Introduction
The ubiquitous fungus Aspergillus fumigatus
is the predominant cause of life-threatening invasive aspergillosis
(IA) in immunocompromised patients but further Aspergillus
species such as A. niger and A. flavus can also cause
invasive disease (1). Over the
last decades, there has been a substantial increase in the number
of immunocompromised persons concomitant with an increase in the
number of IA related deaths (2).
While exposure of immunocompetent individuals to conidia remains
with no further implications, it may become lethal in
immunocompromised patients (3).
Hence, IA is the leading cause of death in patients with acute
leukemia and recipients of allogeneic haematopoietic stem-cell
transplants (2). Furthermore,
patients with advanced HIV infections and patients undergoing
immuno-modulation therapies or chemotherapy are also at risk to
acquire IA (4).
Symptoms which have been linked to IA are
non-specific; this comprises cough, chest pain and fever and
patients might even remain asymptomatic (5). Accordingly, early and accurate
diagnosis of IA is mandatory in order to improve the patient
outcome and restrain utilization of antifungal drugs prescribed for
patients at high risk of IA. Unfortunately, currently applied
diagnostic methods of Aspergillus infections lack
sensitivity and/or specificity.
Diagnostic tools based on ELISA such as the
detection of the cell wall components galactomannan (GM) or
(1–>3)-β-D-glucan (BDG) are widely used in the diagnosis of IA
(6). Nevertheless, many factors
can compromise the results of both assays. The accuracy of GM
testing can be altered by antibiotic (7) or antifungal therapy (8) and inadequate sampling strategies
(6). In addition, both GM and BDG
are not characteristic for fungi of the genus Aspergillus
and are also produced from other species in the fungal kingdom
(9,10). False positive BDG results have also
been reported in patients treated with immunoglobulins or
antibiotics such as amoxicillin (11). PCR based diagnosis is a promising
method for detecting Aspergillus DNA, due to high
sensitivity and specificity. Nevertheless, there are some
constrains for routine laboratory use including lack of
standardized protocols, reproducibility and risk of contamination
(12).
We hypothesized, that the monitoring of
Aspergillus specific proteolytic acitivity in serum
specimens might be an alternative approach for diagnosis of IA. In
immunocompromised patients the airborne conidia can germinate and
produce a branched mycelium that penetrates the lung tissue
invading blood vessels and spreading hematogenously. During
invasion several toxic molecules as well as enzymes including a
variety of proteases are secreted. These proteases play a crucial
role in the breakdown of the extracellular matrix barrier of the
host (13). Recently,
Aspergillus associated protease activity has been
characterized in bronchoalveolar lavage (BAL) using fluorogenic
substrates (14). Due to the high
pulmonary blood flow it is most likely that secreted proteases are
also present in serum specimens that can be obtained much easier
than BAL fluid. In a previous study we have already demonstrated
the feasibility of protease profiling for the detection of disease
associated proteolytic activity in serum specimens. Synthetic
peptides designated as reporter peptides (RP) are added to serum
specimens and the accumulation of cleaved products is indicating
disease associated proteolytic activity. The main advantages of RP
spiking is the ease of standardization making this procedure highly
reproducible (15). Furthermore,
the enzymatic cleavage of RPs in combination with mass spectrometry
based quantitation of RP-fragments leads to high diagnostic
sensitivity (16,17). Here, we selected five different
reporter peptides that are exclusively cleaved by proteases
secreted from Aspergillus species. These RPs were added to
serum specimens of patients with invasive aspergillosis (IA),
possible invasive aspergillosis (PIA) and healthy controls (HC) and
incubated under standardized conditions prior to MALDI-TOF MS
analysis. As expected, the specimens from patients with IA showed
highest concentrations of RP-fragments indicating increased
activity of Aspergillus specific proteases. This functional
protease profiling might be a promising approach for laboratory
based diagnosis of IA. However, our preliminary results of this
proof of concept experiment have to be validated thoroughly in
future studies.
Materials and methods
Aspergillus culture
Clinical isolates of A. fumigatus were grown
on Sabouraud dextrose agar (Oxoid Ltd., Hampshire, UK) at 30°C for
at least 7 days. After spore formation had occurred, conidia were
harvested manually using a pipette tip and inoculated in 50 ml of
Aspergillus minimal medium (AMM) containing 500 μl of a 100X
penicillin/streptomycin solution (final concentration: 0.1 mg/ml
each) and liquid cultures were incubated at 37°C. Supernatant of
cultures was harvested after 4 days, filtered through a syringe
filter (Fisher Scientific GmbH, Schwerte, Germany; size cut-off
0.22 μm) and stored directly at −80°C.
Patients and blood collection
Patients were classified based on the EORTC/MSG
consensus criteria (18). Nine
serum samples from five patients with proven invasive aspergillosis
(IA) were obtained from the Department of Internal Medicine C of
the University of Greifswald, Germany. Furthermore, ten serum
specimens of these five patients were obtained prior to the
diagnosis of IA when patients were classified as having a possible
invasive aspergillosis. The number of blood specimens per patient
ranged from 1 to 6 and longitudinal blood sampling was
consecutively performed within a time range of up to 240 days prior
to diagnosis of IA. Blood from 101 healthy control individuals (HC)
was taken from voluntary employees of the University Hospital
Mannheim during routine laboratory testing at the occupational
medicine. Furthermore, 144 blood samples of 16 immunosuppressed
patients with haematological malignancies that were classified as
having a possible invasive aspergillosis (PIA) were also integrated
into the study. The number of blood specimens per patient ranged
from 1 to 22 and longitudinal blood sampling was consecutively
performed within a time range of up to 185 days. The
characteristics of patients are summarized in Table I.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| EORTC/MSG | Gender | Age (years) | Diagnosis |
|---|
| IA | Male | 47 | NHL |
| Female | 59 | AML |
| Male | 50 | AML |
| Male | 27 | CML |
| Female | 25 | CML |
| PIA | Male | 74 | AML |
| Male | 55 | AML |
| Female | 62 | CML |
| Male | 45 | AML |
| Female | 68 | AML |
| Male | 50 | AML |
| Female | 58 | AML |
| Female | 51 | AML |
| Female | 67 | AML |
| Female | 69 | AML |
| Female | 44 | AML |
| Female | 40 | AML |
| Female | 25 | AML |
| Female | 44 | AML |
| Male | 56 | AML |
| Female | 51 | AML |
Blood collection was performed after we obtained
institutional review board approval from the local ethics committee
and patients’ written informed consent. Whole blood (10 ml) was
collected in serum tubes (Sarstedt AG & Co.,
Nümbrecht-Rommelsdorf, Germany). Samples were kept for 30 min at
room temperature prior to centrifugation at 3,000 × g (10 min,
20°C). Finally, the serum was removed, divided in aliquots and
stored at −80°C until further use.
Galactomannan assay
Serum galactomannan (GM) levels were measured using
the Platelia Aspergillus enzyme immunoassay test (Bio-Rad
Laboratories GmbH, Munich, Germany) according to the manufacturer’s
instructions. Results were recorded as the ratio of optical density
of the sample to that of threshold control samples. Cut-off value
of GM was 0.5 of optical density index (ODI) as approved by FDA
(19).
Reporter peptide synthesis and
spiking
Peptide sequences were selected from our recent
study (20,21) as well as publicly available data
(22,23) and were synthesized by the Peptide
Synthesis Facility of the German Cancer Research Center
(Heidelberg, Germany). The reporter peptides (RP) and respective
reporter peptide fragments (RPF) are shown in Table II. Any RP was prepared as a 4
mmol/l stock solution in DMSO. RPs were diluted to 100 μmol/l in
PBS pH 7.4 (PAA Laboratories). Each serum sample was diluted 1:3 in
PBS. For the proteolytic profiling, 50 μl of diluted serum sample
was mixed with 50 μl RP solution and incubated at 37°C for 24
h.
 | Table IIReporter peptide sequences. |
Table II
Reporter peptide sequences.
| Name | Aa sequence |
[M+H]+ |
|---|
| RP1 |
Bio-Abu-Ahx-RPKPVE-Nva-WREAKdHHHHHH | 2,858.4 |
| RPF1 | KdHHHHHH | 1,084.5 |
| RP2 |
Bio-Abu-Ahx-WKPYDAADLtnHHHHHHt | 2,640.8 |
| RPF2 | DAADLtnHHHHHHt | 1,642.7 |
| RP3 |
Bio-Abu-Ahx-CGSHLVEAttHHHHHH | 2,264.8 |
| RPF3 | AttHHHHHH | 1,114.5 |
| RP4 |
Bio-Abu-Ahx-DGVDLKTQyHHHHHHn | 2,399.9 |
| RPF4 | DLKTQyHHHHHHn | 1,703.8 |
| RP5 |
Bio-Abu-Ahx-GFFYTPKAvHHHHHHn | 2,391.3 |
| RPF5 | AvHHHHHHn | 1,125.5 |
Purification of proteolytic
fragments
As shown in a previous study, the analytical
sensitivity of protease profiling can substantially be enhanced
using serial affinity chromatography of doubly tagged RPs (16). Accordingly, the full length RP was
depleted prior to enrichment of His-tagged C-terminal part of
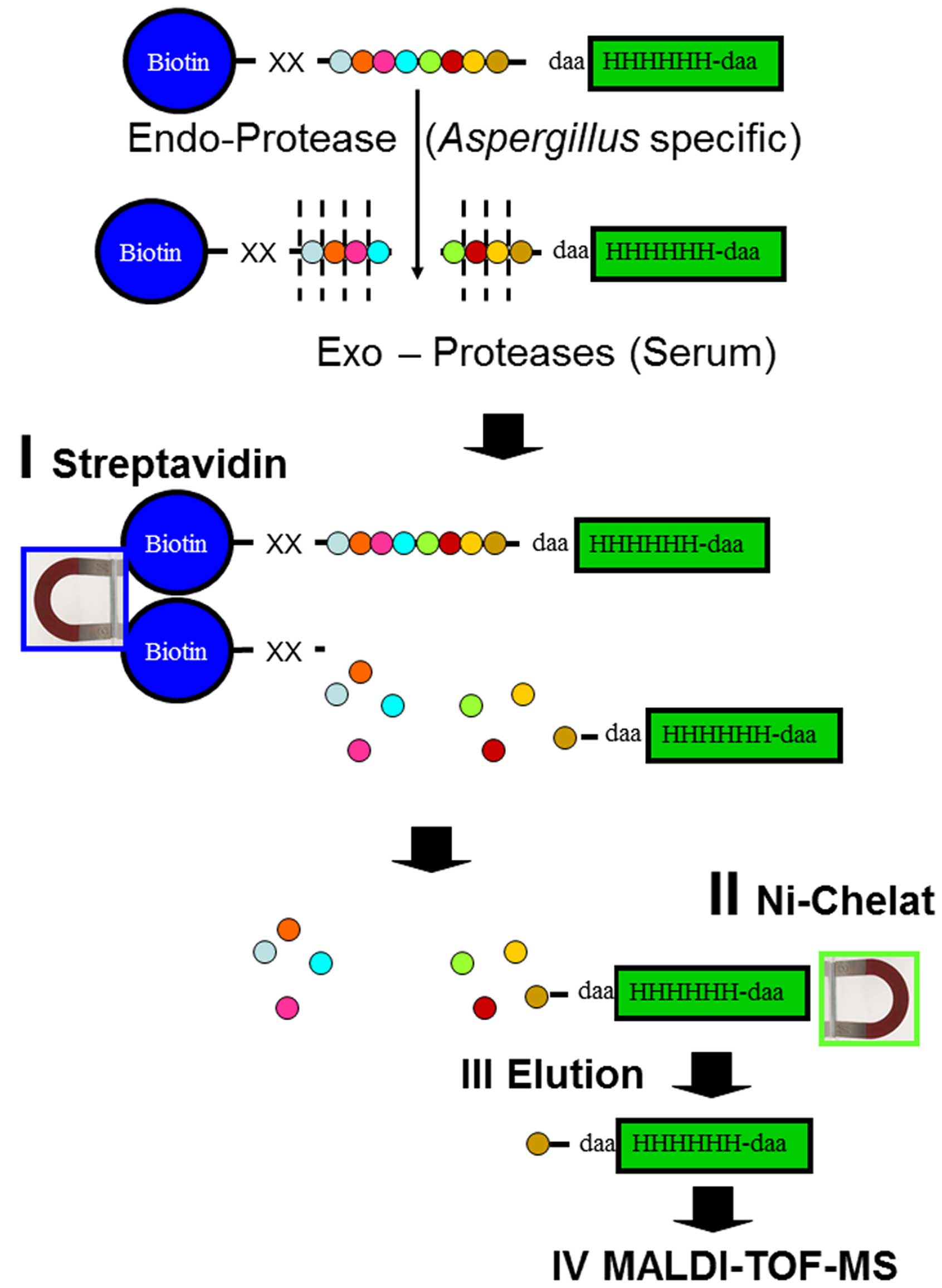
cleaved RPs and subsequent MALDI-TOF MS (Fig. 1). Streptavidin sepharose
high-performance slurry (GE Healthcare, Uppsala, Sweden) was washed
twice and resuspended in PBS pH 7.4 to yield a 50% slurry. We then
added 50 μl streptavidin slurry to each spiked sample and incubated
the mixture for 15 min at room temperature. After binding, the
streptavidin sepharose was centrifuged at 1,100 × g for 5 min and
discarded. A 20-μl aliquot of the supernatant was mixed with 60 μl
binding buffer and 10 μl IMAC SL (immobilized metal ion affinity
chromatography on self-loading) magnetic beads (Bruker Daltonik
GmbH, Bremen, Germany) that were loaded with Ni2+ ions,
and then further processed according to the manufacturer’s
instructions. Briefly, the samples were incubated for 30 min at
room temperature, washed 2 times each with 3 different buffers (100
μl per wash step), and eluted with 10 μl elution buffer for 15 min
at room temperature.
MALDI TOF-mass spectrometry
We diluted 1 μl of each eluted sample with matrix
solution (1:10) and 1 μl of the resulting mixture was spotted onto
a SCOUT 600-μm AnchorChip target (Bruker Daltonik GmbH). The matrix
solution was prepared by dissolving 0.3 g/l of
α-cyano-4-hydroxycinnamic acid solution (Bruker Daltonik GmbH) in
ethanol/acetone (2:1, vol/vol), and external mass calibration was
then performed with a peptide calibration standard (Bruker Daltonik
GmbH). MALDI-TOF MS measurements were performed using an Autoflex
II (Bruker Daltonik GmbH) operating in the positive mode. The MS
spectra for peaks in the range of 1–3 kDa were generated by
summarizing 500 laser shots (50 laser shots at 10 different spot
positions). Peak detection was performed using the centroid
algorithm. The peak intensities were normalized to the total ion
count of the respective mass spectra. For data acquisition and
analysis, the software packages FlexControl™ 2.2 and FlexAnalysis™
2.2 (both from Bruker Daltonik GmbH) were used.
Reproducibility
The 1:3 mixtures of Aspergillus fumigatus
cell culture supernatant and healthy control serum was aliquoted to
generate the specimens designated as ‘quality control’ (QC). All
aliquots were kept at −80°C until further use. Quality control
samples were further on handled similarly to the patient specimens
and regularly interspersed into analytical runs to determine
reproducibility of reporter peptide spiking. All samples were
strictly randomized prior to further handling, processing and
analysis.
Statistical analysis
Coefficients of variation (CV) and standard
deviations (SD) were calculated using Microsoft Excel 2010
(Microsoft software). The data visualization using whisker-box
plots was performed with MedCalc™ software, version 13.1.1 (MedCalc
Software, Ostend, Belgium).
Results
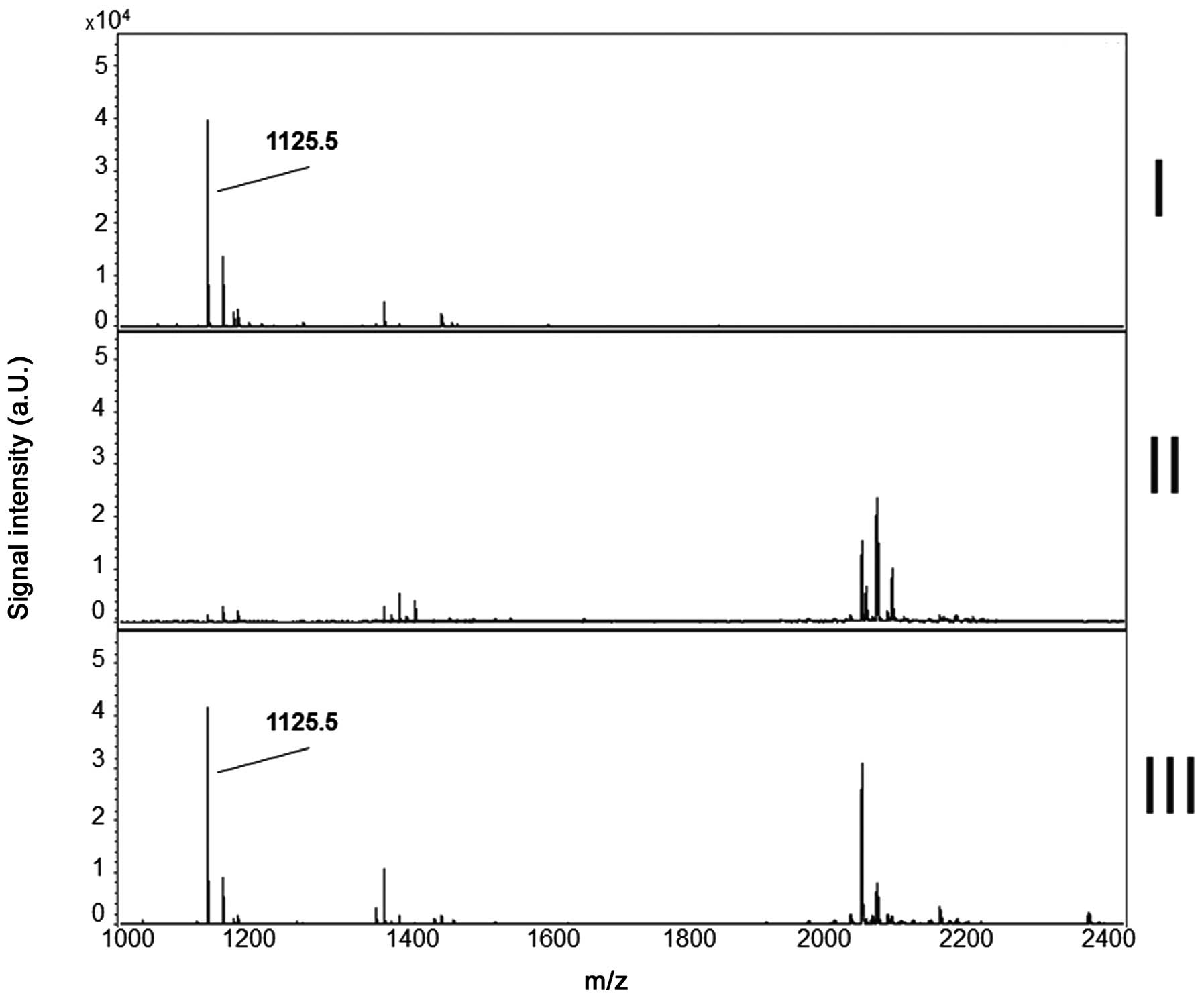
Specificity of RP spiking
Reporter peptides were added to specimens and
incubated prior to MALDI-TOF MS as described in Materials and
methods. The cleavage pattern of Aspergillus fumigatus cell
culture supernatant (ACCS), serum of healthy controls (HC) and the
mixture of ACCS and HC were compared to each other. Any of the five
RPs was cleaved in ACCS and the mixture of ACCS and HC and
respective proteolytic fragments (Table II) were prominent in subsequent
MALDI-TOF MS analyses. In contrast, after incubation of RPs in HC
serum the respective proteolytic fragments were almost
undetectable. This indicates that RPs are selectively cleaved by
secreted proteases of Aspergillus fumigatus whereas they are
stable in serum specimens of healthy controls. The representative
MS-profiles of RP5 are exemplarily shown in Fig. 2.
Reproducibility
The reproducibility of RP spiking was tested with
ten quality control specimens (QC) that were randomly interspersed
into measurements of serum from patients and controls. As expected,
the coefficient of variation (CV) was inversely correlated to the
signal intensity of reporter peptide fragments (RPFs) (24). Lowest CV of 8% was found for RPF5
with median signal intensity of 55.040 a.U. (SD 4.132). Highest CV
of 31% was found for RPF2 with median signal intensity of 20.150
a.U. (SD 6.292).
Reporter peptide processing in serum
specimens from patients with invasive aspergillosis and
controls
Any of the five RPs was incubated in serum specimens
of healthy controls (HC, n=101) and patients with proven invasive
aspergillosis (IA, n=9) as described in Materials and methods. The
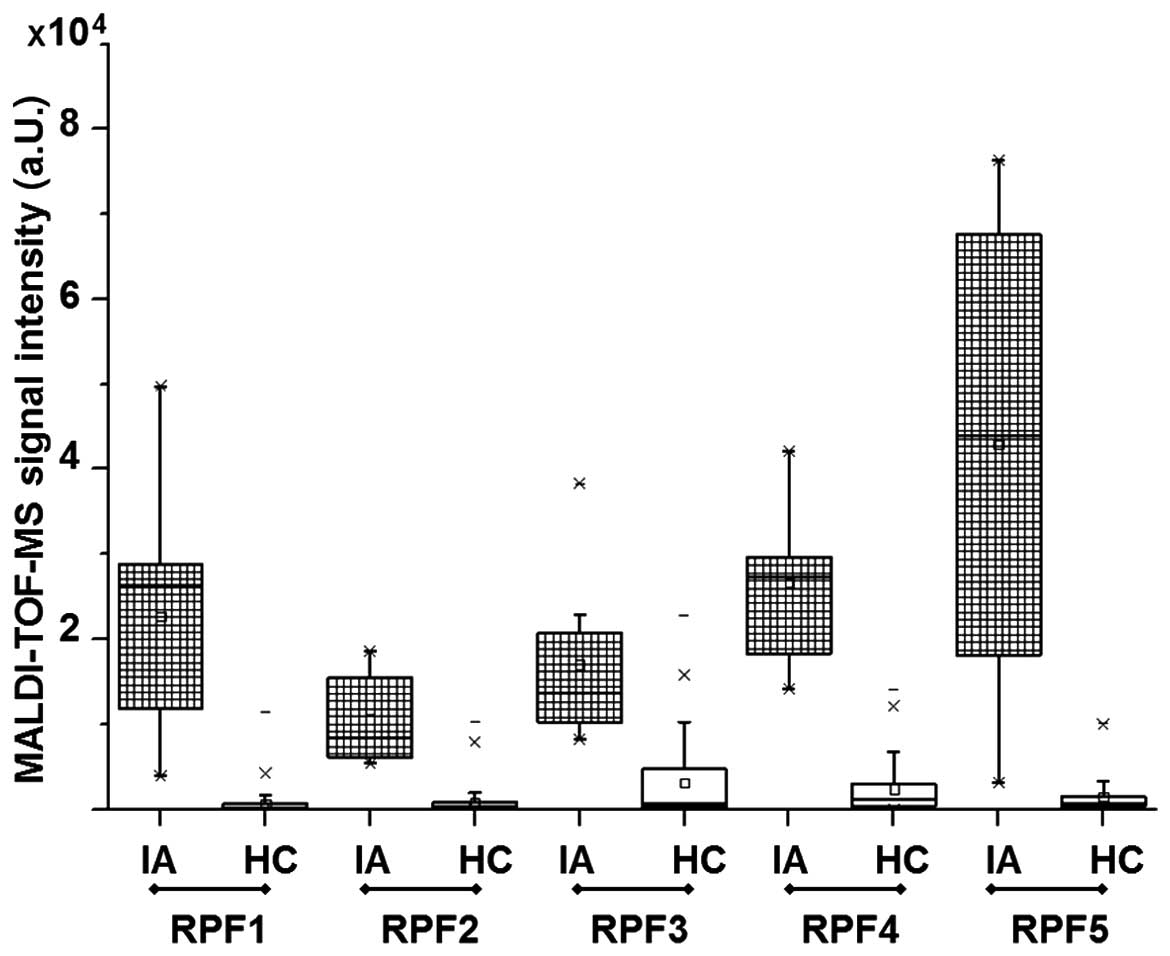
signal intensities of reporter peptide fragments 1–5 were
significantly higher in IA serum specimens when compared to HC
(Fig. 3).
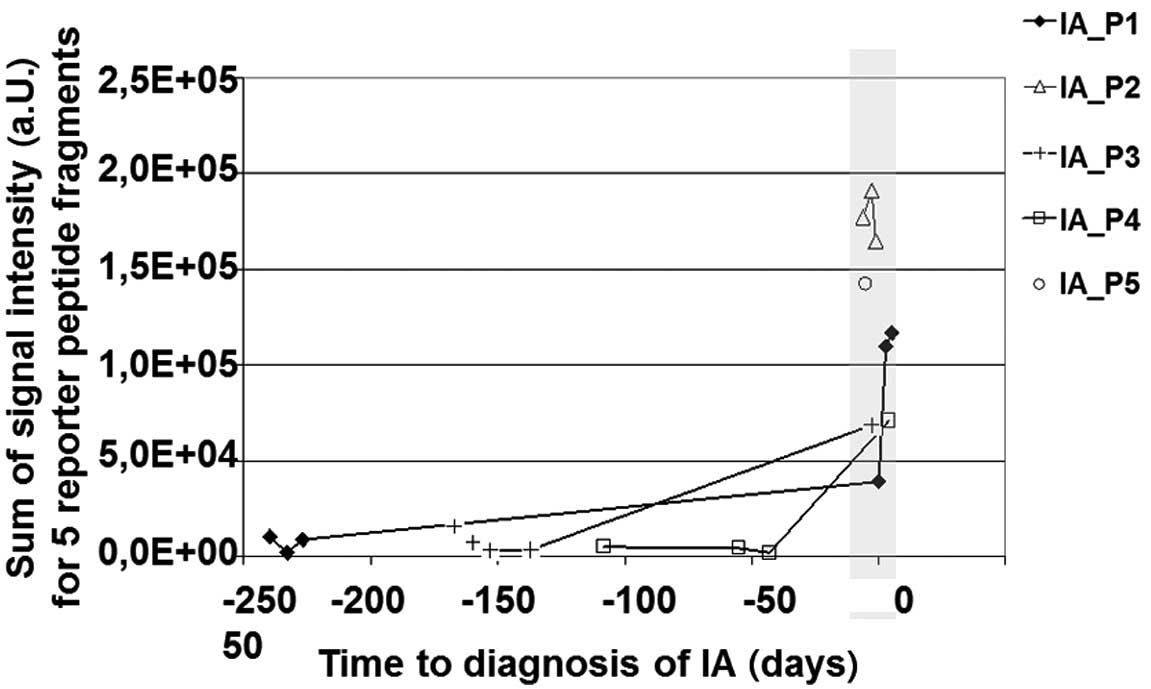
For reason of simplification the signal intensities
of RPFs were summarized and plotted over time to demonstrate that
longitudinal sampling is essential for reporter peptide spiking. As
shown in Fig. 4 there is a strong
accumulation of RPFs next to the time-point (−6 to +4 days) of
diagnosis of proven IA (time-point zero in Fig. 4). In contrast, specimens that were
obtained in a time period ranging from 43 to 240 days prior to
diagnosis of IA showed low signal intensities for RPFs.
Furthermore, these specimens showed no statistically relevant
difference in RPF signal intensities when compared to healthy
controls (data not shown).
Cut-off calculation
The median signal intensity of IA specimens was
116.546 (SD, 53.063) with maximal and minimal values of 190.943 and
39.197, respectively. The median signal intensity of HC specimens
was 5.009 (SD, 8.432) with maximal and minimal values of 36.910 and
210, respectively. As there is no overlap of values between the two
collectives a cut-off >36.910 was chosen that performed with
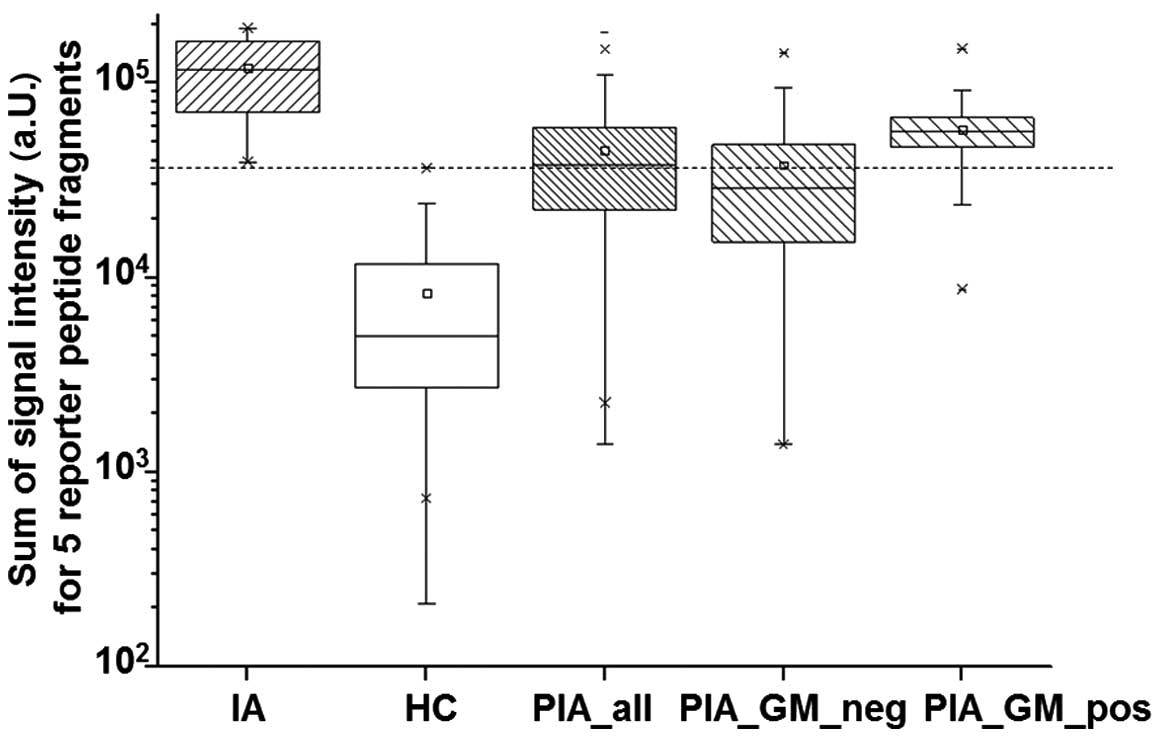
100% diagnostic specificity and sensitivity.
Reporter peptide processing in serum
specimens from patients with possible IA
Reporter peptides were added to serum specimens
(n=144) of 16 patients that were classified as having a possible
invasive aspergillosis (PIA) at the time-point of blood withdrawal.
Samples were processed as described in Materials and methods prior
to MALDI-TOF mass spectrometry. From a total of 144 PIA samples 18%
(26/144) were positive and 65% (93/144) were negative for
galactomannan (GM). For 17% (25/144) of serum specimens no GM
values were available. PIA samples with positive GM values had
significantly higher mass spectrometric signal intensities of
reporter peptide fragments (median, 56.235; SD, 26.298) when
compared to those that were negative for GM (median, 28.781; SD,
30.678). As already mentioned the signal intensities of reporter
peptide fragments showed highest values in IA specimens and lowest
values in HC specimens with no overlap. In contrast, signal
intensity of PIA specimens was between these two collectives and
has overlap with IA and HC collectives, respectively (Fig. 5). With reporter peptide spiking 53%
of specimens (76/144) could be classified as positive and 47%
(68/144) as negative when the cut-off of 36.910 a.U. for sum of RPF
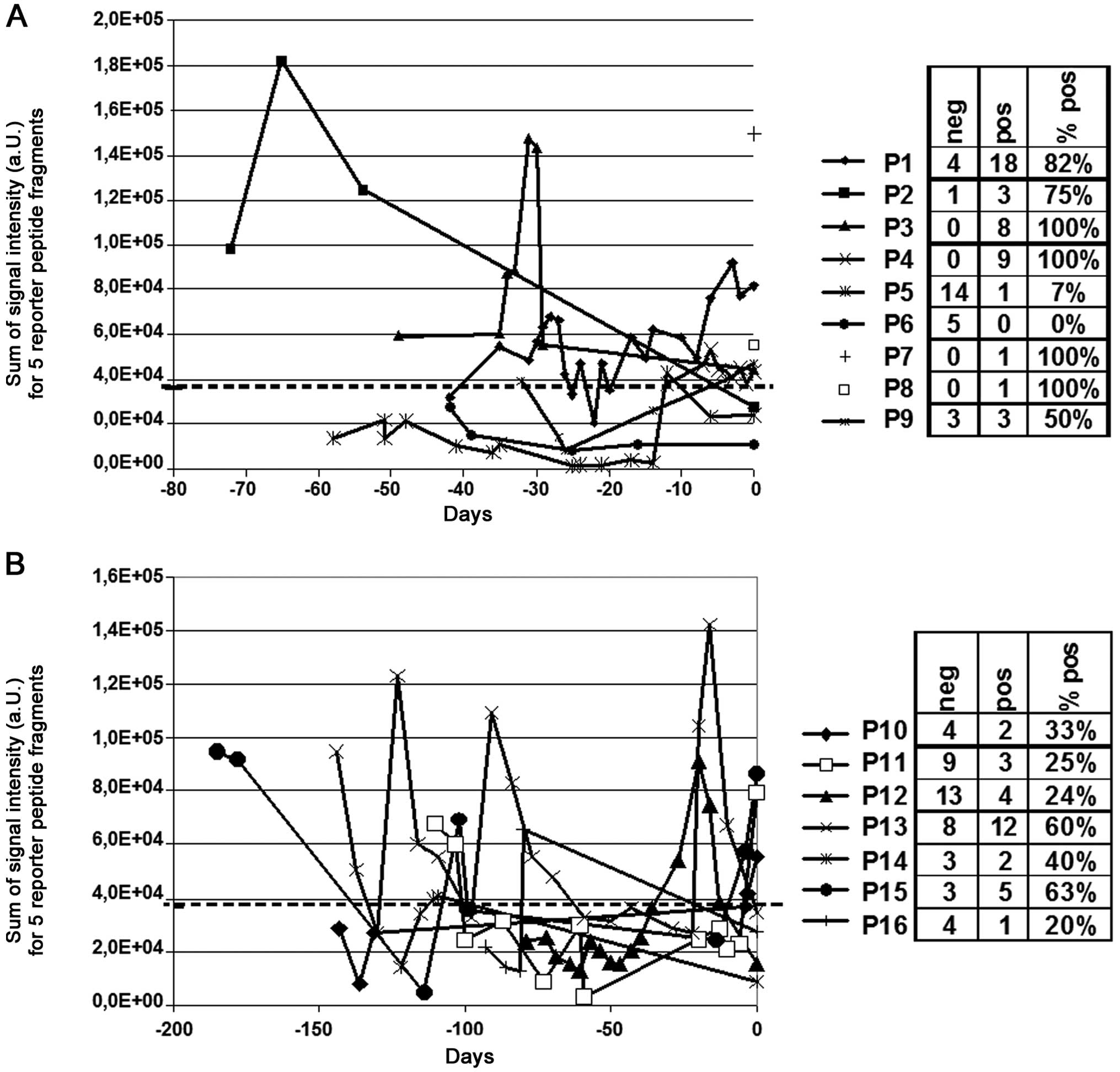
signals was applied. However, the longitudinal monitoring of these
16 patients with RP spiking revealed a heterogeneous pattern of
proteolytic activity (Fig. 6). One
patient had completely negative values over time (patient 6,
Fig. 6A) for the chosen cut-off.
In contrast few other patients had completely positive values over
time (e.g. patient 3, Fig. 6A).
Most patients showed undulating graphs with positive and negative
values and clear maxima with signal intensities >100.000 a.U.
were observed (e.g. patient 13, Fig.
6B). However, for in-depth interpretation of longitudinal
changes additional data, specifically prompt chest CT scan or
Aspergillus antigen testing from BAL would be necessary, but
were not available for this cohort of patients.
Discussion
Since diagnostic tools lack sensitivity or
specificity, the final diagnosis of IA is rather frequently
confirmed only during autopsy (25). Accordingly, there is pressing need
for the development of rapid and accurate diagnostic tests. In the
present study we applied a mass spectrometry based method using
reporter peptide spiking for monitoring of Aspergillus
specific proteolytic activity in serum specimens of healthy
controls and patients with proven or possible IA according to the
EORTC/MSG criteria.
Multiple proteases are secreted from
Aspergillus species during the course of infection (13). We previously demonstrated that
disease specific proteolytic activity can be detected in serum
specimens of respective patients (reviewed in ref. 26). Accordingly, proteases have been
proposed as novel biomarkers for diagnosis of IA (14,20,22).
Spiking of clinical specimens with synthetic reporter peptides
offers an efficient diagnostic tool for monitoring distinct
proteolytic activity in clinical specimens. For example, the
activity of multiple coagulation factors can be quantitatively
assessed by the use of chromogenic substrates (27). These are cleaved by the protease of
interest e.g. the serine protease coagulation factor X that has
high affinity towards a synthetic substrate (S-2222). The cleavage
of S-2222 is inversely correlated to the heparin concentration that
inhibits factor X activity (27).
Mass spectrometry (MS)-based proteomic profiling can
easily be applied to a variety of clinical specimens such as serum,
plasma or tissue samples. MALDI-TOF MS has been typically performed
due to its high sample throughput capacity, robustness and ease of
use. However, the combination of different RPs for multiplex
analysis has limitations due to ion suppression which could hamper
the detection of peptides and lead to an incomplete recovery of
proteolytic fragments (26). In
order to minimize negative effects we applied a tandem affinity
purification protocol for extraction of RPFs as recently described
(16). Furthermore, the synthetic
reporter peptides were stabilized by introducing D-amino acids to
avoid unwanted processing by exoproteases that contribute to the
intrinsic proteolytic activity of serum specimens (28,29).
In the present study, we validated a set of five RPs
for laboratory-based diagnosis of IA. We used a relatively small
number of serum specimens for this proof-of-concept experiment,
because obtaining serum specimens from patients with proven IA is
difficult. We were able to obtain only 9 serum specimens from 5
patients with proven IA. The numerical imbalance of IA specimens
(n=9) compared to HC (n=101) and PIA (n=144) samples is an obvious
limitation of the present study.
The EORTC/MSG criteria were defined to increase
standardization and allowed results of studies of invasive fungal
disease to be interpreted and compared more uniformly (30). However, one of the major hurdles
when performing these criteria is that most patients are classified
as possible cases, whereas probable or proven IA cases are rare
(31–33). For example, Subirà et al
(32) performed a diagnostic study
on 22 patients in order to test the applicability of the EORTC/MSG
criteria. All of the patients in the present study had
haematological malignancies and IA at autopsy. However, results
showed large differences between ante- and post-mortem
classifications. Only 2 patients had been classified as proven
while they were alive, 6 as probable, 13 as possible and 1 remained
unclassified. In addition, 64% of these patients had no
micro-biological or major clinical criteria. Hence, even the 2008
EORTC/MSG criteria still have a low sensitivity rendering the
correct classification of patients an unsolved and serious problem
for clinical studies (32,33).
Accordingly, it may be presumed, that our collective
of possible IA (n=144) also comprises cases of probable or proven
IA as well. Our results for functional protease profiling are
indicating that 53% (76/144) have values above the chosen cut-off
that separates proven IA cases from healthy controls. Furthermore,
higher concentrations of RPFs as surrogate marker for
Aspergillus specific protease activity are associated with
positive galactomannan assays as another surrogate marker for IA
(Fig. 5). In this retrospective
study the patients’ EORTC/MSG status cannot be elucidated
definitely and reclassification attempts are highly speculative.
However, functional protease profiling can be performed from a
simple blood specimen and longitudinal assaying in a future
prospective study seems to be feasible. Laboratory results should
be combined with other diagnostic approaches such as imaging (chest
CT scan) or Aspergillus antigen testing from BAL to enable a
more profound interpretation of the data (34). Taken together, MS-based functional
protease profiling is promising for IA diagnosis and could foster
the diagnostic yield of IA when it is combined with other existing
approaches, and diagnostic accuracy should be evaluated in further
prospective studies.
Acknowledgements
The present study was supported by a grant from the
Deutsche José Carreras Leukämiestiftung (Proposal no. DJCLS R
11/11). We thank Dr William Krüger from the Department of Internal
Medicine C of the Ernst-Moritz-Arndt-University Greifswald, Germany
for providing specimens from patients with proven IA. Furthermore,
we thank Dr Klaus Becker from the Institute for Microbiology of the
Medical Faculty Mannheim of the University of Heidelberg, for
providing cell culture supernatants of Aspergillus species
and results of Galactomannan assays.
Abbreviations:
|
IA
|
invasive aspergillosis
|
|
PIA
|
possible invasive aspergillosis
|
|
HC
|
healthy controls
|
|
MALDI-TOF MS
|
matrix assisted laser desorption
ionisation time of flight mass spectrometry
|
|
RP
|
reporter peptide
|
|
RPF
|
reporter peptide fragment
|
|
CV
|
coefficient of variation
|
|
SD
|
standard deviation
|
|
ACCS
|
Aspergillus fumigatus cell
culture supernatant
|
|
EORTC/MSG
|
European Organization for Research and
Treatment of Cancer/Invasive Fungal Infections Cooperative Group
and the National Institute of Allergy and Infectious Diseases
Mycoses Study Group
|
References
|
1
|
Denning DW: Invasive aspergillosis. Clin
Infect Dis. 26:781–803; quiz 804–805. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Segal BH: Aspergillosis. N Engl J Med.
360:1870–1884. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hohl TM and Feldmesser M: Aspergillus
fumigatus: Principles of pathogenesis and host defense. Eukaryot
Cell. 6:1953–1963. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thompson GR III and Patterson TF:
Pulmonary aspergillosis: Recent advances. Semin Respir Crit Care
Med. 32:673–681. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Latgé JP: Aspergillus fumigatus and
aspergillosis. Clin Microbiol Rev. 12:310–350. 1999.PubMed/NCBI
|
|
6
|
Hope WW, Walsh TJ and Denning DW:
Laboratory diagnosis of invasive aspergillosis. Lancet Infect Dis.
5:609–622. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adam O, Aupérin A, Wilquin F, Bourhis JH,
Gachot B and Chachaty E: Treatment with piperacillin-tazobactam and
false-positive Aspergillus galactomannan antigen test results for
patients with hematological malignancies. Clin Infect Dis.
38:917–920. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
McCulloch E, Ramage G, Rajendran R, Lappin
DF, Jones B, Warn P, Shrief R, Kirkpatrick WR, Patterson TF and
Williams C: Antifungal treatment affects the laboratory diagnosis
of invasive aspergillosis. J Clin Pathol. 65:83–86. 2012.
View Article : Google Scholar
|
|
9
|
Barton RC: Laboratory diagnosis of
invasive aspergillosis: From diagnosis to prediction of outcome.
Scientifica Cairo. 2013:4594052013.PubMed/NCBI
|
|
10
|
Pickering JW, Sant HW, Bowles CA, Roberts
WL and Woods GL: Evaluation of a (1–>3)-beta-D-glucan assay for
diagnosis of invasive fungal infections. J Clin Microbiol.
43:5957–5962. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mennink-Kersten MASH, Warris A and Verweij
PE: 1,3-β-D-glucan in patients receiving intravenous
amoxicillin-clavulanic acid. N Engl J Med. 354:2834–2835. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Einsele H and Loeffler J: Contribution of
new diagnostic approaches to antifungal treatment plans in
high-risk haematology patients. Clin Microbiol Infect. 14(Suppl 4):
37–45. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kogan TV, Jadoun J, Mittelman L,
Hirschberg K and Osherov N: Involvement of secreted Aspergillus
fumigatus proteases in disruption of the actin fiber cytoskeleton
and loss of focal adhesion sites in infected A549 lung pneumocytes.
J Infect Dis. 189:1965–1973. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jambunathan K, Watson DS, Najvar LK,
Wiederhold NP, Kirkpatrick WR, Patterson TF, Askew DS, Kodukula K
and Galande AK: Prolyl endopeptidase activity in bronchoalveolar
lavage fluid: A novel diagnostic biomarker in a guinea pig model of
invasive pulmonary aspergillosis. Med Mycol. 51:592–602. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Findeisen P, Post S, Wenz F and Neumaier
M: Addition of exogenous reporter peptides to serum samples before
mass spectrometry-based protease profiling provides advantages over
profiling of endogenous peptides. Clin Chem. 53:1864–1866. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peccerella T, Lukan N, Hofheinz R,
Schadendorf D, Kostrezewa M, Neumaier M and Findeisen P:
Endoprotease profiling with double-tagged peptide substrates: A new
diagnostic approach in oncology. Clin Chem. 56:272–280. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Findeisen P, Costina V, Yepes D, Hofheinz
R and Neumaier M: Functional protease profiling with reporter
peptides in serum specimens of colorectal cancer patients:
Demonstration of its routine diagnostic applicability. J Exp Clin
Cancer Res. 31:562012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
De Pauw B, Walsh TJ, Donnelly JP, Stevens
DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O,
Kauffman CA, et al: European Organization for Research and
Treatment of Cancer/Invasive Fungal Infections Cooperative Group;
National Institute of Allergy and Infectious Diseases Mycoses Study
Group (EORTC/MSG) Consensus Group: Revised definitions of invasive
fungal disease from the European Organization for Research and
Treatment of Cancer/Invasive Fungal Infections Cooperative Group
and the National Institute of Allergy and Infectious Diseases
Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis.
46:1813–1821. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ostrosky-Zeichner L: Invasive mycoses:
Diagnostic challenges. Am J Med. 125(Suppl): S14–S24. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schaal R, Kupfahl C, Buchheidt D, Neumaier
M and Findeisen P: Systematic identification of substrates for
profiling of secreted proteases from Aspergillus species. J
Microbiol Methods. 71:93–100. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Neustadt M, Costina V, Kupfahl C,
Buchheidt D, Eckerskorn C, Neumaier M and Findeisen P:
Characterization and identification of proteases secreted by
Aspergillus fumigatus using free flow electrophoresis and MS.
Electrophoresis. 30:2142–2150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Watson DS, Feng X, Askew DS, Jambunathan
K, Kodukula K and Galande AK: Substrate specifity profiling of the
Aspergillus fumigatus proteolytic secretome reveals consensus
motifs with predominance of Ile/Leu and Phe/Tyr. PLoS One.
6:e210012011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rawlings ND, Morton FR, Kok CY, Kong J and
Barrett AJ: MEROPS: The peptidase database. Nucleic Acids Res.
36:Database. D320–D325. 2008. View Article : Google Scholar :
|
|
24
|
Findeisen P, Costina V, Yepes D, Hofheinz
R and Neumaier M: Functional protease profiling with reporter
peptides in serum specimens of colorectal cancer patients:
Demonstration of its routine diagnostic applicability. J Exp Clin
Cancer Res. 31:562012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vaideeswar P, Prasad S, Deshpande JR and
Pandit SP: Invasive pulmonary aspergillosis: A study of 39 cases at
autopsy. J Postgrad Med. 50:21–26. 2004.PubMed/NCBI
|
|
26
|
Findeisen P and Neumaier M: Functional
protease profiling for diagnosis of malignant disease. Proteomics
Clin Appl. 6:60–78. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Witt I: Test systems with synthetic
peptide substrates in haemostaseology. Eur J Clin Chem Clin
Biochem. 29:355–374. 1991.PubMed/NCBI
|
|
28
|
Powell MF, Grey H, Gaeta F, Sette A and
Colón S: Peptide stability in drug development: A comparison of
peptide reactivity in different biological media. J Pharm Sci.
81:731–735. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Walker JR, Altman RK, Warren JW and Altman
E: Using protein-based motifs to stabilize peptides. J Pept Res.
62:214–226. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Donnelly JP: Consensus definitions for
invasive fungal disease: Strengths, limitations, and revisions. Med
Mycol. 44(s1): 285–288. 2006. View Article : Google Scholar
|
|
31
|
Subirà M, Martino R, Franquet T, Puzo C,
Altés A, Sureda A, Brunet S and Sierra J: Invasive pulmonary
aspergillosis in patients with hematologic malignancies: Survival
and prognostic factors. Haematologica. 87:528–534. 2002.PubMed/NCBI
|
|
32
|
Subirà M, Martino R, Rovira M, Vazquez L,
Serrano D and De La Cámara R: Clinical applicability of the new
EORTC/MSG classification for invasive pulmonary aspergillosis in
patients with hematological malignancies and autopsy-confirmed
invasive aspergillosis. Ann Hematol. 82:80–82. 2003.PubMed/NCBI
|
|
33
|
Tsitsikas DA, Morin A, Araf S, Murtagh B,
Johnson G, Vinnicombe S, Ellis S, Suaris T, Wilks M, Doffman S, et
al: Impact of the revised (2008) EORTC/MSG definitions for invasive
fungal disease on the rates of diagnosis of invasive aspergillosis.
Med Mycol. 50:538–542. 2012. View Article : Google Scholar
|
|
34
|
Johnson G, Ferrini A, Dolan SK, Nolan T,
Agrawal S, Doyle S and Bustin SA: Biomarkers for invasive
aspergillosis: The challenges continue. Biomarkers Med. 8:429–451.
2014. View Article : Google Scholar
|