Introduction
In recent years, the mortality rate of pancreatic
cancer has leapt to the fifth-highest position among all malignant
tumors in developed countries, and the incidence of pancreatic
cancer is still rising (1).
Pancreatic ductal adenocarcinoma (PDA) that originates in glandular
epithelium accounts for more than 90% of all malignant pancreatic
tumors and exhibits a high degree of malignancy with a poor
prognosis (2). To date, reliable
biomarkers for the early diagnosis and effective treatment of PDA
are lacking. Approximately 60% of PDA patients have advanced-stage
disease at the time of diagnosis, and most patients die within a
year after diagnosis (2). Although
studies have identified the abnormal expression of certain
oncogenes and tumor suppressor genes in PDA, such as the Kirsten
rat sarcoma viral oncogene homolog (KRAS), epidermal growth factor
receptor (EGFR), tumor protein 53 (TP53), Sma and Mad related
family member 4/deleted in pancreatic cancer locus 4 (SMAD4/DPC4)
and P16/cyclin-dependent kinase inhibitor 2A (P16/CDKN2A) (3), the mechanisms of PDA genesis and
development remain unclear. A more profound understanding of the
molecular mechanisms that underlie the biological behaviour of PDA
is crucial to improve the prognosis of PDA patients.
As a newly discovered member of the ubiquitin
hydrolase family, USP22 possesses deubiquitinating activity
(4). USP22 is a key subunit of the
human Spt-Ada-Gcn5-acetyltransferase (hSAGA) transcriptional
coactivator complex and can interact within the hSAGA complex to
hydrolyses the ubiquitin conjugated to histones H2A and H2B, thus
activating target gene transcription as mediated by alterations in
levels of histone ubiquitylation (5). USP22 plays a key role in cell cycle
regulation, embryo development and telomere homeostasis (5–7).
Normal tissues express low levels of USP22, whereas UPS22
expression is significantly elevated in tumor tissues (4). Studies have shown that interfering
with USP22 expression in tumor cells can arrest cell cycle
progression in the G1 phase and inhibit tumor cell proliferation,
leading to tumor cell apoptosis (5,8–10).
Additionally, USP22 is closely related to metastatic potential,
therapy resistance and solid tumor prognosis (11). Furthermore, USP22 is one of the
genes comprising the 11-gene Polycomb/cancer stem cell signature
(12). To date, the mechanism of
the abnormal expression and regulation of USP22 remains unclear,
and the relationship between UPS22 and pancreatic cancer has not
been reported.
FoxM1 is a key member of the forkhead box family of
transcription factors (13), which
is characterised by a DNA-binding domain with a wing-shaped spiral
structure (winged helix domain) (14). FoxM1 plays a critical role in
regulating cell cycle and mitosis, and it is a key regulator of the
G1/S and G2/M cell cycle transitions (15). FoxM1 regulates the expression of
cell division cycle 25 homolog A (cdc25A) at the G1/S cell cycle
checkpoint and controls the transcription of S-phase
kinase-associated protein 2 (skp2) and cyclin-dependent kinase
subunit 1 (cks1) (16,17). Additionally, FoxM1 regulates the
expression of G2/M checkpoint genes such as cdc25B, cyclin B,
aurora-B, survivin, polo-like kinase 1 (PLK1) and the centromere
proteins A/B/F (CENP A/B/F) (14,18,19).
FoxM1 is widely expressed in proliferating cells and is
significantly upregulated in a variety of malignant tumors,
including gastric cancer, non-small cell lung cancer, brain glioma,
prostate cancer, cervical cancer and pancreatic cancer (20). Recent studies have found that FoxM1
is closely related to the occurrence and development of pancreatic
cancer (21). Interference in
FoxM1 expression inhibits the proliferation, invasion and
metastasis of pancreatic cancer cells through the downregulation of
cyclin B, cyclin D1, cyclin-dependent kinase 2 (Cdk2), matrix
metalloproteinase-2 (MMP-2), MMP-9 and vascular endothelial growth
factor (VEGF) (22).
Overexpression of FoxM1 activates mesenchymal cell markers and
promotes the epithelial-mesenchymal transition (EMT) (23). Additionally, FoxM1 is associated
with poor prognosis and therapy resistance in a variety of solid
tumors, including pancreatic cancer (24–26).
The classical Wnt/β-catenin signalling pathway is
among the most widely studied signal transduction pathways in
recent years. Upon abnormal Wnt pathway activation, Wnt ligands
bind to cell surface frizzled/low density lipoprotein
receptor-related protein (Fz/LRP) receptors, and β-catenin is then
released from the degradation complex comprising Axin/denomatous
polyposis coli (Axin/APC) and glycogen synthase kinase-3β (GSK-3β),
resulting in its cellular accumulation and nuclear translocation.
In the nucleus, β-catenin interacts with the T cell factor/lymphoid
enhancer factor (TCF/LEF) and regulates the expression of the
downstream target genes c-Myc and cyclin D1, thus controlling cell
cycle progression (27). Abnormal
activation of the Wnt/β-catenin signalling pathway plays an
essential role in PDA tumorigenesis and metastasis (28,29).
Pasca et al observed varying degrees of Wnt/β-catenin
signalling pathway activation in 26 human pancreatic cancer cell
lines and 3 different animal models of pancreatic cancer. Blockade
of the Wnt/β-catenin signalling pathway inhibits cell proliferation
and promotes cell apoptosis (30).
Zhang et al found that FoxM1 promoted the transfer of
β-catenin to the nucleus and activated the Wnt pathway in glioma,
thereby activating the expression of downstream target genes such
as cyclin D1 and c-Myc and participating in tumorigenesis (31).
Although USP22 has been confirmed to be highly
expressed in a variety of tumors and to be related to cell cycle
regulation, the mechanism that underlies the role of USP22 has not
been clearly reported. As a typical cell proliferation-associated
transcription factor, the role of FoxM1 in cell cycle regulation
has been acknowledged in recent years. However, whether FoxM1 is
involved in USP22-mediated cell cycle regulation, as well as the
relationship between USP22 and FoxM1, remains to be elucidated.
In the present study, we performed a systematic
immunohistochemical analysis of USP22 and FoxM1 expression in PDA.
Associations between USP22/FoxM1 expression, clinicopathological
features and clinical outcome were investigated, respectively. As a
correlation was found between UPS22 and FoxM1 expression in PDA
tissues, the functional studies were carried out in PDA cell lines.
We provide the first evidence that USP22 promoted the G1/S phase
cell cycle transition and accelerated cell proliferation via
upregulated FoxM1 expression. The β-catenin nuclear localization is
required for USP22-mediated FoxM1 expression.
Materials and methods
Cell culture
The poorly differentiated human PDA cell line PANC-1
and the highly differentiated human PDA cell line CFPAC-1 were
purchased from the cell bank of the Committee on Type Culture
Collection of the Chinese Academy of Sciences (Shanghai, China)
(32). PANC-1 cells were cultured
in a 1:1 mixture of Dulbecco’s modified Eagle’s medium (DMEM) and
F12 medium supplemented with 10% fetal bovine serum (FBS;
Invitrogen, Tokyo, Japan). CFPAC-1 cells were maintained in
Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10%
FBS (Invitrogen). All cells were cultured in cell-culture flasks or
Petri dishes in a humidified incubator at 37°C in an atmosphere of
5% CO2.
Patients and tissue specimens
Paraffin-embedded specimens were obtained from 136
patients with stage II PDA who underwent surgical resection from
January, 2002 to January, 2013 at the Department of General Surgery
at the First Affiliated Hospital of Dalian Medical University.
Patients with stage I, III and stage IV disease were excluded from
this study because the case number was too small to be
representative for these disease stages. One hundred and nine
patients (80.1%) underwent pancreaticoduodenectomy, 25 patients
(18.4%) underwent distal pancreatectomy, and 2 patients (1.5%)
received total pancreatectomy. Information on patient demographics
(gender and age) and clinicopathologic features (tumor location,
tumor size, resection margin, histological differentiation, lymph
node metastasis and TNM stage) were obtained from clinical and
pathological records (Table I).
Histological PDA grading was based upon the World Health
Organization grading system.
 | Table ICorrelation between USP22 and FoxM1
protein expression and clinicopathologic features in patients with
stage II pancreatic ductal adenocarcinoma. |
Table I
Correlation between USP22 and FoxM1
protein expression and clinicopathologic features in patients with
stage II pancreatic ductal adenocarcinoma.
| | USP22 | FoxM1 | USP22+FoxM1 |
|---|
| |
|
|
|
|---|
| Variables | N | Positive | Negative | χ2 | P-value | Positive | Negative | χ2 | P-value | Positive | Negative | χ2 | P-value |
|---|
| Gender |
| Male | 74 | 39 | 35 | 0.191 | 0.662 | 51 | 23 | 2.255 | 0.133 | 30 | 44 | 0.168 | 0.682 |
| Female | 62 | 35 | 27 | | | 35 | 27 | | | 23 | 39 | | |
| Age (years) |
| <60 | 51 | 26 | 25 | 0.387 | 0.534 | 29 | 22 | 1.425 | 0.233 | 16 | 35 | 1.981 | 0.159 |
| ≥60 | 85 | 48 | 37 | | | 57 | 28 | | | 37 | 48 | | |
| Tumor location |
| Head | 67 | 39 | 28 | 0.768 | 0.381 | 39 | 28 | 1.135 | 0.231 | 27 | 40 | 0.098 | 0.754 |
| Body/tail | 69 | 35 | 34 | | | 47 | 22 | | | 26 | 43 | | |
| Tumor size |
| ≤2 cm | 61 | 26 | 35 | 6.197 | 0.013 | 32 | 29 | 5.525 | 0.019 | 17 | 44 | 5.732 | 0.017 |
| >2 cm | 75 | 48 | 27 | | | 54 | 21 | | | 36 | 39 | | |
| Resection
margin |
| Negative | 25 | 14 | 11 | 0.031 | 0.860 | 18 | 7 | 1.012 | 0.314 | 10 | 15 | 0.014 | 0.907 |
| Positive | 111 | 60 | 51 | | | 68 | 43 | | | 43 | 68 | | |
| Histological
differentiation |
|
Well/moderately | 79 | 38 | 41 | 3.026 | 0.082 | 45 | 34 | 3.191 | 0.074 | 28 | 51 | 0.986 | 0.321 |
| Poorly | 57 | 36 | 21 | | | 41 | 16 | | | 25 | 32 | | |
| Lymph node
metastasis |
| Negative | 49 | 20 | 28 | 4.858 | 0.028 | 24 | 24 | 5.590 | 0.018 | 11 | 37 | 8.039 | 0.005 |
| Positive | 87 | 54 | 34 | | | 62 | 26 | | | 42 | 46 | | |
| Post operative
treatment |
| No | 31 | 13 | 18 | 2.520 | 0.112 | 19 | 12 | 0.065 | 0.798 | 11 | 20 | 0.205 | 0.651 |
| Yes | 105 | 61 | 44 | | | 67 | 38 | | | 42 | 63 | | |
TNM stages were classified according to the criteria
proposed by UICC/AJCC (2002). There were 74 male and 62 female
patients with age ranging from 34 to 80 years (median age, 67
years). No neo-adjuvant radio- or chemotherapy was applied before
surgical resection to any patient. Post-operatively, 82 patients
(60.3%) received adjuvant chemotherapy alone, 23 patients (16.9%)
received combined chemoradiation therapy and 31 patients (22.8%)
did not receive adjuvant therapy. All patients were followed up
postoperatively until May, 2013 or death. Overall survival (OS) was
defined as the interval between the dates of surgery and death.
Ethical approval for the project was obtained from the First
Affiliated Hospital of Dalian Medical University Research Ethics
Committee. All fresh samples were confirmed by hematoxylin and
eosin staining in frozen sections with histopathologic analysis; 5
pairs of PDA tissues and the paired normal carcinoma-adjacent
tissues were dissected and separated to 2 parts for mRNA and
protein studies.
Immunohistochemistry
USP22 and FoxM1 expression in the paraffin-embedded
specimens was examined according to standard immunohistochemical
methods (33). Anti-USP22 (ab4812;
1:200 dilution; Abcam, Cambridge, UK) and anti-FoxM1 (SC-502; 1:50;
Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibodies were used
as the primary antibodies. The percentage of positive cells was
determined by counting 500 cells in five random areas per section.
Nuclear immunostaining were evaluated using a semiquantitative
assessment, which calculated the staining intensity and the
percentage of positive cells. IHC staining was scored according to
the following criteria: 0, none of the cells stained; 1+, 1–20% of
the cells stained; 2+, 20–50% of the cells stained; 3+, 50–100% of
the cells stained. Furthermore, the expression levels of USP22 and
FoxM1 were divided into the following two groups according to
score: negative (0, 1+, 2+) and positive (3+).
Plasmid preparation
The USP22 and β-catenin overexpression plasmids were
designed and synthesised by Shanghai GenePharma Co., Ltd (Shanghai,
China). Cells were cultured in Petri dishes to 80–90% confluence
and then transfected with Lipofectamine 2000 (Invitrogen) in strict
accordance with the manufacturer’s instructions. After
transfection, the cells were cultured for 48 or 72 h before use in
subsequent experiments.
siRNA analyses
Small interfering RNA (siRNA) for FoxM1
(5′-GCCGGAACAUGACCAUCAATT-3′), USP22 (5′-GCAG
CTCACTATGAAGAAACT-3′), β-catenin (5′-AACAGTCTT ACCTGGACTCTG-3′) and
a negative control (NC) sequence (5′-GTTCTCCGAACGTGTCACGT-3′) were
purchased from Genepharma (Shanghai, China). According to the
manufacturer’s protocol, cells were transfected using Lipofectamine
2000 (Invitrogen). After transfection, the cells were cultured for
48 or 72 h before use in subsequent experiments.
CCK-8 assays
Cell viability in the treated cells was determined
by using Cell Counting Kit-8 (CCK-8) kit (Dojindo Laboratories,
Kumamoto, Japan) following the instructions outlined by the
manufacturer. Cells were plated at a density of 3–5×103
cells/well with 200 μl of medium in 96-well microtiter plates.
After treatment, CCK-8 solution (10 μl) was added to each well and
the plates were incubated at 37°C for 90 min. The absorbance of the
cell suspension was measured with a microplate reader at a
wavelength of 450 nm.
Flow cytometry analysis
Cells pellets were washed with phosphate buffered
saline and fixed/permeabilized with 50% ice-cold ethanol. Pellets
were washed and resuspended in 50 μg/ml ribonuclease A and 62.5
μg/ml propidium iodide. Samples were analysed on the
Becton-Dickinson FACSCalibur (Becton-Dickinson, Franklin Lakes, NJ,
USA). The percentages of cells in various phases of the cell cycle
were quantified using the ModFit LT Version 3.0 program (Verity
Software House, Topsham, ME, USA). The error bars were derived from
the SD of multiple experiments.
RT-PCR
Total RNA was extracted with the TRIzol reagent
according to the manufacturer’s protocol and then it was reverse
transcribed into cDNA using RNA PCR kit (AMV) version 3.0. cDNA was
amplified by PCR using the specific primer for USP22, FoxM1 or
β-actin as an internal control. The sequences of the upstream and
downstream primers were as follows: USP22 forward,
5′-CATGACCCCTTTCATGGCCT-3′ and reverse, 5′-GATGTTCTGGTGACGGGTGT-3′;
FoxM1 forward, 5′-GGC TCCCGCAGCATCAAGCA-3′ and reverse,
5′-TGTTCCGGC GGAGCTCTGGA-3′; β-actin forward, 5′-ATCTGGCACCAC
ACCTTCACAATGAGCTGCG-3′ and reverse, 5′-CGTCATAC
TCCTGCTTGCTGATCCACATCTGC-3′, respectively. PCR analysis was
performed under the following conditions: denaturation at 94°C for
5 min, followed by 30 cycles of denaturation for 40 sec at 94°C,
annealing for 30 sec at 52°C for USP22, 57°C for FoxM1, 58°C for
β-actin and extension for 40 sec at 72°C. The amplified products
were analyzed by 1.0% agarose gel electrophoresis, followed by
ethidium bromide staining. Band intensities were measured using
BioImaging systems (UVP, Labworks™, ver 4.6).
Western blot analysis
Proteins were separated by SDS-PAGE and transferred
to nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). Membranes
were blocked in a buffer (TBS: 50 mM Tris-HCl, 150 mM NaCl, pH 7.4)
containing 5% bovine serum albumin and 0.1% Tween-20, followed by
incubation with the primary antibodys USP22 (ab4812, 1:2,000,
Abcam), FoxM1 (SC-502, 1:200, Santa Cruz Biotechnology), cyclin D1
(60186-1-lg, 1:100), p21 (10355-1-AP, 1:100), p27 (10567-1-AP,
1:100), cdk4 (11026-1-AP, 1:500), cdk6 (14052-1-AP, 1:500, all from
Proteintech, Chicago, IL, USA), β-catenin (SC-7963, 1:500, Santa
Cruz Biotechnology), Axin2 (EPR2005-2, 1:2,000), LEF (EP2030Y,
1:2,000, both from Abcam), c-Myc (10057-1-AP, 1:200) and LMNB1
(12987-1-AP, 1:1,000) or β-actin (20536-1-AP, 1:2,000, all from
Proteintech) diluted in the same buffer. The immunoreactive
proteins were visualized using the ECL western blot analysis system
(Bio-Rad), and densitometric analysis was performed using the Image
Pro-Plus software.
β-catenin/TCF transcription reporter
assay
Briefly, 1×105 cells were seeded per well
in a 24-well plate before transient transfection with the construct
TOPflash or FOPflash reporter plasmid (Millipore, Billerica, MA,
USA). TOPflash comprised three copies of the TCF/LEF sites upstream
of a thymidine kinase (TK) promoter and the Firefly luciferase
gene. FOPflash comprised three mutated copies of TCF/LEF sites and
were used as a control for measuring non-specific activation of the
reporter. All transfections were performed using 0.8 μg of TOPflash
or FOPflash plasmid and 2 μl lipofectamine 2000. To normalise the
transfection efficiency in reporter assays, the cells were
cotransfected with 0.02 μg of an internal control reporter plasmid,
containing Renilla reniformis luciferase driven by the TK
promoter. At 24 h after TOPflash or FOPflash transfection, the
luciferase assay was performed with the Dual Luciferase Assay
System kit (Promega Corp., Madison, WI, USA). Relative luciferase
activity was reported as the fold induction after normalization for
transfection efficiency.
Immunofluorescence
Pancreatic cancer cells were seeded onto cover
slips, fixed with 4% paraformaldehyde and permeabilized with 0.3%
Triton X-100 for 10 min. Slides were blocked with 1% bovine serum
albumin and incubated with USP22, FoxM1 or β-catenin antibodies
overnight at 4°C. After washing in PBS, the cells were stained with
secondary antibodies and incubated for 1 h at room temperature,
followed by nuclear counterstaining with DAPI.
Statistical analysis
Statistical analyses were performed using SPSS
software package version 17.0. The Pearson χ2 test or
Fisher’s exact test was used to compare qualitative variables. OS
was estimated by Kaplan-Meier curves and the curves were compared
using the log-rank test. OS was analysed using the Cox proportional
hazards model for univariate and multivariate analyses. The hazard
ratios (HRs) between prognostic groups and the 95% confidence
intervals were analysed. The quantitative data derived from three
independent experiments are expressed as means (± SD). Unpaired
Student’s t-tests were used to analyze between repeated group
differences. P<0.05 was considered statistically
significant.
Results
USP22 and FoxM1 expression in PDA tissues
and the paired normal carcinoma-adjacent tissues
The expression of USP22 and FoxM1 was analysed by
immunohistochemistry in PDA tissues and the paired normal
carcinoma-adjacent tissues (n=136). USP22 and FoxM1 staining in PDA
tissues appeared as brown particles which were mainly localized
within the nuclei and cytoplasm, with minute staining in the cell
membrane as well as some scattered infiltrated lymphocytes
(Fig. 1B and C). No or minimal
fluorescent staining showed in the normal carcinoma-adjacent
tissues (Fig. 1A). This finding is
consistent with those of earlier reports. The incidence of positive
expression was 54.4% (74/136) for USP22 and 63.2% (86/136) for
FoxM1 in PDA tissues and 8.8% (12/136) for USP22 and 7.4% (10/136)
for FoxM1 in the normal tissues. In addition, 53 (39.0%) PDA
tissues showed a co-positive expression of USP22/FoxM1. Compared
with the normal tissues, PDA tissues expressed significantly high
USP22 and FoxM1 (P<0.001, Fig.
1D). Additionally, a statistical correlation was observed
between USP22 and FoxM1 expression (r=0.221, P=0.01).
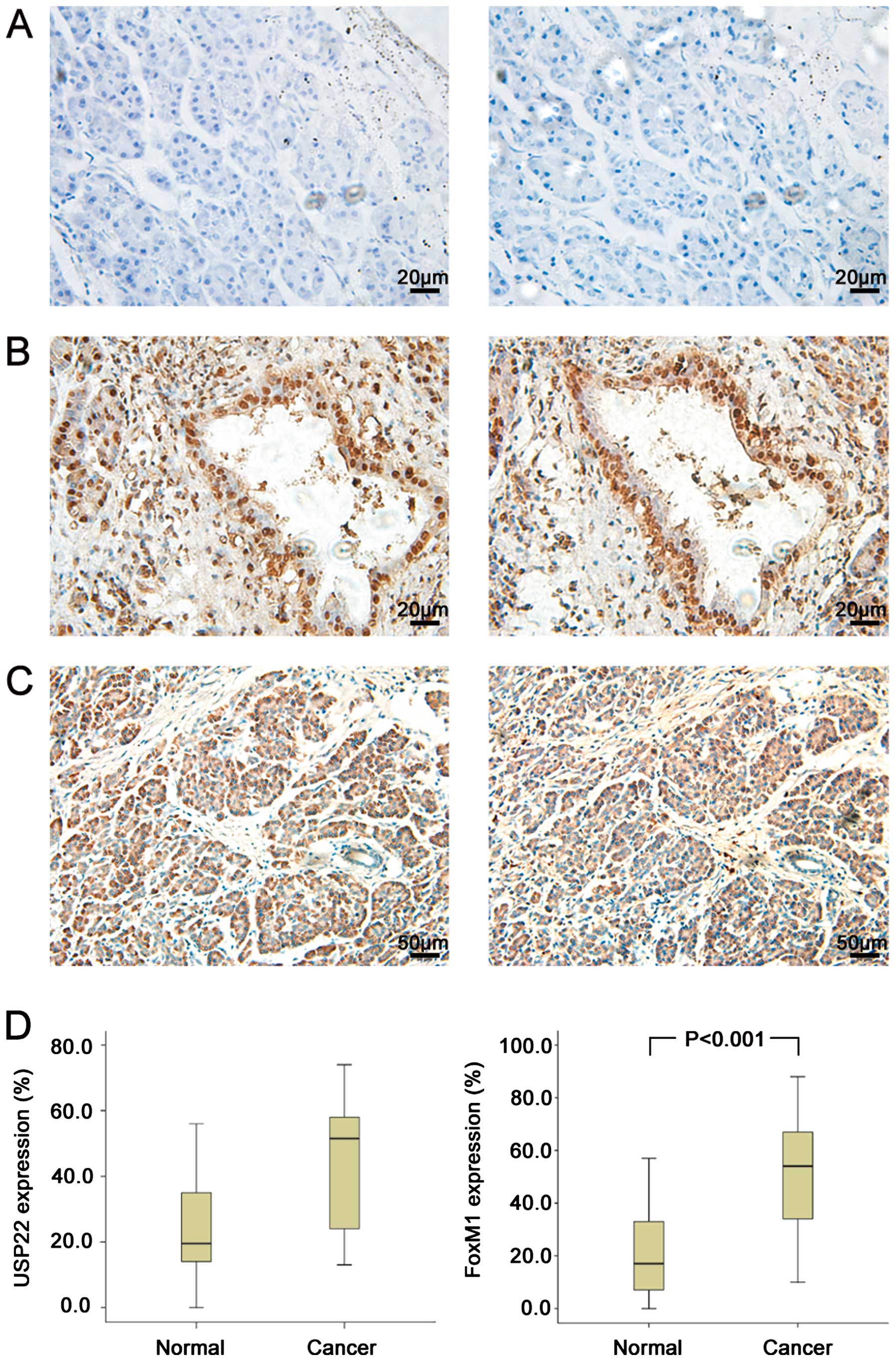 | Figure 1Detection of USP22 and FoxM1
immunoreactivity in normal carcinoma-adjacent tissues and PDA. (A)
Normal carcinoma-adjacent tissues showed no or weak immunoreactions
for USP22 (left) and FoxM1 (right), which was identified as
negative expression in this study. Original magnification, ×400.
(B) In contrast to the situation in the normal carcinoma-adjacent
tissues, strong immunostaining of USP22 (left) and FoxM1 (right),
which were primarily localized in the nuclei, were observed in the
cancer cells. Original magnification, ×400. (C) Strong
immunostaining of USP22 (left) and FoxM1 (right), which were
localized in the cytoplasm, were observed in the cancer cells.
Original magnification, ×200. (D) IHC analysis of proteins
expression in normal tissues and cancer tissues (n=136). Compared
with the normal tissues, cancer tissues expressed significantly
high USP22 (P<0.001) and FoxM1 (P<0.001). |
Correlation between USP22/FoxM1
expression and clinicopathologic features
The co-distribution of PDA with a positive or
negative USP22/FoxM1 expression in relation to clinicopathologic
features is shown in Table I.
USP22 expression was significantly associated with tumor size
(0.013) and lymph node metastasis (P=0.028). FoxM1 expression was
significantly associated with tumor size (P=0.019) and lymph node
metastasis (P=0.018). Furthermore, a co-positive expression of
USP22/FoxM1 was significantly associated with lymph node metastasis
(P=0.017) and TNM stage (P=0.005).
Correlation between USP22/FoxM1
expression and patient survival
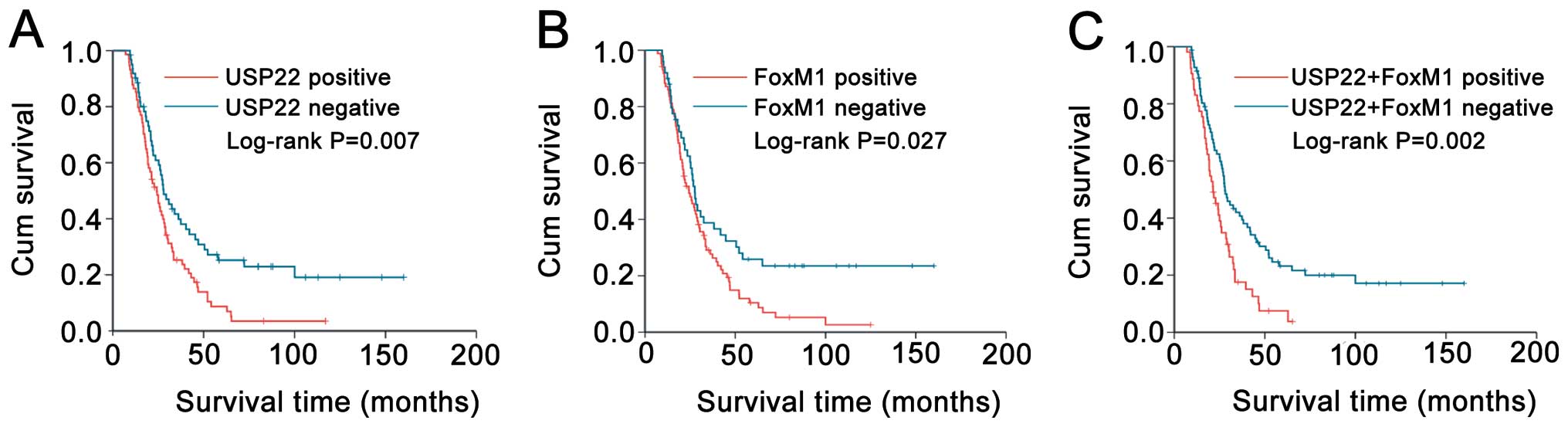
We assessed the prognostic value of USP22 and FoxM1
expression on survival in PDA patients. Kaplan-Meier analysis
demonstrated that PDA patients with USP22, FoxM1 and co-expression
USP22/FoxM1 had a significantly poorer OS compared to the patients
with negative expression (log-rank P=0.007; P=0.027; and P=0.002,
respectively, Fig. 2). In
univariate analysis, USP22 (HR, 3.227; 95% CI, 1.877–6.979;
P=0.007, Table II), FoxM1 (HR,
1.837; 95% CI, 1.185–4.993; P=0.027; Table II), co-expression USP22/FoxM1 (HR,
2.608; 95% CI, 1.659–5.959; P=0.002, Table II), tumor size, margin and lymph
node metastasis were significant prognostic factors of PDA. In
multivariate analysis of Table
III model A, USP22 (adjusted HR, 1.944; 95% CI, 1.492–2.998;
P=0.044), margin (adjusted HR, 1.331; 95% CI, 1.171–1.994; P=0.033)
and lymph node metastasis (adjusted HR, 3.229; 95% CI, 1.573–6.988;
P=0.020) were independent prognostic factors of survival. Table III model B showed that positive
co-expression of USP22/FoxM1 (adjusted HR, 1.728; 95% CI,
1.275–2.684; P=0.012), margin (adjusted HR, 1.827; 95% CI,
1.167–2.192; P=0.027) and lymph node metastasis (adjusted HR,
2.931; 95% CI, 1.476–4.990; P=0.023) were determined as independent
prognostic factors.
 | Table IIUnivariate analyses for survival in
patients with stage II pancreatic ductal adenocarcinoma. |
Table II
Univariate analyses for survival in
patients with stage II pancreatic ductal adenocarcinoma.
| | 95% CI | |
|---|
| |
| |
|---|
| Variables | Hazard ratio | Lower | Upper | P-value |
|---|
| Gender | 1.206 | 0.831 | 1.750 | 0.324 |
| Age | 0.094 | 0.791 | 1.618 | 0.439 |
| Tumor location | 0.825 | 0.569 | 2.195 | 0.309 |
| Tumor size | 2.317 | 1.099 | 4.380 | 0.015 |
| Margin | 1.924 | 1.266 | 2.987 | 0.018 |
| Histological
differentiation | 1.402 | 0.863 | 3.041 | 0.078 |
| Lymph node
metastasis | 5.905 | 1.853 | 9.959 | 0.001 |
| Post operative
treatment | 0.753 | 0.591 | 1.920 | 0.097 |
| USP22 | 3.227 | 1.877 | 6.979 | 0.007 |
| FoxM1 | 1.737 | 1.185 | 4.993 | 0.027 |
| USP22+FoxM1 | 2.608 | 1.659 | 5.959 | 0.002 |
 | Table IIIMultivariate analyses for survival in
patients with stage II pancreatic ductal adenocarcinoma. |
Table III
Multivariate analyses for survival in
patients with stage II pancreatic ductal adenocarcinoma.
| | 95% CI | |
|---|
| |
| |
|---|
| Variables | Hazard ratio | Lower | Upper | P-value |
|---|
| Model A |
| Tumor size | 1.348 | 0.797 | 2.004 | 0.138 |
| Margin | 1.331 | 1.171 | 1.994 | 0.033 |
| Lymph node
metastasis | 3.229 | 1.573 | 6.988 | 0.020 |
| USP22 | 1.944 | 1.492 | 2.998 | 0.044 |
| FoxM1 | 1.270 | 0.713 | 2.830 | 0.316 |
| Model B |
| Tumor size | 1.369 | 0.623 | 1.930 | 0.118 |
| Margin | 1.827 | 1.167 | 2.192 | 0.027 |
| Lymph node
metastasis | 2.931 | 1.476 | 4.990 | 0.023 |
| USP22+FoxM1 | 1.728 | 1.275 | 2.684 | 0.012 |
USP22/FoxM1 expression in fresh-frozen
PDA tissues and cell lines
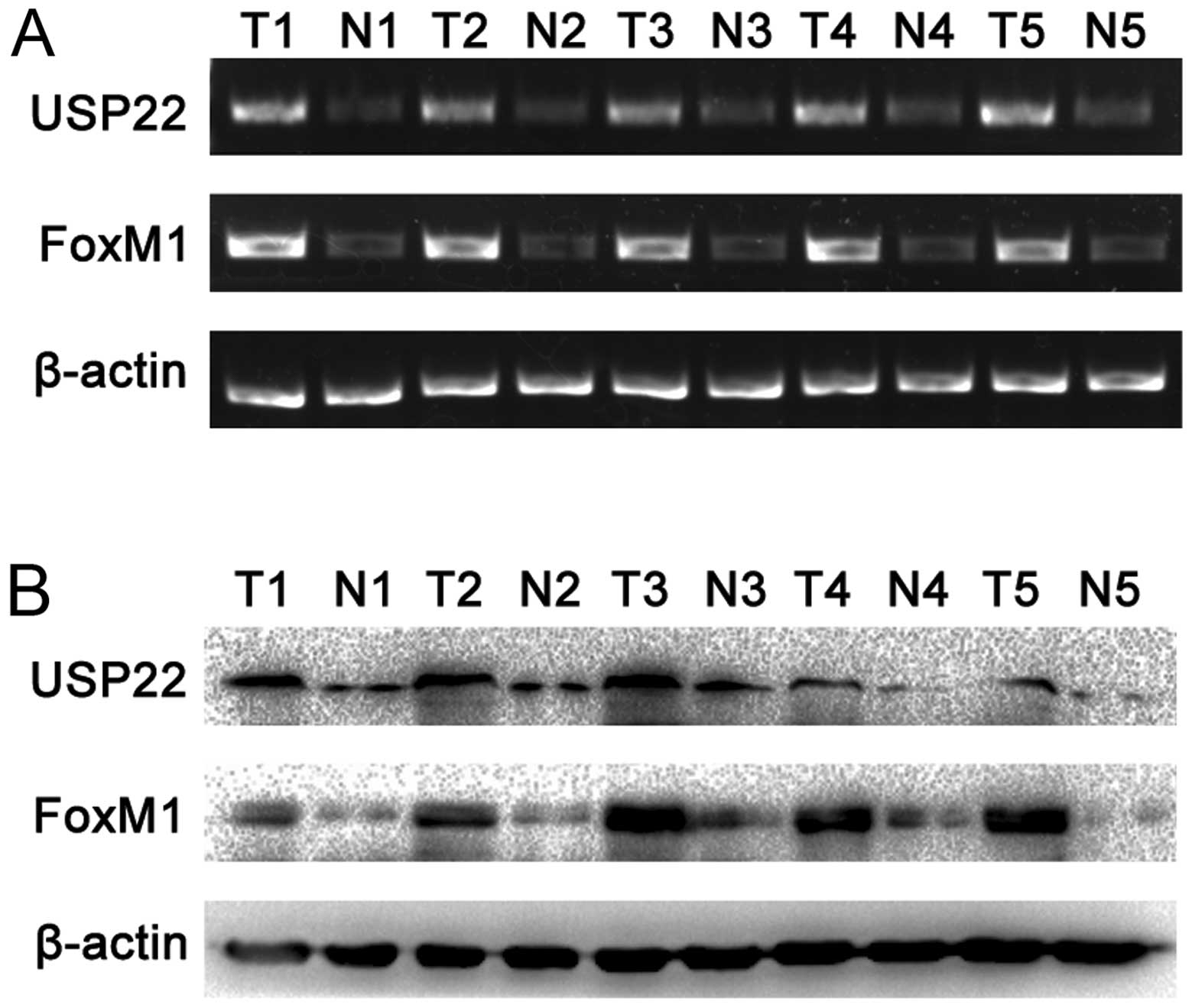
To further confirm the afore-mentioned results,
RT-PCR and western blot analyses were performed on 5 pairs of PDA
fresh-frozen tumor tissues and the paired normal carcinoma-adjacent
tissues. The results showed that both USP22 and FoxM1 expression
were significantly higher in PDA tissues than the paired normal
carcinoma-adjacent tissues (Fig. 3A
and B). USP22 and FoxM1 expression were also examined in PDA
cell lines PANC-1 and CFPAC-1. The results showed that expression
of USP22 and FoxM1 was significantly elevated in the poorly
differentiated PANC-1 cell line in comparison with the highly
differentiated CFPAC-1 cell line (Fig.
3C and D).
USP22 regulates the G1/S phase transition
and affects the expression of G1 phase-related proteins through
FoxM1 in PDA cell lines
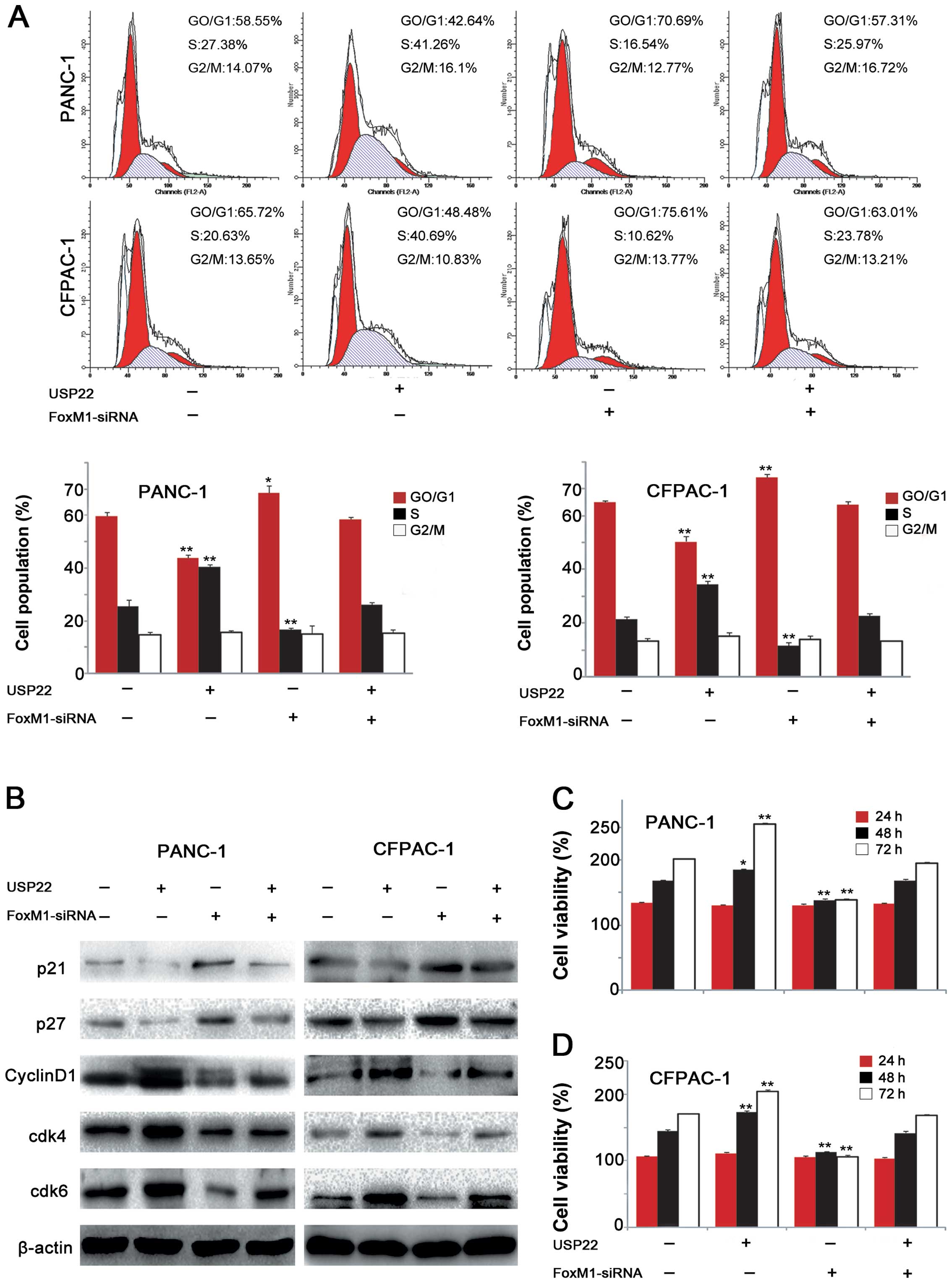
To investigate the role of USP22 and its possible
underlying mechanism in PDA tumorigenesis, USP22 plasmid or
FoxM1-siRNA was transfected in both the poorly differentiated
PANC-1 cell line and highly differentiated CFPAC-1 cell line. The
CCK-8 proliferation assay showed that USP22 overexpression
significantly stimulated the proliferation of both cell lines and
the interference of FoxM1 expression produced significant
inhibition of the proliferation of both cell lines. However, the
depletion of FoxM1 expression, caused by FoxM1-siRNA interference,
in the USP22-overexpressing PANC-1 and CFPAC-1 cell lines abolished
the USP22 overexpression-stimulated cell proliferation (Fig. 4C and D). Additionally, flow
cytometry revealed that USP22 overexpression increased the G1/S
transition rate, resulting in a significant decrease in the
proportion of G1 phase cells and an increase in the proportions of
S phase. Upon FoxM1 downregulation by RNAi, USP22 overexpression no
longer elicited significant changes in cell cycle progression in
either cell line (Fig. 4A).
Further studies showed that USP22 overexpression resulted in
decreased p21 and p27 expression in both cell lines, which were
accompanied by the increased cyclin D1 levels. Similarly, the
expression levels of cdk4 and cdk6 were also increased. FoxM1
depletion abolished the USP22-induced changes in G1 phase-related
protein expression in both cell lines (Fig. 4B). These results suggested that
USP22 promoted the G1/S phase transition and affected G1
phase-related protein expression in PDCA cell lines via the
upregulation of FoxM1 expression.
USP22 expression affects FoxM1 expression
and Wnt/β-catenin pathway activation
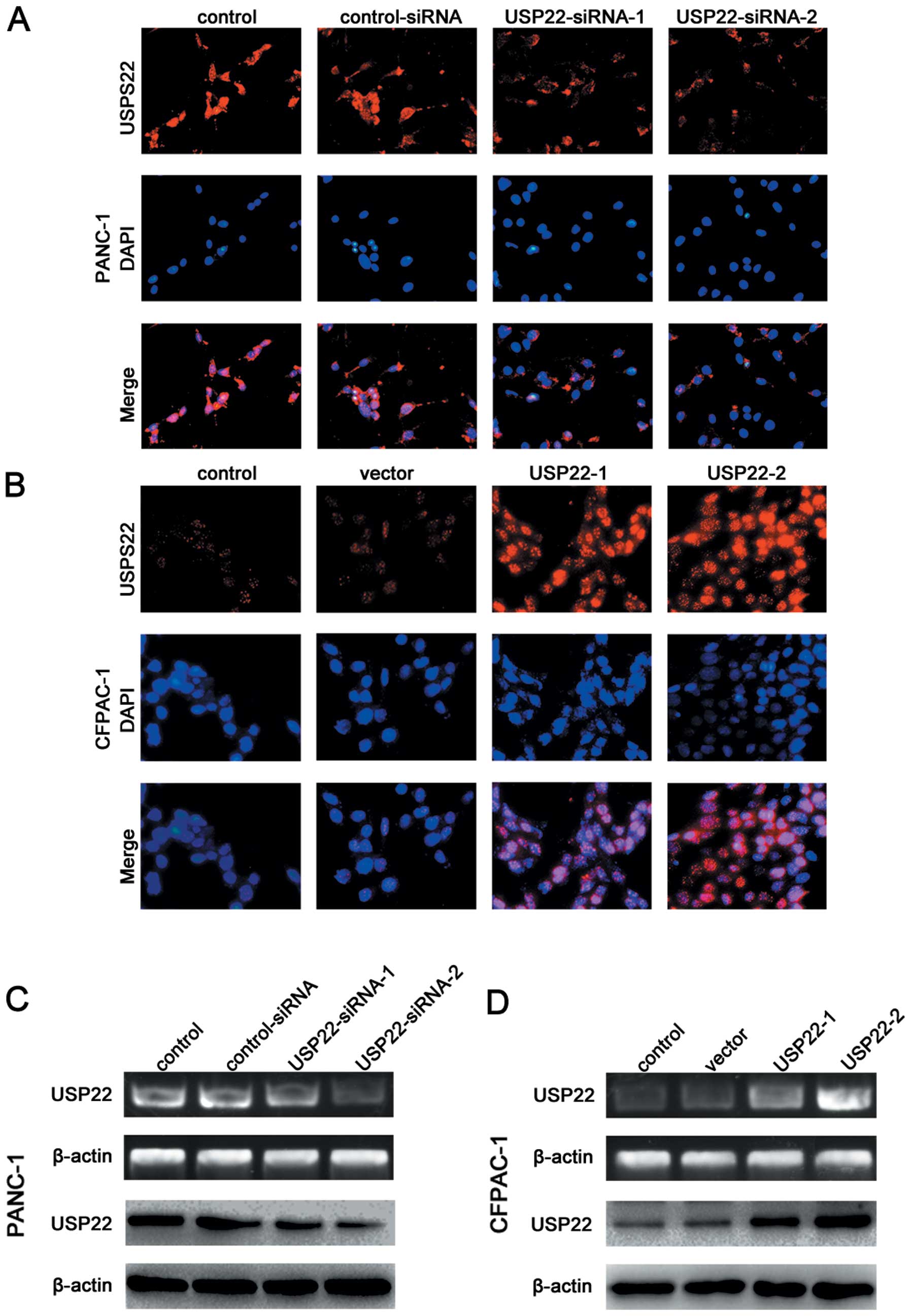
We used different concentrations of the USP22
plasmids or USP22-siRNA to achieve the different degrees of
downregulation of USP22 in the PANC-1 cell line which originally
expressed rather high levels of USP22, as well as the different
degrees of upregulation of USP22 in the CFPAC-1 cell line which
expressed relatively low levels of endogenous USP22. The results of
RT-PCR, western blot and immunofluorescence staining analyses
confirmed the success of the above-described transfection efforts
(Fig. 5). The immunofluorescence
experiments also showed that USP22 expression was mainly nuclear
(Fig. 5A and B). The USP22
downregulation in the PANC-1 cell line resulted in reduction of the
expression of 2 Wnt/β-catenin signalling pathway marker proteins,
Axin2 and lymphoid enhancer-binding factor-1 (LEF-1), as well as
the downstream target protein c-Myc (Fig. 6E). Using a luciferase reporter
plasmid, we also confirmed a decrease in Wnt/β-catenin signalling
activity (Fig. 6F). In the CFPAC-1
cell line, the USP22 upregulation led to increases in the
expression of Axin2, LEF-1 and the downstream target protein c-Myc
(Fig. 6E). The luciferase reporter
assay also confirmed an increase in Wnt/β-catenin signalling
activity (Fig. 6F).
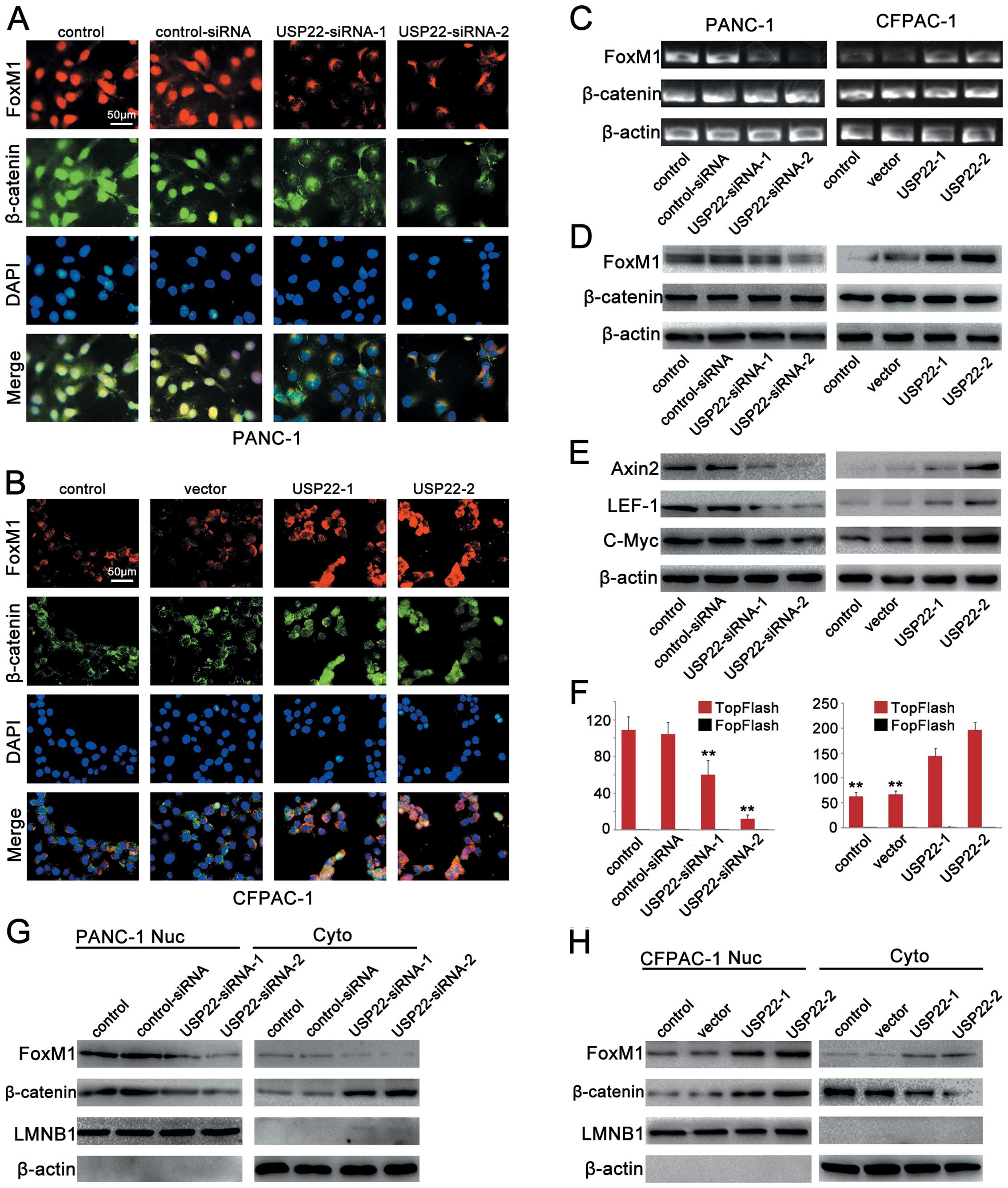 | Figure 6USP22 expression affects FoxM1
expression and Wnt/β-catenin pathway activation. (A) Triple IF
staining for FoxM1 (red), β-catenin (green) and nuclei (DAPI, blue)
was performed on PANC-1 cells that were transfected by
control-siRNA, USP22-siRNA-1 (50 nM) or USP22-siRNA-2 (150 nM). (B)
Triple IF staining for FoxM1 (red), β-catenin (green) and nuclei
(DAPI, blue) was performed on CFPAC-1 cells that were transfected
by vector USP22-1 (2 μg) or USP22-2 (10 μg). (C and D) RT-PCR and
western blot analysis of FoxM1 and β-catenin in PANC-1 cells that
were transfected by control-siRNA, USP22-siRNA-1 (50 nM) or
USP22-siRNA-2 (150 nM) (left panel) and CFPAC-1 cells that were
transfected by vector, USP22-1 (2 μg) or USP22-2 (10 μg) (right
panel). (E) Cellular levels of Axin2, c-Myc and LEF-1 in PANC-1
cells that were transfected by control-siRNA, USP22-siRNA-1 (50 nM)
or USP22-siRNA-2 (150 nM) (left panel) and CFPAC-1 cells that were
transfected by vector, USP22-1 (2 μg) or USP22-2 (10 μg) (right
panel). (F) Activities of TOP-Flash and FOP-Flash in PANC-1 cells
that were transfected by control-siRNA, USP22-siRNA-1 (50 nM) or
USP22-siRNA-2 (150 nM) (left panel) and CFPAC-1 cells that were
transfected by vector, USP22-1 (2 μg) or USP22-2 (10 μg) (right
panel). (G) Cytoplasmic and nuclear levels of FoxM1 and β-catenin
in PANC-1 cells that were transfected by control-siRNA,
USP22-siRNA-1 (50 nM) or USP22-siRNA-2 (150 nM). (H) Cytoplasmic
and nuclear levels of FoxM1 and β-catenin in CFPAC-1 cells that
were transfected by vector USP22-1 (2 μg) or USP22-2 (10 μg). |
To prove that USP22 regulates FoxM1 expression via
the Wnt/β-catenin signalling pathway, western blot and
immunofluorescence analyses were performed. The results showed that
the USP22 downregulation in PANC-1 cells led to a reduction in
nuclear β-catenin expression, as well as an elevation in
cytoplasmic β-catenin expression. Reduction was observed in both
nuclear and cytoplasmic FoxM1 expression (Fig. 6A, C, D and G). In the CFPAC-1 cell
line, the USP22 upregulation led to an increase in nuclear
β-catenin expression and a reduction in cytoplasmic β-catenin
expression. Concurrently, increases were observed in both nuclear
and cytoplasmic FoxM1 expression (Fig.
6B, C, D and H). These results indicated that in PDA cell
lines, USP22 expression significantly altered FoxM1 expression via
the Wnt/β-catenin signalling pathway.
β-catenin nuclear localization is
required for USP22-mediated FoxM1 expression
The nuclear transfer of β-catenin is believed to be
directly responsible for elevated Wnt/β-catenin signalling pathway
activity. The β-catenin expression levels in the nucleus directly
represent the level of Wnt/β-catenin signalling pathway activity.
β-catenin depletion or overexpression has been widely used to
trigger corresponding changes in Wnt/β-catenin signalling pathway
activity. In additional experiments on the PANC-1 cell line, we
found that when β-catenin expression was upregulated with the
β-catenin overexpressing plasmid, USP22 knockdown with USP22-siRNA
failed to induce significant changes in either nuclear or
cytoplasmic FoxM1 expression (Fig. 7A,
C and D). In the CFPAC-1 cell line, when β-catenin expression
was disrupted with β-catenin-siRNA, USP22 overexpression failed to
trigger significant changes in either nuclear or cytoplasmic FoxM1
expression (Fig. 7B, C and E).
These results strongly suggest that in PDA cell lines,
USP22-mediated FoxM1 expression depends on the nuclear localization
of β-catenin.
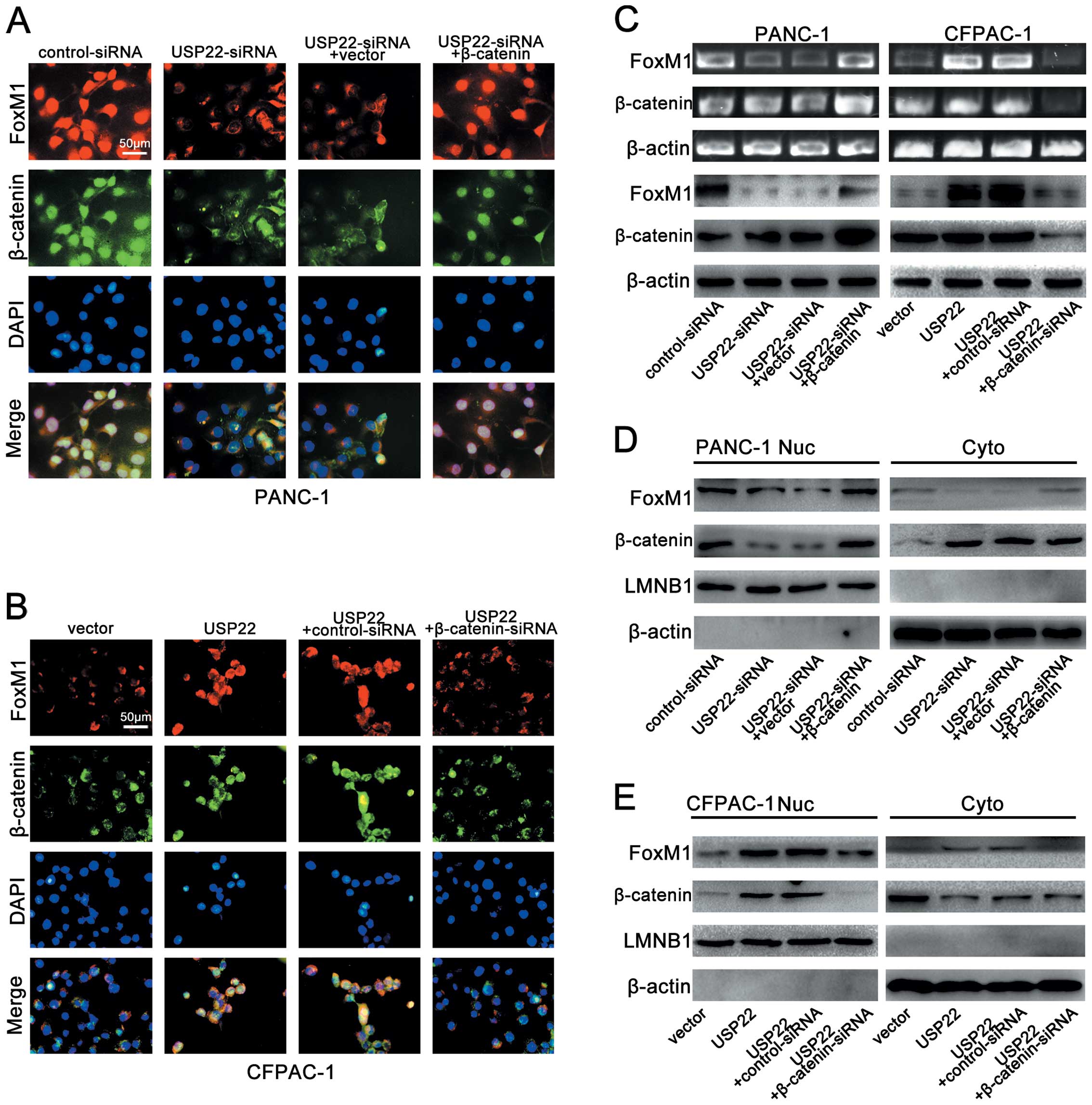 | Figure 7β-catenin is required for
USP22-mediated FoxM1 expression. (A) Triple IF staining for FoxM1
(red), β-catenin (green) and nuclei (DAPI, blue) was performed in
PANC-1 cells that were transfected by control-siRNA, USP22-siRNA,
USP22-siRNA+vector or USP22-siRNA+β-catenin. (B) Triple IF staining
for FoxM1 (red), β-catenin (green) and nuclei (DAPI, blue) was
performed in CFPAC-1 cells that were transfected by vector, USP22,
USP22+control-siRNA or USP22+β-catenin-siRNA. (C) RT-PCR and
western blot analysis of FoxM1 and β-catenin in PANC-1 cells that
were transfected by control-siRNA, USP22-siRNA, USP22-siRNA+vector
or USP22-siRNA+β-catenin (left panel) and CFPAC-1 cells that were
transfected by vector, USP22, USP22+control-siRNA or
USP22+β-catenin-siRNA (right panel). (D) Cytoplasmic and nuclear
levels of FoxM1 and β-catenin in PANC-1 cells that were transfected
by control-siRNA, USP22-siRNA, USP22-siRNA+vector or
USP22-siRNA+β-catenin. (E) Cytoplasmic and nuclear levels of FoxM1
and β-catenin in CFPAC-1 cells that were transfected by vector
USP22, USP22+control-siRNA or USP22+β-catenin-siRNA. |
Discussion
In an analysis of mRNA levels in a variety of tumor
tissues in 2005, Glinsky et al identified 11 overexpressed
genes in tumor tissues that can serve as death-from-cancer
signature genes, including USP22, BUB1,
HEC1/KNTC2, CCNB1, Ki67, Gbx2,
FGFR2, ANK3, CES, BMI-1 and RNF2
(12). The study suggests that
USP22 overexpression may be correlated with short-term tumor
recurrence, distant metastasis and treatment resistance (11). A number of studies have confirmed
that USP22 overexpression is present in various solid tumors such
as papillary thyroid, breast, non-small cell lung, esophageal,
gastric and colon cancer, and is closely related to the metastatic
potential and prognosis of tumors (33–38).
However, no study has reported its expression in pancreatic
cancer.
USP22, one of the 11 death-from-cancer signature
genes, functions as a cell cycle regulator and plays an important
role in cell cycle progression (11). hSAGA is a transcription cofactor
complex that recruits the transcriptional complex to the promoter
of its target genes to activate gene transcription through specific
amino acid covalent modifications of histone tails, such as
acetylation, methylation and deubiquitination (39,40).
USP22, a subunit of hSAGA, can cause deubiquitination of histone
H2A and H2B and acetylation of histone H4 (41). hSAGA is involved in the
transcription of Myc target genes. The amino-terminal of c-Myc
protein carries two conserved domains, MbI and MbII, which combine
with hSAGA to initiate the transcription of c-Myc target genes.
Studies have shown that USP22 knockdown using shRNA resulted in a
decline in the expression of Myc target genes including CDCA7,
cyclin D2, ODC, CAD and MTA1. ChIP assays also confirm that USP22
binds to the promoter regions of Myc target genes and triggers the
transcription of these genes. In addition, downregulation of USP22
expression slows the proliferation of Myc-overexpressing cells and
arrests the cells in the G1 phase, leading to a lower cloning
efficiency (5). These results
suggest that USP22 can promote cell proliferation and mediate
malignant cell behavior via Myc-regulated target genes. Moreover,
USP22 can also modulate the expression of the tumor suppressor gene
p21 by altering the ubiquitination level of its upstream element
FBP1, thereby affecting cell cycle progression (42).
A member of the Fox transcription factor family,
FoxM1 is specifically expressed in proliferating cells and is
absent in terminally differentiated cells. FoxM1 can initiate
target gene transcription, mediate the transcription of cell
cycle-related genes, and promote cell proliferation by suppressing
Cdk inhibitors, such as p21 and p27, and the phosphatase activity
of some cyclins (e.g., cyclin A1, cyclin B1 and cyclin D1) and Cdk
activators Cdc25a and Cdc25b (17–19,43–45).
First, FoxM1 upregulates the expression of CdK inhibitors p21 and
p27 in the nucleus by controlling the transcription of two
important subunits Skp2 and Cks1 in the ubiquitin ligase complex
SCF, thereby promoting DNA replication in S phase (46). Current studies have shown that
numerous cell cycle regulators, such as E2F, cyclin D1, cyclin E,
cyclin A, cyclin B, CDC25B, p27, p21 and p53, are all substrates of
the ubiquitin proteasome pathway (47–52).
Wang et al showed that FoxM1 activation mediates the
degradation of G1/S transition regulators p27 and p21 by activating
Skp2-Cksl ubiquitin ligase. As p27 and p21 are CDK inhibitors,
decreased expression of p27 and p21 will inevitably lead to reduced
inhibition of cell cycle, which helps cells move beyond the G1/S
phase checkpoint and enter S phase to synthesize DNA and make
preparations for cell proliferation (46,53).
In addition, FoxM1 also regulates the transcription of many
mitosis-related genes, including cyclin B1, Cdc25B, polo-like
kinase 1 (PLK1), aurora B kinase, survivin, centromere protein A
(CENPA) and CENPB (54). Previous
studies have shown that FoxM1 expression is abnormally high in a
variety of malignancies such as gastric, non-small cell lung
cancer, glioma and cervical cancer and can be used as a prognostic
marker (24,25,55,56).
Xia et al also confirmed that FoxM1 is overexpressed in
pancreatic cancer and can be used as an independent prognostic
factor (26), but the exact
mechanism has not been examined.
Currently, no study has reported the role of USP22
in pancreatic cancer. Given that both USP22 and FoxM1 are involved
in cell cycle progression, this leads us to question whether USP22
and FoxM1 interact in cell cycle regulation and whether the two
factors are involved in the development and progression of
pancreatic cancer.
To address these issues, we determined the
expression of USP22 and FoxM1 in 136 PDA tissues and 42 PDA
adjacent normal tissues using immunohistochemistry. The results
showed that the expression of USP22 and FoxM1 was abnormally
upregulated in the PDA tissue and that up to 39.0% of patients with
high expression of USP22 also had FoxM1 overexpression, exhibiting
a significant correlation between the expression levels of USP22
and FoxM1. Detection in fresh tissues also supported the above
results. This suggests that there may be some interplay between
USP22 and FoxM1 in pancreatic cancer. Further analysis revealed
that USP22/FoxM1 overexpression is closely associated with the
clinical and pathological features of PDA patients and can be used
as an independent prognostic factor for PDA patients. In
particular, patients with positive co-expression of USP22/FoxM1
have a worse prognosis. These results indicate that USP22 and FoxM1
may be jointly involved in the development and progression of PDA.
However, it remains unknown whether and how USP22 interacts with
FoxM1 in pancreatic cancer.
With the above question in mind, we selected two PDA
cell lines for study. The results showed that the expression of
USP22 and FoxM1 was significantly higher in poorly differentiated
PANC-1 cell line than in highly differentiated CFPAC-1 cell line.
To determine the role of USP22 in the proliferation and cell cycle
of PDA cells, we upregulated USP22 expression in both cell lines
and found faster G1/S phase transition and significantly
accelerated cell proliferation. Next, we upregulated USP22
expression and knocked down FoxM1 expression in both cell lines.
The results showed that with FoxM1 downregulation, USP22
upregulation alone caused no significant change in G1/S transition
and cell proliferation. This indicates that FoxM1 plays a key role
in the regulation by USP22 of G1/S transition and cell
proliferation. Moreover, we found that USP22 upregulation was
accompanied by p21 and p27 downregulation as well as upregulation
of cell cycle-related proteins cyclin D1, cdk4 and cdk6; USP22
upregulation combined with FoxM1 knockdown did not cause
significant change in the expression of the above proteins. This
suggests that FoxM1 plays an important role in the regulation by
USP22 of G1 phase related proteins. Therefore, our study indicates
that FoxM1 is a key factor essential for the regulation by USP22 of
G1/S transition.
USP22 and FoxM1 play a role in the transcriptional
regulation of a variety of oncogenes and tumor suppressor genes
(e.g., c-Myc, SIRT1, BMI-1). Previous studies have shown that USP22
promotes tumor growth by enhancing c-Myc-mediated gene
transcription (5). FoxM1 can also
help tumor cells pass through checkpoints and promote tumor cell
proliferation through direct transcriptional activation of the TATA
box in human c-Myc transcription promoters P1 and P2 (57). SIRT1 upregulation is involved in
tumorigenesis in a variety of tumor cells by promoting tumor cell
proliferation and inhibiting apoptosis (58). Lin et al revealed that as
part of the SAGA complex, USP22 can directly regulate the
expression of SIRT1 via deubiquitination and affect the activity of
SIRT1 target genes, thereby regulating DNA repair and apoptosis
(6). Zhu et al demonstrated
that FoxM1 directly associates with SIRT1 promoter, increases the
transcriptional activity of SIRT1, and promote glioma cell
proliferation (59). As a
component of PcG protein, BMI-1 determines to a certain extent the
proliferation potential of cancer stem cells (60). Previous research showed high
expression of USP22 and BMI-1 in gastric and colorectal cancer
(33,38). Liu et al found that USP22
can regulate cell cycle in colon cancer through BMI-1-mediated pRb,
P53 and Akt/GSK3β pathways (61).
Li et al showed that BMI-1 is one of the downstream target
genes of FoxM1. FoxM1 overexpression can attenuate the activity of
p19 (Arf)-p53 pathway, downregulate p53 and p21 gene expression,
and activate c-Myc-mediated BMI-1 gene expression (62). These studies suggest that USP22 and
FoxM1 share common target genes and may work in synergy to
participate in development and progression of tumors.
The Wnt signaling pathway plays an important role in
embryonic development and tumorigenesis. β-catenin is an important
component in the classical Wnt signaling pathway. c-Myc is the
second target in the Wnt signaling pathway. Upon its translocation
to the nucleus, β-catenin reacts with transcription factors in the
Tcf/Lef family to form β-catenin/Tcf complexes, which then activate
target genes such as c-Myc, c-jun and cyclin D1, leading to
abnormal cell proliferation and differentiation and tumor formation
(63). Heiser et al found
that sustained activation of β-catenin in mice can lead to
pancreatic intraepithelial neoplasia and eventual tumor formation
(64–66). Zhang et al showed that
inhibition of the Wnt pathway can significantly delay the
development of pancreatic intraepithelial neoplasia (67). Ripka et al and Griesmann
et al suggest that the Wnt pathway enhances the
proliferation, invasion and metastasis activity of pancreatic
cancer cells and is involved in epithelial-to-mesenchymal
transformation of pancreatic cancer cells (68,69).
Accordingly, substantial evidence indicates that the Wnt/β-catenin
signaling pathway plays an important role in the development and
progression of pancreatic cancer (30). Furthermore, Zhang et al
showed that FoxM1 binds to β-catenin and promotes the nuclear
translocation of the latter, hence increasing the activity of the
Wnt/β-catenin pathway (31). This
finding is corroborated by Bowman et al (70). These studies also strongly suggest
that the activation of the Wnt/β-catenin signaling pathway,
especially β-catenin nuclear translocation, may play an important
role in the regulation by USP22 of FoxM1.
To test the above hypothesis, we used different
plasmid concentrations to create a descending gradient of USP22
expression in the PANC-1 cell line and an ascending gradient of
USP22 expression in the CFPAC-1 cell line. With the gradient change
in USP22 expression, a similar trend change also occurred in
Wnt/β-catenin signaling pathway activity and FoxM1 expression in
the nucleus and cytoplasm in the two cell lines. Further analysis
showed that after β-catenin expression was upregulated in the
PANC-1 cell line, even USP22 expression was significantly
decreased, there was no significant change in the expression of
FoxM1. After the β-catenin expression was knocked down in the
CFPAC-1 cell line, even significant USP22 upregulation failed to
cause a significant increase in FoxM1 expression. These results
confirm our hypothesis that USP22 regulates FoxM1 expression
through the Wnt/β-catenin signaling pathway, especially β-catenin
nuclear translocation.
In summary, our immunohistochemical analysis
demonstrated that varying levels of USP22 and FoxM1 overexpression
are present in PDA tissue and that the expression of both factors,
especially the co-expression of the factors, is an independent
prognostic factor of PDA. In vitro investigation showed that
USP22 overexpression is accompanied by an increase in FoxM1
expression and that USP22 increases FoxM1 expression to promote
G1/S transition and cell proliferation through promoting β-catenin
nuclear translocation in PDA cell lines. These results demonstrate
for the first time that USP22 and FoxM1 are jointly involved in the
development and progression of pancreatic cancer and are promising
new targets for pancreatic cancer targeted therapy.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China Research grant (no. 30870719 to Z.W.,
no. 30672753 to J.L.) and China 973 grant (no. 2012CB822100 to
Q.Y.).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar
|
|
3
|
Singh P, Srinivasan R and Wig JD: Major
molecular markers in pancreatic ductal adenocarcinoma and their
roles in screening, diagnosis, prognosis, and treatment. Pancreas.
40:644–652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM
and Baek KH: The expression patterns of deubiquitinating enzymes,
USP22 and Usp22. Gene Expr Patterns. 6:277–284. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang XY, Varthi M, Sykes SM, et al: The
putative cancer stem cell marker USP22 is a subunit of the human
SAGA complex required for activated transcription and cell-cycle
progression. Mol Cell. 29:102–111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin Z, Yang H, Kong Q, et al: USP22
antagonizes p53 transcriptional activation by deubiquitinating
Sirt1 to suppress cell apoptosis and is required for mouse
embryonic development. Mol Cell. 46:484–494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Atanassov BS, Evrard YA, Multani AS, et
al: Gcn5 and SAGA regulate shelterin protein turnover and telomere
maintenance. Mol Cell. 35:352–364. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu H, Liu YL, Yang YM and Dong XS:
Knock-down of ubiquitin-specific protease 22 by micro-RNA
interference inhibits colorectal cancer growth. Int J Colorectal
Dis. 27:21–30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lv L, Xiao XY, Gu ZH, Zeng FQ, Huang LQ
and Jiang GS: Silencing USP22 by asymmetric structure of
interfering RNA inhibits proliferation and induces cell cycle
arrest in bladder cancer cells. Mol Cell Biochem. 346:11–21. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ling SB, Sun DG, Tang B, et al: Knock-down
of USP22 by small interfering RNA interference inhibits HepG2 cell
proliferation and induces cell cycle arrest. Cell Mol Biol
(Noisy-le-grand). 58:L1803–L1808. 2012.PubMed/NCBI
|
|
11
|
Glinsky GV: Genomic models of metastatic
cancer: functional analysis of death-from-cancer signature genes
reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype
with altered cell cycle control and activated Polycomb Group (PcG)
protein chromatin silencing pathway. Cell Cycle. 5:1208–1216.
2006.
|
|
12
|
Glinsky GV, Berezovska O and Glinskii AB:
Microarray analysis identifies a death-from-cancer signature
predicting therapy failure in patients with multiple types of
cancer. J Clin Invest. 115:1503–1521. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jackson BC, Carpenter C, Nebert DW and
Vasiliou V: Update of human and mouse forkhead box (FOX) gene
families. Hum Genomics. 4:345–352. 2010.PubMed/NCBI
|
|
14
|
Laoukili J, Kooistra MR, Bras A, et al:
FoxM1 is required for execution of the mitotic programme and
chromosome stability. Nat Cell Biol. 7:126–136. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wierstra I and Alves J: FOXM1, a typical
proliferation-associated transcription factor. Biol Chem.
388:1257–1274. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luscher-Firzlaff JM, Lilischkis R and
Luscher B: Regulation of the transcription factor FOXM1c by Cyclin
E/CDK2. FEBS Lett. 580:1716–1722. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Major ML, Lepe R and Costa RH: Forkhead
box M1B transcriptional activity requires binding of Cdk-cyclin
complexes for phosphorylation-dependent recruitment of p300/CBP
coactivators. Mol Cell Biol. 24:2649–2661. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang X, Hung NJ and Costa RH: Earlier
expression of the transcription factor HFH-11B diminishes induction
of p21(CIP1/WAF1) levels and accelerates mouse hepatocyte entry
into S-phase following carbon tetrachloride liver injury.
Hepatology. 33:1404–1414. 2001. View Article : Google Scholar
|
|
19
|
Wang X, Krupczak-Hollis K, Tan Y,
Dennewitz MB, Adami GR and Costa RH: Increased hepatic Forkhead Box
M1B (FoxM1B) levels in old-aged mice stimulated liver regeneration
through diminished p27Kip1 protein levels and increased Cdc25B
expression. J Biol Chem. 277:44310–44316. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laoukili J, Stahl M and Medema RH: FoxM1:
at the crossroads of ageing and cancer. Biochim Biophys Acta.
1775:92–102. 2007.PubMed/NCBI
|
|
21
|
Wang Z, Ahmad A, Banerjee S, et al: FoxM1
is a novel target of a natural agent in pancreatic cancer. Pharm
Res. 27:1159–1168. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z, Banerjee S, Kong D, Li Y and
Sarkar FH: Down-regulation of Forkhead Box M1 transcription factor
leads to the inhibition of invasion and angiogenesis of pancreatic
cancer cells. Cancer Res. 67:8293–8300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bao B, Wang Z, Ali S, et al:
Over-expression of FoxM1 leads to epithelial-mesenchymal transition
and cancer stem cell phenotype in pancreatic cancer cells. J Cell
Biochem. 112:2296–2306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Qiu W, Liu B, et al: Forkhead box
transcription factor 1 expression in gastric cancer: FOXM1 is a
poor prognostic factor and mediates resistance to docetaxel. J
Transl Med. 11:2042013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Wen L, Zhao SH, Ai ZH, Guo JZ and
Liu WC: FoxM1 expression is significantly associated with
cisplatin-based chemotherapy resistance and poor prognosis in
advanced non-small cell lung cancer patients. Lung Cancer.
79:173–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia JT, Wang H, Liang LJ, et al:
Overexpression of FOXM1 is associated with poor prognosis and
clinicopathologic stage of pancreatic ductal adenocarcinoma.
Pancreas. 41:629–635. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang H and He X: Wnt/beta-catenin
signaling: new (and old) players and new insights. Curr Opin Cell
Biol. 20:119–125. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Karayiannakis AJ, Syrigos KN,
Polychronidis A and Simopoulos C: Expression patterns of alpha-,
beta- and gamma-catenin in pancreatic cancer: correlation with
E-cadherin expression, pathological features and prognosis.
Anticancer Res. 21:4127–4134. 2001.PubMed/NCBI
|
|
29
|
Lowy AM, Fenoglio-Preiser C, Kim OJ, et
al: Dysregulation of beta-catenin expression correlates with tumor
differentiation in pancreatic duct adenocarcinoma. Ann Surg Oncol.
10:284–290. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pasca DMM, Biankin AV, Heiser PW, et al:
Common activation of canonical Wnt signaling in pancreatic
adenocarcinoma. PLoS One. 2:e11552007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang N, Wei P, Gong A, et al: FoxM1
promotes beta-catenin nuclear localization and controls Wnt
target-gene expression and glioma tumorigenesis. Cancer Cell.
20:427–442. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Deer EL, Gonzalez-Hernandez J, Coursen JD,
et al: Phenotype and genotype of pancreatic cancer cell lines.
Pancreas. 39:425–435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang DD, Cui BB, Sun LY, et al: The
co-expression of USP22 and BMI-1 may promote cancer progression and
predict therapy failure in gastric carcinoma. Cell Biochem Biophys.
61:703–710. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang H, Li YP, Chen JH, et al: Prognostic
significance of USP22 as an oncogene in papillary thyroid
carcinoma. Tumour Biol. 34:1635–1639. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Yao L, Zhang X, et al: Elevated
expression of USP22 in correlation with poor prognosis in patients
with invasive breast cancer. J Cancer Res Clin Oncol.
137:1245–1253. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hu J, Liu YL, Piao SL, Yang DD, Yang YM
and Cai L: Expression patterns of USP22 and potential targets
BMI-1, PTEN, p-AKT in non-small-cell lung cancer. Lung Cancer.
77:593–599. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li J, Wang Z and Li Y: USP22 nuclear
expression is significantly associated with progression and
unfavorable clinical outcome in human esophageal squamous cell
carcinoma. J Cancer Res Clin Oncol. 138:1291–1297. 2012. View Article : Google Scholar
|
|
38
|
Liu Y, Yang Y, Xu H and Dong X:
Implication of USP22 in the regulation of BMI-1, c-Myc, p16INK4a,
p14ARF, and cyclin D2 expression in primary colorectal carcinomas.
Diagn Mol Pathol. 19:194–200. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nagy Z, Riss A, Romier C, et al: The human
SPT20-containing SAGA complex plays a direct role in the regulation
of endoplasmic reticulum stress-induced genes. Mol Cell Biol.
29:1649–1660. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nagy Z and Tora L: Distinct
GCN5/PCAF-containing complexes function as co-activators and are
involved in transcription factor and global histone acetylation.
Oncogene. 26:5341–5357. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang XY, Pfeiffer HK, Thorne AW and
McMahon SB: USP22, an hSAGA subunit and potential cancer stem cell
marker, reverses the polycomb-catalyzed ubiquitylation of histone
H2A. Cell Cycle. 7:1522–1524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Atanassov BS and Dent SY: USP22 regulates
cell proliferation by deubiquitinating the transcriptional
regulator FBP1. EMBO Rep. 12:924–930. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fu Z, Malureanu L, Huang J, et al:
Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional
programme required for mitotic progression. Nat Cell Biol.
10:1076–1082. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Anders L, Ke N, Hydbring P, et al: A
systematic screen for CDK4/6 substrates links FOXM1 phosphorylation
to senescence suppression in cancer cells. Cancer Cell. 20:620–634.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang X, Bhattacharyya D, Dennewitz MB, et
al: Rapid hepatocyte nuclear translocation of the Forkhead Box M1B
(FoxM1B) transcription factor caused a transient increase in size
of regenerating transgenic hepatocytes. Gene Expr. 11:149–162.
2003. View Article : Google Scholar
|
|
46
|
Wang IC, Chen YJ, Hughes D, et al:
Forkhead box M1 regulates the transcriptional network of genes
essential for mitotic progression and genes encoding the SCF
(Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 25:10875–10894. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Allende-Vega N and Saville MK: Targeting
the ubiquitin-proteasome system to activate wild-type p53 for
cancer therapy. Semin Cancer Biol. 20:29–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Clurman BE, Sheaff RJ, Thress K, Groudine
M and Roberts JM: Turnover of cyclin E by the ubiquitin-proteasome
pathway is regulated by cdk2 binding and cyclin phosphorylation.
Genes Dev. 10:1979–1990. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bae Y, Choi D, Rhim H and Kang S: Hip2
interacts with cyclin B1 and promotes its degradation through the
ubiquitin proteasome pathway. FEBS Lett. 584:4505–4510. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pagano M, Tam SW, Theodoras AM, et al:
Role of the ubiquitin-proteasome pathway in regulating abundance of
the cyclin-dependent kinase inhibitor p27. Science. 269:682–685.
1995. View Article : Google Scholar
|
|
51
|
Ding GX, Liu J, Feng CC, Jiang HW, Xu JF
and Ding Q: Slug regulates Cyclin D1 expression by
ubiquitin-proteasome pathway in prostate cancer cells. Panminerva
Med. 54:219–223. 2012.PubMed/NCBI
|
|
52
|
Mandal S, Freije WA, Guptan P and Banerjee
U: Metabolic control of G1-S transition: cyclin E degradation by
p53-induced activation of the ubiquitin-proteasome system. J Cell
Biol. 188:473–479. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang IC, Chen YJ, Hughes DE, et al: FoxM1
regulates transcription of JNK1 to promote the G1/S transition and
tumor cell invasiveness. J Biol Chem. 283:20770–20778. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nakamura S, Hirano I, Okinaka K, et al:
The FOXM1 transcriptional factor promotes the proliferation of
leukemia cells through modulation of cell cycle progression in
acute myeloid leukemia. Carcinogenesis. 31:2012–2021. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Priller M, Poschl J, Abrao L, et al:
Expression of FoxM1 is required for the proliferation of
medulloblastoma cells and indicates worse survival of patients.
Clin Cancer Res. 17:6791–6801. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
He SY, Shen HW, Xu L, et al: FOXM1
promotes tumor cell invasion and correlates with poor prognosis in
early-stage cervical cancer. Gynecol Oncol. 127:601–610. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wierstra I and Alves J: FOXM1c
transactivates the human c-myc promoter directly via the two TATA
boxes P1 and P2. FEBS J. 273:4645–4667. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
De Boer VC, de Goffau MC, Arts IC, Hollman
PC and Keijer J: SIRT1 stimulation by polyphenols is affected by
their stability and metabolism. Mech Ageing Dev. 127:618–627.
2006.PubMed/NCBI
|
|
59
|
Zhu GY, Shi BZ and Li Y: FoxM1 regulates
Sirt1 expression in glioma cells. Eur Rev Med Pharmacol Sci.
18:205–211. 2014.PubMed/NCBI
|
|
60
|
Jiang L, Li J and Song L: Bmi-1, stem
cells and cancer. Acta Biochim Biophys Sin (Shanghai). 41:527–534.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu YL, Jiang SX, Yang YM, Xu H, Liu JL
and Wang XS: USP22 acts as an oncogene by the activation of
BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem
Biophys. 62:229–235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Li SK, Smith DK, Leung WY, et al: FoxM1c
counteracts oxidative stress-induced senescence and stimulates
Bmi-1 expression. J Biol Chem. 283:16545–16553. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Katoh M and Katoh M: WNT signaling pathway
and stem cell signaling network. Clin Cancer Res. 13:4042–4045.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Heiser PW, Lau J, Taketo MM, Herrera PL
and Hebrok M: Stabilization of beta-catenin impacts pancreas
growth. Development. 133:2023–2032. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Rulifson IC, Karnik SK, Heiser PW, et al:
Wnt signaling regulates pancreatic beta cell proliferation. Proc
Natl Acad Sci USA. 104:6247–6252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Heiser PW, Cano DA, Landsman L, et al:
Stabilization of beta-catenin induces pancreas tumor formation.
Gastroenterology. 135:1288–1300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang Y, Morris JT, Yan W, et al:
Canonical wnt signaling is required for pancreatic carcinogenesis.
Cancer Res. 73:4909–4922. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ripka S, Konig A, Buchholz M, et al: WNT5A
- target of CUTL1 and potent modulator of tumor cell migration and
invasion in pancreatic cancer. Carcinogenesis. 28:1178–1187. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Griesmann H, Ripka S, Pralle M, et al:
WNT5A-NFAT signaling mediates resistance to apoptosis in pancreatic
cancer. Neoplasia. 15:11–22. 2013.PubMed/NCBI
|
|
70
|
Bowman A and Nusse R: Location, location,
location: FoxM1 mediates beta-catenin nuclear translocation and
promotes glioma tumorigenesis. Cancer Cell. 20:415–416. 2011.
View Article : Google Scholar : PubMed/NCBI
|