF7 and SKBR3 were PI3K and
E-cadherin positive, but vimentin negative. Compared to the
epithelial cells, MDA-MB-231 was shown to be HER2, ER, PI3K
negative while expressing a low level of E-cadherin and high level
of vimentin, a mesenchymal cell-specific marker.
Similar analysis of the expression has been
performed on the captured cells using spike-in cells into
peripheral blood. To distinguish background hematologic cells from
the captured cancer cells, we used CD45 as a marker for leukocyte
staining. Examples of the stained captured cancer cells and
leucocytes are shown in Fig. 5.
The results are highly consistent with those from the cell lines.
MCF7 cells were PanCK, ER, PI3K, E-cadherin positive, but HER2 and
vimentin negative. MDA-MB-231 cells possessed high level of
vimentin and PanCK expression and low level of E-cadherin
expression, but no expression on HER2, ER and PI3K. Our results
suggested that the microchannel device capture both epithelial
cancer cells such as MCF7 and SKBR3 and EMT-like cells such as
MDA-MB-231. Furthermore, the microchannel device is able to
identify differential expression and phenotype of capture cells
using panel of epithelial and mesenchymal breast cancer biomarkers.
The data of the molecular characterization in spike-in cells is
summarized in Table IV.
 | Table IV.Differential expression of cancer
biomarkers in spike-in breast cancer cells. |
Table IV.
Differential expression of cancer
biomarkers in spike-in breast cancer cells.
| MCF7 | MDA-MB-231 |
|---|
| PanCK | + | + |
| HER-2 | − | − |
| ER | + | − |
| PI3K | + | − |
| E-cadherin | + | + (low) |
| Vimentin | − | + |
Capture of CTCs in patient clinical
samples
To test the clinical application of the microfluidic
device, blood samples from metastatic breast cancer patients were
processed. CTCs have been captured and enumerated using the
antibodies against PanCK and CD45 (Fig. 6). From 2 ml of blood, 1 to >600
CTCs have been counted from the metastatic breast cancer samples.
Interestingly, the device also captured clusters of cancer cells,
which have been implicated as micrometastases and probably
represent more aggressive tumor cells than individual CTCs.
Detailed clinical data and further analysis of the study is being
carried out with the aim towards demonstrating the clinical use of
the platform.
Discussion
We investigated a novel technology of capturing and
characterizing CTCs by using a microchannel device. Different
breast cancer cells including MCF7, MDA-MB-231 and SKBR3, as well
as a panel of breast cancer biomarkers were used to test the
device. The device can capture cells in a range of 20–2,000 with
high reproducibility. The capturing efficiency of the cells is
greater than 80% with a minimum background of leukocyte
contamination in the captured cell population. Furthermore, it
captured both epithelial cancer cells such as MCF7 and SKBR3 and
mesenchymal cells such as MDA-MB-231. Immunostaining of the
captured cells on the microchannel device suggested that a panel of
breast cancer biomarkers can be used to characterize differential
expression of the captured cells. This device is unique in its
ability to segregate cancer cells in their individual chambers thus
separating them from contaminating leukocytes and also allowing for
on chip molecular analysis at the single cell level. This study is
laying the foundation for future studies that will test the
clinical validity and utility of this CTC technology.
Breast cancer represents a heterogeneous group of
diseases. Cell lines derived from primary tumors can reflect the
molecular diversity of the disease. One objective of this study was
to investigate the expression patterns of those clinically relevant
biomarkers for breast cancer (ER, HER2, PI3K, vimentin and
E-cadherin) in commonly used breast cancer cells. The panel of
breast cancer markers selected for the study has been implicated to
be specific for breast epithelial cells and/or mesenchymal cells.
The detection of the markers in the captured cells not only
confirmed that the cells originated from subtypes of breast cancer,
but also revealed that the majority of captured cells kept the
properties of breast cancer cells. Among the three breast cancer
cell lines, MCF7 resembles the Luminal A subtype because it is ER
positive and HER2 negative. SKBR3 with high HER2 expression and no
ER expression belongs to HER2 subtype. In contrast, MDA-MB-231 with
vimentin positive, HER2 negative, and ER negative resembles within
the basal-like subtype. This demonstrated the feasibility of using
the biomarkers to classify different types of breast cancer cells
using the microchannel platform.
We observed that CD45, a leukocyte specific marker
was expressed in the majority of the background leukocytes. This
level of the background leukocytes was consistent with the
observation of leukocytes presence in the CTC-enriched populations
with other CTC capturing technologies. Although the background
leukocytes create a challenge for detecting and analyzing CTCs, the
level of leukocyte background observed with this technology kept
leukocytes in separate microchambers and did not seem to affect the
analytical sensitivity of immunostaining of the captured cells.
In summary, clinical oncology is challenged by a
lack of predictive tests for therapy choice and therapy response
that are simple, non-invasive and inexpensive. CTC technologies
provide a great promise of delivering such a tool that enables
enumeration and molecular characterization of metastatic cancer
cells and estimate prognosis and therapeutic response of the
patient. Fundamental research continues to increase our knowledge
of molecular and cellular processes that contribute to the clinical
behavior of cancer. Further development of the technology could
potentially lead to benefits of the patients through personalized
treatment strategies to improve patient management and
outcomes.
Acknowledgements
We would like to thank Dr Pak Kin Wong
at the University of Arizona for his support to the study and
critical review of the manuscript, Dr Wenjun Zhang and Noah Theiss
for their technical assistance.
References
|
1.
|
Weigelt B, Peterse JL and van’t Veer LJ:
Breast cancer metastasis: markers and models. Nature Rev Cancer.
5:591–602. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Maheswaran S and Haber DA: Circulating
tumor cells: a window into cancer biology and metastasis. Curr Opin
Genet Dev. 20:96–99. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Pantel K and Alix-Panabieres C:
Circulating tumour cells in cancer patients: challenges and
perspectives. Trends Mol Med. 16:398–406. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Cristofanilli M, Budd GT, Ellis MJ,
Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ,
Terstappen LW and Hayes DF: Circulating tumor cells, disease
progression, and survival in metastatic breast cancer. N Engl J
Med. 351:781–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Cristofanilli M, Hayes DF, Budd GT, Ellis
MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC,
Fritsche HA, Hortobagyi GN and Terstappen LW: Circulating tumor
cells: a novel prognostic factor for newly diagnosed metastatic
breast cancer. J Clin Oncol. 23:1420–1430. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Pantel K, Brakenhoff RH and Brandt B:
Detection, clinical relevance and specific biological properties of
disseminating cancer cells in breast cancer patients. Nature Rev
Cancer. 8:329–340. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Punnoose EA, Atwal SK, Spoerke JM, Savage
H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS and
Lackner MR: Molecular biomarker analyses using circulating tumor
cells. PLoS One. 5:e125712010. View Article : Google Scholar
|
|
8.
|
Kirby BJ, Jodari M, Loftus, Gakhar G,
Pratt ED, Chanel-Vos C, Gleghorn JP, Santana SM, Liu H, Smith JP,
Navarro VN, Tagawa ST, Bander NH, Nanus DM and Giannakakou P:
Functional characterization of circulating tumor cells with a
prostate-cancer-specific microfluidic device. PLoS One.
7:e359162012. View Article : Google Scholar
|
|
9.
|
Riethdorf S, Fritsche H, Muller V, Rau T,
Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, Jackson
S, Gornet T, Cristofanilli M and Pantel K: Detection of circulating
tumor cells in peripheral blood of patients with metastatic breast
cancer: a validation study of the CellSearch system. Clin Cancer
Res. 13:920–928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Reinholz MM, Nibbe A, Jonart LM, Kitzmann
K, Suman VJ, Ingle JN, Houghton R, Zehentner B, Roche PC and Lingle
WL: Evaluation of a panel of tumor markers for molecular detection
of circulating cancer cells in women with suspected breast cancer.
Clin Cancer Res. 11:3722–3732. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Warner EA, Kotz KT, Ungaro RF, Abouhamze
AS, Lopez MC, Cuenca AG, Kelly-Scumpia KM, Moreno C, O’Malley KA,
Lanz JD, Baker HV, Martin LC, Toner M, Tompkins RG, Efron PA and
Moldawer LL: Microfluidics-based capture of human neutrophils for
expression analysis in blood and bronchoalveolar lavage. Lab
Invest. 91:1787–1795. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Dong Y, Skelley AM, Merdek KD, Sprott KM,
Jiang C, Pierceall WE, Lin J, Stocum M, Carney WP and Smirnov DA:
Microfluidics and circulating tumor cells. J Mol Diagn. 15:149–157.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Zhang Z and Nagrath S: Microfluidics and
cancer: are we there yet? Biomed Microdevices. 15:595–609. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Nagrath S, Sequist LV, Maheswaran S,
Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L,
Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ,
Tompkins RG, Haber DA and Toner M: Isolation of rare circulating
tumor cells in cancer patients by microchip technology. Nature.
450:1235–1239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Stott SL, Hsu CH, Tsukrov DI, Yu M,
Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK,
Floyd FP Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D,
Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber
DA and Toner M: Isolation of circulating tumor cells using a
microvortex-generating herringbone-chip. Proc Natl Acad Sci USA.
107:18392–18397. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Li P, Stratton ZS, Dao M, Ritz J and Huang
TJ: Probing circulating tumor cells in microfluidics. Lab Chip.
13:602–609. 2013. View Article : Google Scholar
|
|
17.
|
Saliba AE, Saias L, Psychari E, Minc N,
Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J,
Saada V, Farace F, Vielh P, Malaquin L and Viovy JL: Microfluidic
sorting and multimodal typing of cancer cells in self-assembled
magnetic arrays. Proc Natl Acad Sci USA. 107:14524–14529. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Warkiani ME, Guan G, Luan KB, Lee WC,
Bhagat AA, Kant Chaudhuri P, Tan DS, Lim WT, Lee SC, Chen PC, Lim
CT and Han J: Slanted spiral microfluidics for the ultra-fast,
label-free isolation of circulating tumor cells. Lab Chip.
14:128–137. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Ring AE, Zabaglo L, Ormerod MG, Smith IE
and Dowsett M: Detection of circulating epithelial cells in the
blood of patients with breast cancer: comparison of three
techniques. Br J Cancer. 92:906–912. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wicha MS and Hayes DF: Circulating tumor
cells: not all detected cells are bad and not all bad cells are
detected. J Clin Oncol. 29:1508–1511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Smirnov DA, Zweitzig DR, Foulk BW, Miller
MC, Doyle GV, Pienta KJ, Meropol NJ, Weiner LM, Cohen SJ, Moreno
JG, Connelly MC, Terstappen LW and O’Hara SM: Global gene
expression profiling of circulating tumor cells. Cancer Res.
65:4993–4997. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Sieuwerts AM, Mostert B, Bolt-de Vries J,
Peeters D, de Jongh FE, Stouthard JM, Dirix LY, van Dam PA, Van
Galen A, de Weerd V, Kraan J, van der Spoel P, Ramírez-Moreno R,
van Deurzen CH, Smid M, Yu JX, Jiang J, Wang Y, Gratama JW,
Sleijfer S, Foekens JA and Martens JW: mRNA and microRNA expression
profiles in circulating tumor cells and primary tumors of
metastatic breast cancer patients. Clin Cancer Res. 17:3600–3618.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Tang P, Skinner KA and Hicks DG: Molecular
Classification of breast carcinomas by immunohistochemical
analysis: are we ready? Diagn Mol Pathol. 18:125–132. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Subik K, Lee JF, Baxter L, Strzepek T,
Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG and
Tang P: The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67
and AR by immunohistochemical analysis in breast cancer cell lines.
Breast Cancer. 4:35–41. 2010.PubMed/NCBI
|
|
25.
|
Ignatiadis M, Kallergi G, Ntoulia M,
Perraki M, Apostolaki S, Kafousi M, Chlouverakis G, Stathopoulos E,
Lianidou E, Georgoulias V and Mavroudis D: Prognostic value of the
molecular detection of circulating tumor cells using a multi-marker
reverse transcription-PCR assay for cytokeratin 19, mammaglobin A,
and HER2 in early breast cancer. Clin Cancer Res. 14:2593–2600.
2008. View Article : Google Scholar
|
|
26.
|
US Patent Application # US 2013/0190212 A1
(DeNovo Sciences).
|
|
27.
|
Allan AL and Keeney M: Circulating tumor
cell analysis: technical and statistical considerations for
application to the clinic. J Oncol. 426218:e1–e10. 2010. View Article : Google Scholar : PubMed/NCBI
|