Introduction
The assessment of B-cell clonality by analysis of
immunoglobulin (Ig) gene rearrangements is an important tool in the
diagnosis of suspected B-cell proliferations, for which the results
of cyto/histopathological and immunophenotyping analysis are
inconclusive (1–5). The immunoglobulin heavy chain gene
(IGH) rearrangements have been the most frequent targets for
clonality analysis by polymerase chain reaction (PCR) in mature
B-cell proliferations (5).
However, this method fails to detect clonal rearrangements of the
IGH gene in a significant proportion of B-cell lymphomas.
The most possible reason for these false negative results could be
the process of somatic hypermutation (SHM) (4–8). SHM
is a cellular mechanism by which the immune system adapts B cell
receptors to recognize foreign antigens and to respond by
production of specific immunoglobulins. The SHM process takes place
in germinal center (GC) cells in the secondary lymphoid organs. As
a result of SHM, variable (VH) and joining
(JH) sequences of the rearranged VDJ exon of the
IGH gene are altered by single-nucleotide mutations or small
deletions or insertions of nucleotides. Thus, SHM can be
responsible for preventing primer annealing, which leads to
false-negative IGH-PCR results. The majority of B-cell
lymphomas arise from GC and post-GC cells potentially carrying the
risk of somatic mutations and lack of monoclonal IGH-PCR
result (1–3,5).
The immunoglobulin light chain genes represent an
alternative target for B-cell clonality assessment. It has been
shown that detection of clonal rearrangements in the immunoglobulin
κ light chain gene (IGK) can improve clonality detection
rates in mature B-cell lymphomas that are heavily somatically
mutated (4,5,9,10).
During normal B-cell differentiation, rearrangements of the
IGK gene start soon after the IGH gene
rearrangements, and are followed by rearrangements of the
immunoglobulin λ light chain gene (IGL). A functional
IGK rearrangement produces Vκ-Jκ product, and generates an
IGK+ B-cell. If a non-functional Vκ-Jκ product is
generated, IGK allele is inactivated through recombination
of the κ-deleting element (Kde). In a case of non-functional Vκ-Jκ
rearrangements on both alleles, rearrangements of the IGL gene take
place and generate an IGL+ B-cell. Thus, clonal Vκ-Jκ
rearrangements should be detected in IGK+ B-cell
lymphomas, and at least one clonal Kde rearrangement should be
detected in IGL+ B-cell lymphomas (5,11,12).
The applicative value of the IGK gene
rearrangement analysis in suspected B-cell lymphomas, particularly
in cases of germinal center (GC) and post-GC lymphomas has been
reported in a number of studies (4–8,13,14).
In our previous study, we evaluated the utility of standardized
BIOMED-2 clonality assay protocols for clonality analysis in a
routine diagnostical setting of non-Hodgkin lymphomas (15). In the aforementioned study, we used
only the assay protocol for the detection of clonal rearrangements
in the IGH gene for assessment of B-cell clonality (15).
The aim of the present study was to evaluate the
added value of standardized BIOMED-2 assay for detection of clonal
IGK gene rearrangements in the diagnostic setting of
suspected B-cell lymphomas using fresh and formalin-fixed
diagnostic specimens.
Materials and methods
Study group
Ninety-two specimens from 80 patients submitted for
routine diagnostics were evaluated in the present study. The study
group included 37 specimens from 32 patients with B-cell lymphoma.
Twenty-seven specimens were previously evaluated in our recently
published study (15) and 10
specimens of suspected B-cell lymphoma were analyzed during routine
diagnostic assessment from October through December 2013. In
addition to B-cell lymphomas, 26 specimens of T-cell lymphomas
(T-NHLs) and 29 specimens of reactive lymphoid proliferations were
also included in the study. Different types of diagnostic samples
were analyzed including 41 bone marrow (BM) aspirates, 25
fine-needle aspiration specimens (FNA), 22 formalin-fixed,
paraffin-embedded tissue specimens (FFPE), 3 pleural fluid and 1
ascites.
All specimens were subjected to the
cyto/histomorphological and immunophenotyping examination as well
as to the PCR-based clonality analysis of B-cell populations during
routine diagnostic assessment.
Final diagnosis
The final diagnosis of each lymphoid proliferation
was set upon careful evaluation of clinical, morphological,
immunophenotyping and molecular data. The diagnosis of lymphoma
subtype was made according to the WHO Classification of Tumours of
Hematopoietic and Lymphoid Tissues (16).
DNA isolation
DNA was isolated using commercial DNA isolation kits
according to the manufacturers’ protocols. The QIAamp FFPE Tissue
kit (Qiagen GmbH, Hilden, Germany) was used for FFPE tissue
specimens and High Pure PCR Template Preparation kit (Roche Applied
Science, Penzberg, Germany) was used for other types of specimens.
The concentration and the purity of DNA
(A260nm/A280nm) were determined using the
NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE,
USA).
B-cell clonality analysis
BIOMED-2 clonality assays
B-cell clonality was assessed using the BIOMED-2
clonality assays-ABI Fluorescence Detection (IdentiClone; InVivo
Scribe Technologies, San Diego, CA, USA) according to the
manufacturer’s instructions. Rearrangements in the immunoglobulin
heavy chain gene (IGH) were analyzed using the IdentiClone
IGH Gene clonality assay and rearrangements in the immunoglobulin κ
light chain gene (IGK) using the IdentiClone IGK Gene
clonality assay.
The DNA quality was checked for all samples using
the control gene PCR (Specimen Control Size Ladder Master Mix) and
only samples yielding control gene PCR products of ≥ 300 bp were
included in the study.
Based on the results of our previous study (15) the IGH clonality was
evaluated with three IGH multiplex PCR reactions for detection of
the complete rearrangements (VH-JH) in the
IGH gene. The IGK clonality was evaluated with two
IGK multiplex PCR reactions, one for the detection of the complete
rearrangements (Vκ-Jκ) and one for the
detection of Kde (κ deleting element) rearrangements in the
IGK gene. Each run included monoclonal and polyclonal
control DNAs for particular primer Master Mix, supplied with each
BIOMED-2 clonality assay, and a contamination control (no template
DNA in a reaction).
Analysis of IGH and IGK amplification
products
The fluorescently labeled PCR products were detected
by capillary gel electrophoresis using the ABI 3500 Genetic
Analyzer (Applied Biosystems, Foster City, CA, USA) and analyzed by
fragment analysis (GeneScan), which discriminates amplification
products according to their size (in bp). Fragment analysis was
performed using GeneScan™ 600LIZ® Size Standard v2.0
(Applied Biosystems). In case of the IGK clonality assay, PCR
products were also analyzed by heteroduplex analysis and
electrophoresis in non-denaturing polyacrylamide gels (HD-PAGE),
stained with ethidium bromide and visualized under UV light. The
HD-PAGE method discriminates amplification products according to
their size and nucleotide composition, and is useful in cases with
borderline (doubtful) GS results. Amplified products from
diagnostic samples were interpreted according to the manufacturer’s
instructions. Samples that failed to amplify following repeated
testing were reported as ‘not detected’ (i.e. clonality could not
be detected due to insufficient quality or quantity of DNA for
analysis) and were excluded from the study.
Results
Ninety-two specimens from 80 patients with the final
diagnosis of mature B-cell lymphoma (37 specimens), T-cell lymphoma
(26 specimens) and reactive lymphoid proliferation (29 specimens)
were analyzed for B-cell clonality. According to the WHO
classification, our series of B-cell lymphomas comprised 2
specimens of mantle cell lymphoma (MCL), 8 specimens of follicular
lymphoma (FL), 11 specimens of nodal marginal zone B-cell lymphoma
(MZL), 2 specimens of extranodal marginal zone lymphoma of
mucosa-associated tissue (MALT lymphoma), 3 specimens of diffuse
large B-cell lymphoma (DLBCL), 1 specimen of B-cell lymphoma with
features between DLBCL and Burkitt lymphoma (BL), 1 specimen of
plasmablastic lymphoma, 3 specimens of lymphoplasmacytic lymphoma
and 6 specimens of unclassified B-cell lymphoma (B-cell lymphoma,
unclassified). All specimens were analyzed for both, IGH and
IGK clonality and detection rates of the IGH and the IGK
clonality assays were compared. The results of clonality analysis
using the IGH, the IGK and combined IGH+IGK clonality assays are
presented in Table I. The
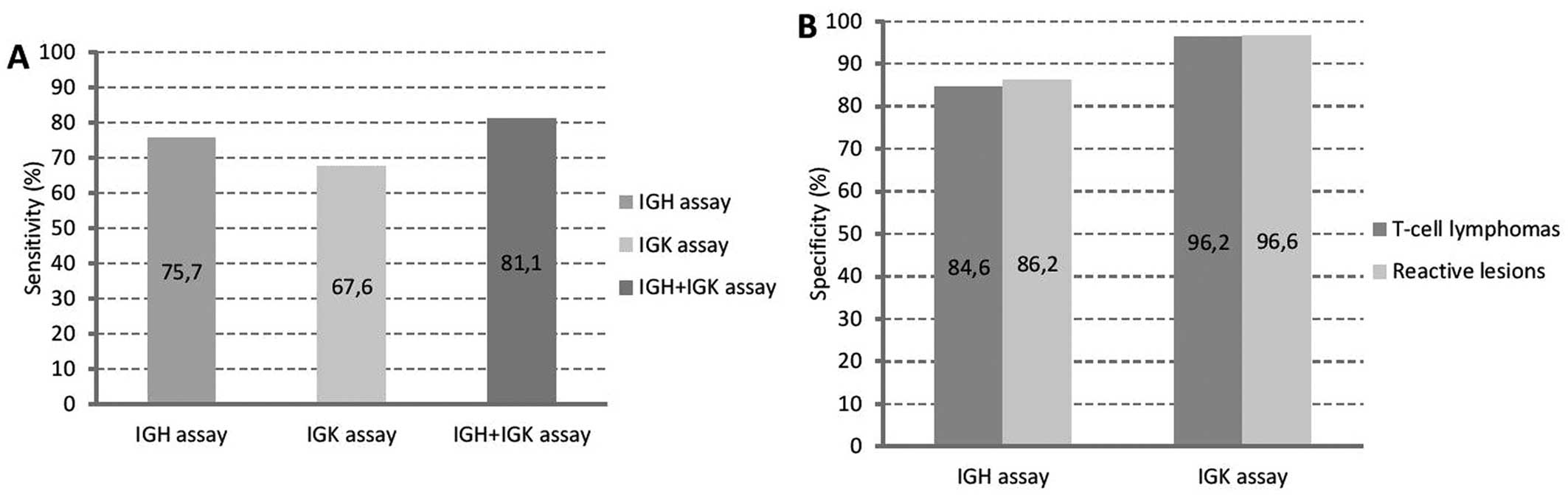
calculated sensitivity and specificity of the IGH/IGK clonality
assays are shown in Fig. 1.
 | Table IResults of clonality analysis using
the IGH, IGK and combined IGH+IGK clonality assays in 92 specimens
of lymphoid proliferations (LP). |
Table I
Results of clonality analysis using
the IGH, IGK and combined IGH+IGK clonality assays in 92 specimens
of lymphoid proliferations (LP).
| Diagnosis | No. of IGH
monoclonal/no. of tested specimens (%) | No. of IGK
monoclonal/no. of tested specimens (%) | No. of IGH+K
monoclonal/no. of tested specimens (%) |
|---|
| B-cell
lymphoma |
| MALT lymphoma | 2/2 (100.0) | 2/2 (100.0) | 2/2 (100.0) |
| MCL | 2/2 (100.0) | 2/2 (100.0) | 2/2 (100.0) |
| FL | 6/8 (75.0) | 5/8 (62.5) | 7/8 (87.5) |
| DLBCL | 3/3 (100.0) | 2/3 (66.7) | 3/3 (100.0) |
| DLBCL/BL | 0/1 (0.0) | 0/1 (0.0) | 0/1 (0.0) |
| MZL | 7/11 (63.6) | 6/11 (54.5) | 7/11 (63.6) |
| Plasmablastic
lymphoma | 1/1 (100.0) | 1/1 (100.0) | 1/1 (100.0) |
| Lymphoplasmacytic
lymphoma | 3/3 (100.0) | 3/3 (100.0) | 3/3 (100.0) |
| B-cell lymphoma,
unclassified | 4/6 (66.7) | 4/6 (66.7) | 5/6 (83.3) |
| Total B-cell
lymphomas | 28/37 (75.7) | 25/37 (67.6) | 30/37 (81.1) |
| T-cell
lymphomas | 4/26 (15.4) | 1/26 (3.8) | 4/26 (15.4) |
| Reactive
specimens | 4/29 (13.8) | 1/29 (3.4) | 5/29 (17.3) |
| Total LP | 36/92 (39.1) | 27/92 (29.3) | 39/92 (42.4) |
The BIOMED-2 IGH clonality assay
Monoclonal IGH rearrangements corresponding
to monoclonal B-cell proliferations were detected in 27 of 37
B-cell lymphoma specimens (73.0%). Polyclonal IGH
rearrangements corresponding to polyclonal B-cell proliferations
were detected in 9 B-cell lymphoma specimens (24.3%). In one
specimen of unclassified B-cell lymphoma monoclonal IGH
products in a background of polyclonal products were detected and
this specimen was concluded as borderline (monoclonal in a
polyclonal background). Polyclonal IGH rearrangements were
detected in MZL (4 specimens), FL (2 specimens), unclassified
B-cell lymphoma (2 specimens) and B-cell lymphoma with features
between DLBCL and BL (1 specimen). In 22 of 27 IGH
monoclonal B-cell lymphoma specimens also monoclonal IGK
rearrangements were detected. Four specimens with monoclonal
IGH rearrangements were polyclonal by the IGK assay and one
specimen with monoclonal IGH rearrangements was IGK
borderline. The IGH borderline specimen was polyclonal by
the IGK assay.
In the T-cell lymphoma group, polyclonal IGH
rearrangements were detected in 22 of 26 analyzed specimens (84.6%)
and monoclonal IGH rearrangements were detected in 4
specimens (15.4%). In the group of reactive specimens, polyclonal
IGH rearrangements were detected in 25 of 29 cases (86.2%)
and monoclonal IGH rearrangements were detected in 4 cases
(13.8%).
The BIOMED-2 IGK clonality assay
The detection rate of the BIOMED-2 IGK clonality
assay was lower than that of the IGH assay. Monoclonal IGK
rearrangements were detected in 23 of 37 analyzed B-cell lymphoma
specimens (62.2%). Of these, 22 specimens were monoclonal also by
the IGH assay and one specimen was IGH polyclonal. Polyclonal
IGK rearrangements were detected in 12 specimens (32.4%).
Two specimens of B-cell lymphoma were borderline for IGK
rearrangements (monoclonal in a polyclonal background) (two BM
aspirates with minimal infiltration with lymphoma cells from
patients with FL and lymphoplasmacytic lymphoma, respectively).
Polyclonal IGK rearrangements were detected in MZL (5
specimens), FL (3 specimens), unclassified B-cell lymphoma (2
specimens), DLBCL (1 specimen) and B-cell lymphoma with features
between DLBCL and BL (1 specimen).
Analysis of IGK amplification
products
We also compared the efficacy of GeneScan (GS) and
HD-PAGE analysis for the discrimination between monoclonal and
polyclonal IGK rearrangements in a series of 27 B-cell
lymphomas from 2011. In this regard, we evaluated only Vκ-Jκ
amplification products (tube IGK-A). The results of GS and HD-PAGE
analysis in a group of 27 B-cell lymphomas are shown in Table II.
 | Table IIComparison of GeneScan (GS) and
heteroduplex pretreatment-polyacrylamide gel electrophoresis
(HD-PAGE) methods for analysis of IGK gene rearrangements in
a group of 27 B-cell lymphomas. |
Table II
Comparison of GeneScan (GS) and
heteroduplex pretreatment-polyacrylamide gel electrophoresis
(HD-PAGE) methods for analysis of IGK gene rearrangements in
a group of 27 B-cell lymphomas.
| A, The results of
GS and HD-PAGE analysis in 27 cases of B-cell lymphomas |
|---|
|
|---|
| Sample no. | Diagnosis | IGK-A-GS | IGK-A-HD | IGK-A-final | IGK-B | IGK-final |
|---|
| 1 | MZL | P | P | P | P | P |
| 2 | DLBCL/BL-like | P | P | P | P | P |
| 3 | MZL | M | M/P | M | P | M |
| 4 |
Lymphoplasmacytic | M | M/P | M | M | M |
| 5 | FL | P | P | P | P | P |
| 6 |
Lymphoplasmacytic | P | M/P | M/P | M/P | M/P |
| 7 |
Lymphoplasmacytic | M | M | M | M | M |
| 8 | FL | M | M | M | M | M |
| 9 | MZL | P | M/P | M/P | M | M |
| 10 | MCL | P | M/P | M/P | M | M |
| 11 | FL | M/P | P | M/P | P | M/P |
| 12 | MZL | M | P | M | P | M |
| 13 | MZL | M | M/P | M | P | M |
| 14 | Plasmablastic | M | M | M | M | M |
| 15 | B-cell lymphoma,
unspecified | P | P | P | P | P |
| 16 | DLBCL | P | P | P | P | P |
| 17 | FL | P | P | P | M | M |
| 18 | MALT lymphoma | M | M/P | M | P | M |
| 19 | MZL | P | P | P | P | P |
| 20 | B-cell lymphoma,
unspecified | M | M/P | M | P | M |
| 21 | MZL | P | P | P | P | P |
| 22 | MZL | M | M/P | M | P | M |
| 23 | DLBCL | M | M | M | M | M |
| 24 | DLBCL | M | M | M | M | M |
| 25 | B-cell lymphoma,
unspecified | M | M | M | P | M |
| 26 | FL | P | P | P | P | P |
| 27 | FL | P | P | P | ND | P |
|
| B, The summary of
results obtained by GS and HD-PAGE analysis of tube A (Vκ-Jκ)
amplification products in 27 cases of B-cell lymphomas |
|
| Summary | Data |
|
| No. of IGK
monoclonal by both methods/no. of tested specimens (%) | 6/27 (22.2) |
| No. of IGK
polyclonal by both methods/no. of tested specimens (%) | 10/27 (37.0) |
| No. of IGK
borderline by both methods/no. of tested specimens (%) | 0/27 (0.0) |
| No. of discordant
results between GS and HD-PAGE methods/no. of tested specimens
(%) | 11/27 (40.7) |
Using the GS analysis we were able to discriminate
between monoclonal and polyclonal Vκ-Jκ rearrangements in 26
specimens (96.3%); 13 specimens had monoclonal and 13 specimens had
polyclonal Vκ-Jκ rearrangements. One specimen was borderline
(monoclonal in a polyclonal background) by GS analysis, while
HD-PAGE detection showed polyclonal ‘smear’ (Table II).
The interpretation of HD-PAGE results was
straightforward in 18 of 27 analyzed specimens (66.7%), in which
clear monoclonal bands (6 specimens) or polyclonal ‘smears’ (12
specimens) were detected. On the contrary, 9 of 27 (33.3%) of
analyzed specimens showed ‘monoclonal in a polyclonal background’
pattern by HD-PAGE analysis (6 specimens were monoclonal and 3
specimens were polyclonal by GS analysis) and were difficult to
interpret (Table II). HD-PAGE
detection of Vκ-Jκ amplification products in 17 representative
B-cell lymphoma specimens are shown in Fig. 2A.
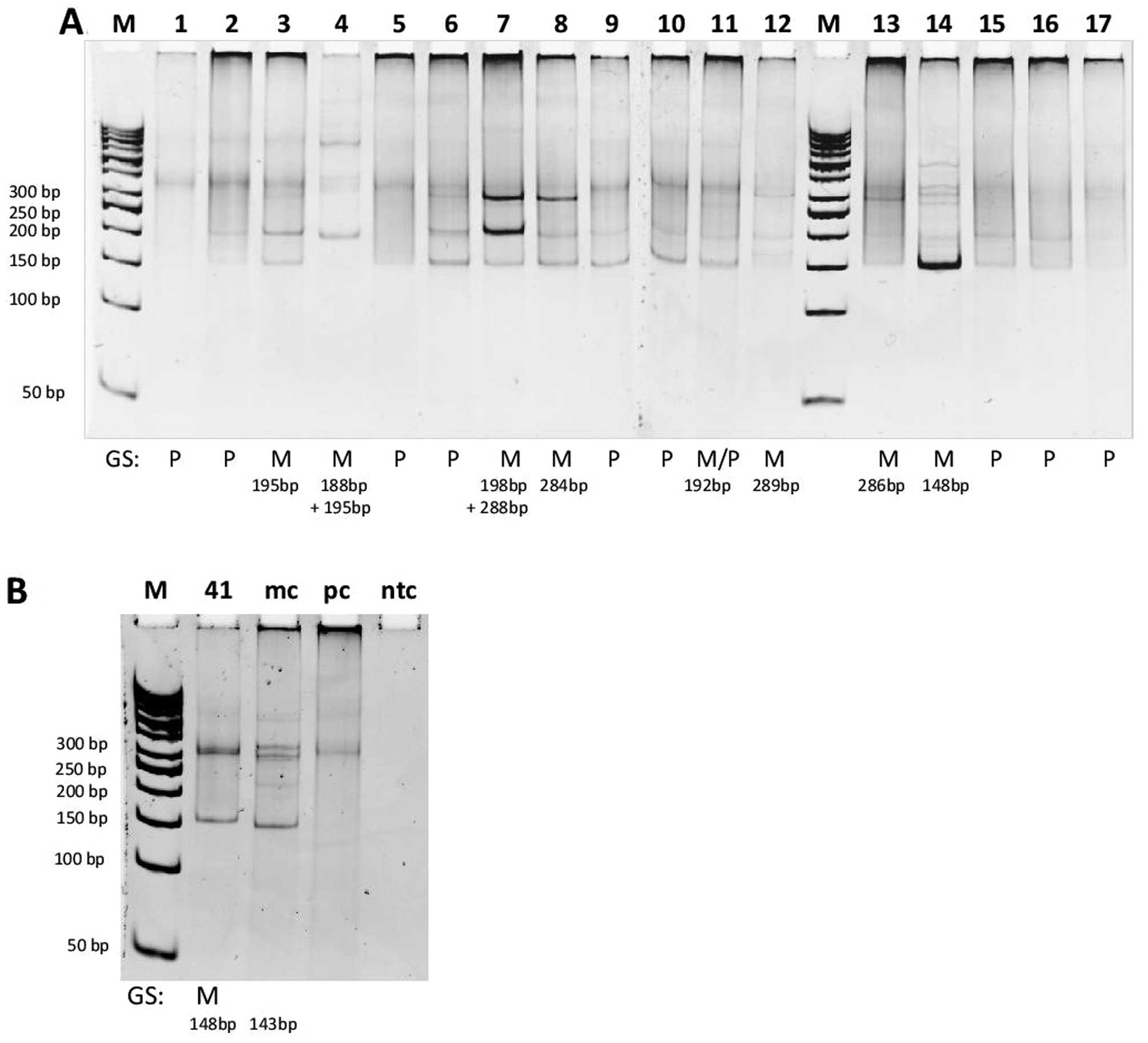 | Figure 2HD-PAGE detection of Vκ-Jκ
rearrangements (tube IGK-A) in representative B-cell lymphoma
cases. M, molecular weight marker (50 bp DNA ladder); lanes 1–17
(A) and 41 (B) represent amplifications from B-cell lymphoma cases;
mc, monoclonal control; pc, polyclonal control; ntc, no template
control (water instead of DNA); the presence of one or two distinct
bands in the expected size range (120–160, 190–210 and 260–300 bp)
indicates monoclonality; ‘smear’ in the expected size range
indicates polyclonality; results of GeneScan analysis (GS) of the
same cases are presented on the bottom of the gel; M, monoclonal;
P, polyclonal; M/P, ‘monoclonal in a polyclonal background’. In
cases 3, 4 and 13 monoclonal Vκ-Jκ amplified products were observed
by GS, but after HD-PAGE analysis ‘monoclonal in a polyclonal
background’ pattern was observed (A). Conversely, in cases 6, 9 and
10 polyclonal Vκ-Jκ products were detected by GS analysis, but
HD-PAGE analysis showed ‘monoclonal in a polyclonal background’
pattern (A). Monoclonal Vκ-Jκ product of 148 bp detected by GS in a
case of suspected IGL+ B-cell lymphoma was confirmed by
HD-PAGE analysis (B). |
In routine diagnostic series, the HD-PAGE analysis
was performed only in specimens with doubtful results of the GS
analysis. Monoclonal Vκ-Jκ amplification product by GS analysis in
the case of suspected B-cell lymphoma with the expression of λ
light chains (IGL+ B-cell lymphoma) was confirmed by
HD-PAGE method (Fig. 2B).
Among 26 analyzed T-cell lymphoma specimens,
polyclonal IGK rearrangements were detected in 25 specimens
(96.2%) and monoclonal IGK rearrangements in 1 specimen
(3.8%). Twenty-eight of 29 reactive lymphoid proliferations were
polyclonal for IGK rearrangements (96.5%). In one reactive
specimen monoclonal IGK rearrangement was detected.
The combined IGH and IGK assay
By the combination of the IGH and the IGK clonality
assay, monoclonal B-cell populations were detected in 28 specimens
of B-cell lymphoma (75.7%). Of these, 22 specimens were monoclonal
for both the IGH and the IGK clonality. Two specimens with
polyclonal Ig rearrangements detected by one of the clonality
assays (IGH or IGK) and borderline result by the other assay, were
concluded as borderline (monoclonal in a polyclonal background).
Polyclonal rearrangements in both genes, the IGH and the
IGK, were detected in 7 B-cell lymphoma specimens (18.9%).
Considering monoclonal and borderline cases as ‘true positives’ the
detection rate of B-cell clonality determined by the combination of
IGH and IGK was 81.1% (30/37).
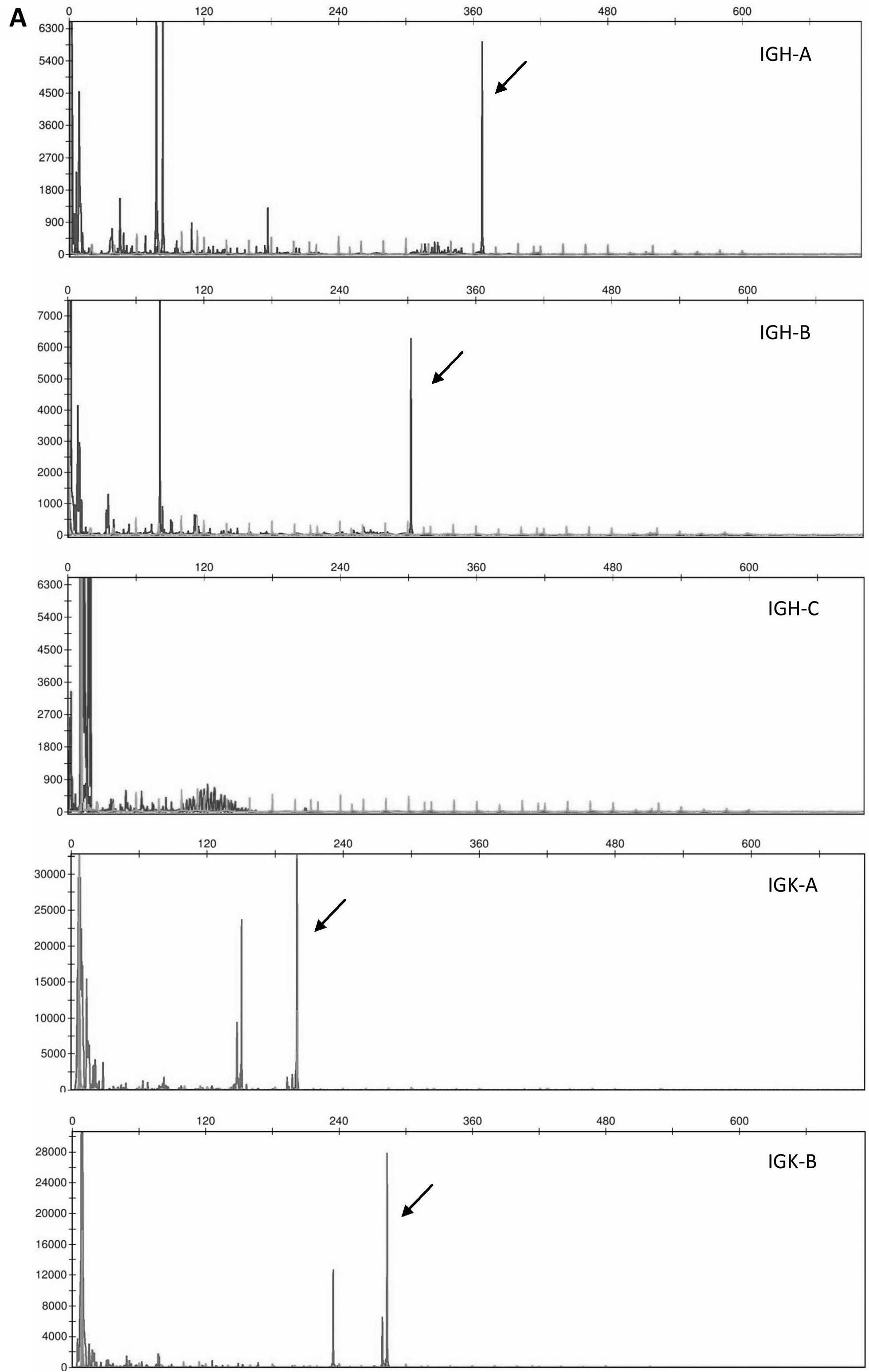
Three representative specimens with monoclonal
rearrangements in both genes, IGH and IGK, monoclonal
IGH and polyclonal IGK rearrangement, and polyclonal
IGH and monoclonal IGK rearrangement, respectively,
are presented in Fig. 3.
Discussion
In the present study, we evaluated the added value
of standardized BIOMED-2 assay for the detection of clonal
IGK gene rearrangements in the diagnostic setting of
suspected B-cell lymphomas. The sensitivity of the IGK assay
(67.6%) in our diagnostic series of B-cell lymphomas was lower than
expected from the literature (6–8,17).
The sensitivity of the IGH assay in our series was 75.7%, which is
also lower than some of the authors reported (10,18),
but higher than the obtained sensitivity of the IGK assay. With the
combination of both assays (IGH+IGK) the sensitivity of the B-cell
clonality detection was improved to 81.1% (Table I, Fig.
1). Nevertheless, we were not able to achieve the B-cell
clonality detection rates reported in most studies using the
BIOMED-2 protocols, even with the combination of the IGH and the
IGK assay (6–8,17).
The lower sensitivity of the IGH assay in our series
is probably related to a rather high percentage of included
GC/post-GC B-cell lymphomas (29 of 37), for which it has been well
documented that frequent somatic hypermutations contribute to a
lower IGH monoclonality rate (6–8,18).
Indeed, we detected monoclonal IGH rearrangements in only 22
of 29 (75.9%) GC/post-GC B-cell lymphomas (Table I), which is in agreement with other
studies (54–85%) (6,7,10,14,18).
Therefore, we expected a higher clonality detection rate by the IGK
assay in this group of B-cell lymphomas, somewhere in the range of
74–86% (6,7,10,14,18).
Surprisingly, monoclonal IGK rearrangements were detected in
only 19 of 29 (65.5%) GC/post-GC B-cell lymphoma cases (Table I). The detection rates of the IGK
assay in the most common GC/post-GC B-cell lymphoma entities,
follicular lymphoma and diffuse large B-cell lymphoma were 62.5% (5
of 8) and 66.7% (2 of 3), respectively (Table I). Causes for the low sensitivity
of the IGK assay in our hands, especially in histologically and
immunophenotypically confirmed cases of GC/post-GC B-cell lymphoma
subtypes, are not clear.
The comparison of GeneScan (GS) and heteroduplex
pretreatment-polyacrylamide gel electrophoresis (HD-PAGE) methods
for analysis of IGK gene rearrangements showed a better
performance of the GS analysis in our series of 27 B-cell lymphomas
analyzed by both methods. Namely, we were able to discriminate
between monoclonal and polyclonal Vκ-Jκ rearrangements (tube IGK-A)
in 26 cases (96.3%) using the GS analysis and in only 18 cases
(66.7%) using HD-PAGE analysis (Table
II). One case was borderline (monoclonal in a polyclonal
background) with the GS analysis, whereas 9 cases were borderline
with the HD-PAGE method. According to the EuroClonality/BIOMED-2
guidelines for interpretation and reporting of Ig/TCR clonality
testing in suspected lymphoid proliferations, GS and HD-PAGE
methods have complementary value for analysis of IGK
rearrangements (19). The HD-PAGE
analysis is recommended in cases with doubtful GS results, in
particular when a prominent peak of ~148 bp is detected in tube
IGK-A (Vκ-Jκ rearrangements). It is well known that in polyclonal
samples this peak represents rearrangements of different sequences
with the same size due to limited junction diversity of the
rearranged IGK gene (19,20).
Indeed, the monoclonal Vκ-Jκ product of 148 bp detected by GS
analysis in a case of suspected IGL+ B-cell lymphoma was
confirmed by the HD-PAGE method (Fig.
2B).
The specificity of the IGK assay was higher than the
specificity of the IGH assay (Fig.
1). It was 96.2% in the group of mature T-cell lymphomas and
96.6% in the group of reactive lesions, which is comparable to
other studies using the BIOMED-2 protocols (17,21,22).
The specificity of the IGH assay in the group of T-cell lymphomas
(84.6%) and in the group of reactive lesions (86.2%) is comparable
to the one determined in our previous study and does not need
further discussion (15).
In conclusion, the present study was applied to
demonstrate the utility of combined IGH+IGK clonality assay for the
assessment of B-cell clonality in suspected B-cell lymphomas with
inconclusive clinical and cyto/histological diagnosis. Using the
combined IGH+IGK clonality assay, the overall detection rate of
B-cell clonality was increased by 5.4% (from 75.7% using only the
IGH assay to 81.1% using both, IGH and IGK assays). Thus, we
confirmed the added value of standardized BIOMED-2 IGK assay for
the assessment of B-cell clonality. However, the sensitivity of
standardized BIOMED-2 IGK assay in the present study was lower than
expected, even in GC/post-GC B-cell lymphoma cases. Furthermore,
care should be taken with the interpretation of the IGK
amplification products due to restricted IGK junctional
diversity.
We are aware that our conclusions derive from a
rather small diagnostic series with only 37 specimens of confirmed
B-cell lymphomas. Certainly, the evaluation of a larger series of
B-cell lymphomas is needed for firmer conclusions. With these
considerations in mind, we believe that the assessment of
IGK gene rearrangements is valuable in demonstrating the
B-cell clonality in the diagnostic setting, in particular in the
absence of clonal IGH rearrangements.
References
|
1
|
Diss TC, Peng H, Wotherspoon AC, Isaacson
PG and Pan L: Detection of monoclonality in low-grade B-cell
lymphomas using the polymerase chain reaction is dependent on
primer selection and lymphoma type. J Pathol. 169:291–295. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McCarthy KP, Sloane JP and Wiedemann LM:
Rapid method for distinguishing clonal from polyclonal B cell
populations in surgical biopsy specimens. J Clin Pathol.
43:429–432. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trainor KJ, Brisco MJ, Wan JH, Neoh S,
Grist S and Morley AA: Gene rearrangement in B- and
T-lymphoproliferative disease detected by the polymerase chain
reaction. Blood. 78:192–195. 1991.PubMed/NCBI
|
|
4
|
Amara K, Trimeche M, Ziadi S, Sriha B,
Mokni M and Korbi S: PCR-based clonality analysis of B-cell
lymphomas in paraffin-embedded tissues: diagnostic value of
immunoglobulin k and l light chain gene rearrangement
investigation. Pathol Res Pract. 202:425–431. 2006. View Article : Google Scholar
|
|
5
|
Gameiro P, Sebastião M, Spetalen S, Gomes
da Silva M and Cabeçadas J: The added value of immunoglobulin Kappa
light chain gene (IGK) rearrangement analysis in suspected B-cell
lymphomas: three illustrative cases. J Hematopathol. 5:45–56. 2012.
View Article : Google Scholar
|
|
6
|
Liu H, Bench AJ, Bacon CM, Payne K, Huang
Y, Scott MA, et al: A practical strategy for the routine use of
BIOMED-2 PCR assays for detection of B- and T-cell clonality in
diagnostic haematopathology. Br J Haematol. 138:31–43. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Payne K, Wright P, Grant JW, Huang Y,
Hamoudi J, Bacon CM, et al: BIOMED-2 PCR assays for IGK gene
rearrangements are essential for B-cell clonality analysis in
follicular lymphoma. Br J Haematol. 155:84–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berget E, Helgeland L, Molven A and
Vintermyr OK: Detection of clonality in follicular lymphoma using
formalin-fixed, paraffin-embedded tissue samples and BIOMED-2
immunoglobulin primers. J Clin Pathol. 64:37–41. 2011. View Article : Google Scholar
|
|
9
|
Diss TC, Liu HX, Du MQ and Isaacson PG:
Improvements to B cell clonality analysis using PCR amplification
of immunoglobulin light chain genes. Mol Pathol. 55:98–101. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Evans PA, Pott C, Groenen PJ, Salles G,
Davi F, Berger F, et al: Significantly improved PCR-based clonality
testing in B-cell malignancies by use of multiple immunoglobulin
gene targets. Report of the BIOMED-2 Concerted Action
BHM4-CT98-3936. Leukemia. 21:207–214. 2007. View Article : Google Scholar
|
|
11
|
Langerak AW, Nadel B, De Torbal A,
Wolvers-Tettero IL, van Gastel-Mol EJ, Verhaaf B, et al: Unraveling
the consecutive recombination events in the human IGK locus. J
Immunol. 173:3878–3888. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
van der Burg M, Tumkaya T, Boerma M, de
Bruin-Versteeg S, Langerak AW and van Dongen JJ: Ordered
recombination of immunoglobulin light chain genes occurs at the IGK
locus but seems less strict at the IGL locus. Blood. 97:1001–1008.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pai RK, Chakerian AE, Binder JM, Amin M
and Viswanatha DS: B-cell clonality determination using an
immunoglobulin k light chain polymerase chain reaction method. J
Mol Diagn. 7:300–307. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Catherwood MA, Gonzales D, Patton C,
Dobbin E, Venkatraman L and Alexander HD: Improved clonality
assessment in germinal centre/post germinal centre non-Hodgkin’s
lymphomas with high rates of somatic hypermutation. J Clin Pathol.
60:524–528. 2007. View Article : Google Scholar
|
|
15
|
Kokovic I, Jezersek Novakovic B, Cerkovnik
P and Novakovic S: Clonality analysis of lymphoid proliferations
using the BIOMED-2 clonality assays: a single institution
experience. Radiol Oncol. 48:155–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Swerdlow SH, Campo E, Harris NL, Jaffe ES,
Pileri SA, Stein H, et al: WHO Classification of Tumors of
Haematopoietic and Lymphoid Tissues. IARC Press; Lyon: 2008
|
|
17
|
Mannu C, Gazzola A, Bacci F, Sabattini E,
Sagramoso C and Roncolato F: Use of IGK gene rearrangement analysis
for clonality assessment of lymphoid malignancies: a single center
experience. Am J Blood Res. 1:167–174. 2011.PubMed/NCBI
|
|
18
|
van Krieken JH, Langerak AW, Macintyre EA,
Kneba M, Hodges E, Garcia Sanz R, et al: Improved reliability of
lymphoma diagnostics via PCR-based clonality testing: report of the
BIOMED-2 concerted action BHM4-CT98-3936. Leukemia. 21:201–206.
2007. View Article : Google Scholar
|
|
19
|
Langerak AW, Groenen PJ, Brüggemann M,
Beldjord K, Bellan C, Bonello L, et al: EuroClonality/BIOMED-2
guidelines for interpretation and reporting of Ig/TCR clonality
testing in suspected lymphoproliferations. Leukemia. 26:2159–2171.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van Dongen JJ, Langerak AW, Brüggemann M,
Evans PA, Hummel M, Lavender FL, et al: Design and standardization
of PCR primers and protocols for detection of clonal immunoglobulin
and T-cell receptore gene recombinations in suspect
lymphoproliferations: report of the BIOMED-2 Concerted Action
BHM4-CT98-3936. Leukemia. 17:2257–2317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brüggemann M, White H, Gaulard P,
Garcia-Sanz R, Gameiro P, Oeschger S, et al: Powerful strategy for
polymerase chain reaction-based clonality assessment in T-cell
malignancies. Report of the BIOMED-2 Concerted Action
BHM4-CT98-3936. Leukemia. 21:215–221. 2007. View Article : Google Scholar
|
|
22
|
Langerak AW, Molina TJ, Lavender FL,
Pearson D, Flohr T, Sambade C, et al: Polymerase chain
reaction-based clonality testing in tissue samples with reactive
lymphoproliferations: usefulness and pitfalls. A report of the
BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 21:222–229.
2007. View Article : Google Scholar
|