Introduction
Concurrent irinotecan and fluorinated-pyrimidine is
a common first-line therapy for metastatic colorectal cancer (mCRC)
(1–6). Although prolonged survival is
associated with regimens involving irinotecan, severe neutropenia
occurs in 20–35% of mCRC cases treated with irinotecan regimens.
Carboxylesterases catabolized irinotecan to
7-ethyl-10-hydroxycamptothecin (SN-38), which is a potent
topoisomerase I inhibitor (7,8).
SN-38 is then further catabolized by hepatic uridin
diphosphate-glucuronosyltransferase (UGT) 1A enzymes to an inactive
SN-38 glucuronide (SN-38G) (9).
Many mCRC patients with a genetic variant (UGT1A1*28)
experience severe irinotecan toxicity; UGT1A1*28 is a
variation in the number (seven vs. six) of TA repeats in the
promoter region of UGT1A1 (10,11).
Interestingly, the toxicity and tumor response of concurrent
leucovorin, 5-fluorouracil, and irinotecan (FOLFIRI) reportedly
also correlate with UGT1A variants (UGT1A1,
UGT1A7 and UGT1A9) and haplotypes including these
variants (12–18). There are differences between
Caucasian and Asian populations in frequencies of UGT1A
variants, and UGT1A1*6 reportedly associates strongly with
severe neutropenia especially among Asian patients (12,17).
To predict the risk of irinotecan toxicity for
individual patients, it is important that determining the relative
contributions of UGT1A variants other than UGT1A1*28
and UGT1A1*6 is important to the development of any system
designed to predict irinotecan toxicity for individual patients
because patients without UGT1A1*28 or *6 do
experience severe irinotecan toxicity. Several studies have
examined associations between irinotecan toxicity and UGT1A
haplotypes in addition to each genotype of UGT1A (17–19).
However, determining the haplotype or diplotype for each patient is
difficult; moreover, most haplotypes and diplotypes are too rare to
constitute a group large enough for meaningful statistical
analysis. Moreover, gender and age of patients each reportedly have
an impact on irinotecan toxicity (20–22).
Hence, these factors should be also taken into consideration when
developing a system designed to predict irinotecan toxicity.
The aim of this study was to evaluate whether the
combinations of UGT1A genotypes, but not haplotypes,
together with patient characteristics might be useful in predicting
the risk to patients with mCRC treated of irinotecan-containing
regimens. Here, we investigated the genotypes of 123 patients at
six loci: UGT1A1*6 (211G>A, rs4148323), UGT1A1*28
(TA6>TA7, rs8175347), UGT1A1*60
(−3279T>G, rs4124874), UGT1A7 (387T>G, rs17868323),
UGT1A7 (622T>C, rs11692021), and UGT1A9*1b
(−118T9>T10, rs35426722, also called
UGT1A9*22) (23). Next, we
evaluated the contribution of each UGT1A genotype,
haplotype, and diplotype to the risk of irinotecan toxicity.
Furthermore, we developed a new system for predicting the risk that
a patient will experience irinotecan toxicity; this system uses
sequential forward floating selection (SFFS) algorithm based on
statistical pattern recognition to select the combinations of
UGT1A genotypes, gender and age. SFFS is a sequential search
method characterized by a dynamically changing number of features
included or eliminated at each step of an individual analysis
(24). This is the first study
conducted to assess the role of the combination of genotypes at six
polymorphic sites in UGT1A and clinical features constructed
by SFFS on the risk of irinotecan toxicity.
Materials and methods
Patients
In this study, 123 mCRC patients were examined for
association between UGT1A genotypes and irinotecan toxicity
(Table I). This study was
performed as an ancillary investigation; data collected from three
prospective studies [FLIGHT1 (5),
FLIGHT2 (5) and FRUTIRI (6)] and from consecutive patients who
received FOLFIRI at the Department of Digestive Surgery and
Surgical Oncology, Yamaguchi University Graduate School of
Medicine, Japan. Each participant received irinotecan at the dose
of 150 mg/m2, which has been approved in Japan.
 | Table ICharacteristics of the patients. |
Table I
Characteristics of the patients.
| | Sub-population
(treatment regimen) | |
|---|
| |
| |
|---|
| Clinical features
and genotypes | Total (n=123) | FLIGHT1a (n=38) | FLIGHT2a (n=35) | FRUTIRIb (n=22) | 2nd-line
FOLFILIc (n=28) | |
|---|
| Toxicity of
irinotecan |
| No | 72 | 20 | 19 | 16 | 17 | NSd |
| Yes | 51 | 18 | 16 | 6 | 11 | |
| Gender |
| Male | 78 | 24 | 24 | 17 | 13 | NSd |
| Female | 45 | 14 | 11 | 5 | 15 | |
| Age |
| ≤60 | 50 | 14 | 14 | 9 | 13 | NSd |
| >60 | 73 | 24 | 21 | 13 | 15 | |
|
UGT1A1*6 |
| −/− | 84 | 25 | 23 | 15 | 21 | NSd |
| −/*6 | 36 | 12 | 11 | 6 | 7 | |
| *6/*6 | 3 | 1 | 1 | 1 | 0c | |
|
UGT1A1*28 |
| −/− | 103 | 32 | 27 | 22 | 22 | NSd |
| −/*28 | 20 | 6 | 8 | 0b | 6 | |
|
*28/*28 | 0 | 0a | 0a | 0b | 0c | |
|
UGT1A1*60 |
| −/− | 71 | 19 | 21 | 15 | 16 | NSd |
| −/*60 | 46 | 17 | 12 | 6 | 11 | |
|
*60/*60 | 6 | 2 | 2 | 1 | 1 | |
| UGT1A7 |
| 387T/T | 41 | 13 | 12 | 8 | 8 | NSd |
| 387T/G | 69 | 18 | 18 | 13 | 20 | |
| 387G/G | 13 | 7 | 5 | 1 | 0 | |
| UGT1A7 |
| 387T/T | 70 | 21 | 19 | 14 | 16 | NSd |
| 387T/G | 48 | 15 | 13 | 8 | 12 | |
| 387G/G | 5 | 2 | 3 | 0 | 0 | |
|
UGT1A9*1b |
|
*1b/*1b | 43 | 14 | 12 | 9 | 8 | NSd |
| −/*1b | 67 | 17 | 18 | 12 | 20 | |
| −/− | 13 | 7 | 5 | 1 | 0 | |
FLIGHT1 (UMIN000002388) and FLIGHT2 (UMIN000002476)
were phase II studies of first line and second line chemotherapy,
respectively, for mCRC. Study designs and key eligibility and
exclusion criteria have been described in detail (5,25,26).
Briefly, each regimen consisted of irinotecan on day 1 +400
mg/m2 fluorouracil bolus followed by 2,400
mg/m2 fluorouracil continuous infusion during 46 h + 200
mg/m2 leucovorin on day 1 every 2 weeks. Of all patients
from the FLIGHT1 and FLIGHT2 studies, 38 and 35, respectively,
participated in this ancillary investigation and use; these 73
patients constituted the training population. FLIGHT1 or FLIGHT2
patients homozygous for UGT1A1*28 were excluded from the
training population because these patients received a lower
starting dose of irinotecan (100 mg/m2) (5).
The validation population comprised 50 patients from
two different study groups: 22 patients who participated in FRUTIRI
(UMIN000005011), a phase II study of a combination therapy
comprised irinotecan and 5′-deoxy-5-fluorouridine (5′-DFUR)
(6) and 28 consecutive patients
who underwent second-line FOLFILI treatment between October, 2008
and July, 2012 in the Department of Digestive Surgery and Surgical
Oncology, Yamaguchi University Graduate School of Medicine, Japan.
Detail treatment regimen tested in FRUTIRI was described previously
(6). Briefly, irinotecan was
administered every two weeks, and 400 mg 5′-DFUR was administered
every week orally twice a day on five consecutive days that were
followed by a weekly 2-day washout. The 28 consecutive patients
undergoing FOLFIRI treatment were following the protocol used in
FLIGHT2 (26). In a validation
population, patients with UGT1A1*28 homozygous were not
found in the FRUTIRI study (n=28). Additionally, patients
heterozygous for UGT1A1*28 (n=6) were excluded from the
FRUTIRI study because these patients received lower starting dose
of irinotecan 70 mg/m2. Among the 28 consecutive
patients who received second-line FOLFILI therapy, homozygous for
UGT1A1*6 or *28 and those compound heterozygous for
UGT1A1*6 and UGT1A1*28 been excluded from this
ancillary study. The training (n=73) and validation (n=50)
populations did not differ significantly with regard to the
distribution of any clinical feature or genotype that is listed in
Table I except for the
distributions of the UGT1A7 (387T>G) and
UGT1A9*1b alleles (data not shown).
In this study, we defined patients who exhibited
hematologic toxicity greater than grade 3 during the entire course
of therapy as experiencing irinotecan toxicity. The study protocols
were approved by the Institutional Review Board at Yamaguchi
University Graduate School of Medicine, and were carried out in
accordance with the Helsinki declaration on experimentation on
human subjects. Each patient gave written, informed consent for
their participation in this study.
Genotyping of UGT1A and haplotype
construction
A conventional sodium iodide (NaI) method was used
to extract genomic DNA from peripheral blood samples (27). The number of TA repeats in the
UGT1A1 promoter region was determined by the fragment size
analysis followed by direct sequencing as described previously
(4). The TaqMan technique with a
hydrolysis probe was used to determine the UGT1A1*6 genotype
as described previously (28);
similarly, hydrolysis probes were used to determine the genotypes
at UGT1A1*60; a direct sequencing method was also used to
determine the genotypes at UGT1A7 (387T>G and 622T>C)
and UGT1A9*1b.
Each nucleotide variant was evaluated to determine
whether it was in Hardy-Weinberg equilibrium; Haploview 4.2
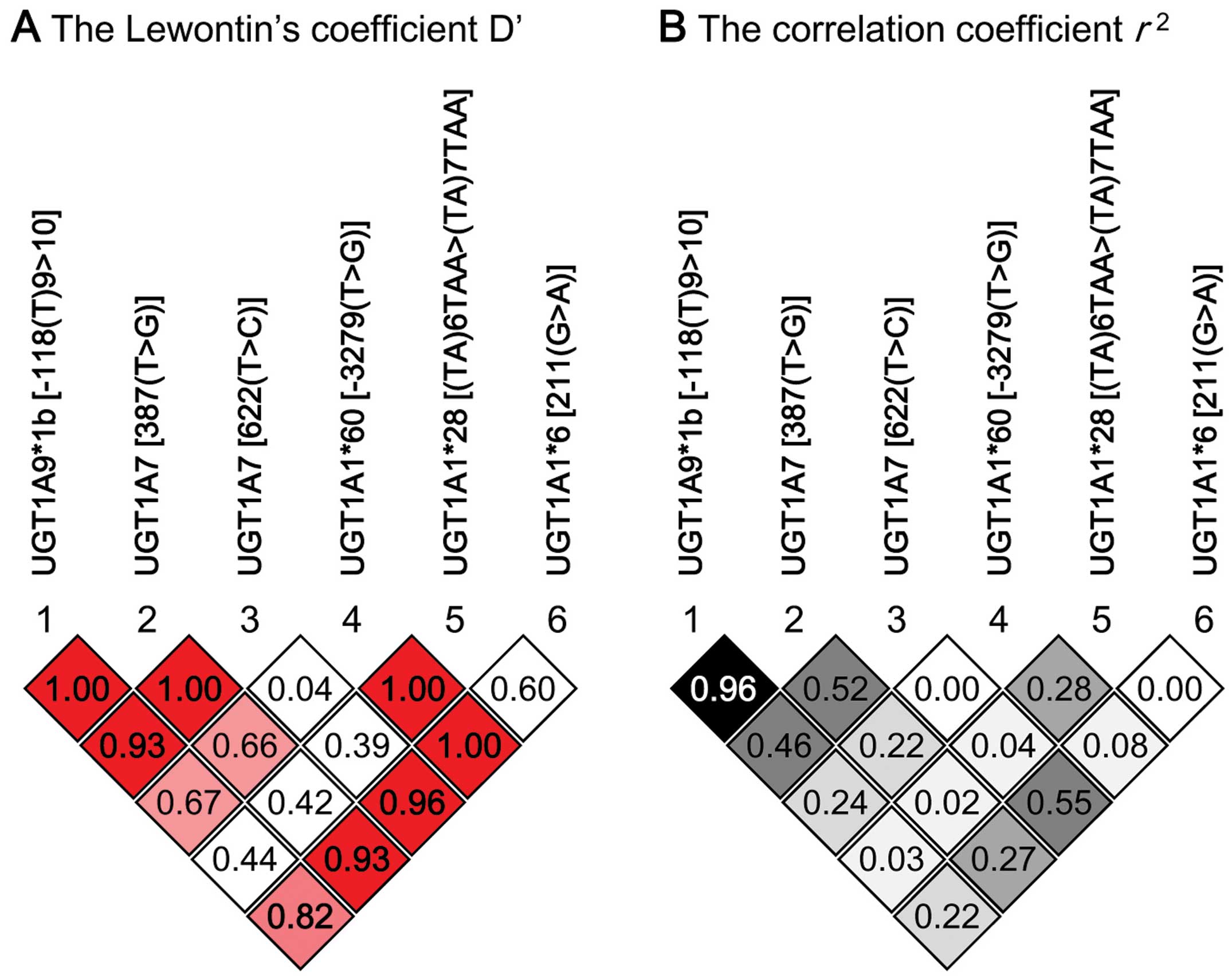
software was used to perform the linkage disequilibrium (LD) and
case-control haplotype analyses (29). Lewontin’s coefficient D’ and
correlation coefficient r2 were calculated as
measures of LD.
Construction of toxicity prediction
system by genotype combinations
To predict severe toxicities of irinotecan, the age,
the gender and a comprehensive 6-site UGT1A genotype were
determined for each of the 73 patients in the training population.
SFFS, a method of statistical pattern recognition, was then used to
determine the optimal genotype combinations for predicting the risk
of irinotecan toxicity. The statistical pattern recognition, SFFS,
identified the genotype combinations with the ‘maximum number of
cases’ and ‘maximum prediction rate’ to maximize overall diagnostic
accuracy (24). Briefly, the
algorithm of the SFFS used in this study was as follows: i) Suppose
that at stage k we have a set of
X1, …, Xk of
sizes 1 to k, respectively. ii) Let the corresponding values
of the feature selection criteria be J1 to
Jk, where Ji =
J(Xi), for the feature selection
criterion J(.). iii) Let the total set of features be
X. Then at the kth stage of the SFFS procedure follow
these steps: Step 1, select the feature xj
from X-Xk that increases the value
of J to the greatest degree and add it to the current set:
X(k + 1) =
Xk + xj. Step 2,
find the feature xr in the current set
X(k + 1) that
reduces the value of J the least; if this feature is the
same as xj then set
J(k + 1) =
J(X(k + 1));
increment k; go to step 1; otherwise remove it from the set
to from X′k =
X(k + 1) -
xr. Step 3, continue removing features
from the set X′k to form reduced sets
X′ (k − 1) while
J(X′ (k − 1))
> J(k − 1);
k = k − 1; until k = 2; then continue with
step 1. The algorithm is initialized by setting k = 0 and
X0 = Ø.
Statistical analysis
Fisher’s exact test was used to assess the
relationship between toxicity and each UGT1A variant. The
Cochran-Armitage trend test was used to examine the linearity of
the relationship between UGT1A genotypes and irinotecan
toxicity. SPSS Statics 17.0 software (IBM, Tokyo, Japan) and R
version 2.13.0 software were used to perform the calculations
(30). p<0.05 was considered
statistically significant.
Results
UGT1A allele and haplotype
frequencies
The minor allele frequencies (MAF) of each
UGT1A allele among the 103 patients without genetic bias;
all patients regardless of the starting dose of irinotecan enrolled
in FLIGHT1, FLIGHT2, and FRUTIRI studies, and 123 patients received
a starting dose of 150 mg/m2 for case-control study
participating in this study are listed in Table II. In this study, the MAFs of
UGT1A1*28 and UGT1A1*6 were approximately 0.117 and
0.184, respectively. The MAF for each other UGT1A SNP
examined in this study was greater than 0.20. Among all patients,
the Hardy-Weinberg equilibrium p-value for each locus examined in
this study was higher than 0.05. LD analysis with 103 patients
showed that high LD (r2>0.9) was evident
between UGT1A7 (387T>G) and
UGT1A9*1b (Fig. 1). We
found 12 UGT1A haplotypes (Hp-I to Hp-XII)
using 6 loci in 103 patients: UGT1A1*6, *28,
*60, UGT1A7 (387T>G), UGT1A7
(622T>C), and UGT1A9*1b (Table III). Three common haplotypes
(Hp-I, Hp-II and Hp-III) accounted for 82.5%
of all haplotypes identified in this study.
 | Table IIMinor allele frequency and
Hardy-Weinberg equilibrium in 123 patients. |
Table II
Minor allele frequency and
Hardy-Weinberg equilibrium in 123 patients.
| 103
patientsa | 123
patientsb |
|---|
|
|
|
|---|
| MAF | HWp | MAF | HWp |
|---|
| UGT1A1*6
[211 (G>A)] | 0.18 | 1.00 | 0.17 | 1.00 |
| UGT1A1*28
[(TA)6>(TA)7] | 0.12 | 0.80 | 0.08 | 0.86 |
| UGT1A1*60
[−3279 (T>G)] | 0.27 | 0.99 | 0.24 | 0.92 |
| UGT1A7 [387
(T>G)] | 0.42 | 1.00 | 0.39 | 0.07 |
| UGT1A7 [622
(T>C)] | 0.27 | 0.61 | 0.24 | 0.54 |
| UGT1A9*1b
[−118 (T9>T10)] | 0.41 | 0.84 | 0.38 | 0.13 |
 | Table IIIHaplotype frequency. |
Table III
Haplotype frequency.
| Haplotypes | UGT1A
alleles | Allele
frequencies |
|---|
|
|
|---|
| UGT1A9 | UGT1A7 | UGT1A1 | (n=103)b | (n=123)c |
|---|
| *1b | 387T>G | 622T>C | *60 | *28 | *6 |
|---|
| Hp-I | T10 | T | T | T | TA6 | G | 0.524 | 0.573 |
| Hp-II |
T9a | Ga | Ca | T | TA6 | Aa | 0.170 | 0.159 |
| Hp-III |
T9a | Ga | T | Ga | TA6 | G | 0.131 | 0.134 |
| Hp-IV |
T9a | Ga | Ca | Ga |
TA7a | G | 0.063 | 0.041 |
| Hp-V | T10 | T | T | Ga |
TA7a | G | 0.044 | 0.028 |
| Hp-VI |
T9a | Ga | Ca | T | TA6 | G | 0.015 | 0.016 |
| Hp-VII |
T9a | Ga | Ca | Ga | TA6 | G | 0.015 | 0.012 |
| Hp-VIII |
T9a | Ga | T | Ga |
TA7a | G | 0.010 | 0.012 |
| Hp-IX | T10 | T | T | Ga | TA6 | G | 0.010 | 0.008 |
| Hp-X | T10 | Ga | Ca | T | TA6 | Aa | 0.010 | 0.008 |
| Hp-XI |
T9a | Ga | T | T | TA6 | G | 0.005 | 0.004 |
| Hp-XII | T10 | T | T | T | TA6 | Aa | 0.005 | 0.004 |
Associations between UGT1A
genotypes/haplotypes and irinotecan toxicity
We examined associations between individual
UGT1A genotypes or haplotypes and severe irinotecan toxicity
among 123 patients with mCRC who receive chemotherapy that included
irinotecan (Table IV). Each of
four UGT1A genotypes [UGT1A1*6, UGT1A7
(387T>G), UGT1A7 (622T>C)
and UGT1A9*1b] showed a significant association to
irinotecan toxicity and linear trend (p<0.05). Similarly, two
haplotypes (Hp-I and Hp-II) each showed a significant
association to and linear trend with irinotecan toxicity
(p<0.05). Among two patients received a starting dose of 100
mg/m2 irinotecan, diplotype of Hp-IV/V did not
show toxicity and diplotype of Hp-V/V showed toxicity. Six
patients excluded from FRUTIRI study did not show toxicity of
irinotecan (a starting dose of 70 mg/m2; UGT1A
diplotypes of Hp-I/V, II/IV and III/IV were
found in 2, 3 and 1 patients). Regarding non-hematological
toxicities, only 5 patients developed grade 3 diarrhea
(UGT1A diplotype of these 5 patients consists of 4
Hp-I/II and 1 Hp-II/XII).
 | Table IVAssociations between UGT1A
genotypes/haplotypes and irinotecan toxicity. |
Table IV
Associations between UGT1A
genotypes/haplotypes and irinotecan toxicity.
| | Toxicity | p-value |
|---|
| |
|
|
|---|
| | Yes | No | (% of yes) | Fisher’s exact | CA trend |
|---|
| Genotypes |
|
UGT1A1*6 | −/− | 27 | 57 | (32.1) | 0.002 | 0.001 |
| −/*6 | 21 | 15 | (58.3) | | |
| *6/*6 | 3 | 0 | (100.0) | | |
|
UGT1A1*28 | −/− | 40 | 63 | (38.8) | 0.218 | - |
| −/1*28 | 11 | 9 | (55.0) | | |
|
1*28/1*28 | - | - | - | | |
|
UGT1A1*60 | −/− | 27 | 44 | (38.0) | 0.349 | 0.219 |
| −/1*60 | 20 | 26 | (43.5) | | |
|
1*60/1*60 | 4 | 2 | (66.7) | | |
| UGT1A7
(387T>G) | 387T/T | 9 | 32 | (22.0) | 0.005 | 0.002 |
| 387T/G | 34 | 35 | (49.3) | | |
| 387G/G | 8 | 5 | (61.5) | | |
| UGT1A7
(622T>C) | 622T/T | 18 | 52 | (25.7) | <0.001 | <0.001 |
| 622T/C | 31 | 17 | (64.6) | | |
| 622C/C | 2 | 3 | (40.0) | | |
|
UGT1A9*1b |
9*1b/9*1b | 9 | 34 | (20.9) | 0.003 | 0.001 |
| −/9*1b | 34 | 33 | (50.7) | | |
| −/− | 8 | 5 | (61.5) | | |
| Haplotypes |
| Hp-I | 0a | 12 | 6 | (66.7) | 0.002 | <0.001 |
| 1a | 32 | 37 | (46.4) | | |
| 2a | 7 | 29 | (19.4) | | |
| Hp-II | 0a | 27 | 59 | (31.4) | 0.001 | <0.001 |
| 1a | 22 | 13 | (62.9) | | |
| 2a | 2 | 0 | (100.0) | | |
| Hp-III | 0a | 38 | 53 | (41.8) | 0.517 | 0.900 |
| 1a | 12 | 19 | (38.7) | | |
| 2a | 1 | 0 | (100.0) | | |
| Clinical
features |
| Gender | Male | 31 | 47 | (39.7) | 0.705 | - |
| Female | 20 | 25 | (44.4) | | |
| Age | ≤60 | 15 | 35 | (30.0) | 0.027 | - |
| >60 | 36 | 37 | (49.3) | | |
Performances of the toxicity prediction
system by genotype combination
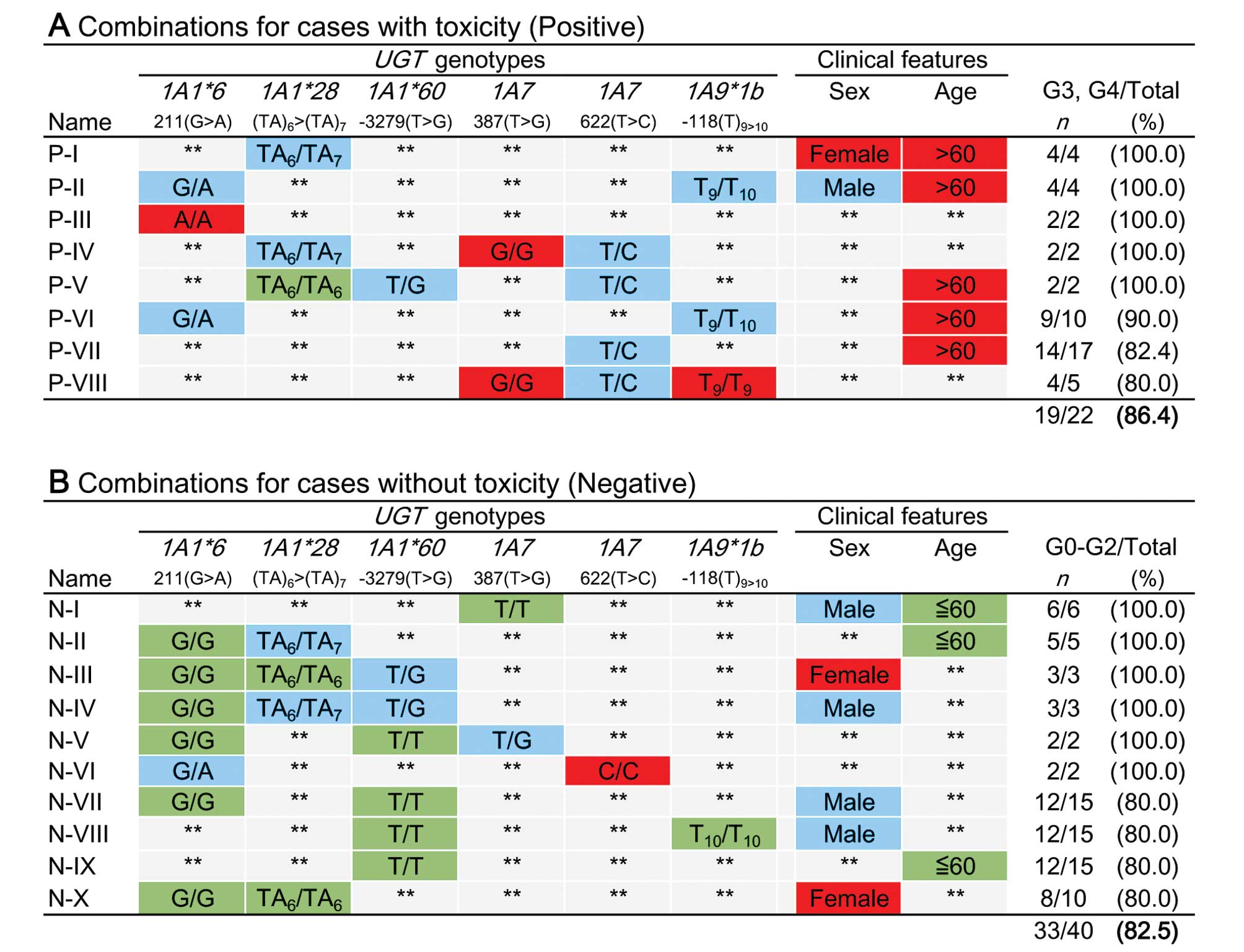
To construct a system for predicting the risk of
severe irinotecan toxicity, genetic data from 73 patients that
constituted the training population were analyzed exhaustively;
specifically, SFFS was used to assess gender, age and the
individual genotypes at six polymorphic UGT1A sites
(Fig. 2). In addition to the three
possible genotypes (wild-type homozygous, heterozygous, variant
homozygous), a fourth option for each site (designated ‘unspecified
genotype’) was included into the algorithm. Similarly, patient
gender (male, female, regardless of gender) and age (≤60, >60
years old, regardless of age) were assessed. The cutoff value for
age (60 years) was determined by Youden index obtained by the
receiver operating characteristic (ROC) curve analysis with the
training population. Among possible combinations (46 ×
32 − 1 = 36,863), the following cases were excluded:
cases not found, single cases, and cases that represented positive
or negative predictive values <80%. In order to optimize the
combinations, categorization according to predictive value and
exclusion of redundant combinations in each category were
performed. As a result, 8 combinations (P-I to P-VIII, Fig. 1A) appeared to predict an increased
risk of toxicity, and 10 combinations (N-I to N-X, Fig. 1B) appeared to predict a lack of
toxicity.
The system for predicting irinotecan toxicity based
on combinations of 8 factors (6 genotypes, gender and age) was
generated using data from of all 73 patients in the training
population. The system was then applied to data from 84.9 and 86.0%
of the patients in the training and validation populations,
respectively (Table V). This
prediction system showed 83.9% accuracy (positive predictive value,
86.4%; negative predictive value, 82.5%) for the training
population (n=62) and 72.1% accuracy (positive predictive value,
70.0%; negative predictive value, 72.7%) for the validation
population (n=43). When patients who were not applied to the
combinations were included, the performance of the system was 71.2%
accuracy (sensitivity, 55.9%; specificity, 84.6%) in training
population (n=73) and 62.0% accuracy (sensitivity, 41.2%;
specificity, 72.7%) in validation population (n=50). Odds ratios of
positive prediction for irinotecan toxicity for this prediction
system were 8.0 (95% CI, 1.5–42.5) and 16.3 (95% CI, 2.2–121.4) in
training and validation populations, respectively (p<0.05,
Table VI).
 | Table VPredicitive performance for
irinotecan toxicity by the genotype combinations. |
Table V
Predicitive performance for
irinotecan toxicity by the genotype combinations.
| Training
(n=73) | Validation
(n=50) |
|---|
|
|
|
|---|
| n | (%) | n | (%) |
|---|
| Matched with the
combinationa | 62/73 | (84.9) | 43/50 | (86.0) |
| Accuracy in
applied patients | 52/62 | (83.9) | 31/43 | (72.1) |
| Positive
predictive valueb | 19/22 | (86.4) | 7/10 | (70.0) |
| Negative
predictive valueb | 33/40 | (82.5) | 24/33 | (72.7) |
| Accuracy | 52/73 | (71.2) | 31/50 | (62.0) |
| Sensitivity | 19/34 | (55.9) | 7/17 | (41.2) |
| Specificity | 33/39 | (84.6) | 24/33 | (72.7) |
 | Table VIAssociations between UGT1A
genotypes/haplotypes and irinotecan toxicity in training and
validation sub-populations. |
Table VI
Associations between UGT1A
genotypes/haplotypes and irinotecan toxicity in training and
validation sub-populations.
| Training
(n=73) | Validation
(n=50) |
|---|
| Toxicity | Fisher’s exact
test | Toxicity | Fisher’s exact
test |
|---|
|
|
|
|
|
|---|
| Yes | No | (% of yes) | OR | (95% CI) | p-value | Yes | No | (% of yes) | OR | (95% CI) | p-value |
|---|
| Haplotypes |
| Hp-I
(+/+) | 5 | 15 | (25.0) | 1c | | | 2 | 14 | (12.5) | 1c | | |
| Hp-I (−/−,
−/+) | 29 | 24 | (54.7) | 3.63 | (1.15–11.42) | 0.035 | 15 | 19 | (44.1) | 5.53 | (1.08–28.18) | 0.053 |
| Hp-II
(+/+,−/+) | 16 | 8 | (66.7) | 0.64 | (1.60–22.48) | 0.008 | 8 | 5 | (61.5) | 11.20 | (1.75–71.64) | 0.016 |
| The predicition
systema |
| Negative for
toxicity | 7 | 33 | (17.5) | 0.64 | (0.17–2.34) | 0.511 | 9 | 24 | (27.3) | 2.63 | (0.50–13.92) | 0.300 |
| Positive for
toxicity | 19 | 3 | (86.4) | 8.00 | (1.51–42.45) | 0.021 | 7 | 3 | (70.0) | 16.33 | (2.20–121.43) | 0.009 |
| Not
matchedb | 8 | 3 | (72.7) | | | | 1 | 6 | (14.3) | | | |
| Genotypes |
| UGT1A1*6
(+/+,−/+) | 16 | 9 | (64.0) | 5.33 | (1.45–19.58) | 0.016 | 8 | 6 | (57.1) | 9.33 | (1.51–57.65) | 0.019 |
| UGT1A1*28
(−/+) | 7 | 7 | (50.0) | 3.00 | (0.70–12.88) | 0.163 | 4 | 2 | (66.7) | 14.00 | (1.47–133.23) | 0.025 |
| UGT1A1*60
(+/+,−/+) | 18 | 15 | (54.5) | 3.60 | (1.06–12.22) | 0.048 | 6 | 13 | (31.6) | 3.23 | (0.55–18.96) | 0.244 |
| UGT1A7
(387G/G, T/G) | 27 | 21 | (56.3) | 3.86 | (1.21–12.33) | 0.032 | 15 | 19 | (44.1) | 5.53 | (1.08–28.18) | 0.053 |
| UGT1A7
(622C/C, T/C) | 20 | 13 | (60.6) | 4.62 | (1.35–15.78) | 0.022 | 13 | 7 | (65.0) | 13.00 | (2.27–74.32) | 0.002 |
| UGT1A9*1b
(−/−, −/+) | 27 | 20 | (57.4) | 4.05 | (1.26–12.99) | 0.018 | 15 | 18 | (45.5) | 5.83 | (1.14–29.84) | 0.028 |
Patients with either of three UGT1A alleles
[UGT1A1*6, UGT1A7 (622T>C) or
UGT1A9*1b], UGT1A haplotype-I or haplotype-II
showed significant association to severe irinotecan toxicity
(p<0.05) in both the training and validation populations (data
not shown).
Discussion
The novel system for predicting severe irinotecan
toxicity described here was based on genotypes at 6 polymorphic
sites in UGT1A and 2 basic clinical features; notably, it
showed high predictive performance even though the treatment
regimens differed among the training and validation patients
(Tables V and VI). The odds ratio of positive
prediction for severe irinotecan toxicity was higher for this
prediction system than for that of any other haplotype or for that
of any genotype (Table VI). The
performance of this prediction system was reduced from the 83.9%
accuracy seen with applied patients to this system in the training
population to 72.1% accuracy in the validation. With regard to
positive prediction, the inconsistency in accuracy between training
and validation populations was seen when the combinations included
the UGT1A9*1b site and patient age (P-II, VI and VII in
Fig. 2). The frequencies of
UGT1A9*1b genotype differed between the training and
validation populations; moreover, the UGT1A9*1b alleles were
not in Hardy-Weinberg equilibrium in the validation population
(data not shown). The cutoff value for patient age (60 years old)
was determined by a ROC curve generated with data from the training
population; however, previous studies used a cutoff age of 65 years
(20,21). Indeed, one patient without
toxicity, but predicted as presence of toxicity in this system, was
aged 63 years.
Some genotypic combinations decreased the
performance of negative prediction for sever irinotecan toxicity in
the validation population relative to the training population
(N-II, IV, and V in Fig. 2).
Specifically, 36.4% (n=4/11) of patients in training population
with a combined genotype that included heterozygous for
UGT1A1*28 alleles and UGT1A1*6 (−/−)
experienced severe irinotecan toxicity, but 66.7% (n=4/6) of the
patients in validation population with the same genotype
combinations (UGT1A1*6, −/− and UGT1A1*28,
−/+) showed severe toxicity. Of the 73 patients in the training
population and the 50 in the validation population, 11 (15.1%) and
7 (14.0%), respectively, were matched with neither of the
combination in our prediction system. Interestingly, the incidence
of severe toxicity among patients who were not matched with either
combination identified by this prediction system was 72.7%
(training population) and 14.3% (validation population) (Table VI). Therefore, the frequency of
the irinotecan toxicity among patients who do not have any
combination of UGT1A variants identified by this novel
prediction system might be due to factors other than UGT1A
polymorphisms.
Many published studies have focused on associations
between irinotecan toxicity, irinotecan efficacy, or both and any
one or more of each UGT1A variants examined here (10–19,31,32).
Patients, especially Asian patients, homozygous for UGT1A1*6
or *28 or compound heterozygous for these variants are at
high risk for hematologic toxicity (13,33,34).
In this study, each patient homozygous for UGT1A1*6 (n=3)
and those compound heterozygous for UGT1A1*6 and *28
(n=3) showed severe hematologic toxicity; however, 45 patients of
the remaining 117 patients still exhibited severe irinotecan
toxicity. UGT1A1*6 and *28 each have strong effects
on UGT1A1 activity and expression, but frequency of each allele is
low; moreover, the frequencies of each allele differ between races
(11,14,35–37).
Among the patients that lacked these rare, highly effective
variants, this novel prediction system could accurately predict
whether there is severe irinotecan toxicity.
Here, as in previous studies, each identified
UGT1A haplotypes was useful for precisely predicting the
presence or absence of severe irinotecan toxicity (14,18,38–40).
Consistent with our study, Cecchin et al reported that a
haplotype comprising UGT1A1*28 (−), UGT1A1*60 (−),
UGT1A7 (387T and 622T), and UGT1A9*1b
(+) was a predictor of severe hematologic toxicity during the
entire course of therapy (18).
However, determining the haplotypes for any one patient is a
difficult clinical measurement. Therefore, the genotypes at each of
the 6 sites (rather than the haplotype or diplotype) could be used
for clinical assessments.
Our prediction system depend not only on
UGT1A genotypes but also on patient gender and age. Previous
studies showed that patient gender and age were related to the risk
of irinotecan toxicity (20–22).
In the training population, patient age was associated with severe
irinotecan toxicity, but patient gender was not (Table IV). Interestingly, when patient
age, patient gender or both the patient age and gender were
excluded from the factors used by the prediction system, the number
of patients that matched with the prediction system decreased,
although the system maintained the high positive and negative
predictive values (data not shown).
The SFFS algorithm could be modified to include
other factors (e.g., mutations in the tumor, patients’ clinical
characteristics, additional genetic variants, etc.) to improve the
prediction performance. Such modifications may result in a system
that could meaningfully predict clinical outcomes, including tumor
response. Recent advances in technology for sequencing whole
genomes of individuals may lead to substantial increases in
information that might be useful for personalized therapy. However,
such complicated information could not be efficiently or fully
utilized in the currently available formats. SFFS could easily
construct a system that can utilize huge data sets such as
whole-genome sequences. Our strategy for developing SFFS-based
systems for clinical use could serve as a powerful tool for
advancing personalized therapy, although additional prospective
study of this prediction system is needed.
Acknowledgements
This study was supported in part by a non-profit
organization Epidemiological and Clinical Research Information
Network (ECRIN) and by a Grant-in-Aid for Scientific Research from
the Ministry of Education, Culture, Sports, Science and Technology
of Japan (JSPS KAKENHI grant nos. 21591725 and 19591545). We thank
Ms. Mai Hatta for clinical research coordination.
References
|
1
|
Douillard JY, Cunningham D, Roth AD,
Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J,
Alakl M, Gruia G, Awad L and Rougier P: Irinotecan combined with
fluorouracil compared with fluorouracil alone as first-line
treatment for metastatic colorectal cancer: a multicentre
randomised trial. Lancet. 355:1041–1047. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saltz LB, Cox JV, Blanke C, Rosen LS,
Fehrenbacher L, Moore MJ, Maroun JA, Ackland SP, Locker PK, Pirotta
N, Elfring GL and Miller LL: Irinotecan plus fluorouracil and
leucovorin for metastatic colorectal cancer. Irinotecan Study
Group. N Engl J Med. 343:905–914. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tournigand C, André T, Achille E, Lledo G,
Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G,
Landi B, Colin P, Louvet C and de Gramont A: FOLFIRI followed by
FOLFOX6 or the reverse sequence in advanced colorectal cancer: a
randomized GERCOR study. J Clin Oncol. 22:229–237. 2004. View Article : Google Scholar
|
|
4
|
Hazama S, Nagashima A, Kondo H, Yoshida S,
Shimizu R, Araki A, Yoshino S, Okayama N, Hinoda Y and Oka M: Phase
I study of irinotecan and doxifluridine for metastatic colorectal
cancer focusing on the UGT1A1*28 polymorphism. Cancer Sci.
101:722–727. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hazama S, Mishima H, Tsunedomi R, Okuyama
Y, Kato T, Takahashi K, Nozawa H, Ando H, Kobayashi M, Takemoto H,
Nagata N, Kanekiyo S, Inoue Y, Hamamoto Y, Fujita Y, Hinoda Y,
Okayama N, Oba K, Sakamoto J and Oka M: UGT1A1*6, 1A7*3, and 1A9*22
genotypes predict severe neutropenia in FOLFIRI-treated mCRC in two
prospective studies in Japan. Cancer Sci. 104:1662–1669. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanekiyo S, Hazama S, Kondo H, Nagashima
A, Eto R, Yoshida S, Shimizu R, Araki A, Yamamoto T, Uchiyama T,
Yoshino S, Okayama N, Hinoda Y and Oka M:
UDP-glucuronosyltransferase (UGT) 1A1*28 polymorphism-directed
phase II study of irinotecan with 5′-deoxy-5-fluorouridine
(5′-DFUR) for metastatic colorectal cancer. Anticancer Res.
33:3423–3430. 2013.PubMed/NCBI
|
|
7
|
Kawato Y, Aonuma M, Hirota Y, Kuga H and
Sato K: Intracellular roles of SN-38, a metabolite of the
camptothecin derivative CPT-11, in the antitumor effect of CPT-11.
Cancer Res. 51:4187–4191. 1991.PubMed/NCBI
|
|
8
|
Rivory LP, Bowles MR, Robert J and Pond
SM: Conversion of irinotecan (CPT-11) to its active metabolite,
7-ethyl-10-hydroxycamptothecin (SN-38), by human liver
carboxylesterase. Biochem Pharmacol. 52:1103–1111. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mathijssen RH, van Alphen RJ, Verweij J,
Loos WJ, Nooter K, Stoter G and Sparreboom A: Clinical
pharmacokinetics and metabolism of irinotecan (CPT-11). Clin Cancer
Res. 7:2182–2194. 2001.PubMed/NCBI
|
|
10
|
Iyer L, Das S, Janisch L, Wen M, Ramírez
J, Karrison T, Fleming GF, Vokes EE, Schilsky RL and Ratain MJ:
UGT1A1*28 polymorphism as a determinant of irinotecan disposition
and toxicity. Pharmacogenomics J. 2:43–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Toffoli G, Cecchin E, Corona G, Russo A,
Buonadonna A, D’Andrea M, Pasetto LM, Pessa S, Errante D, De
Pangher V, Giusto M, Medici M, Gaion F, Sandri P, Galligioni E,
Bonura S, Boccalon M, Biason P and Frustaci S: The role of
UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics
of irinotecan in patients with metastatic colorectal cancer. J Clin
Oncol. 24:3061–3068. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guillemette C, Ritter JK, Auyeung DJ,
Kessler FK and Housman DE: Structural heterogeneity at the
UDP-glucuronosyltransferase 1 locus: functional consequences of
three novel missense mutations in the human UGT1A7 gene.
Pharmacogenetics. 10:629–644. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ando Y, Saka H, Ando M, Sawa T, Muro K,
Ueoka H, Yokoyama A, Saitoh S, Shimokata K and Hasegawa Y:
Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan
toxicity: a pharmacogenetic analysis. Cancer Res. 60:6921–6926.
2000.PubMed/NCBI
|
|
14
|
Ando M, Ando Y, Sekido Y, Ando M,
Shimokata K and Hasegawa Y: Genetic polymorphisms of the
UDP-glucuronosyltransferase 1A7 gene and irinotecan toxicity in
Japanese cancer patients. Jpn J Cancer Res. 93:591–597. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamanaka H, Nakajima M, Katoh M, Hara Y,
Tachibana O, Yamashita J, McLeod HL and Yokoi T: A novel
polymorphism in the promoter region of human UGT1A9 gene
(UGT1A9*22) and its effects on the transcriptional activity.
Pharmacogenetics. 14:329–332. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sai K, Saeki M, Saito Y, Ozawa S, Katori
N, Jinno H, Hasegawa R, Kaniwa N, Sawada J, Komamura K, Ueno K,
Kamakura S, Kitakaze M, Kitamura Y, Kamatani N, Minami H, Ohtsu A,
Shirao K, Yoshida T and Saijo N: UGT1A1 haplotypes associated with
reduced glucuronidation and increased serum bilirubin in
irinotecan-administered Japanese patients with cancer. Clin
Pharmacol Ther. 75:501–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han JY, Lim HS, Shin ES, Yoo YK, Park YH,
Lee JE, Jang IJ, Lee DH and Lee JS: Comprehensive analysis of UGT1A
polymorphisms predictive for pharmacokinetics and treatment outcome
in patients with non-small-cell lung cancer treated with irinotecan
and cisplatin. J Clin Oncol. 24:2237–2244. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cecchin E, Innocenti F, D’Andrea M, Corona
G, De Mattia E, Biason P, Buonadonna A and Toffoli G: Predictive
role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their
haplotypes on the outcome of metastatic colorectal cancer patients
treated with fluorouracil, leucovorin, and irinotecan. J Clin
Oncol. 27:2457–2465. 2009. View Article : Google Scholar
|
|
19
|
Fujita K, Ando Y, Nagashima F, Yamamoto W,
Eodo H, Araki K, Kodama K, Miya T, Narabayashi M and Sasaki Y:
Genetic linkage of UGT1A7 and UGT1A9 polymorphisms to UGT1A1*6 is
associated with reduced activity for SN-38 in Japanese patients
with cancer. Cancer Chemother Pharmacol. 60:515–522. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roth AD, Yan P, Dietrich D, Fiocca R,
Bodoky G, Labianca R, Cunningham D, Van Cutsem E, Bosman F and
Tejpar S: Is UGT1A1*28 homozygosity the strongest predictor for
severe hematotoxicity in patients treated with 5-fluorouracil
(5-FU)-irinotecan (IRI)? Results of the PETACC 3 - EORTC 40993 -
SAKK 60/00 trial comparing IRI/5-FU/folinic acid (FA) to 5-FU/FA in
stage II-III colon cancer (COC) patients. J Clin Oncol. 26(Suppl):
abs 4036. 2008.
|
|
21
|
Kweekel D, Guchelaar HJ and Gelderblom H:
Clinical and pharmacogenetic factors associated with irinotecan
toxicity. Cancer Treat Rev. 34:656–669. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Innocenti F, Kroetz DL, Schuetz E, Dolan
ME, Ramírez J, Relling M, Chen P, Das S, Rosner GL and Ratain MJ:
Comprehensive pharmacogenetic analysis of irinotecan neutropenia
and pharmacokinetics. J Clin Oncol. 27:2604–2614. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
UGT Alleles Nomenclature Home Page. UGT
Nomenclature Committee. June. 2005, http://www.pharmacogenomics.pha.ulaval.ca/cms/site/pharmacogenomics/ugt_alleles.
Accessed 04.01.2013
|
|
24
|
Pudil P, Novovicova J and Kittler J:
Floating search methods in feature selection. Pattern Recognition
Lett. 15:1119–1125. 1994. View Article : Google Scholar
|
|
25
|
Okuyama Y, Hazama S, Nozawa H, Kobayashi
M, Takahashi K, Fujikawa K, Kato T, Nagata N, Kimura H, Oba K,
Sakamoto J and Mishima H: Prospective phase II study of FOLFIRI for
mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms.
Jpn J Clin Oncol. 41:477–482. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hirata K, Nagata N, Kato T, Okuyama Y,
Andoh H, Takahashi K, Oba K, Sakamoto J, Hazama S and Mishima H:
Prospective phase II trial of second-line FOLFIRI in patients with
advanced colorectal cancer including analysis of UGT1A1
polymorphisms: FLIGHT 2 study. Anticancer Res. 34:195–201.
2014.PubMed/NCBI
|
|
27
|
Wang L, Hirayasu K, Ishizawa M and
Kobayashi Y: Purification of genomic DNA from human whole blood by
isopropanol-fractionation with concentrated NaI and SDS. Nucleic
Acids Res. 22:1774–1775. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okayama N, Hamanaka Y, Suehiro Y, Hasui Y,
Nakamura J and Hinoda Y: Association of interleukin-10 promoter
single nucleotide polymorphisms-819 T/C and -592 A/C with aging. J
Gerontol A Biol Sci Med Sci. 60:1525–1529. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
The R project website. http://www.r-project.org/.
|
|
31
|
Smith NF, Figg WD and Sparreboom A:
Pharmacogenetics of irinotecan metabolism and transport: an update.
Toxicol In Vitro. 20:163–175. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lamas MJ, Duran G, Balboa E, Bernardez B,
Candamio S, Vidal Y, Mosquera A, Giraldez JM, Lopez R, Carracedo A
and Barros F: The value of genetic polymorphisms to predict
toxicity in metastatic colorectal patients with irinotecan-based
regimens. Cancer Chemother Pharmacol. 69:1591–1599. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Araki K, Fujita K, Ando Y, Nagashima F,
Yamamoto W, Endo H, Miya T, Kodama K, Narabayashi M and Sasaki Y:
Pharmacogenetic impact of polymorphisms in the coding region of the
UGT1A1 gene on SN-38 glucuronidation in Japanese patients with
cancer. Cancer Sci. 97:1255–1259. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Satoh T, Ura T, Yamada Y, Yamazaki K,
Tsujinaka T, Munakata M, Nishina T, Okamura S, Esaki T, Sasaki Y,
Koizumi W, Kakeji Y, Ishizuka N, Hyodo I and Sakata Y:
Genotype-directed, dose-finding study of irinotecan in cancer
patients with UGT1A1*28 and/or UGT1A1*6 polymorphisms. Cancer Sci.
102:1868–1873. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Innocenti F, Undevia SD, Iyer L, Chen PX,
Das S, Kocherginsky M, Karrison T, Janisch L, Ramírez J, Rudin CM,
Vokes EE and Ratain MJ: Genetic variants in the
UDP-glucuronosyltransferase 1A1 gene predict the risk of severe
neutropenia of irinotecan. J Clin Oncol. 22:1382–1388. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Innocenti F, Liu W, Chen P, Desai AA, Das
S and Ratain MJ: Haplotypes of variants in the
UDP-glucuronosyltransferase1A9 and 1A1 genes. Pharmacogenet
Genomics. 15:295–301. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Massacesi C, Terrazzino S, Marcucci F,
Rocchi MB, Lippe P, Bisonni R, Lombardo M, Pilone A, Mattioli R and
Leon A: Uridine diphosphate glucuronosyl transferase 1A1 promoter
polymorphism predicts the risk of gastrointestinal toxicity and
fatigue induced by irinotecan-based chemotherapy. Cancer.
106:1007–1016. 2006. View Article : Google Scholar
|
|
38
|
Minami H, Sai K, Saeki M, Saito Y, Ozawa
S, Suzuki K, Kaniwa N, Sawada J, Hamaguchi T, Yamamoto N, Shirao K,
Yamada Y, Ohmatsu H, Kubota K, Yoshida T, Ohtsu A and Saijo N:
Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic
polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet
Genomics. 17:497–504. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Saito Y, Sai K, Maekawa K, Kaniwa N,
Shirao K, Hamaguchi T, Yamamoto N, Kunitoh H, Ohe Y, Yamada Y,
Tamura T, Yoshida T, Minami H, Ohtsu A, Matsumura Y, Saijo N and
Sawada J: Close association of UGT1A9 IVS1+399C>T with
UGT1A1*28, *6, or *60 haplotype and its apparent influence on
7-ethyl-10-hydroxycamptothecin (SN-38) glucuronidation in Japanese.
Drug Metab Dispos. 37:272–276. 2009.PubMed/NCBI
|
|
40
|
Martinez-Balibrea E, Abad A,
Martínez-Cardús A, Ginés A, Valladares M, Navarro M, Aranda E,
Marcuello E, Benavides M, Massutí B, Carrato A, Layos L, Manzano JL
and Moreno V: UGT1A and TYMS genetic variants predict toxicity and
response of colorectal cancer patients treated with first-line
irinotecan and fluorouracil combination therapy. Br J Cancer.
103:581–589. 2010. View Article : Google Scholar
|