Introduction
Since conventional approaches cannot detect
micrometastasis with high sensitivity, development of novel and
effective methods is needed. Circulating tumor cells (CTCs) and/or
their specific molecules are important determinants for predicting
poor prognosis in patients with cancer (1–4).
CTCs also are measured to assess the therapeutic effects of
chemotherapy, radiotherapy and chemoradiotherapy (5–8).
CTCs or tumor-associated DNAs also have been detected frequently in
serum samples from poorly diagnosed patients with oral squamous
cell carcinoma (OSCC) (9,10), indicating that this type of blood
test is useful during cancer treatment and follow-up to monitor
patients for recurrent or metastatic lesions.
Due to low cellular copy numbers of genomic DNAs in
serum samples, isolating sufficient DNA for molecular analyses can
be difficult. Considering this, we recently reported the clinical
relevance of detecting tumor-derived mutant mitochondrial DNAs
(mut-mtDNAs) at the regions including the D-loop, 12S-rRNA and
16S-rRNA, the copy numbers of which are much higher than those of
genomic DNAs (11). To examine
whether discrete mutation(s) may exist in the mitochondorial
genome, and moreover, detection of CTCs with the mutant
mitochondrial DNA could be useful to predict micrometastasis of
patients with OSCC with no histologic evidence of cancer cells in
their surgical margins, we used a comprehensive approach for
detecting tumor-derived mut-mtDNAs in the ND2 and ND3 regions by
quantitative real-time polymerase chain reaction combined with
high-resolution melting curve analysis (qRT-PCR-HRMA). This
investigation showed compelling evidence that evaluation of
circulating tumor-derived mitochondrial DNAs with ND2 and/or ND3
mutation may be an additional clinical tool to monitor the
post-operative patients with OSCC.
Materials and methods
Ethical statement
The study protocol was approved by the Ethics
Committee of the Graduate School of Medicine, Chiba University
(approval number, 236) and was performed in accordance with the
ethical standards laid down in the Declaration of Helsinki. Written
informed consent was received from all patients or their
families.
All experimental animals were treated and cared for
in accordance with the guidelines of Chiba University. Experimental
animals were sacrificed by cervical dislocation. We made every
effort to relieve the pain of experimental animals. The protocol
was approved by the Committee on the Ethics of Animal Experiments
of Chiba University (approval number, 25-221).
Mutation detection of mtDNA for OSCC cell
lines in vitro and in vivo
The human OSCC-derived cell lines Sa3 and HSC-4 were
purchased, respectively, from the RIKEN BioResource Center through
the National Bioresource Project of the Ministry of Education,
Culture, Sports, Science and Technology (Tsukuba, Japan) and the
Human Science Research Resources Bank (Osaka, Japan). A DNA
profiling procedure validated the cell lines (11). The cells were cultured in the same
manner as previously reported (12).
Ten sets of specific PCR primers were prepared for
amplification of regions ND2 and ND3 of the human mitochondrial
genome. The primer sequences are shown in Table I. The PCR products were subcloned
into a pCR8/GW/TOPO TA cloning vector (Invitrogen, Carlsbad, CA,
USA), and then sequenced using ABI 3730xl DNA sequencers (Applied
Biosystems, Foster City, CA, USA) to validate the identity of the
amplified products by comparing them with the MITOMAP database
(www.mitomap.org/MITOMAP/HumanMitoSeq).
 | Table IThe primer sequences used for PCR
amplification. |
Table I
The primer sequences used for PCR
amplification.
| Primer sequence
(5′-3′) | Nucleotide
positions | Product size
(bp) |
|---|
| ND2-1F |
CCTATCACACCCCATCCTAAA | 4382–4402 | 157 |
| ND2-1R |
GCTTAGCGCTGTGATGAGTG | 4519–4538 | |
| ND2-2F |
TACCATCTTTGCAGGCACAC | 4502–4521 | 175 |
| ND2-2R |
GATTATGGATGCGGTTGCTT | 4657–4676 | |
| ND2-3F |
GTTCCACAGAAGCTGCCATC | 4621–4640 | 191 |
| ND2-3R |
TCAGAAGTGAAAGGGGGCTA | 4792–4811 | |
| ND2-4F |
GGAATAGCCCCCTTTCACTT | 4788–4807 | 215 |
| ND2-4R |
ATTTTGCGTAGCTGGGTTTG | 4983–5002 | |
| ND2-5F |
CCATCATAGCAGGCAGTTGA | 4951–4970 | 228 |
| ND2-5R |
GCTTGTTTCAGGTGCGAGAT | 5169–5188 | |
| ND2-6F |
TCGCACCTGAAACAAGCTAA | 5162–5181 | 208 |
| ND2-6R |
GGTGGAGTAGATTAGGCGTAGG | 5348–5369 | |
| ND2-7F |
CACCATCACCCTCCTTAACC | 5318–5337 | 248 |
| ND2-7R |
TGCAACTTACTGAGGGCTTTG | 5545–5565 | |
| ND3-1F |
CCGTTAACTTCCAATTAACTAGTTTTG | 10012–10038 | 164 |
| ND3-1R |
GCACTCGTAAGGGGTGGAT | 10157–10175 | |
| ND3-2F |
ACCACAACTCAACGGCTACA | 10130–10149 | 169 |
| ND3-2R |
TTGTAGGGCTCATGGTAGGG | 10279–10298 | |
| ND3-3F |
CCCTCCTTTTACCCCTACCA | 10267–10286 | 219 |
| ND3-3R |
TGTAAATGAGGGGCATTTGG | 10466–10485 | |
We optimized the conditions and examined the
feasibility of using qRT-PCR-HRMA for detecting three discrete
sequence variations (ND2-T5108C, ND3-A10397G and ND3-C10400T) in
Sa3 and HSC-4 cell lines. Using specific PCR primer sets (Table I), qPCR-HRMA was performed using a
LightCycler 480 system (Roche Diagnostics GmbH, Mannheim, Germany)
in a final volume of 20 μl of a reaction mixture comprised of 10 μl
of LightCycler 480 High Resolution Melting Master Mix (Roche), 3 mM
of MgCl2, and 4 μM of the primers, according to the
manufacturer's instructions.
The in vivo experiments, i.e., detecting
Sa3-derived mut-mtDNAs of the ND2 and ND3 regions in serum samples
from BALB/cAnNCrj-nu/nu mice (n=2, Charles River Laboratories,
Yokohama, Japan), were performed according to our previous methods
(11). In brief, to validate
whether mut-mtDNA of the ND2 and/or ND3 regions in human oral
cancer cells were detectable quantitatively from peripheral blood
samples, we transplanted Sa3 cells (2×106) by
subcutaneous injection into BALB/cAnNCrj-nu/nu mice (n=2, Charles
River Laboratories). The mice were sacrificed after 6 weeks as
previously described (11). The
non-Sa3 transplanted mice (n=2) were used as controls and their
serum samples were collected. MtDNA was extracted from them for
qRT-PCR-HRMA. All mice were maintained under specific pathogen-free
conditions. Environmental conditions were a temperature of 24 ±2°C,
humidity of 50±10%, lighting of 300 lux and a 12:12 light:dark
cycle with lights on at 07:00 and off at 19:00. The mice were
housed individually in 210×300×225 mm cages.
The committee of the Chiba University Laboratory
Animal Center reviewed and approved the protocol.
Determination of mut-mtDNAs in patients
with OSCC
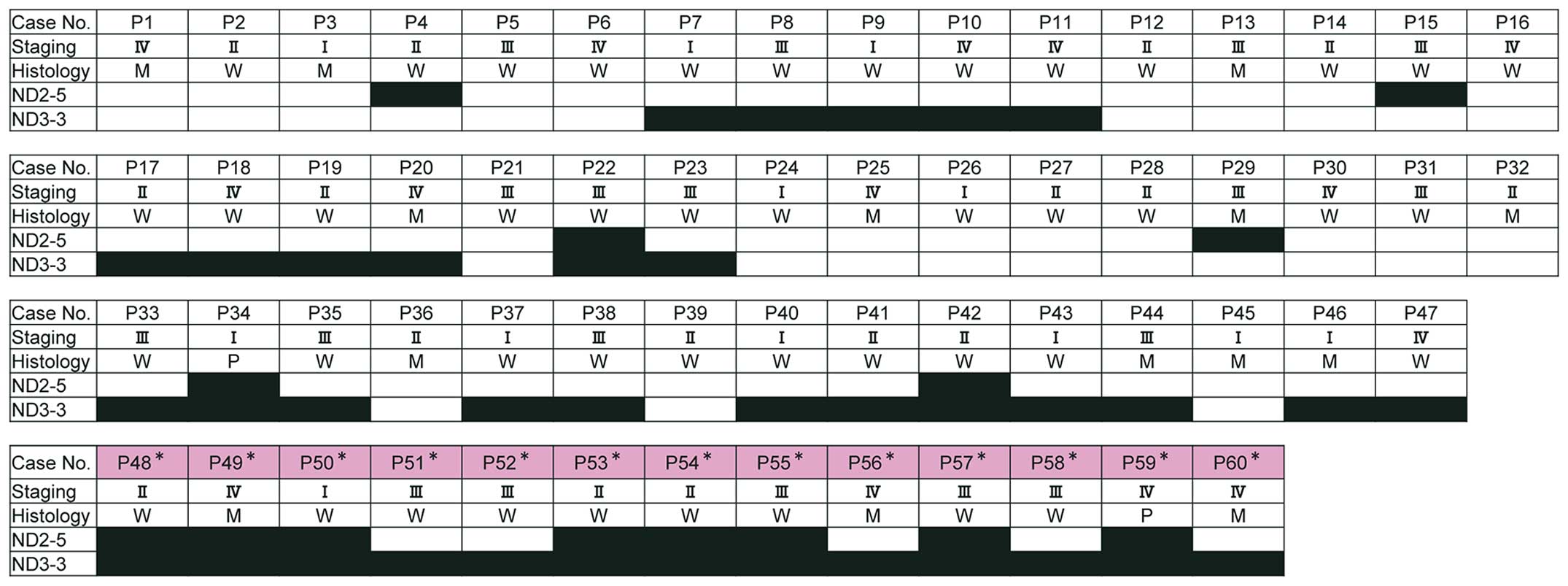
Sixty patients with newly diagnosed OSCC with
surgical malignancy-free margins were included. The patients were
divided into two groups: 47 patients with a good prognosis with no
recurrence and/or metastasis and 13 patients with a poor prognosis
with a recurrence or metastasis 17 months post-operatively.
Additional patient information is shown in Fig. 1.
Overall, we analyzed 240 mtDNAs comprised of normal
tissue, tumoral tissue, pre-operative serum samples, and serum
samples obtained 4 weeks post-operatively from each patient. To
quantify the mut-mtDNAs in each sample, the qRT-PCR-HRMA procedure
was performed as previously described (11) with specific primer sets for the ND2
region and the ND3 region (Table
I). The amount of each mut-mtDNA in the samples was determined
based on the standard curves that were created by diluting
mut-mtDNA from Sa3 or HSC-4 with wild-type mtDNAs to prepare 100,
75, 50, 35, 20 and 0% mutated samples for detecting the ND2 or ND3
region in the mtDNA genome as previously described (11).
All results, expressed as the mean ± standard error
of the mean, were similar among experiments repeated three times.
P-values were analyzed using the Mann-Whitney U-test. P<0.05 was
considered significant. Statistical analyses were performed using
Microsoft Office Excel 2010 (Microsoft, Seattle, WA, USA). For
receiver operating characteristic (ROC) curve analysis, EZR
software (Saitama Medical Center, Jichi Medical University,
Saitama, Japan) (13) was used. We
also utilized the area under the ROC curve (AUC) values with
estimated odds ratios and 95% confidence intervals (CIs) to
evaluate the diagnostic relevance for predicting the serum
mut-mtDNAs in patients with a poor prognosis.
Results
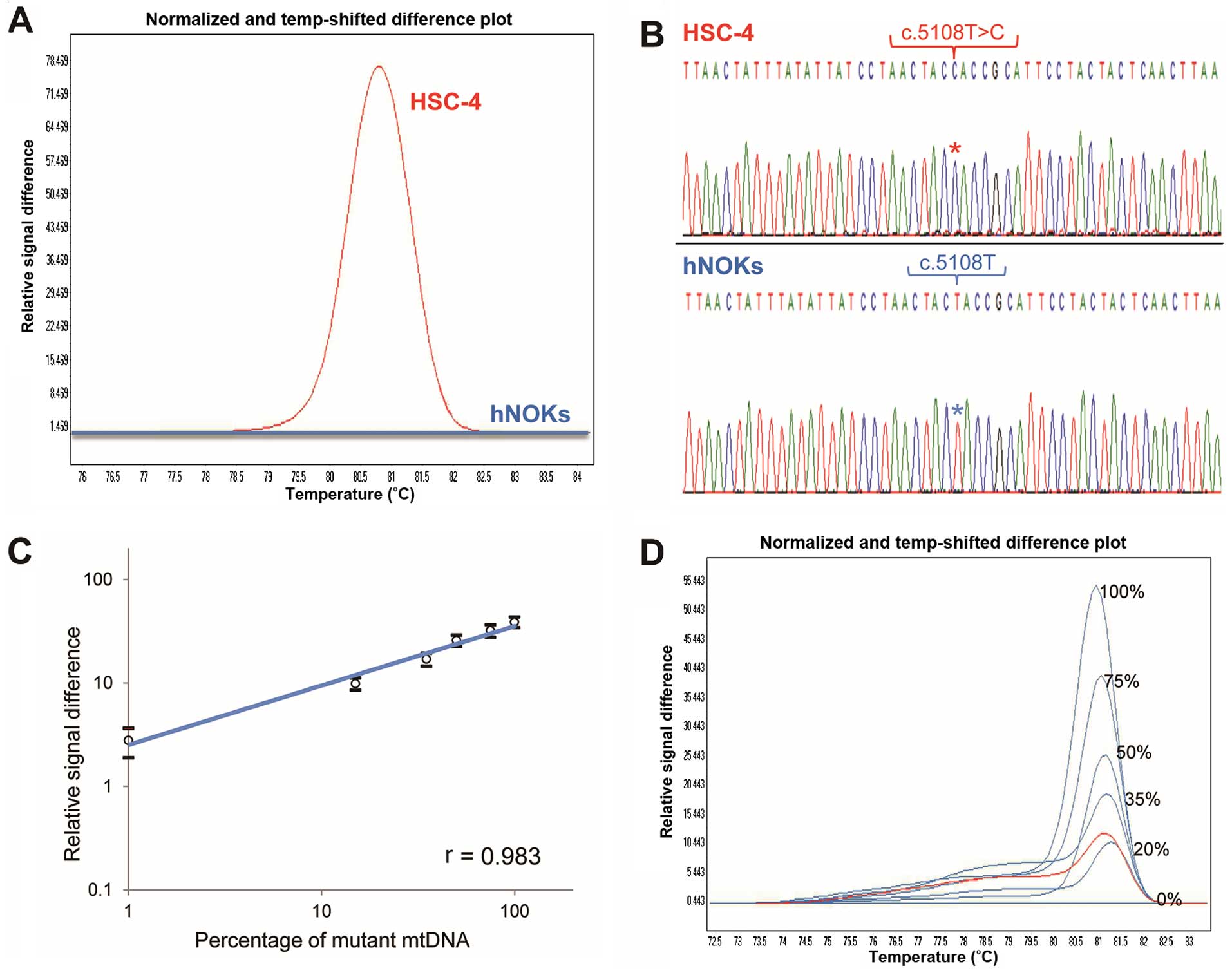
Three homoplasmic nucleotide substitutions defined
as single-nucleotide polymorphisms (SNPs) were identified in the
ND2 region (T:A to C:G at position 5108) in HSC-4 cells and the ND3
region (A:T to G:C at position 10397 and C:G to T:A at position
10400) in Sa3 cells; no mutation was observed in normal control
human normal oral keratinocytes (hNOKs) (Fig. 2A and B). In blood samples from
Sa3-xenografted mice, we detected mut-mtDNAs identical to
Sa3-associated mut-mtDNAs, but control mice did not have mut-mtDNAs
(Table II). The results indicated
that this blood test is clinically useful for detecting
tumor-related mut-mtDNAs. Based on the melting curves separated by
the HRMA chromatogram, we created a standard curve by serial
dilution of the DNA from hNOKs (Fig.
2C and D) and detected the mut-mtDNA amounts in samples from
the mice and humans examined. Typical results are shown in Fig. 2D.
 | Table IIComparison of Sa3 specific mut-mtDNA
levels between xenografted and control mice. |
Table II
Comparison of Sa3 specific mut-mtDNA
levels between xenografted and control mice.
| Mouse | Analyzed
regions | Serum |
|---|
| No. 1 | ND2 | 23% |
| ND3 | 50% |
| No. 2 | ND2 | 17% |
| ND3 | 45% |
| No. 3 | ND2 | 0% |
| ND3 | 0% |
| No. 4 | ND2 | 0% |
| ND3 | 0% |
As previously described (11,14),
we isolated sufficient mtDNAs for analysis from clinical samples
(n=240) from 60 patients with OSCC (746.8±476 ng/μl, tissue
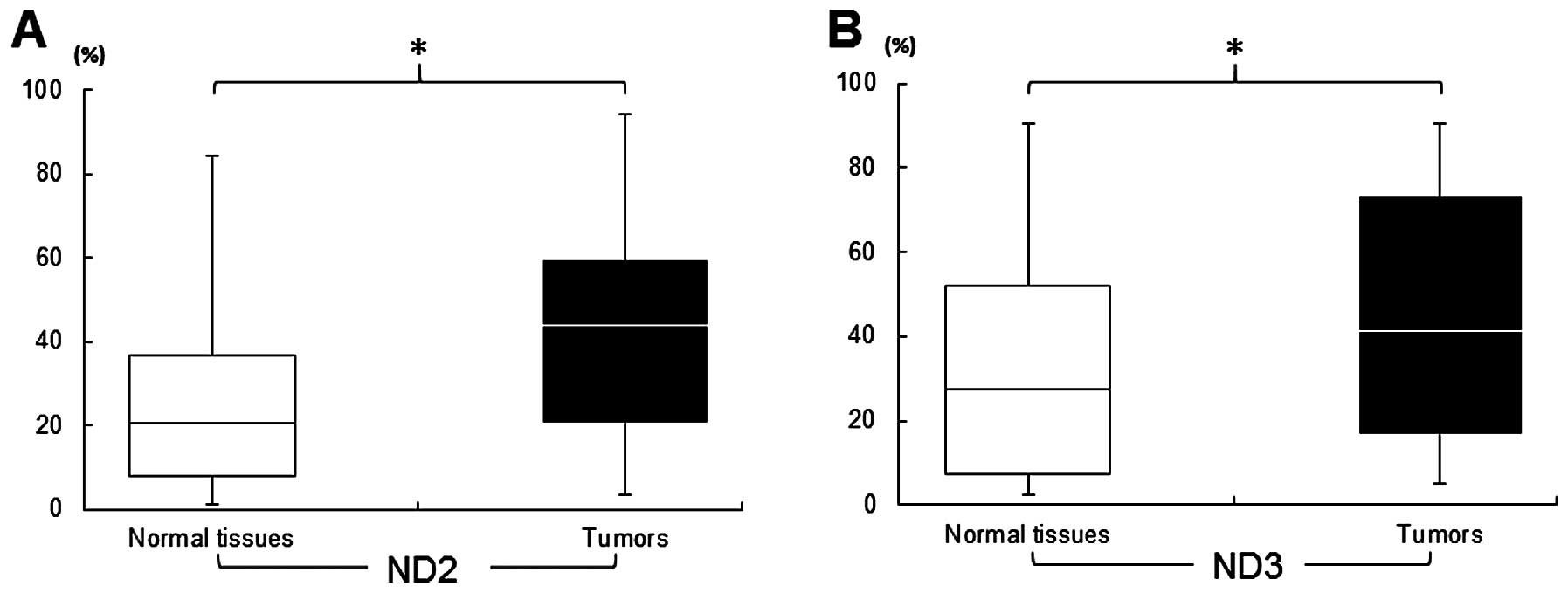
samples; 761.7±340 ng/μl, blood samples). In resected tissues, a
significantly (P<0.05) higher concentration of mut-mtDNA was
detected in tumoral tissues from patients with a good prognosis and
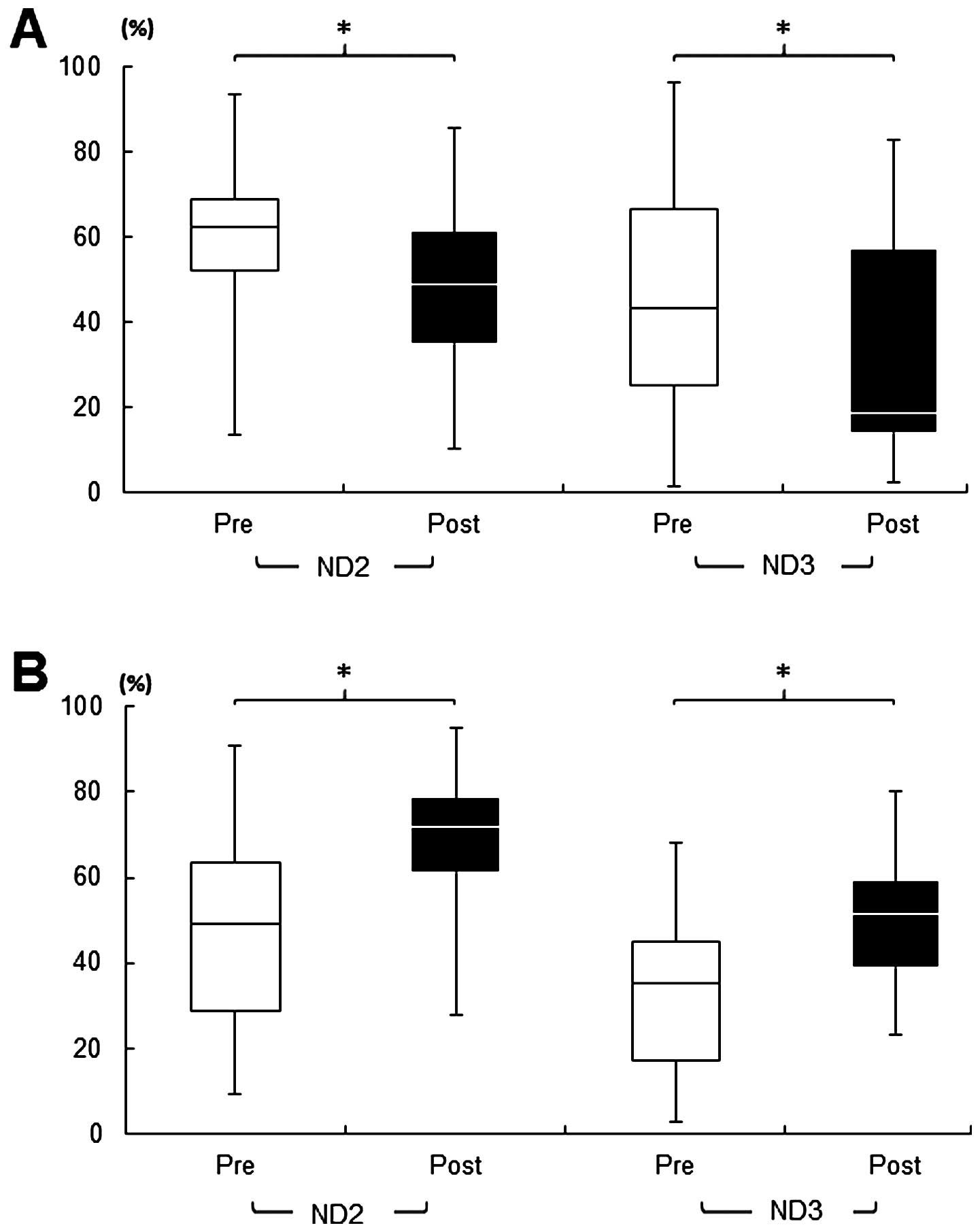
a poor prognosis, compared to each normal counterpart (Fig. 3). The blood test analyzed by
qRT-PCR-HRMA indicated that surgery significantly (P<0.05)
decreased the circulating mut-mtDNAs in patients with OSCC without
recurrence and/or metastasis (Fig.
4A). Compared to the group with a good prognosis, a significant
(P<0.05) increase in the circulating tumor-associated mut-mtDNAs
was confirmed in the blood samples obtained postoperatively from
patients with a poor prognosis (Fig.
4B), all of whom had substantial mut-mtDNA in their serum,
without exception, in at least one region examined (Fig. 1).
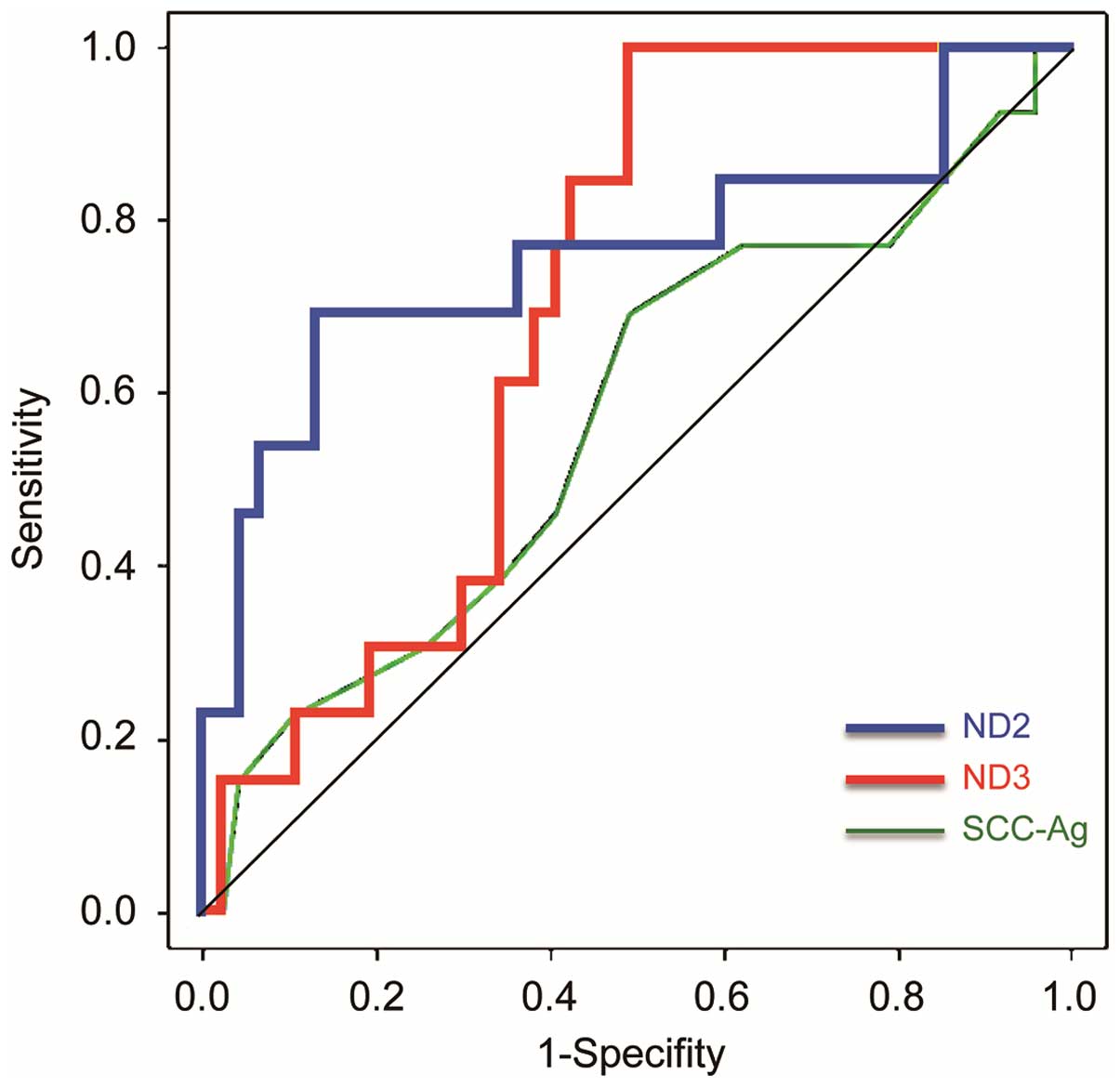
The area under the ROC curve (AUC) values were more
sensitive across a range of mut-mtDNA levels in the sera for the
risk of recurrence/metastasis than serum SCC antigen (SCC-Ag)
levels (Fig. 5). Using the optimal
threshold values of 68% (sensitivity, 61.5%; specificity, 87.2%)
for ND2, 22.9% (sensitivity, 92.3%; specificity, 51.1%) for ND3,
and 1.0 ng/ml (sensitivity, 69.2%; specificity, 51.1%) for SCC-Ag,
each AUC was 0.761 [95% confidence interval (CI), 0.580–0.9421,
P<0.05], 0.704 (95% CI, 0.5696–0.838, P<0.05) and 0.574 (95%
CI, 0.386–0.761, P=0.793), respectively.
Discussion
CTCs are promising clinical tools in many human
cancers (15–17). Evidence indicates that the
epithelial cell adhesion molecule is one of the most useful
molecular markers for detecting CTCs, including human SCCs
(18,19). In contrast, Wirtschafter et
al (20) reported that CTCs
were validated only in a small portion of patients with head and
neck SCC, suggesting limited clinical application.
The present study, in which a unique set of human
OSCC specimens was used, found that circulating mut-mtDNAs at the
ND2 and/or ND3 regions are significant predictive biomarkers for
postoperative recurrence/metastasis in OSCC. Nawroz et al
(21) first reported their
potential clinical use by detecting tumor-derived microsatellite
alterations in serum genomic DNAs in patients with head and neck
cancer. Recently, accumulating data on circulating mtDNAs have been
published on malignant tumors (22–24)
and other human diseases (25–28).
From a clinical standpoint, there are several benefits to adopting
mtDNA for clinical blood tests: DNA, including genomic DNA and
mtDNA, is more stable than RNA, including mRNA and microRNA, and
extracted protein; as He et al (14) and we (29) reported, the copy number of the
mtDNA is hundreds to thousands of times higher than that of genomic
DNA; and a high rate of somatic sequence variations resulting in a
pathogenic state are present in patients with OSCC.
We identified cancer-specific somatic variants in
the ND2 and ND3 regions (Fig. 1B).
These genes encoding ND2 and ND3 are subunits of NADH, which may
act as the rate-limiting enzyme of oxidative phosphorylation
(30). Alterations in these genes
are correlated with human cancers (31–33).
It has been proposed that once these genes function abnormally in
cancer cells, enhanced reactive oxygen species induces HIF1α
stabilization (34,35). Thus, we speculated that genetic
mutations identified in the present study, even in SNPs, may be
linked partly to the above-mentioned mechanisms for oral
tumorigenesis. In this context, several studies have reported an
association between SNPs on mtDNA, especially in the ND3 region,
and the risk of developing breast cancer (36–38).
We previously described the usefulness of
qRT-PCR-HRMA for searching mtDNA mutations with high
sensitivity/specificity. As indicated in the present study, our
method, even in different regions on the mitochondrial genome, is
sufficient for clinical use as well. However, as Kandel (39) pointed out, several issues need
attention such as minimization of cellular contamination,
determination of mut-mtDNA characteristics specific for OSCC, and
elimination of the effect of other diseases.
The study limitations were the small number of OSCC
cases and the absence of other human malignancies. However, our
data were highly significant for early detection of high-risk
individuals with OSCCs, since subjects expected to have a good
prognosis who had recurrence/metastasis postoperatively can be
distinguished by the level of tumor-derived mtDNA in their serum 4
weeks postoperatively. When a more precise approach for mut-mtDNA
detection of CTCs in cancer patients is established, we will
identify earlier the patients with undetectable lesions.
Acknowledgements
We thank L.C. Charters for editing the manuscript.
The present study was supported by a Grant-in-Aid for Exploratory
Research from The Ministry of Education, Culture, Sports, Science
and Technology (MEXT) (no. 50236775).
Abbreviations:
|
CTCs
|
circulating tumor cells
|
|
mut-mtDNA
|
mutant mitochondorial DNAs
|
|
OSCC
|
oral squamous cell carcinoma
|
|
qRT-PCR-HRMA
|
quantitative real-time polymerase
chain reaction combined with high-resolution melting curve
analysis
|
|
hNOKs
|
human normal oral keratinocytes
|
|
ROC
|
receiver operating characteristic
|
|
CI
|
confidence interval
|
|
SNP
|
single-nucleotide polymorphism
|
References
|
1
|
Königsberg R, Gneist M, Jahn-Kuch D,
Pfeiler G, Hager G, Hudec M, Dittrich C and Zeillinger R:
Circulating tumor cells in metastatic colorectal cancer: Efficacy
and feasibility of different enrichment methods. Cancer Lett.
293:117–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mavroudis D: Circulating cancer cells. Ann
Oncol. 21(Suppl 7): vii95–v100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khan MS, Kirkwood A, Tsigani T,
Garcia-Hernandez J, Hartley JA, Caplin ME and Meyer T: Circulating
tumor cells as prognostic markers in neuroendocrine tumors. J Clin
Oncol. 31:365–372. 2013. View Article : Google Scholar
|
|
4
|
Bidard FC, Peeters DJ, Fehm T, Nolé F,
Gisbert-Criado R, Mavroudis D, Grisanti S, Generali D, Garcia-Saenz
JA, Stebbing J, et al: Clinical validity of circulating tumour
cells in patients with metastatic breast cancer: A pooled analysis
of individual patient data. Lancet Oncol. 15:406–414. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Buglione M1, Grisanti S, Almici C, Mangoni
M, Polli C, Consoli F, Verardi R, Costa L, Paiar F, Pasinetti N, et
al: Circulating tumour cells in locally advanced head and neck
cancer: preliminary report about their possible role in predicting
response to non-surgical treatment and survival. Eur J Cancer.
48:3019–3026. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu CY, Tsai HL, Uen YH, Hu HM, Chen CW,
Cheng TL, Lin SR and Wang JY: Circulating tumor cells as a
surrogate marker for determining clinical outcome to mFOLFOX
chemotherapy in patients with stage III colon cancer. Br J Cancer.
108:791–797. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pavese JM and Bergan RC: Circulating tumor
cells exhibit a biologically aggressive cancer phenotype
accompanied by selective resistance to chemotherapy. Cancer Lett.
352:179–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tinhofer I, Konschak R, Stromberger C,
Raguse JD, Dreyer JH, Jöhrens K, Keilholz U and Budach V: Detection
of circulating tumor cells for prediction of recurrence after
adjuvant chemoradiation in locally advanced squamous cell carcinoma
of the head and neck. Ann Oncol. 25:2042–2047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hamana K, Uzawa K, Ogawara K, Shiiba M,
Bukawa H, Yokoe H and Tanzawa H: Monitoring of circulating
tumour-associated DNA as a prognostic tool for oral squamous cell
carcinoma. Br J Cancer. 92:2181–2184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gröbe A, Blessmann M, Hanken H, Friedrich
RE, Schön G, Wikner J, Effenberger KE, Kluwe L, Heiland M, Pantel
K, et al: Prognostic relevance of circulating tumor cells in blood
and disseminated tumor cells in bone marrow of patients with
squamous cell carcinoma of the oral cavity. Clin Cancer Res.
20:425–433. 2014. View Article : Google Scholar
|
|
11
|
Uzawa K, Baba T, Uchida F, Yamatoji M,
Kasamatsu A, Sakamoto Y, Ogawara K, Shiiba M, Bukawa H and Tanzawa
H: Circulating tumor-derived mutant mitochondrial DNA: A predictive
biomarker of clinical prognosis in human squamous cell carcinoma.
Oncotarget. 3:670–677. 2012.PubMed/NCBI
|
|
12
|
Saito K, Uzawa K, Kasamatsu A, Shinozuka
K, Sakuma K, Yamatoji M, Shiiba M, Shino Y, Shirasawa H and Tanzawa
H: Oncolytic activity of Sindbis virus in human oral squamous
carcinoma cells. Br J Cancer. 101:684–690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR' for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar :
|
|
14
|
He Y, Wu J, Dressman DC, Iacobuzio-Donahue
C, Markowitz SD, Velculescu VE, Diaz LA Jr, Kinzler KW, Vogelstein
B and Papadopoulos N: Heteroplasmic mitochondrial DNA mutations in
normal and tumour cells. Nature. 464:610–614. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cen P, Ni X, Yang J, Graham DY and Li M:
Circulating tumor cells in the diagnosis and management of
pancreatic cancer. Biochim Biophys Acta. 1826:350–356.
2012.PubMed/NCBI
|
|
16
|
Lianidou ES, Mavroudis D and Georgoulias
V: Clinical challenges in the molecular characterization of
circulating tumour cells in breast cancer. Br J Cancer.
108:2426–2432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Romero-Laorden N, Olmos D, Fehm T,
Garcia-Donas J and Diaz-Padilla I: Circulating and disseminated
tumor cells in ovarian cancer: A systematic review. Gynecol Oncol.
133:632–639. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bozec A, Ilie M, Dassonville O, Long E,
Poissonnet G, Santini J, Chamorey E, Ettaiche M, Chauvière D,
Peyrade F, et al: Significance of circulating tumor cell detection
using the CellSearch system in patients with locally advanced head
and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol.
270:2745–2749. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Driemel C, Kremling H, Schumacher S, Will
D, Wolters J, Lindenlauf N, Mack B, Baldus SA, Hoya V, Pietsch JM,
et al: Context-dependent adaption of EpCAM expression in early
systemic esophageal cancer. Oncogene. 33:4904–4915. 2014.
View Article : Google Scholar
|
|
20
|
Wirtschafter A, Benninger MS, Moss TJ,
Umiel T, Blazoff K and Worsham MJ: Micrometastatic tumor detection
in patients with head and neck cancer: A preliminary report. Arch
Otolaryngol Head Neck Surg. 128:40–43. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nawroz H, Koch W, Anker P, Stroun M and
Sidransky D: Microsatellite alterations in serum DNA of head and
neck cancer patients. Nat Med. 2:1035–1037. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mehra N, Penning M, Maas J, van Daal N,
Giles RH and Voest EE: Circulating mitochondrial nucleic acids have
prognostic value for survival in patients with advanced prostate
cancer. Clin Cancer Res. 13:421–426. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kohler C, Radpour R, Barekati Z,
Asadollahi R, Bitzer J, Wight E, Bürki N, Diesch C, Holzgreve W and
Zhong XY: Levels of plasma circulating cell free nuclear and
mitochondrial DNA as potential biomarkers for breast tumors. Mol
Cancer. 8:1052009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ellinger J, Müller DC, Müller SC, Hauser
S, Heukamp LC, von Ruecker A, Bastian PJ and Walgenbach-Brunagel G:
Circulating mitochondrial DNA in serum: A universal diagnostic
biomarker for patients with urological malignancies. Urol Oncol.
30:509–515. 2012. View Article : Google Scholar
|
|
25
|
Tsai NW, Lin TK, Chen SD, Chang WN, Wang
HC, Yang TM, Lin YJ, Jan CR, Huang CR, Liou CW, et al: The value of
serial plasma nuclear and mitochondrial DNA levels in patients with
acute ischemic stroke. Clin Chim Acta. 412:476–479. 2011.
View Article : Google Scholar
|
|
26
|
Bliksøen M, Mariero LH, Ohm IK, Haugen F,
Yndestad A, Solheim S, Seljeflot I, Ranheim T, Andersen GØ, Aukrust
P, et al: Increased circulating mitochondrial DNA after myocardial
infarction. Int J Cardiol. 158:132–134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kung CT, Hsiao SY, Tsai TC, Su CM, Chang
WN, Huang CR, Wang HC, Lin WC, Chang HW, Lin YJ, et al: Plasma
nuclear and mitochondrial DNA levels as predictors of outcome in
severe sepsis patients in the emergency room. J Transl Med.
10:1302012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qiu C, Hevner K, Enquobahrie DA and
Williams MA: A case-control study of maternal blood mitochondrial
DNA copy number and preeclampsia risk. Int J Mol Epidemiol Genet.
3:237–244. 2012.PubMed/NCBI
|
|
29
|
Lai CH, Huang SF, Liao CT, Chen IH, Wang
HM and Hsieh LL: Clinical significance in oral cavity squamous cell
carcinoma of pathogenic somatic mitochondrial mutations. PLoS One.
8:e655782013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kumazaki T, Sakano T, Yoshida T, Hamada K,
Sumida H, Teranishi Y, Nishiyama M and Mitsui Y: Enhanced
expression of mitochondrial genes in senescent endothelial cells
and fibroblasts. Mech Ageing Dev. 101:91–99. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu VW, Shi HH, Cheung AN, Chiu PM, Leung
TW, Nagley P, Wong LC and Ngan HY: High incidence of somatic
mitochondrial DNA mutations in human ovarian carcinomas. Cancer
Res. 61:5998–6001. 2001.PubMed/NCBI
|
|
32
|
Kumimoto H1, Yamane Y, Nishimoto Y, Fukami
H, Shinoda M, Hatooka S and Ishizaki K: Frequent somatic mutations
of mitochondrial DNA in esophageal squamous cell carcinoma. Int J
Cancer. 108:228–231. 2004. View Article : Google Scholar
|
|
33
|
Prior SL, Griffiths AP, Baxter JM, Baxter
PW, Hodder SC, Silvester KC and Lewis PD: Mitochondrial DNA
mutations in oral squamous cell carcinoma. Carcinogenesis.
27:945–950. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun W, Zhou S, Chang SS, McFate T, Verma A
and Califano JA: Mitochondrial mutations contribute to HIF1alpha
accumulation via increased reactive oxygen species and up-regulated
pyruvate dehydrogenease kinase 2 in head and neck squamous cell
carcinoma. Clin Cancer Res. 15:476–484. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Singh RK, Srivastava A, Kalaiarasan P,
Manvati S, Chopra R and Bamezai RN: mtDNA germ line variation
mediated ROS generates retrograde signaling and induces
pro-cancerous metabolic features. Sci Rep. 4:65712014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bai RK, Leal SM, Covarrubias D, Liu A and
Wong LJ: Mitochondrial genetic background modifies breast cancer
risk. Cancer Res. 67:4687–4694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Covarrubias D, Bai RK, Wong LJ and Leal
SM: Mitochondrial DNA variant interactions modify breast cancer
risk. J Hum Genet. 53:924–928. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Czarnecka AM, Krawczyk T, Zdrozny M,
Lubiński J, Arnold RS, Kukwa W, Scińska A, Golik P, Bartnik E and
Petros JA: Mitochondrial NADH-dehydrogenase subunit 3 (ND3)
polymorphism (A10398G) and sporadic breast cancer in Poland. Breast
Cancer Res Treat. 121:511–518. 2010. View Article : Google Scholar
|
|
39
|
Kandel ES: Mutations in circulating
mitochondrial DNA: Cassandra of oral cancer? Oncotarget. 3:664–665.
2012.PubMed/NCBI
|