Introduction
Multipotent human mesenchymal stroma/stem cells
(MSC) are characterized as a heterogeneous cell population which
can be found in nearly all vascularized organs and tissues. The
stem cell properties include self-renewal and regenerative
potential (1). Moreover, MSC
display at least a tri-lineage differentiation capacity along the
osteogenic, chondrogenic and adipogenic phenotype (2). Certain heterogeneity in morphology
and cell fate as demonstrated by isolation of MSC subpopulations
(3) may result partially from the
cellular microenvironment by neighboring cells, altered trophic
factors, pH or hypoxic conditions. Consequently, a broad range of
simultaneous properties including the capacity for plastic
adherence, paralleled by expression of the CD73, CD90 and CD105
surface molecules with concomitant absence of other cell
type-specific markers including CD14, CD31, CD34 CD45 and HLA-DR
can be identified for MSC (2,4).
MSC are recruited during tissue damage to support
wound repair and tissue regeneration. Likewise, invasive tumor
growth also causes tissue injuries and inflammatory processes with
the consequence of MSC attraction and cellular crosstalk. MSC can
interact with tumor cells via exchange of soluble factors like
chemokines, cytokines and further trophic molecules as well as
various microvesicles including exosomes. These types of
interaction enable multiple pathways for MSC to communicate with
neighboring tumor cells whereby release of exosomes can also affect
more distant tumor cells. Exosomes represent small membrane
particles of endocytic origin which are released into the
extracellular compartment (5) and
contain a large panel of proteins, mRNAs and regulatory microRNAs
(miRs) which can alter the functionality of recipient cells
(6). According to the
heterogeneity of MSC populations, exosomes from MSC of different
tissue origin contain a variety of unique proteins together with
some common exosomal marker proteins such as CD29 or CD63 (7,8).
Several effects of MSC-derived exosomes on tumor cells have been
demonstrated including suppression of angiogenic potential by
down-modulation of VEGF in breast cancer cells via
exosome-associated miR-16 (9).
Moreover, human umbilical cord MSC-derived exosomes can protect
against cisplatin-induced nephrotoxicity and promote cell
proliferation (10) and other
research has demonstrated that human bone marrow MSC-derived
exosomes increase tumor growth in vivo (11), however, mechanisms for these
findings remain unclear.
In the present study, we investigated the effects of
MSC-derived exosomes on different tumor types including breast
cancer and a rare type of ovarian carcinoma cells. The data
demonstrated a variety of functional changes including MMP-2 and
ecto-5′-nucleotidase acquisition by different tumor cells following
internalization of exosomes.
Materials and methods
Cell culture of mesenchymal stem/stroma
cells (MSC)
Primary human mesenchymal stem cells were obtained
after explant culture of umbilical cord tissue; the procedure was
approved by the Ethics Committee of Hannover Medical School,
Project no. 443, February 26, 2009, respectively, following
informed written consent by the patient.
MSC-like cells were isolated from human umbilical
cords as reported previously (12). The cells were obtained from
different patients following delivery of full-term (38–40 weeks)
infants either spontaneously or by cesarean section. In brief,
umbilical cord tissue was washed with PBS to remove blood cells,
cut into ~0.5 cm3 large pieces and incubated in αMEM
(Sigma Chemie GmbH, Steinheim, Germany) supplemented with 15% of
allogeneic human AB-serum (HS; commercially obtained from blood
bank, University Campus Lübeck, Germany), 100 U/ml penicillin, 100
μg/ml streptomycin and 2 mM L-glutamine (Sigma) at 37°C in a
humidified atmosphere with 5% CO2. After ~14 days of
explant culture, the umbilical cord tissue pieces were removed and
the adherent cells were harvested by accutase (Sigma) treatment for
3 min at 37°C. The obtained cell suspension was centrifuged at 320
× g for 5 min and the cells were resuspended in MSC culture medium
(αMEM supplemented with 10% HS, 100 U/ml penicillin, 100 μg/ml
streptomycin and 2 mM L-glutamine) and subcultured in the
appropriate passage. For the experiments, MSC primary cultures from
5 different donors in different passages (P1 to P5) were used
(MSC241111 in P3, MSC131113 in P2, MSC101213 in P1 and P3,
MSC180314 in P5 and MSC270114 in P1), respectively.
Cell culture of tumor cells
Human MDA-MB-231 and MCF-7 breast carcinoma cell
lines were obtained from the American Type Culture Collection
(Rockville, MD, USA). MCF-7 cells were grown in Dulbecco’s modified
Eagle’s medium and MDA-MB-231 cells were cultured in Leibovitz
medium supplemented with [10% (v/v) fetal calf serum, 2 mM
L-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin] (all
from Sigma Chemie GmbH, Steinheim, Germany), respectively.
Subculture was performed by trypsin/EDTA (Biochrom GmbH, Berlin,
Germany) treatment for 5 min at 37°C.
SCCOHT-1 cells represent a spontaneously
proliferating population derived from a patient with recurrent
small cell carcinoma of the ovary hypercalcemic type (SCCOHT) and
were maintained in RPMI-1640 with medium supplements as described
previously (13).
Cells were cultivated at 37°C in a humidified
atmosphere containing 5% CO2 and tested for mycoplasma
by the luminometric MycoAlert Plus mycoplasma detection kit (Lonza
Inc., Rockland, ME, USA) according to the manufacturer’s
recommendations. Moreover, authentication of the cell lines was
performed by short tandem repeat (STR) fragment analysis using the
GenomeLab human STR primer set (Beckman Coulter Inc., Fullerton,
CA, USA) demonstrating similar STR pattern according to the STR
database provided by the Deutsche Sammlung von Mikroorganismen und
Zellkulturen (DSMZ, Braunschweig, Germany).
Labeling of MSC by lentiviral
transduction
For discrimination of the different tumor cells in
co-culture with MSC and for proliferation measurements, all tumor
cell populations were transduced with a 3rd generation lentiviral
SIN vector containing the mcherry gene. Likewise, the different MSC
populations were transduced with a 3rd generation lentiviral SIN
vector containing the eGFP gene according to a labeling technique
previously described (14).
Analysis of surface markers by flow
cytometry
Characterization of the MSC immunophenotype was
performed as described previously (15). Briefly, continuously proliferating
MSC were harvested and analyzed for cell surface marker expression
by flow cytometry. After blocking non-specific binding to
Fc-receptors by incubation of 106 cells with 2% FCS for
30 min at 4°C and washing with PBS-BSA, the cells were incubated
with the following appropriately-labeled monoclonal anti-human
antibodies, respectively: CD73-PE (clone AD2) (BD Bioscience);
CD90-PE (clone 5E10, IgG1, BioLegend Inc., San Diego, CA, USA);
CD105-PE (clone 43A3, IgG1, BioLegend Inc.); CD31-FITC (Miltenyi
Biotec, Bergisch-Gladbach, Germany); CD34-PE and CD45-PE (BD
Biosciences). Following antibody staining, all samples were washed
twice with PBS-BSA. Positive staining was obtained according to
control measurements of the different populations with
isotype-matching IgG control antibodies. Flow cytometry analysis
and histograms were performed in a Galaxy FACSan (Partec) using
FloMax analysis software (Partec).
Preparation of exosomes
Exosomes were isolated using the total exosome
isolation kit reagent (Invitrogen, USA). After culture in
serum-free conditions for 24 h, cell media were harvested from
MSCGFP, MCF-7cherry, and
MDA-MB231cherry mono-cultures and from co-cultures of
MSC with the two breast cancer cell lines, respectively. Co-culture
of MSC and the tumor cells was performed at a cell ratio of 60:40.
The cells were cultured at an initial density of 2,000
cells/cm2 for 7–15 days and following medium exchange
with serum-free media, the released exosomes were isolated after
subsequent 24 h.
The serum-free cell media samples were centrifuged
at 2,000 × g for 30 min to remove cell debris. The supernatant was
stored on ice and supplemented with the half volume of the total
exosome isolation kit reagent (Invitrogen) according to the
manufacturer’s instructions. After thorough mixing, the samples
were incubated at 4°C overnight and centrifuged at 100,000 × g/4°C
for 1 h. The supernatant was aspirated and discarded, and the
exosome pellet was resuspended for protein and enzymatic
analyses.
For incorporation of MSCGFP-derived
exosomes into tumor cells, equal aliquots of exosomes were added to
104 MCF-7 or SCCOHT-1 cells in a 6-well plate (Nunc).
The tumor cell cultures were then incubated for 24 h in the
presence of MSCGFP-derived exosomes followed by 5
extensive washes of the wells with PBS to remove free and loosely
cell-attached exosomes. Thereafter, the cells were detached by
trypsin/EDTA treatment, homogenized in appropriate buffer and cell
homogenates were analyzed by western blot analysis or zymography
assay. Alternatively, analysis of exosomes and of the tumor cells
with incorporated GFP-labeled exosomes was performed after lysis in
10% (w/v) SDS. Relative fluorescence intensities of the homogenates
were quantified for GFP (excitation 485 nm/emission 520 nm) using
the Fluoroscan Ascent Fl (Thermo Fisher Scientific).
2D-gel analysis and mass spectrometry
identification
The proteins of the different exosome preparations
were separated by isoelectric focusing (IEF) followed by SDS-PAGE
in the second dimension, respectively. Thus, aliquots of exosomes
from each preparation were incubated in reswelling buffer [8 M
urea, 1% CHAPS (v/v), 0.5% pharmalytes 3–10 (v/v), 0.002%
bromophenol blue (w/v), 0.4% DTT (w/v); according to the GE
Healthcare protocol] on an 18-cm IPG Immobiline Dry Strip (pH
3.0–10.0; NL) (Amersham Biosciences GmbH, Freiburg, Germany) and
separated for 18 h at 150 V in the first dimension using the
IPGphor isoelectric focusing system (Amersham). Thereafter, the IPG
strips were incubated in two subsequent equilibration buffers for
15 min, respectively (according to the GE Healthcare protocol), and
polymerized on a 10% SDS-PAGE separation gel using 0.5% (w/v) low
melting point agarose. Electrophoresis was standardized using
appropriate molecular-weight markers (Amersham). Following staining
of the gels with Coomassie brilliant blue appropriate protein spots
were cut and analyzed by liquid chromatography coupled with tandem
mass spectrometry (LC-MS/MS) using the AB5800 TOF/TOF (ABSys GmbH,
Darmstadt, Germany).
Western blot analysis
Exosome aliquots from the different cell cultures
were homogenized in RIPA buffer containing 0.3 M NaCl, 1% (w/v)
sodium desoxycholate, 0.1% (w/v) sodium dodecyl sulfate (SDS), 1%
(v/v) Triton X-100, 20 mM Tris-HCl (pH 8.0), 1 mM EDTA supplemented
with 1 mM phenylmethylsulfonylfluoride (PMSF). Approximately 15 μg
of exosomal protein was separated by electrophoresis on a 15%
SDS-polyacrylamide gel and transferred to a PVDF membrane
(Millipore Corp., Bedford, MA, USA). Immunoblotting was performed
with the following antibodies: polyclonal anti-CD63; polyclonal
anti-collagen αI; polyclonal anti-collagen αII, polyclonal
anti-CD73 and anti-MMP2 (each 1:500 dilution; all from Abcam,
Cambrige, UK); monoclonal anti-CD90 (1:500 dilution; Dianova,
Hamburg, Germany). Odyssey 680/800 nm secondary conjugates were
used for the quantification of protein expression levels and
signals were visualized using the Odyssey Infra-Red Imaging System
and software (Li-Cor BioSciences, Lincoln, NE, USA).
Transcript analysis by RT-PCR
Total RNA was isolated from MSC and MSC-derived
exosomes using RNeasy Mini kit (Qiagen, Hilden, Germany) according
to the manufacturer’s instructions. One microgram of RNA was
reverse transcribed into cDNA using 500 μM of dNTP (R0193), 5 μM
Oligo(dT)18 primer (S0132), 5 μM Random Hexan primer (S0142), 1 U
RiboLock™ RNase inhibitor (E00381) and 5 U RevertAid™ M-MuLV
reverse transcriptase (EP0441) in the supplied reaction buffer (all
reagents from Thermo Scientific, Schwerte, Germany). The cDNA
reactions were performed for 10 min/25°C, 1 h/37°C and stopped at
72°C for 10 min. As a template 2.5 μl of cDNA was used with primers
specific for: CD73 (forward, 5′-CGCAACAATGGCACAATTAC-3′; reverse,
5′-CTCGACA CTTGGTGCAAAGA-3′; amplification product 241 bp); CD90
(forward, 5′-GGACTGAGATCCCAGAACCA-3′; reverse,
5′-ACGAAGGCTCTGGTCCACTA-3′; amplification product 124 bp); CD105
(forward, 5′-TGTCTCACTTCATGCCTCC AGCT-3′; reverse,
5′-AGGCTGTCCATGTTGAGGCAGT-3′; amplification product 378 bp); MMP-2
(forward, 5′-TTTTCT CGAATCCATGATGG-3′; reverse, 5′-CTGGTGCAGCTCT
CATATTT-3′; amplification product 619 bp); fibronectin (forward,
5′-AGCCGCCACGTGCCAGGATTAC-3′; reverse,
5′-CTTATGGGGGTGGCCGTTGTGG-3′; amplification product 439 bp); (all
primers customized by Eurofins, MWG GmbH, Ebersberg, Germany). PCR
reactions included 0.2 μM of each primer, 200 μM of dNTP (R0193,
Thermo Scientific) and 0.03 U One Taq Hot Start DNA polymerase (New
England Biolabs GmbH, Frankfurt am Main, Germany) in the supplied
reaction buffer. PCR cycling conditions were performed 30 sec at
94°C, 1 min at 60 and 68°C for 1 min, respectively, including an
initial 30-sec denaturation step at 94°C and a final 5-min
extension step at 68°C (35 cycles). Aliquots of 25 μl of each
RT-PCR product were separated on a 2% agarose gel and visualized by
GelRed™ (Biotium Inc., Hayward, CA, USA) staining.
MMP-2 zymography assay
Exosome aliquots from the MSCGFP,
MDA-MB-231cherry cells, and from co-cultures of
MSCGFP/MDA-MB-231cherry cells were used in a
zymographic assay. Moreover, conditioned media was prepared from
106 MSCGFP or MCF-7cherry cells or
MCF-7cherry cells with incorporated
MSCGFP-derived exosomes after 24-h culture in 0.1% serum
and concentrated ~20-fold using Amicon Ultra-4 Centrifugal Filter
Devices (10 kDa; Millipore, Carrigtwohill, Ireland) according to
the manufacturer’s instructions. In the following MMP-2 zymographic
assay, aliquots were mixed 2:1 (v/v) with non-reducing sample
buffer [10 mM Tris (pH 6.0–8.0), 1% SDS, 10% glycerol and 0.02%
bromophenol blue] and subjected to SDS-PAGE containing 2 mg/ml of
gelatine (Sigma). Electrophoresis was performed for 30 min at 60 V
followed by 120 min at 125 V. The gels were washed twice in 2.5%
Triton X-100 on a vertical shaker and five times with
H2O. Thereafter, the gels were incubated with fresh MMP
enzyme buffer (50 mM Tris-HCl, pH 7.0, 5 mM CaCl2)
overnight at 37°C. Finally, the gels were stained with 0.4%
Coomassie blue whereby the proteolytic activity was detectable by
the appearance of light bands.
CD73 activity by analysis of 5′-AMP and
adenosine
Exosomes were isolated from steady-state SCCOHT-1
mono-cultures after 7 days, from steady-state MSC101213 P3
mono-cultures after 7 days, and from 7-day co-culture of these MSC
with SCCOHT-1 (ratio 60:40; initial seeding of 2,000
cells/cm2). The appropriate exosomal fractions as well
as SCCOHT-1 control cells and SCCOHT-1 cells with 24 h incorporated
MSC-derived exosomes were cultivated in PBS with 20 μM 5′-AMP
(Sigma, Schnelldorf, Germany) as a substrate for 30 min at 37°C. An
exosome-free and a cell-free PBS incubation served as a negative
control, respectively. Supernatants were collected and centrifuged
(14.000 × g/5 min) to remove debris and supernatants were analyzed
by HPLC-MS/MS using a Shimadzu HPLC-system (Shimadzu, Duisburg,
Germany) coupled with a QTRAP5500™ triple quadrupole mass
spectrometer (ABSCIEX, Foster City, CA, USA) operating in positive
ionization mode. Reversed phase chromatographic separation of
adenosine and 5′-AMP was performed on a Hypercarb column (30×4.6
mm; 5 μm; Thermo Scientific, Dreieich, Germany) using a linear
organic gradient. Data were collected and analyzed with Analyst
1.5.1 software (ABSCIEX).
Results
Direct cellular interaction of MSCGFP
with MDA-MB-231cherry breast cancer cells was associated
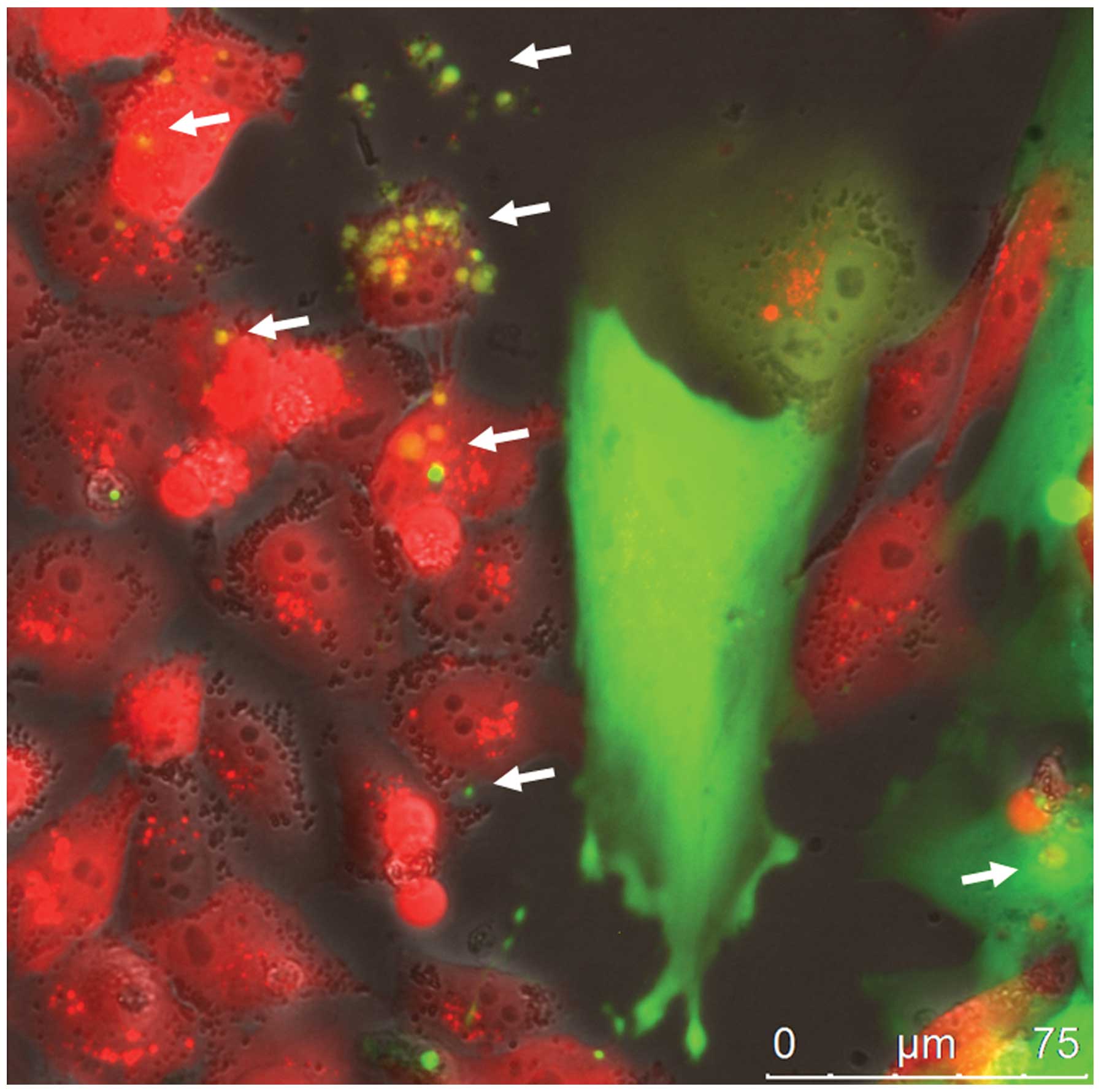
with the exchange of exosome-like micovesicles (Fig. 1). GFP-labeled exosomes derived from
MSC241111GFP P3 were detectable within the extracellular
space and were incorporated into MDA-MB-231cherry breast
cancer cells as indicated by the white arrows (Fig. 1). While these cellular interactions
and the exchange of exosomes suggested an intercellular transfer of
biological material between MSC and the tumor cells, a 24-h
production of exosomes was isolated from the mono-cultures and the
co-cultures for further analysis. The protein amount of isolated
exosomes from MDA-MB-231cherry mono-cultures was 108 μg,
from MSC241111GFP was 90 μg, and the co-culture of
MSCGFP with MDA-MB-231cherry yielded 144 μg
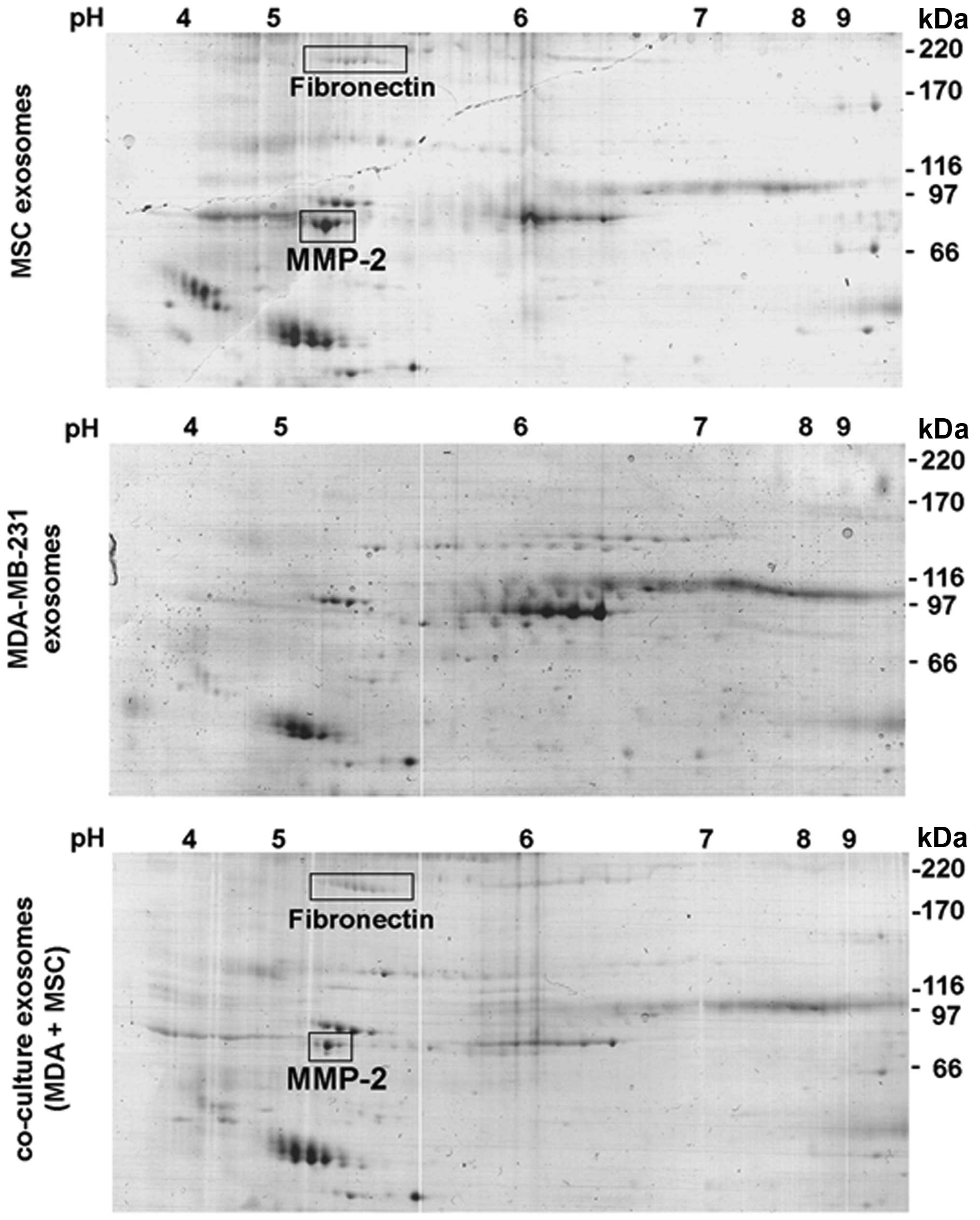
within 24 h. Comparative protein analysis was performed by 2D-gel
separation of 35 μg of the exosomal protein fraction of each
preparation (Fig. 2). A variety of
common protein spots were detectable in all 3 preparations,
however, at least two additional proteins were identified in MSC
exosomes (Fig. 2, upper panel) and
co-culture exosomes (Fig. 2, lower
panel) in contrast to MDA-MB-231 exosomes (Fig. 2, middle panel). Mass spectrometric
analysis revealed fibronectin and matrix metalloproteinase-2
(MMP-2) as those protein spots which were undetectable in exosomes
from MDA-MB-231 cells (Fig. 2).
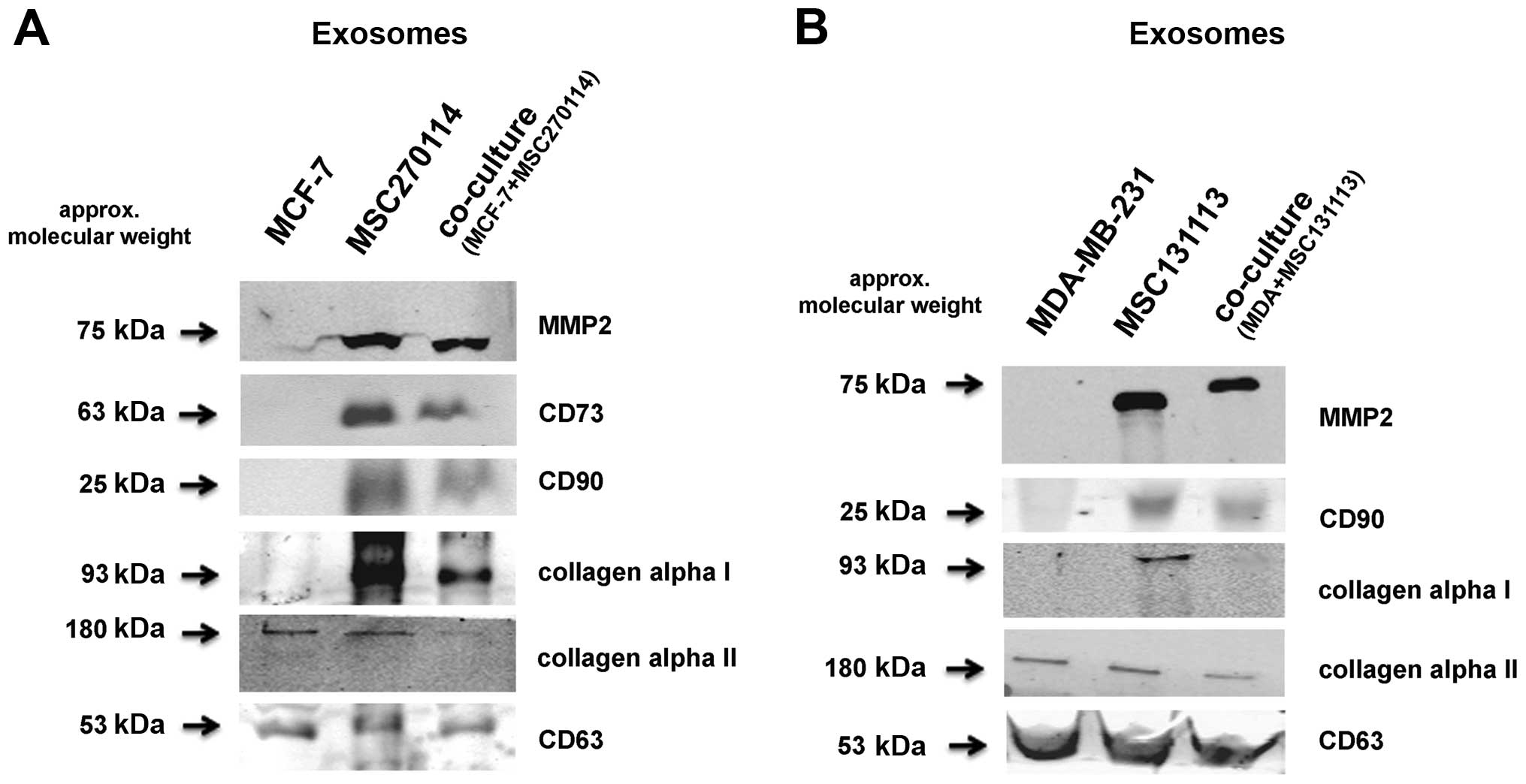
Western blot analysis of exosomal preparation confirmed these
findings (Fig. 3A). Moreover,
similar data were also obtained from exosomes after co-culture of
MSC with MCF-7 breast cancer cells (Fig. 3B). In addition to the presence of
MMP-2 exclusively in exosomes from MSC and the co-culture, the
GPI-anchored CD90 antigen was similarly detectable and likewise the
ecto-5′-nucleotidase CD73, all of which remain absent in the two
breast cancer cell lines (Fig. 3).
Whereas fibronectin can associate with certain types of collagen,
the isoform collagen α1 was also detectable in exosomes from MSC
mono-cultures and was reduced in the co-culture exosomes, however,
it was undetectable in the exosome preparation of MDA-MB-231 and
MCF-7 cells, respectively (Fig.
3). In contrast, collagen α2 appeared in all exosome
preparations and likewise, CD63 as an exosomal marker protein was
uniformly present (Fig. 3).
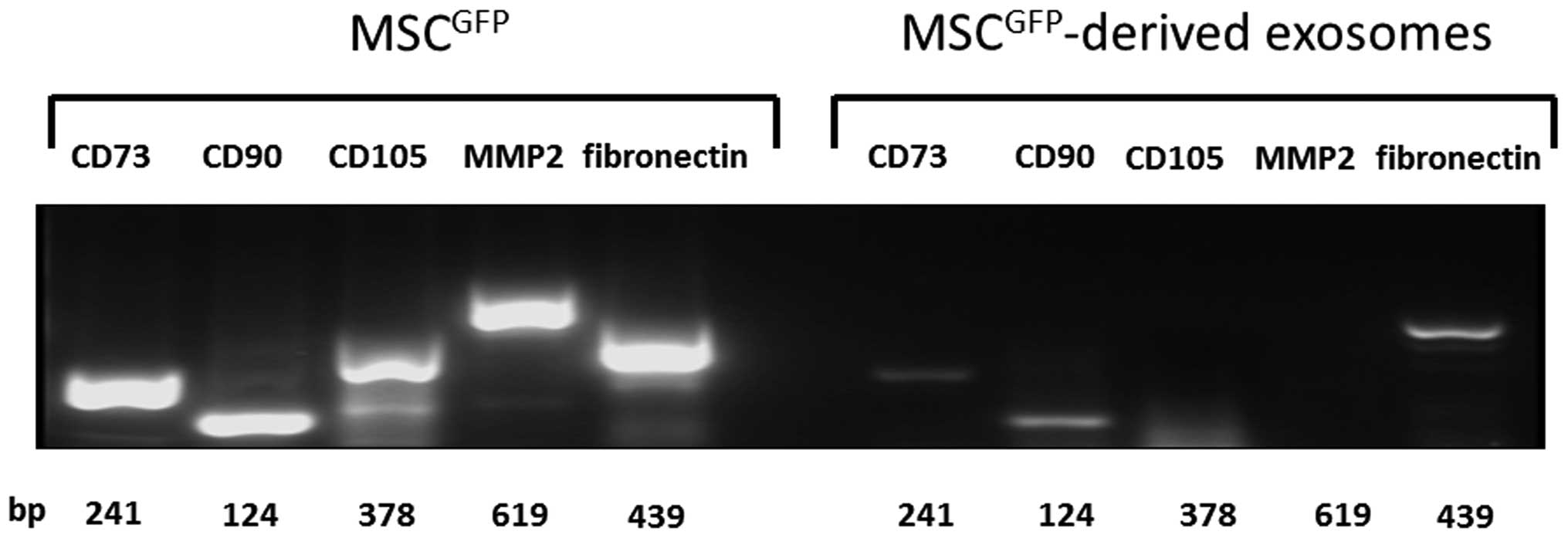
Furthermore, transcripts of the typical MSC marker
proteins CD73, CD90, and CD105 were detectable in MSC lysates as
well as MMP2 and fibronectin in accordance with the mass
spectrometric analysis of the 2D gels (Fig. 4). Likewise, mRNAs of CD73, CD90,
and fibronectin also appeared in MSC-derived exosomes although in
reduced quantities as compared to the cells. However, PCR products
of CD105 and MMP-2 remained undetectable in these vesicles
(Fig. 4) suggesting that only the
proteins are carried.
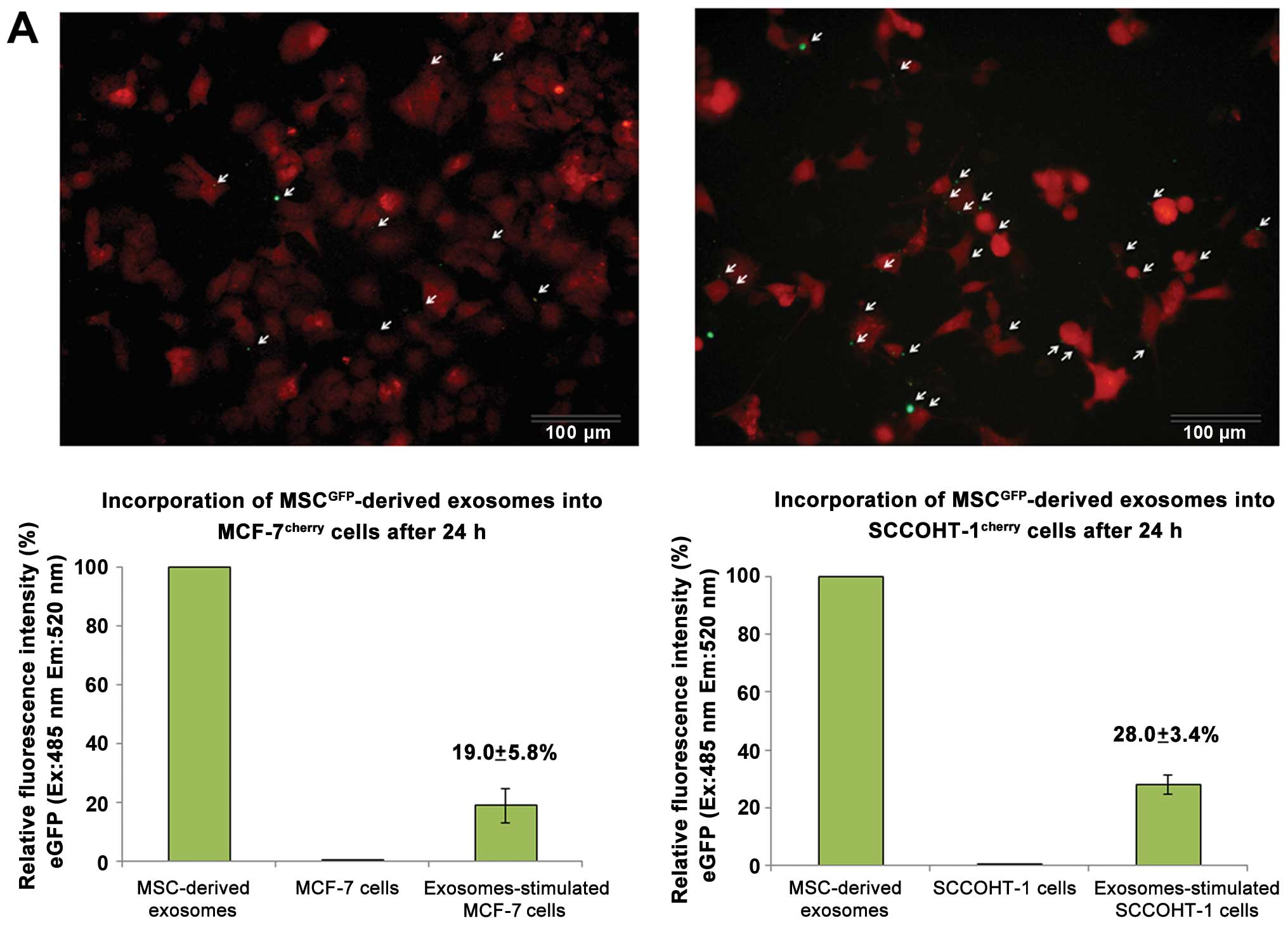
Incubation of MSCGFP-derived exosomes
with tumor cells was associated with an uptake of exosomes by
MCF-7cherry cells (Fig.
5A, upper left) and by SCCOHT-1cherry cells
(Fig. 5A, upper right). The tumor
cell cultures were incubated with the exosomes for 24 h and
analysis of incorporated exosomes was performed after 5 extensive
washes with PBS to remove free and loosely cell-attached exosomes.
Quantification of incorporated exosomes into the cells was
performed by fluorescence measurement of the GFP-labeled vesicles
and revealed a relative uptake of 19.0±5.8% (n=3) by MCF-7 cells
(Fig. 5A, lower left) and 28.0±3.4
% (n=3) by SCCOHT-1 cells (Fig.
5A, lower right) after 24 h compared to the total amount of
available exosomes.
To test, whether the assimilation of MSC-derived
exosomes by tumor cells includes transfer of biologically active
proteins, gelatinase activity was measured for MMP-2 in an
appropriate gelatin zymography assay. In contrast to exosomes from
MDA-MB-231, MSC-derived exosomes and exosomes isolated from the
MSC/breast cancer cell co-culture exhibited a marked gelatinase
activity at ~63 kDa, which corresponded to the active form of MMP-2
(Fig. 5B). A potential
MSC-mediated transfer of MMP-2 was also tested for MCF-7 breast
cancer cells. MSC cell homogenates demonstrated MMP-2 protein
expression in contrast to undetectable MMP-2 levels in MCF-7 cells.
However, incubation of MCF-7 cells in the presence of MSC-derived
exosomes and subsequent removal of non-incorporated exosomes by
extensive washes of the cells was associated with MMP-2 protein
expression in the breast cancer cells (Fig. 5C). Moreover, the acquired MMP-2
expression in MCF-7 cells by MSC-derived exosomes was also tested
for enzymatic activity. Whereas MSC cell homogenates exhibited
MMP-2 gelatinase in-gel activity at ~59 and 63 kDa, there was
little if any MMP-2 activity detectable in MCF-7 cell homogenates.
In contrast, MMP-2 activity was displayed in MCF-7 cells after
incorporation of MSC-derived exosomes in the zymography assay
(Fig. 5D).
Further acquisition of enzymatic activity from
MSC-derived exosomes was evaluated for the CD73
ecto-5′-nucleotidase which associates to the external face of the
plasma membrane via a GPI-anchor. The exosomal transfer of
biologically active CD73 was tested by the capability to metabolize
5′-AMP into adenosine using a tumor cell culture model of primary
cells from a small cell hypercalcemic ovarian carcinoma (SCCOHT-1).
The preparation of exosomes from SCCOHT-1 cells demonstrated no
detectable adenosine synthesis similar to PBS control incubation
without any cells (Fig. 5E). In
contrast, LC/MS-MS analysis of exosomes from MSC mono-culture and
from MSC/SCCOHT-1 co-culture revealed a marked reduction of the
substrate 5′-AMP paralleled by a significantly increased level of
the product adenosine (Fig. 5E)
suggesting CD73 activity exclusively in MSC-derived exosomes.
Transfer of CD73 became obvious when a markedly elevated
ecto-5′-nucleotidase activity was detectable after incorporation of
MSC-derived exosomes into SCCOHT-1 cells compared to SCCOHT-1
control cells (Fig. 5F).
Discussion
Previous research has demonstrated that mesenchymal
stem/stroma cells contribute to a direct interaction with tumor
cells and promote mutual exchange/induction of cellular markers
(14,16,17).
Alternatively, MSC interaction can be mediated indirectly by the
release of soluble biological factors (18) and/or vesicles such as exosomes
whereby MSC can affect cellular functionality of distant cell
populations in a paracrine manner. These effects can be mediated
both, by proteins and RNAs including mRNAs and miRs.
The appearance of the MSC marker proteins CD73, CD90
and CD105 in MSC-derived exosomes has been confirmed by previous
studies (19). In addition,
MSC-derived exosomes also contain transcripts for CD73, CD90, and
fibronectin, whereas CD105 expression remained undetectable.
Likewise, MMP-2 mRNAs were not observed in MSC-derived exosomes,
suggesting appropriate protein transport in the microvesicles or
regulators that induced expression in the target cells. Previous
research has demonstrated that cancer cell-associated fibronectin
induces release of matrix metalloproteinase-2 from normal
fibroblasts (20). Consequently,
MMP-2 protein appearance in MCF-7 and MDA-MB-231 breast cancer cell
populations with corresponding enzymatic activity after
incorporation of MSC-derived exosomes enables degradation of
certain collagens as structural component of basement membranes.
Therefore, the acquisition of distinct matrix metalloproteinase
activities suggested new properties of the tumor cells with the
capability to restructure the tumor microenvironment. Indeed,
transfection of MDA-MB-231 human breast cancer cells with
pro-matrix metalloproteinase-2 increased growth and metastasis in
nude mice (21). Moreover,
supportive evidence demonstrated that human adipose MSC-derived
exosomes in conditioned medium promote migratory activity of MCF-7
cells accompanied by an upregulation of several cancer-related
pathways such as Wnt signaling (22).
The incorporation of MSC-derived exosomes was also
associated with acquired ecto-5′-nucleotidase activity by SCCOHT-1
tumor cells whereby this interaction included both, protein and
corresponding mRNA assimilation. Previous findings substantiated
acquisition of 5′-nucleotidase activity by SCCOHT-1 cells (17) and by normal natural killer cells
(23) following direct co-culture
with MSC which suggested a transfer by the cells and/or by the
exosomes. With this new capability of metabolizing 5′-AMP into
adenosine, SCCOHT-1 cells can suppress and modulate
pro-inflammatory activities via activation of adenosine receptor
signaling present on the surface of most immune cells (24). Indeed, previous reports
demonstrated that exosomal conversion of 5′-AMP to adenosine can
inhibit T cell activation in a tumor microenvironment (25).
Together, these findings suggested that MSC-derived
exosomes can change the cellular functionality of tumor cells by
induction of MMP-2 and ecto-5′-nucleotidase activity and thereby
contribute to an altered tumor microenvironment and increased tumor
heterogeneity (26,27). Further factors including certain
microRNAs in MSC-derived exosomes can also hide metastatic breast
cancer cells by inducing dormancy (28). Thus, functional changes by
MSC-derived exosomes can support a protection of tumor cells
against chemotherapeutic approaches and consequently promote tumor
cell resistance. Alternatively, MSC-derived exosomes may represent
a useful carrier to deliver antitumor cargo.
Acknowledgements
Yuanyuan Yang is a visiting research fellow from
Tongji University, Shanghai, China.
References
|
1
|
Caplan AI: Mesenchymal stem cells. J
Orthop Res. 9:641–650. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Majore I, Moretti P, Hass R and Kasper C:
Identification of subpopulations in mesenchymal stem cell-like
cultures from human umbilical cord. Cell Commun Signal. 7:62009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop DJ and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The International Society for Cellular
Therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Théry C, Regnault A, Garin J, Wolfers J,
Zitvogel L, Ricciardi-Castagnoli P, Raposo G and Amigorena S:
Molecular characterization of dendritic cell-derived exosomes.
Selective accumulation of the heat shock protein hsc73. J Cell
Biol. 147:599–610. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lai RC, Tan SS, Teh BJ, Sze SK, Arslan F,
de Kleijn DP, Choo A and Lim SK: Proteolytic potential of the MSC
exosome proteome: Implications for an exosome-mediated delivery of
therapeutic proteasome. Int J Proteomics. 2012:9719072012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu B, Zhang X and Li X: Exosomes derived
from mesenchymal stem cells. Int J Mol Sci. 15:4142–4157. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee JK, Park SR, Jung BK, Jeon YK, Lee YS,
Kim MK, Kim YG, Jang JY and Kim CW: Exosomes derived from
mesenchymal stem cells suppress angiogenesis by down-regulating
VEGF expression in breast cancer cells. PLoS One. 8:e842562013.
View Article : Google Scholar
|
|
10
|
Zhou Y, Xu H, Xu W, Wang B, Wu H, Tao Y,
Zhang B, Wang M, Mao F, Yan Y, et al: Exosomes released by human
umbilical cord mesenchymal stem cells protect against
cisplatin-induced renal oxidative stress and apoptosis in vivo and
in vitro. Stem Cell Res Ther. 4:342013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu W, Huang L, Li Y, Zhang X, Gu J, Yan
Y, Xu X, Wang M, Qian H and Xu W: Exosomes derived from human bone
marrow mesenchymal stem cells promote tumor growth in vivo. Cancer
Lett. 315:28–37. 2012. View Article : Google Scholar
|
|
12
|
Lavrentieva A, Majore I, Kasper C and Hass
R: Effects of hypoxic culture conditions on umbilical cord-derived
human mesenchymal stem cells. Cell Commun Signal. 8:182010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Otte A, Göhring G, Steinemann D,
Schlegelberger B, Groos S, Länger F, Kreipe HH, Schambach A,
Neumann T, Hillemanns P, et al: A tumor-derived population
(SCCOHT-1) as cellular model for a small cell ovarian carcinoma of
the hypercalcemic type. Int J Oncol. 41:765–775. 2012.PubMed/NCBI
|
|
14
|
Mandel K, Yang Y, Schambach A, Glage S,
Otte A and Hass R: Mesenchymal stem cells directly interact with
breast cancer cells and promote tumor cell growth in vitro and in
vivo. Stem Cells Dev. 22:3114–3127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Otte A, Bucan V, Reimers K and Hass R:
Mesenchymal stem cells maintain long-term in vitro stemness during
explant culture. Tissue Eng Part C Methods. 19:937–948. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hass R and Otte A: Mesenchymal stem cells
as all-round supporters in a normal and neoplastic
microenvironment. Cell Commun Signal. 10:262012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Y, Otte A and Hass R: Human
mesenchymal stroma/stem cells exchange membrane proteins and alter
functionality during interaction with different tumor cell lines.
Stem Cells Dev. Jan 26–2015.(Epub ahead of print). View Article : Google Scholar
|
|
18
|
Karnoub AE, Dash AB, Vo AP, Sullivan A,
Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R and Weinberg
RA: Mesenchymal stem cells within tumour stroma promote breast
cancer metastasis. Nature. 449:557–563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim HS, Choi DY, Yun SJ, Choi SM, Kang JW,
Jung JW, Hwang D, Kim KP and Kim DW: Proteomic analysis of
microvesicles derived from human mesenchymal stem cells. J Proteome
Res. 11:839–849. 2012. View Article : Google Scholar
|
|
20
|
Saad S, Gottlieb DJ, Bradstock KF, Overall
CM and Bendall LJ: Cancer cell-associated fibronectin induces
release of matrix metalloproteinase-2 from normal fibroblasts.
Cancer Res. 62:283–289. 2002.PubMed/NCBI
|
|
21
|
Tester AM, Waltham M, Oh SJ, Bae SN, Bills
MM, Walker EC, Kern FG, Stetler-Stevenson WG, Lippman ME and
Thompson EW: Pro-matrix metalloproteinase-2 transfection increases
orthotopic primary growth and experimental metastasis of MDA-MB-231
human breast cancer cells in nude mice. Cancer Res. 64:652–658.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin R, Wang S and Zhao RC: Exosomes from
human adipose-derived mesenchymal stem cells promote migration
through Wnt signaling pathway in a breast cancer cell model. Mol
Cell Biochem. 383:13–20. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chatterjee D, Tufa DM, Baehre H, Hass R,
Schmidt RE and Jacobs R: Natural killer cells acquire CD73
expression upon exposure to mesenchymal stem cells. Blood.
123:594–595. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ohta A and Sitkovsky M: Extracellular
adenosine-mediated modulation of regulatory T cells. Front Immunol.
5:3042014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Clayton A, Al-Taei S, Webber J, Mason MD
and Tabi Z: Cancer exosomes express CD39 and CD73, which suppress T
cells through adenosine production. J Immunol. 187:676–683. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ungefroren H, Sebens S, Seidl D, Lehnert H
and Hass R: Interaction of tumor cells with the microenvironment.
Cell Commun Signal. 9:182011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Friedl P and Alexander S: Cancer invasion
and the micro-environment: Plasticity and reciprocity. Cell.
147:992–1009. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ono M, Kosaka N, Tominaga N, Yoshioka Y,
Takeshita F, Takahashi RU, Yoshida M, Tsuda H, Tamura K and Ochiya
T: Exosomes from bone marrow mesenchymal stem cells contain a
microRNA that promotes dormancy in metastatic breast cancer cells.
Sci Signal. 7:ra632014. View Article : Google Scholar : PubMed/NCBI
|