Introduction
Lung cancer is the leading cause of cancer death in
Japan and worldwide (1). The
majority of lung cancers are non-small cell lung cancers (NSCLC)
that include adenocarcinomas (ADC) and squamous cell carcinomas
(SQCC). Recently, oncogenic driver mutations including EGFR gene
mutation and ALK fusion gene have been found in the majority of ADC
(2–4). Recent randomized trials of gefitinib,
erlotinib and crizotinib have demonstrated the significant
superiority of these molecular targeted drugs on progression-free
survival compared with standard chemotherapies as the key agents
for the treatment of advanced ADC with driver mutations (5–7).
However, oncogenic driver mutations are not typically present in
SQCC. Molecular targeted agents developed for lung ADC are largely
ineffective against lung SQCC. The standard of care for advanced
SQCC patients is still cytotoxic chemotherapy. Unfortunately,
modern cytotoxic and molecular targeted agents, pemetrexed and
bevacizumab, are not recommended for patients with SQCC (8,9).
Therefore, identification of biomarkers predictive of drug
sensitivity and personalized therapies using the biomarkers could
have a clinically significant impact on treatment strategies for
SQCC.
Surgical resection is the standard treatment for
early-stage NSCLC. Adjuvant chemotherapy after surgery is
recognized as a standard therapy for resected stage IB, II and IIIA
NSCLC. Three large randomized clinical trials have demonstrated a
survival benefit for adjuvant cisplatin (CDDP)-based chemotherapy
compared with surgery alone in patients who underwent surgery for
NSCLC (10–12). While adjuvant chemotherapy has
improved survival for patients with early-stage NSCLC, the
prognosis of NSCLC after recurrence remains poor, in particular
SQCC. Numerous promising biomarkers are currently being evaluated
in the adjuvant setting. Among the molecular markers used as
potential predictors of a survival benefit from adjuvant
chemotherapy, ERCC1 and RRM1 have been reported (13,14).
However, it is controversial whether ERCC1 predicts the prognosis
of lung cancer treated with CDDP-based chemotherapy (15). Molecular biomarkers for adjuvant
therapy decision should be identified in NSCLC patients, in
particular SQCC patients. Patient selection based on the presence
of prognostic or predictive biomarkers offers the potential to
improve survival in SQCC patients.
Recent human genome studies have demonstrated the
possibility of alteration of several genes as oncogenic driver
mutations of SQCC (16–22). Multiplex testing for driver
mutations in SQCC of the lung (SQ-MAP) using specimens from 40 SQCC
patients was reported in 2012 (17). FGFR1 amplification was found in 25%
and PIK3CA mutation was observed in 11% of SQCC tumors by SQ-MAP.
Based on the SQ-MAP, two approved clinical trials using FGFR1
inhibition and PI3K inhibition have begun. In addition, the Cancer
Genome Atlas Research reported that overexpression and
amplification of SOX2 were observed in 21% and PIK3CA gene
alteration was found in 16% of 178 lung SQCC samples (18). Chromosome 3q26 is frequently
amplified in lung SQCC, showing the most frequent overexpression of
two known oncogenes on 3q26: SOX2 and PIK3CA (20,21).
Based on these findings, FGFR1, PIK3CA and SOX2 have been
recognized as candidate driver genes of SQCC. However, the
association between these genes including FGFR1, PIK3CA and SOX2
and patients’ prognosis has not been clarified.
In the present study, we evaluated whether three
proteins, FGFR1, PIK3CA and SOX2, could be used as prognostic
factors in SQCC patients from three country cohorts by
immunohistochemical analysis (IHC). We ultimately found that the
combination of PIK3CA and SOX2 protein expression could be useful
for the prognosis of Asian patients with SQCC, in particular stage
I patients. Our prediction criteria may be applicable to selection
of SQCC patients who would benefit from adjuvant chemotherapy.
Materials and methods
Tissue microarray and clinical
samples
Commercially available paraffin-embedded sections of
the tissue microarray (TMA) of Chinese patients (OD-CT-RsLug01-009;
Shanghai Outdo Biotech Co., Ltd., Shanghai, China) was used as the
training set (Table I).
Fifty-seven SQCC specimens were available in this array. Another
TMA from American TA116 (TriStar, Rockville, MD, USA) was used as
the validation set. Fifty-two SQCC specimens were available in this
array.
 | Table IImmunohistochemical analysis of
Chinese SQCC specimens. |
Table I
Immunohistochemical analysis of
Chinese SQCC specimens.
| | | FGFR1
expression | | PIK3CA
expression | | SOX2
expression | |
|---|
| | |
| |
| |
| |
|---|
| | | Positive | Negative | | Positive | Negative | | Positive | Negative | |
|---|
| | |
|
| |
|
| |
|
| |
|---|
| Variables | N | % | N | % | N | % | P-value | Nl | % | N | % | P-value | N | % | N | % | P-value |
|---|
| Total | 57 | | 6 | | 51 | | | 45 | | 12 | | | 40 | | 17 | | |
| Gender |
| Male | 53 | 93 | 3 | 50 | 50 | 98 | | 41 | 91 | 12 | 100 | | 37 | 93 | 16 | 94 | |
| Female | 4 | 7 | 3 | 50 | 1 | 2 | 0.003 | 4 | 9 | 0 | 0 | 1 | 3 | 8 | 1 | 6 | 1.00 |
| Age (years) |
| <65 | 23 | 40 | 4 | 67 | 19 | 37 | | 18 | 40 | 5 | 42 | | 16 | 40 | 7 | 41 | |
| ≥65 | 34 | 60 | 2 | 33 | 32 | 63 | 0.21 | 27 | 60 | 7 | 58 | 1 | 24 | 60 | 10 | 59 | 1.00 |
| Stage |
| I | 24 | 42 | 4 | 67 | 20 | 39 | | 19 | 42 | 5 | 42 | | 16 | 40 | 8 | 47 | |
| II+III | 33 | 58 | 2 | 33 | 31 | 61 | 0.23 | 26 | 58 | 7 | 58 | 1 | 24 | 60 | 9 | 53 | 0.77 |
| Grade |
| G1 | 7 | 12 | 2 | 33 | 5 | 10 | | 7 | 16 | 0 | 0 | | 4 | 10 | 3 | 18 | |
| G2+G3 | 50 | 88 | 4 | 67 | 46 | 90 | 0.15 | 38 | 84 | 12 | 100 | 0 | 36 | 90 | 14 | 82 | 0.42 |
| T factor |
| T1 | 5 | 9 | 3 | 50 | 2 | 4 | | 5 | 11 | 0 | 0 | | 5 | 13 | 0 | 0 | |
| T2+T3 | 52 | 91 | 3 | 50 | 49 | 96 | 0.006 | 40 | 89 | 12 | 100 | 1 | 35 | 88 | 17 | 100 | 0.31 |
| N factor |
| N0 | 34 | 60 | 5 | 83 | 29 | 57 | | 26 | 58 | 8 | 67 | | 25 | 63 | 9 | 53 | |
| N1+N2 | 23 | 40 | 1 | 17 | 22 | 43 | 0.39 | 19 | 42 | 4 | 33 | 1 | 15 | 38 | 8 | 47 | 0.56 |
We used a validation set of lung tumor tissue
samples from 66 Japanese patients with stage I–III SQCC who had
undergone surgical resection at the Nippon Medical School Hospital
from 2001 to 2008. All tissues were freshly collected during
surgery, snap-frozen and stored at −80°C. TNM stage, T factor, N
factor, Grade, Ly factor and V factor were classified according to
the World Health Organization TNM staging 7th edition. Information
on patient survival and recurrence during 5 years of follow-up was
available for all 66 cases (Table
II). Twenty-eight patients received adjuvant chemotherapy with
uracil-tegafur (UFT). The lung tissues from lung SQCC patients were
used only for immunohistochemical analysis. Immunohistochemical
staining of the lung cancer tissue samples was carried out in
accordance with the principles embodied in the Declaration of
Helsinki, 2008. All included patients provided written informed
consent for the use of their tissue specimens for medical
research.
 | Table IIAssociations between PIK3CA and SOX2
expression levels and the patient characteristics. |
Table II
Associations between PIK3CA and SOX2
expression levels and the patient characteristics.
| | | PIK3CA
expression | | SOX2
expression | |
|---|
| | |
| |
| |
|---|
| | | Positive | Negative | | Positive | Negative | |
|---|
| | |
|
| |
|
| |
|---|
| Variables | N | % | N | % | N | % | P-value | N | % | N | % | P-value |
|---|
| Total | 66 | | 10 | | 56 | | | 45 | | 21 | | |
| Age (years) | | | | | | | | | | | | |
| <65 | 13 | 20 | 2 | 20 | 11 | 20 | | 10 | 22 | 3 | 14 | |
| ≥65 | 53 | 80 | 8 | 80 | 45 | 80 | 0.71 | 35 | 78 | 18 | 86 | 1.00 |
| Gender | | | | | | | | | | | | |
| Male | 58 | 88 | 9 | 90 | 49 | 88 | | 42 | 93 | 16 | 76 | |
| Female | 8 | 12 | 1 | 10 | 7 | 13 | 1.00 | 3 | 7 | 5 | 24 | 0.10 |
| Smoking status | | | | | | | | | | | | |
| Ever-smoker | 60 | 91 | 9 | 90 | 51 | 91 | | 44 | 98 | 16 | 76 | |
| Never smoker | 6 | 9 | 1 | 10 | 5 | 9 | 1.00 | 1 | 2 | 5 | 24 | 0.01 |
| T factor | | | | | | | | | | | | |
| T1 | 17 | 26 | 3 | 30 | 14 | 25 | | 14 | 31 | 3 | 14 | |
| T2–4 | 49 | 74 | 7 | 70 | 42 | 75 | 0.71 | 31 | 69 | 18 | 86 | 0.23 |
| N factor | | | | | | | | | | | | |
| N0 | 41 | 62 | 7 | 70 | 34 | 61 | | 27 | 60 | 14 | 67 | |
| N1–2 | 25 | 38 | 3 | 30 | 22 | 39 | 0.73 | 18 | 40 | 7 | 33 | 0.79 |
| Stage | | | | | | | | | | | | |
| I | 32 | 48 | 6 | 60 | 26 | 46 | | 21 | 47 | 11 | 52 | |
| II–III | 36 | 55 | 4 | 40 | 30 | 54 | 0.51 | 24 | 53 | 10 | 48 | 0.79 |
| Grade | | | | | | | | | | | | |
| G1 | 10 | 15 | 2 | 20 | 8 | 14 | | 9 | 20 | 1 | 5 | |
| G2–3 | 56 | 85 | 8 | 80 | 48 | 86 | 0.64 | 36 | 80 | 20 | 95 | 0.15 |
| ly factor | | | | | | | | | | | | |
| ly0 | 17 | 26 | 3 | 30 | 14 | 25 | | 15 | 33 | 2 | 10 | |
| Ly1–3 | 49 | 74 | 7 | 70 | 42 | 75 | 0.71 | 30 | 67 | 19 | 90 | 0.07 |
| v factor | | | | | | | | | | | | |
| v0 | 26 | 39 | 3 | 30 | 23 | 41 | | 20 | 44 | 6 | 29 | |
| v1–3 | 40 | 61 | 7 | 70 | 33 | 59 | 0.73 | 25 | 56 | 15 | 71 | 0.28 |
| Adjuvant
chemotherapy (UFT) | | | | | | | | | | | | |
| Yes | 28 | 42 | 3 | 30 | 25 | 45 | 22 | 49 | 6 | 29 | | |
| No | 38 | 58 | 7 | 70 | 31 | 55 | 0.31 | 23 | 51 | 15 | 71 | 0.10 |
Immunohistochemistry
Immunohistochemical staining was performed on
paraffin-embedded sections. After deparaffinization, antigen
retrieval was carried out in 10 mmol/l citrate buffer (pH 6.0) (LSI
Medience Corp., Tokyo, Japan) using an autoclave. After blocking
swine serum albumin (1:50 dilution; Vector Laboratories Inc.,
Burlingame, CA, USA), the sections were washed and incubated with
rabbit anti-human FGFR1 polyclonal antibody (1:250 dilution; Abcam
Biochemicals, Cambridge, MA, USA) or with rabbit anti-human PIK3CA
polyclonal antibody (1:50 dilution; Santa Cruz Biotechnology,
Dallas, TX, USA) or with rabbit anti-human SOX2 polyclonal antibody
(1:500 dilution; Millipore, Bedford, MA, USA) at 4°C overnight.
After washing, they were incubated with biotinylated goat
anti-rabbit IgG (1:200 dilution; Vector Laboratories) for 30 min.
Finally, they were incubated with avidin-biotin complex kit
(Funakoshi, Tokyo, Japan). Negative controls were prepared by
omitting the primary antibody under the same experimental
conditions.
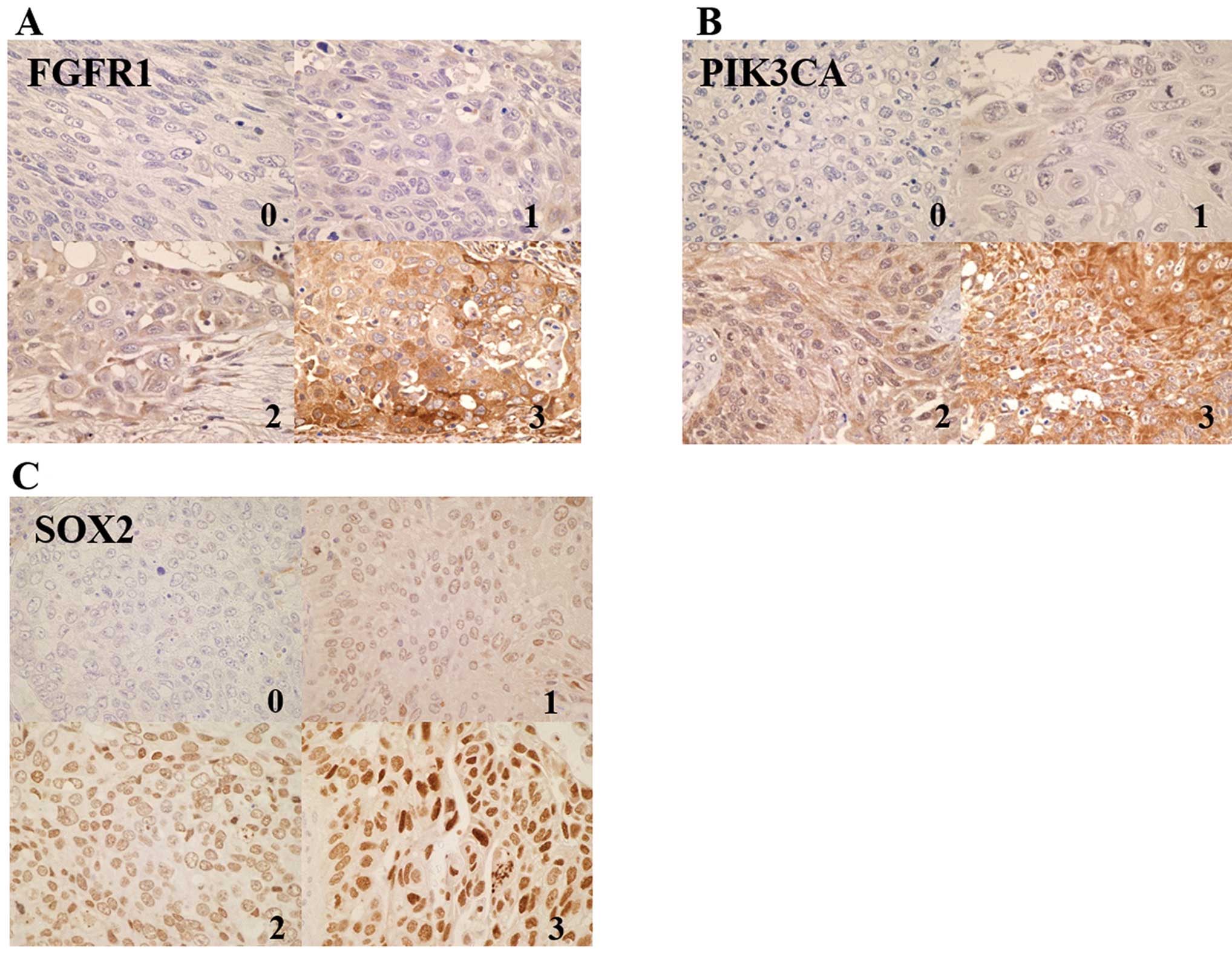
Evaluation of FGFR1, SOX2 and PIK3CA
protein expression
Immunohistochemical scoring was performed using the
Histoscore (H-score) (23). FGFR1
expression level was scored on a scale as follows: no expression
(0), low expression according to a previous study (1+) and high
expression (2+ and 3+). PIK3CA staining was scored on a scale as
follows according to a previous study (24): 0, no staining; 1+, staining
<50%; 2+, ≥50% with weak intensity: 3+, ≥50% with strong
intensity. Nuclear expression of SOX2 protein was scored
semiquantitatively by the combination of intensity (scored as 0, no
staining; 1, weak staining; 2, moderate staining; 3, strong
staining) and proportion of positively stained tumor cells in 5
high power fields (scored as 0, <5%; 1, 5–25%; 2, 26–50%; 3,
51–75%; 4, >75%). The sum of the staining intensity score and
score on the percentage of positive tumor cells was graded as
follows: −, 0–1; 1+, 2–3; 2+, 4–5; and 3+, 6–7. The results of IHC
were judged independently by two investigators (Y.I. and R.N.) who
were unaware of the clinical data, and consensus was reached for
any discordant cases.
Statistical analyses
Correlations between protein expression and
characteristics were assessed by the Fisher’s exact test. Overall
survival (OS) was calculated from the date of surgery. Kaplan-Meier
survival curves were drawn for OS and compared by log-rank test.
Univariate and multivariate analyses were performed using the COX
regression model. Statistical significance was set at P<0.05 for
each analysis. All statistical analyses were carried using the IBM
SPSS Statistics version 21 (IBM SPSS, Inc., Armonk, NY, USA).
Results
TMA analysis of Chinese patients
Fifty-seven Chinese SQCC specimens were available
for IHC analysis. Six specimens (11%) were observed to be
FGFR1-positive (Table I, Fig. 1A). Forty-five (82%) of 57 SQCC
patients were positive for PIK3CA (Table I, Fig.
1B). High SOX2 expression was observed in 40 (70%) of 57 SQCC
specimens (Table I, Fig. 1C). The results of correlation
analysis between FGFR1, PIK3CA and SOX2 protein expression and
patients’ characteristics are summarized in Table I. FGFR1 expression status was found
to be related to gender and T factors.
Association between FGFR1, PIK3CA and
SOX2 expression and prognosis in Chinese SQCC specimens
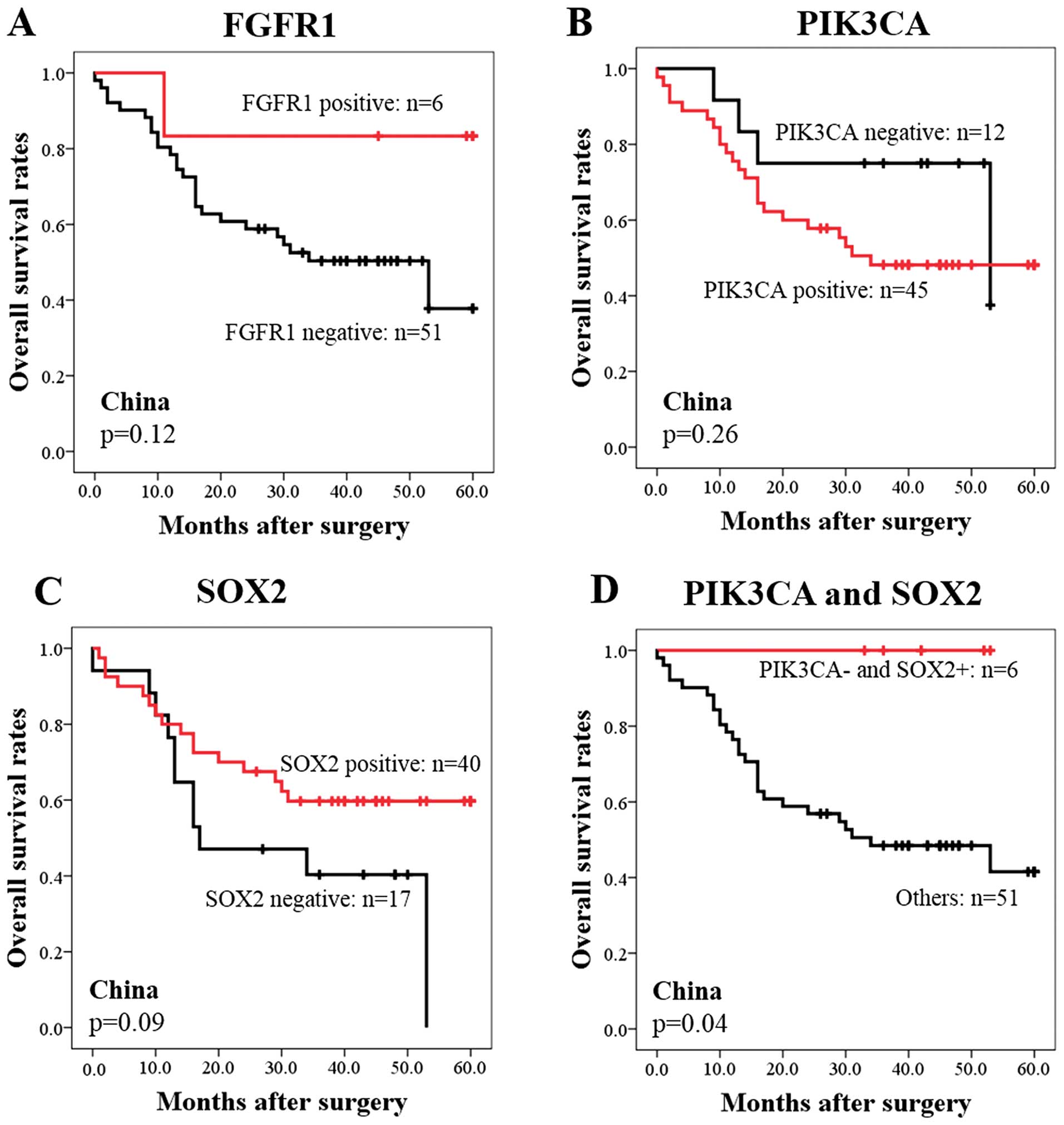
We next evaluated the prognostic significance of the
expression of three proteins by IHC. FGFR1-positive or
PIK3CA-negative or SOX2-positive status showed trends toward better
survival, although the differences were not statistically
significant (P=0.12, 0.26, and 0.09, respectively) (Fig. 2A–C). Therefore, we next used a
combination of expression of two proteins by IHC to improve the
prognostic classification of SQCC patients. Among all combinations,
the combination of PIK3CA and SOX2 was the best classification
distinguishing good prognosis from poor prognosis. Patients with
PIK3CA-negative and SOX2-positive staining
(PIK3CA−/SOX2+) showed good prognosis
compared to those with PIK3CA-positive or SOX2-negative staining
(Fig. 2D). The 5-year survival
rate among the cases was 100% (Fig.
2D). In contrast, those with PIK3CA-positive or SOX2-negative
staining had significantly worse survival than the low-risk
PIK3CA−/SOX2+ group (P=0.04) (Fig. 2D). The 5-year survival rate was 42%
among the cases in the PIK3CA-positive or SOX2-negative group
(Fig. 2D).
Validation of the prognostic significance
of SOX2 and PIK3CA
We next examined the robustness of the
PIK3CA−/SOX2+ immunohistological profile for
classifying patients into prognostic groups in two independent sets
of specimens from American TMA and 66 Japanese SQCC patients who
had undergone surgical resection. Unfortunately, the prognostic
significance of PIK3CA−/SOX2+ could not be
validated in the USA cohort, because only 1 of the 52 USA cases was
PIK3CA−/ SOX2+. In the Japanese cohort,
negative immunoreactions of PIK3CA were found in 56 (85%) of 66
cases and positive immunoreactions of SOX2 were observed in 45
(68%) of 66 cases (Table II). The
associations between PIK3CA and SOX2 expression levels and the
patient characteristics are shown in Table II. SOX2 expression status was
related to smoking status (P=0.01).
The 5-year survival rates were 54% among patients
with PIK3CA-negative and 30% among patients with PIK3CA-positive
status (data not shown). The 5-year survival rates were 56% among
patients with SOX2-positive and 38% among patients with
SOX2-negative status (data not shown). However, these differences
were not statistically significant (P=0.13 and P=0.12,
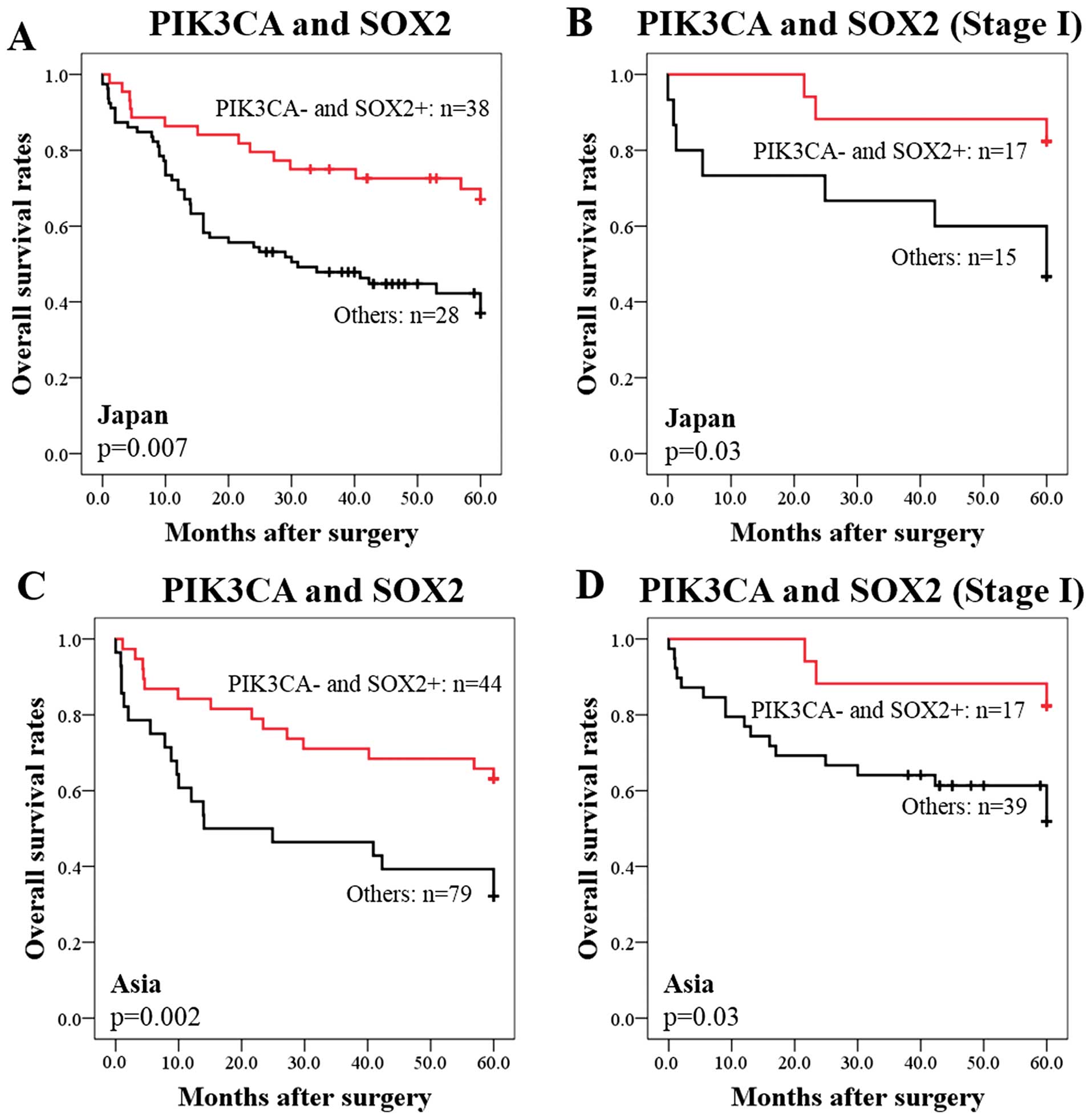
respectively). Kaplan-Meier survival analysis showed that the 38
PIK3CA−/SOX2+ cases had significantly
favorable survival than the other 28 cases (P=0.007) (Fig. 3A). The 5-year survival rates were
63% among patients with PIK3CA−/SOX2+ and 32%
among the other patients (Fig.
3A). Furthermore, among the 32 patients with stage I SQCC, the
PIK3CA−/SOX2+ cases (n=17) had significantly
better survival than the group with PIK3CA-positive or
SOX2-negative status (n=15) (P=0.03) (Fig. 3B). The 5-year survival rates were
82% among the PIK3CA−/SOX2+ cases and 47%
among the group with PIK3CA positive or SOX2-negative status
(Fig. 3B). Among the 123 Asian
(Chinese and Japanese) patients with SQCC, the group with
PIK3CA-positive or SOX2-negative status (n=79) had significantly
poorer survival than the PIK3CA−/SOX2+ cases
(n=44) (P=0.002) (Fig. 3C). The
5-year survival rates were 70% among the
PIK3CA−/SOX2+ cases and 40% among the group
with PIK3CA-positive or SOX2-negative status (Fig. 3C). Furthermore, among 56 Asian
stage I patients, the 5-year survival rates were 82% among the
PIK3CA−/SOX2+ cases and 52% among the group
with PIK3CA-positive or SOX2-negative status (P=0.03) (Fig. 3D). Notably, this classification
correctly predicted poor survival in 16 (89%) of the 18 stage I
cases who had died during the 5-year follow-up period (Fig. 3D).
Univariate analysis and multivariate
analysis
Finally, we investigated whether the prognostic
ability of PIK3CA+/SOX2− expression was
affected by underlying clinical covariates by performing univariate
and multivariable Cox proportional hazards survival analyses.
Univariate analysis of the 123 Asian patients revealed that age, T
factor, N factor, p-stage and PIK3CA−/SOX2+
classification (HR for death in the high-risk PIK3CA-positive or
SOX2-negative group compared with the low-risk reference
PIK3CA−/SOX2+ group = 2.47; 95% CI,
1.35–4.51; P=0.003) were statistically significant predictors of
survival (Table III).
Multivariable analysis of the 123 patients, adjusted for age, T
factor, N-factor, p-stage and PIK3CA/SOX2 classification, showed
that age, N factor and PIK3CA/SOX2 classification were
statistically significant (HR, 1.99, 2.98 and 2.50; 95% CI;
1.05–3.77, 1.23–7.22 and 1.35–4.65; P=0.04, 0.02 and 0.004,
respectively). We also performed univariate and multivariable
survival analyses of the 56 Asian patients with stage I. Univariate
analysis revealed that PIK3CA/SOX2 classification (HR for death in
the PIK3CA-positive or SOX2-negative group compared with the
low-risk reference group = 3.51; 95% CI, 1.02–12.08; P<0.05) was
significantly associated with survival (Table III). In the multivariable Cox
proportional hazards model of Asian stage I patients, the
PIK3CA/SOX2 classification was significantly associated with
survival and was an independent prognostic factor among the stage I
cases (HR for death in the high-risk PIK3CA/SOX2 expression group
compared with the low-risk reference group = 4.23; 95% CI,
1.17–15.4; P=0.03). The combination of PIK3CA and SOX2 expression
identified Asian stage I lung SQCC patients who have a poor
prognosis.
 | Table IIIUnivariate and multivariable Cox
proportional hazards models of factors associated with death for
all Asian patients and for the stage I patients. |
Table III
Univariate and multivariable Cox
proportional hazards models of factors associated with death for
all Asian patients and for the stage I patients.
| | Univariate
analysis | Multivariate
analysis |
|---|
| |
|
|
|---|
|
Characteristics | Comparison | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| All cases
(n=123) |
| Age (years) | <65 vs. ≥65 | 1.93 | 1.02–3.62 | 0.04 | 1.99 | 1.05–3.77 | 0.04 |
| Gender | Male vs.
female | 1.24 | 0.50–3.11 | 0.65 | | | |
| T factor | T1 vs. T2–4 | 2.65 | 1.13–6.20 | 0.03 | | | 0.37 |
| N factor | N0 vs. N1–2 | 3.01 | 1.79–5.07 | <0.001 | 2.98 | 1.23–7.22 | 0.02 |
| p-Stage | I vs. II–III | 2.19 | 1.28–3.76 | 0.005 | | | 0.88 |
| Grade | G1 vs. G2+G3 | 0.76 | 0.39–1.50 | 0.43 | | | |
| SOX2/PIK3CA
expression |
SOX2+/PIK3CA− vs.
others | 2.47 | 1.35–4.51 | 0.003 | 2.50 | 1.35–4.65 | 0.005 |
| Stage I cases
(n=56) |
| Age (years) | <65 vs. ≥65 | 2.09 | 0.70–6.25 | 0.18 | | | 0.08 |
| Gender | Male vs.
female | 1.35 | 0.31–5.83 | 0.69 | | | 0.68 |
| T factor T1
vs. | T2–4 | 1.50 | 0.57–3.94 | 0.41 | | | 0.94 |
| Grade G1 vs. | G2+G3 | 1.67 | 0.39–7.20 | 0.49 | | | 0.57 |
| SOX2/PIK3CA
expression |
SOX2+/PIK3CA− vs.
others | 3.51 | 1.02–12.08 | 0.05 | 4.23 | 1.17–15.37 | 0.03 |
Discussion
In the present study, we examined the prognostic
significance of FGFR1, PIK3CA and SOX2 protein expression by IHC in
SQCC. We ultimately developed a prognostic model which was based on
the combination status of PIK3CA/SOX2 expression to predict the
prognosis of Asian patients with stage I SQCC. PIK3CA and SOX2
genes are located on chromosome 3q26. Recent whole genomic studies
indicated frequent amplification at chromosome 3q26 in SQCC,
suggesting a potential critical role of this chromosome in
carcinogenesis and progression in SQCC (20,21).
PIK3CA regulates the phosphatidylinositol 3-kinase
(PI3K)/Akt signaling pathway which is critical for cell survival of
human cancer (25–27). PIK3CA gene alteration was found in
16% of lung SQCC samples by The Cancer Genome Atlas, resulting in
PI3K pathway alterations (17). In
addition, PIK3CA gene alteration was more frequently observed in
lung SQCC than in ADC (28,29).
In Asian patients, PIK3CA copy number gains by FISH were found in
43% of Japanese SQCC patients (29). PIK3CA amplification was also
observed in 42% of Chinese SQCC patients (30). These results are consistent with
comparative genomic hybridization studies of SQCC showing
consistent gains in chromosome 3q26 including the PIK3CA gene. The
association between PIK3CA gene alteration and the patient
prognosis has not been clarified; however, patients with PIK3CA
gene mutation tended to have an unfavorable prognosis (29). In the present study, patients with
PIK3CA-negative status showed a trend toward better survival
compared with patients with PIK3CA-positive staining. These
findings suggest that PIK3CA alteration may be involved in the
process of carcinogenesis of SQCC and contribute to the prognosis
of SQCC.
SOX2 is a transcription factor essential for early
mammalian development and maintenance of stem cells in multiple
adult tissues (31). SOX2 is also
important for maintenance of lung epithelium cells, Clara cells,
ciliated cells, and goblet cells which have known roles in squamous
cell differentiation (32). SOX2
and TP63 play roles in squamous cell differentiation. SOX2-positive
and p63-negative immunohistochemical staining correlated with
high-grade histology across NSCLC subtypes (33). Recently, SOX2 has been shown as an
oncogene activated by 3q26.3 amplification in lung SQCC (20,34).
The Cancer Genome Atlas Research reported that overexpression and
amplification of SOX2 were observed in 21% of SQCC samples
(17). SOX2 was also highly
overexpressed in lung SQCC when compared with ADC by IHC (31). As to prognostic significance, SOX2
amplification or overexpression showed a trend toward better
survival (31,35). SOX2-positive status showed a trend
toward better prognosis in this study, which is consistent with
findings from previous studies. The association between high SOX2
expression and favorable prognosis is unclear; however, SOX2 might
be implicated in the control of cell differentiation toward the
formation of squamous metaplasia and well-differentiated SQCC
(36,37). Low SOX2 expression tended to be
associated with poorly-differentiated SQCC tumors (38). In our analysis using the Japanese
cohort, 9 of the 10 Grade 1 SQCC cases showed SOX2-positive
expression. Therefore, the role of SOX2 in squamous cell
differentiation might affect the prognosis of SQCC patients.
A limitation of this study is validation of the
American cohort. The prognostic significance of
PIK3CA−/SOX2+ could not be confirmed in the
American cohort, because there was only one patient with
PIK3CA−/SOX2+. The frequency of driver
mutations seems to be different in Asian and Caucasian lung AC
patients. EGFR mutations are found at different frequencies in
Caucasian and East Asian patients with lung AC (39). Although ethnic differences in the
frequency of driver mutations in lung SQCC were not clarified, the
differences seem to contribute to the incidence and prognosis of
lung SQCC.
In conclusion, we found that SQCCs with
PIK3CA−/SOX2+ showed a trend towards better
survival in Japanese and Chinese SQCC patient cohorts. Notably,
PIK3CA/SOX2 classification, which was based on IHC expression, was
useful for identifying stage I SQCC patients who are at high risk
of death. Patients in the high-risk stage I group classified by
PIK3CA/SOX2 expression pattern may be candidates for adjuvant
therapy in SQCC. Therefore, patient selection based on PIK3CA/SOX2
expression offers the potential to improve survival in SQCC
patients. The significance of PIK3CA/ SOX2 classification will be
further validated in a large-scale SQCC cohort.
Acknowledgements
The present study was supported in part by a
grant-in-aid from the Ministry of Education, Culture, Sports,
Science, and Technology of Japan (grant no. 24591179 to M.S; grant
no. 25461172 to A.G), and the Clinical Rebiopy Bank Project for
Comprehensive Cancer Therapy Development in Nippon Medical
School.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soda M, Choi YL, Enomoto M, et al:
Identification of the transforming EML4-ALK fusion gene in
non-small-cell lung cancer. Nature. 448:561–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maemondo M, Inoue A, Kobayashi K, et al:
Gefitinib or chemotherapy for non-small-cell lung cancer with
mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitsudomi T, Morita S, Yatabe Y, et al:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): an open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
7
|
Shaw AT, Kim DW, Nakagawa K, et al:
Crizotinib versus chemotherapy in advanced ALK-positive lung
cancer. N Engl J Med. 368:2385–2394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scagliotti GV, Parikh P, von Pawel J, et
al: Phase III study comparing cisplatin plus gemcitabine with
cisplatin plus pemetrexed in chemotherapy-naive patients with
advanced-stage non-small-cell lung cancer. J Clin Oncol.
26:3543–3551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sandler A, Gray R, Perry MC, et al:
Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell
lung cancer. N Engl J Med. 355:2542–2550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arriagada R, Bergman B, Dunant A, Le
Chevalier T, Pignon JP and Vansteenkiste J: Cisplatin-based
adjuvant chemotherapy in patients with completely resected
non-small-cell lung cancer. N Engl J Med. 350:351–360. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Winton T, Livingston R, Johnson D, et al:
Vinorelbine plus cisplatin vs. observation in resected
non-small-cell lung cancer. N Engl J Med. 352:2589–2597. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Douillard JY, Rosell R, De Lena M, et al:
Adjuvant vinorelbine plus cisplatin versus observation in patients
with completely resected stage IB-IIIA non-small-cell lung cancer
(Adjuvant Navelbine International Trialist Association (ANITA)): a
randomised controlled trial. Lancet Oncol. 7:719–727. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Olaussen KA, Fouret P and Kroemer G:
ERCC1-specific immunostaining in non-small-cell lung cancer. N Engl
J Med. 357:1559–1561. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng Z, Chen T, Li X, Haura E, Sharma A
and Bepler G: DNA synthesis and repair genes RRM1 and ERCC1 in lung
cancer. N Engl J Med. 356:800–808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Friboulet L, Olaussen KA, Pignon JP, et
al: ERCC1 isoform expression and DNA repair in non-small-cell lung
cancer. N Engl J Med. 368:1101–1110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Drilon A, Rekhtman N, Ladanyi M and Paik
P: Squamous-cell carcinomas of the lung: emerging biology,
controversies, and the promise of targeted therapy. Lancet Oncol.
13:e418–e426. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Paul KP, Adnan H, Lu W, Natasha R, Marc L
and Mark GK: Multiplex testing for driver mutations in squamous
cell carcinomas of the lung. J Clin Oncol. 30(Suppl): 75052012.
|
|
18
|
The Cancer Genome Atlas Research Network.
Comprehensive genomic characterization of squamous cell lung
cancers. Nature. 489:519–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim TJ, Lee JW, Song SY, et al: Increased
expression of pAKT is associated with radiation resistance in
cervical cancer. Br J Cancer. 94:1678–1682. 2006.PubMed/NCBI
|
|
20
|
Yamamoto H, Shigematsu H, Nomura M, et al:
PIK3CA mutations and copy number gains in human lung cancers.
Cancer Res. 68:6913–6921. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hussenet T, Dali S, Exinger J, et al: SOX2
is an oncogene activated by recurrent 3q26.3 amplifications in
human lung squamous cell carcinomas. PLoS One. 5:e89602010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hammerman PS, Sos ML, Ramos AH, et al:
Mutations in the DDR2 kinase gene identify a novel therapeutic
target in squamous cell lung cancer. Cancer Discov. 1:78–89. 2011.
View Article : Google Scholar
|
|
23
|
McCarty KS Jr, Miller LS, Cox EB, Konrath
J and McCarty KS Sr: Estrogen receptor analyses. Correlation of
biochemical and immunohistochemical methods using monoclonal
antireceptor antibodies. Arch Pathol Lab Med. 109:716–721.
1985.PubMed/NCBI
|
|
24
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Samuels Y, Wang Z, Bardelli A, et al: High
frequency of mutations of the PIK3CA gene in human cancers.
Science. 304:5542004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Karakas B, Bachman KE and Park BH:
Mutation of the PIK3CA oncogene in human cancers. Br J Cancer.
94:455–459. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Campbell IG, Russell SE, Choong DY, et al:
Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer
Res. 64:7678–7681. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Massion PP, Kuo WL, Stokoe D, et al:
Genomic copy number analysis of non-small cell lung cancer using
array comparative genomic hybridization: implications of the
phosphatidylinositol 3-kinase pathway. Cancer Res. 62:3636–3640.
2002.PubMed/NCBI
|
|
29
|
Kawano O, Sasaki H, Endo K, et al: PIK3CA
mutation status in Japanese lung cancer patients. Lung Cancer.
54:209–215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ji M, Guan H, Gao C, Shi B and Hou P:
Highly frequent promoter methylation and PIK3CA amplification in
non-small cell lung cancer (NSCLC). BMC Cancer. 11:1472011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu Y, Futtner C, Rock JR, et al: Evidence
that SOX2 overexpression is oncogenic in the lung. PLoS One.
5:e110222010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tompkins DH, Besnard V, Lange AW, et al:
Sox2 is required for maintenance and differentiation of bronchiolar
Clara, ciliated, and goblet cells. PLoS One. 4:e82482009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sholl LM, Long KB and Hornick JL: Sox2
expression in pulmonary non-small cell and neuroendocrine
carcinomas. Appl Immunohistochem Mol Morphol. 18:55–61. 2010.
View Article : Google Scholar
|
|
34
|
Bass AJ, Watanabe H, Mermel CH, et al:
SOX2 is an amplified lineage-survival oncogene in lung and
esophageal squamous cell carcinomas. Nat Genet. 41:1238–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Velcheti V, Schalper K, Yao X, et al: High
SOX2 levels predict better outcome in non-small cell lung
carcinomas. PLoS One. 8:e614272013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brcic L, Sherer CK, Shuai Y, Hornick JL,
Chirieac LR and Dacic S: Morphologic and clinicopathologic features
of lung squamous cell carcinomas expressing Sox2. Am J Clin Pathol.
138:712–718. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen S, Xu Y, Chen Y, et al: SOX2 gene
regulates the transcriptional network of oncogenes and affects
tumorigenesis of human lung cancer cells. PLoS One. 7:e363262012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hussenet T and du Manoir S: SOX2 in
squamous cell carcinoma: amplifying a pleiotropic oncogene along
carcinogenesis. Cell Cycle. 9:1480–1486. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Toyooka S, Matsuo K, Shigematsu H, et al:
The impact of sex and smoking status on the mutational spectrum of
epidermal growth factor receptor gene in non small cell lung
cancer. Clin Cancer Res. 13:5763–5768. 2007. View Article : Google Scholar : PubMed/NCBI
|