Introduction
Ovarian cancer remains the most lethal of all
gynaecologic malignancies in the world (1,2).
According to a report of the American Cancer Society, ~22,240 new
cases of ovarian cancer would be diagnosed in the United States in
2013 while nearly 14,030 of those affected would succumb to this
disease (3). Epithelial ovarian
cancer (EOC) accounts for 90% of ovarian cancers; it is not a
single disease as there are several different histological
subtypes, with serous being the most common subtype, yet, its
aetiology remains poorly understood. Most EOC cannot be detected
until peritoneal or distant metastases occur, which are the major
cause of high mortality in ovarian cancer. Despite cytotoxic
therapy, only 30% of patients with advanced ovarian cancer survive
5 years post diagnosis. The metastatic cascade consists of a series
of sequential, interrelated steps that are not yet completely
understood. However, it is known that these metastatic events are
modulated by many factors, including metastasis activators and
suppressors. Metastasis suppressors can inhibit metastasis at any
step of the metastatic cascade without blocking tumorigenicity.
Metastasis suppressor 1 (MTSS1) protein, which is
also known as MIM (missing-in-metastasis) has been recently
characterized as a tumor suppressor protein (4). MTSS1 is mostly expressed in normal
tissues and in some non-metastatic cancer cell lines, however, its
expression is significantly decreased or mostly absent in many
metastatic types of cancer including metastatic bladder (5), prostate (6), kidney (7) and gastric cancer (8), suggesting that MTSS1 could function
as an anti-metastatic protein. Furthermore, an inverse correlation
has also been observed between MTSS1 expression and poor prognosis
in breast cancer (9). These
findings indicate that MTSS1 might function as a tumor suppressor
and that loss of MTSS1 facilitates the development of human cancers
including breast and prostate cancers. However, in contrast to its
reduced expression in many human cancers, overexpression of MTSS1
has been observed in hepatocellular carcinoma (10) although its physiological
significance to liver cancer remains elusive. MTSS1 is an actin and
membrane binding protein. Functionally, it acts as a cytoskeletal
scaffold protein that regulates cytoskeletal dynamics through
interacting with many different proteins such as Rac, actin and
actin-associated proteins (11–13).
In addition, recent research has shown that anti-microRNA (miR182)
treatment can significantly reduce the expression of miR-182 target
genes including BRCA1, HMGA2 and MTSS1; downregulation of MTSS1 is
strongly associated with aggressive invasion of ovarian cancer
cells (14). There is a wealth of
biochemical data concerning MTSS1, but the physiological roles of
its various activities that regulate plasma membrane dynamics
including actin monomer binding, membrane deformation and
interaction with Gli transcription factors are still not fully
understood.
Studies suggest that further understanding of MTSS1
expression or inactivation in different human malignancies may
define it as a novel candidate to be used as a marker of primary
tumors or metastasis. We sought to determine the relevance of MTSS1
in EOC and provide new insights into its biological functions and
role in EOC. In the present study, expression of MTSS1 was examined
in a cohort of human EOC samples and cell lines. EOC cells were
forced to express this molecule by transfection with a mammalian
expression plasmid containing the full sequence of MTSS1 enabling
further understanding of the functional role of MTSS1 in EOC cell
behaviour.
Materials and methods
Clinical sample collection, processing
and IHC staining
All clinical samples examined in the present study
were obtained from surgically removed ovarian tissues of inpatients
in Wuhan Tongji Hospital of Huazhong, University of Science and
Technology (Wuhan, China) from 2013 to 2014; patients who had
received pre-operative radiotherapy or chemotherapy were excluded.
Immunohistochemistry was performed on 17 epithelial ovarian serous
carcinomas, 10 samples were non-metastatic and 7 had lymph node or
omentum metastases. All of the tumor samples were obtained from the
primary tumor site. Diagnosis was confirmed by histopathology in
all cases. All protocols were reviewed and approved by the Ethics
Committee and all patients gave written informed consent.
Tissue sections (4 μm) were prepared from
formalin-fixed paraffin embedded blocks. IHC was performed using
mouse anti-human MTSS1 antibody (Abnova; Caltag-Medsystems Ltd.,
Buckingham, UK) and the Vectastain® Elite Universal ABC
kit (Vector Laboratories, Ltd., Cambridgeshire, UK). The
de-paraffinized sections were rehydrated in Tris-buffered saline
(TBS). Antigen retrieval was then performed by heating the samples
for 20 min in a microwave in 1 mM EDTA buffer (pH 8.0). The
sections were cooled and washed in tap water (10 min). Non-specific
binding was blocked with 5–10% goat serum (90 min) and slides were
then incubated with the primary MTSS-1 antibody (1:100 in TBS) for
1 h. Following sequential 30-min incubations with mouse
biotinylated secondary and ABC complex respectively, the target
protein was visualised using freshly prepared 3,3-diaminobenzidine
(DAB; Sigma-Aldrich Co., Ltd., Dorset, UK). Slides were rinsed with
water, counterstained with haematoxylin, dehydrated, cleared in
xylene and mounted in DPX. Negative controls were prepared by
substituting the primary antibody with TBS. The sections were then
viewed under the Leica MC120 microscope, photographed and the
intensity and localisation of the staining was analysed.
Cell lines and culture conditions
Human ovarian epithelial-serous carcinoma cell lines
SKOV3 and COV504, human ovarian epithelial-mucinous carcinoma cell
lines COV644 (ECACC; European Collection of Animal Cell Culture,
Salisbury, UK) were routinely maintained in DMEM-F12 medium
supplemented with 10% fetal bovine serum, penicillin (100 U/ml),
streptomycin (100 μg/ml) and amphotericin B (0.25 μg/ml)
(Sigma-Aldrich). Cells were incubated at 37°C with 95% humidity in
5% CO2.
Construction of MTSS1 expression vectors
and transfection
The full coding sequence of MTSS1 was amplified from
cDNA prepared from normal human mammary tissue RNA using the
standard PCR procedure and a Master Mix with a proof-reading enzyme
(sense primer, ATGGAGGCTGTGAT TGAG and antisense,
CTAAGAAAAGCGAGGGG). This MTSS1 sequence was then T-A cloned into
the pEF6/V5/-His-TOPO vector (Invitrogen, Paisley, UK). According
to the manufacturers' protocol, the recombinant plasmid vectors
were transformed into chemically competent OneShot®
TOP10 Escherichia coli (Invitrogen) and bacteria were grown
overnight on agar plates containing ampicillin (100 μg/ml).
Colonies were analysed and those carrying correct recombinant
plasmids were amplified, then extracted and purified (Elute
Miniprep; Sigma-Aldrich). Purified MTSS1 plasmids (10 μg) and
control plasmid vectors were then transfected into SKOV3 (300 V,
1500 μF), COV504 (300 V, 1500 μF) and COV644 (275 V, 100 μF) cells
(1×106/ml) using a Gene Pulser Xcell electroporator
(Bio-Rad Laboratories Ltd., Hemel Hempstead, UK). After ~2 weeks of
selection with blasticidin (2–5 μg/ml) (Melford Laboratories Ltd.,
Ipswich, UK), the transfectants were verified for their expression
of MTSS1 mRNA and protein and successful clones were used in
subsequent studies. Transfected cell cultures were maintained in
medium containing 0.5 μg/ml blasticidin.
RNA extraction and reverse transcription
PCR
Total cellular RNA was isolated from the EOC cells
using Tri Reagent according to the manufacturer's protocol
(Sigma-Aldrich). RNA concentration and quality were determined
through spectrophotometric measurement (NanoPhotometer; Implen
GmbH, Munich, Germany). RNA (500 ng) was reverse transcribed into
cDNA using an Applied Biosystems high capacity reverse
transcription kit (Life Technologies, Paisley, UK). DNA quality was
verified using GAPDH PCR (sense GGCTG CTTTTAACTCTGGTA and antisense
GACTGTGGTCATG AGTCCTT) which was also used as a loading control.
MTSS1 mRNA levels were assessed using primers (sense, TCAAGAA
CAGATGGAAGAATGG and antisense, TGCGGTAGCGGT AATGTG). PCR was
carried out in an Applied Biosystems thermo cycler using a GoTaq
Green PCR reaction mix (Promega UK Ltd., Hampshire, UK). Cycling
conditions were 94°C for 5 min, followed by 25–32 cycles of 94°C
for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. This was followed
by a final 7-min extension period at 72°C. The products were
visualized on 2% agarose gel stained with SYBR® Safe
(Life Technologies).
Immunofluorescence staining
Cells were seeded at a density of 20,000 cells/well
in an 8-well chamber slide (Merck-Millipore, East Midlands, UK).
Following an overnight incubation, the medium was aspirated and the
cells were fixed in 4% formalin (4°C, 20 min). Following fixation,
the cells were rehydrated in phosphate-buffered saline (PBS) for 20
min at room temperature before being permeabilised for 5 min in a
0.1% Triton in PBS. Non-specific binding was blocked by 1-h
incubation in phosphate-buffered saline (PBS) containing 5–10% goat
serum. Cells were incubated for 1 h with MTSS1 antibody (1:100) in
PBS blocking solution (Abnova; Caltag-Medsystems). Slides were
washed 3×5 min in PBS then incubated on a shaker platform in the
dark for 1 h with FITC conjugated anti-mouse secondary antibody
(Insight Biotechnology Ltd., Middlesex, UK) and 1:1,000 DAPI
(Roche, Hertfordshire, UK). Slides were finally washed 3×5min PBS,
mounted with fluorsave (Merk-Millipore) and visualised using an
EVOS fluorescence auto imaging system (Life Technologies).
Western blot analysis
Cell lines were grown to 70% confluence, monolayers
were washed with PBS and lysed in ice cold lysis buffer (50 mm
Tris, 150 mM NaCl, 5 mM EGTA, 1% Triton X-100 pH 7.5) supplemented
with protease inhibitor cocktail (Roche). Lysates were clarified by
centrifugation (12,000 rpm, 15 min, 4°C) and the protein
concentrations in the supernatants were determined using the DC
protein assay kit (Bio-Rad Laboratories). Protein was reduced and
denatured by boiling (5 min) in Laemmli buffer (Sigma-Aldrich) and
20 μg protein samples were resolved by SDS-PAGE and transferred
onto nitrocellulose membrane (GE Healthcare Life Sciences,
Buckinghamshire UK). After blocking for 1 h in 5% skimmed milk
(TBS/Tween: 140 mM NaCl; 50 mM Tris, 0.05% Tween pH 7.4), blots
were incubated overnight at 4°C with primary antibodies MTSS1
(1:300 prepared in TBS/Tween/1% milk) and GAPDH (1:1,000 in
TBS/Tween/1% milk) (Santa Cruz Biotechnology, Heidelberg, Germany)
was used as a loading control. Blots were washed with TBS/Tween and
bound antibodies were detected after 1-h incubation (room
temperature) with appropriate horseradish peroxidase-conjugated
secondary antibody (1:1,000; Sigma-Aldrich). Following 3×5min
TBS/Tween washes, protein bands were visualized using enhanced
chemiluminescence (Luminata Forte; Millipore, Hertfordshire, UK)
and photographed using a UVItec imager (UVItec, Inc., Cambridge,
UK).
Cell proliferation assay
Cells were seeded into 96-well plates at a seeding
density of 3,000 cells/well with 12 replicates/experiment. Cells
were fixed with 4% formalin after 1, 3 and 5 days growth. Fixed
cells were stained with 0.5% crystal violet, washed and dried. Dye
was re-solubilised in 200-μl acetic acid/well and absorbance was
determined at 540 nm using an ELx800 multiplate reader (BioTek UK,
Bedfordshire, UK). Each experiment was repeated at least 3 times.
For each cell line, analysis compared cell number (absorbance) on
day 3 and 5 relative to day 1.
Cell adhesion assay
Cell-matrix adhesion was examined using an in
vitro Matrigel adhesion assay adapted from a previously
described method (15–17). Cells were seeded into 96-well
plates pre-coated with 5 μg/well Matrigel basement membrane matrix
(BD Biosciences, Oxford, UK). After 40 min of incubation (37°C) the
cells were washed with PBS to remove unbound cells. The remaining
adherent cells were fixed with 4% formalin, stained with 0.5%
crystal violet, visualized under a microscope (×20) and cell number
counted per field of view. Four counts were made from each of 6
replicate wells and results were expressed as mean cell
number/well. Each experiment was repeated 3 times.
Cell invasion assay
Cell invasive capability was examined using an in
vitro Matrigel invasion assay. Transwell inserts (Greiner
Bio-One Ltd., Stonehouse, UK) with an 8.0 μm pore size were coated
with 50 μg Matrigel (BD Biosciences), dried at 55°C and rehydrated
with 100-μl serum-free medium before seeding 4,000 cells/insert.
After 48 h of incubation at 37°C, non-invasive cells and Matrigel
were removed from the inside of the inserts with a cotton swab.
Cells that had invaded to the underside of the insert were fixed
(4% formalin), stained with 0.5% crystal violet and washed. Cell
invasion was quantitated by counting the cell number in 4 fields of
view (×20 magnification). Data were analysed as mean cell number
per field of view for 3 independent experiments with 3 replicates
per experiment. Results were confirmed by incubating the stained
inserts in 10% acetic acid. Absorbance of solubilized crystal
violet was determined at 540 nm.
Migration assay
A cellular wounding assay was used to study
directional cell migration in vitro as previously described
(18). In brief, cells were
cultured to confluence in a 24-well plate before scratching the
cell monolayer with a 10-μl pipette tip. The closure of the induced
wound, through the migration of cells, was tracked and recorded
over a 48-h period using an automated cell imaging system EVOS
(Life Technologies). Using ImageJ software, the relative cell
migration distance was calculated using multiple measurements of
the width of wound gap after 6, 12, 24 and 48 h compared to 0
h.
Electric cell-substrate impedance sensing
(ECIS)-based attachment and migration assay
Cell attachment and migration were further studied
using an ECIS ZTheta instrument and 96W1E arrays (Applied
BioPhysics, Inc., Troy, NY, USA) as previously described (19). Briefly, 40,000 cells/well were
added to the ECIS arrays. Impedance and resistance of the cell
layer was immediately recorded for a period of up to 15 h. When
confluence was reached, the monolayer in each well was electrically
wounded at 2,600 μA and 6,0000 Hz for 20 sec to create a 250-μm
wound/well. Impedance and resistance of the wounded cells as they
migrated in the wound was then recorded for a period of up to 20 h.
Data were analysed using the ECIS software, supplied by the
manufacturer.
Statistical analysis
All statistical analysis was performed using the
paired t-test for normally distributed data. Differences were
considered to be statistically significant at P<0.05.
Results
Expression of MTSS1 in human ovarian
tissues and EOC cells
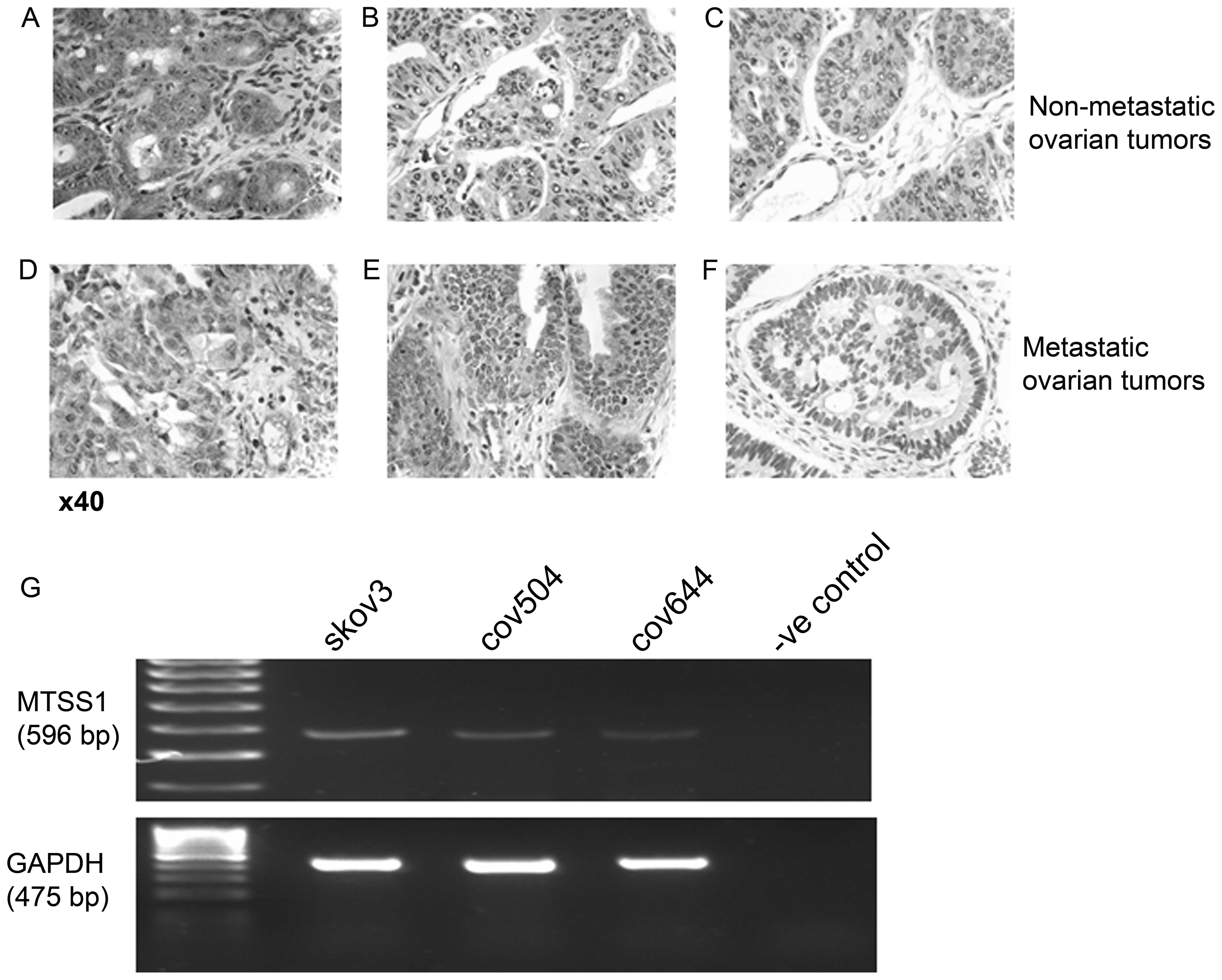
HC staining of 10 sections of non-metastastic and 7
sections of metastatic epithelial cancerous ovarian growths was
used to assess MTSS1 expression pattern in the clinical setting.
Images are shown of both metastatic and non-meta-static samples,
representing the range of staining detected (Fig. 1). Preliminary experiments showed
that MTSS1 protein could also be detected in non-cancerous ovarian
tissue (data not shown). In 6 out of 10 non-metastatic primary
tumors very strong staining for MTSS1 was detected in epithelial
cell cytoplasm (Fig. 1A). Weaker
staining was present in the cytoplasm of connective tissue. In the
other 4 non-metastatic samples, epithelial MTSS1 expression was
either weak (n=2) (Fig. 1B) or
could not be detected (n=2) (Fig.
1C). When tissue from 7 metastatic tumors was examined, MTTS1
epithelial staining was detected in 4 samples but the MTSS1
staining was typically weaker than the very strong epithelial
staining MTSS1 detected in most of the non-metastatic samples
(Fig. 1D and E). In the remaining
3 metastatic samples, MTSS1 staining could not be detected
(Fig. 1F), suggesting some
reduction of MTSS1 expression in metastatic compared to
non-metastatic ovarian cancer.
The mRNA expression of MTSS1 was also examined in
three EOC cell lines using RT-PCR. MTSS1 mRNA was expressed at
relatively low levels in all cell lines, with SKOV3 cells
expressing a slightly increased amount compared to the COV504 and
COV644 cells (Fig. 1G).
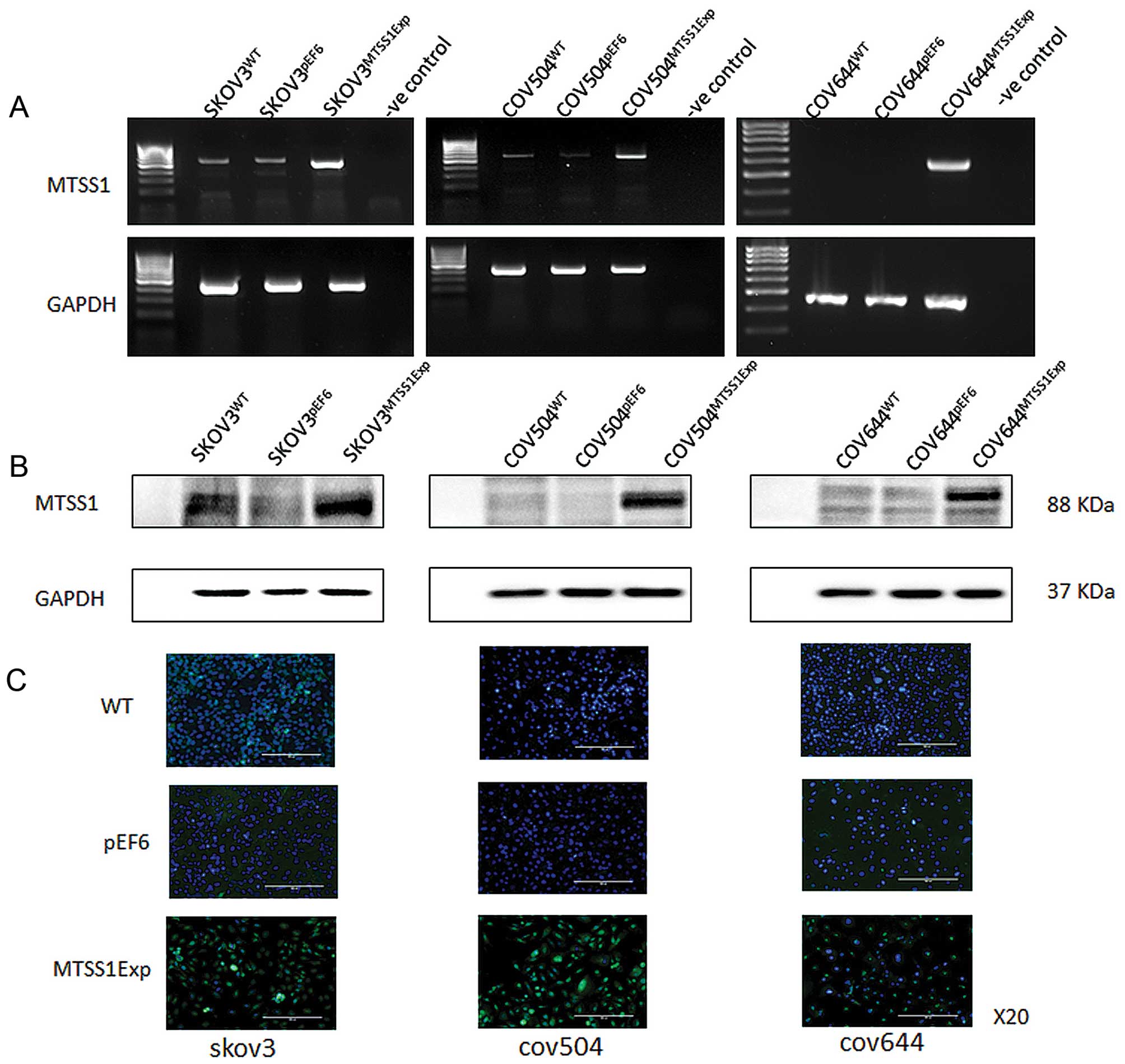
Overexpression of MTSS1 in EOC cells
To investigate the impact of MTSS1 on functions of
ovarian cancer cells, MTSS1 expression vectors were utilised to
overexpress MTSS1. After selection using blasticidin, the
expression of MTSS1 in the transfected cells was verified using
RT-PCR, immunofluorescent staining and western blotting (Fig. 2). Increased expression of both mRNA
(Fig. 2A) and protein (Fig. 2B) of MTSS1 was seen in
SKOV3MTSS1Exp, in comparison with the controls,
wild-type SKOV3WT and empty plasmid
SKOV3pEF6. Overexpression of MTSS1 was also confirmed in
COV504MTSS1Exp cells, in comparison with
COV504WT and COV504pEF6 control cells.
Similarly, overexpression of MTSS1 was confirmed in
COV644MTSS1Exp cells, in comparison with control
COV644WT and COV644pEF6 control cells.
Immunofluorescent staining was carried out to examine the
expression and localisation of the MTSS1 in the transfected cells
(Fig. 2C). MTSS1 staining (green),
was predominantly associated with the cytoplasm. Control cells,
both wild-type and empty vector transfectants, had weak staining
intensity in all three cell lines, with the majority of the cells
in the microscopic fields showing minimal staining levels. However,
in the transfected cells overexpressing MTSS1 had a more intense
and frequently observed staining.
Regulation of MTSS1 expression affects
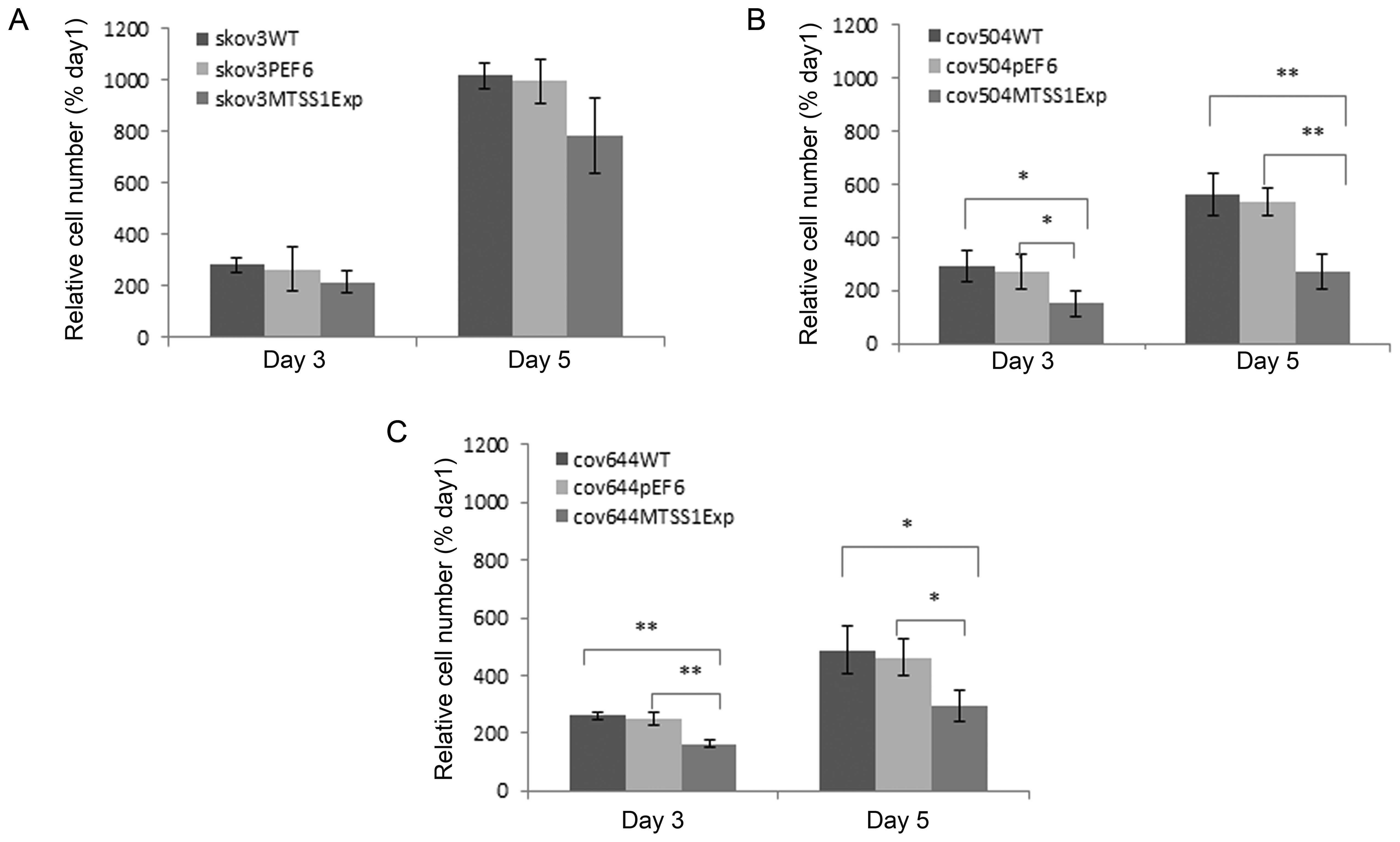
the rate of cell growth of EOC cells
The growth capacity of the EOC cells following MTSS1
overexpression was examined and compared to the wild-type and empty
vector control cells using an in vitro cell growth assay.
The growth rates of the three wild-type ovarian cell lines was
notably different, with SKVO3 cells growing twice faster than
(P<0.05) than either COV504 or COV644 cells (Fig. 3). COV644 were the slowest growing
cell line. In all transfected cells, overexpression of MTSS1
protein reduced growth rate by both day 3 and day 5. In
SKOV3MTSS1Exp cells, the mean cell number at day 5 was
decreased by 21% (not significant) compared to pEF6 control, in
COV644MTSS1Exp cell numbers significantly decreased by
36% (P<0.05) and in COV504MTSS1Exp cells, growth rate
decreased by 50% (P<0.001).
Effect of MTSS1 overexpression on
cell-matrix adhesion in EOC cells
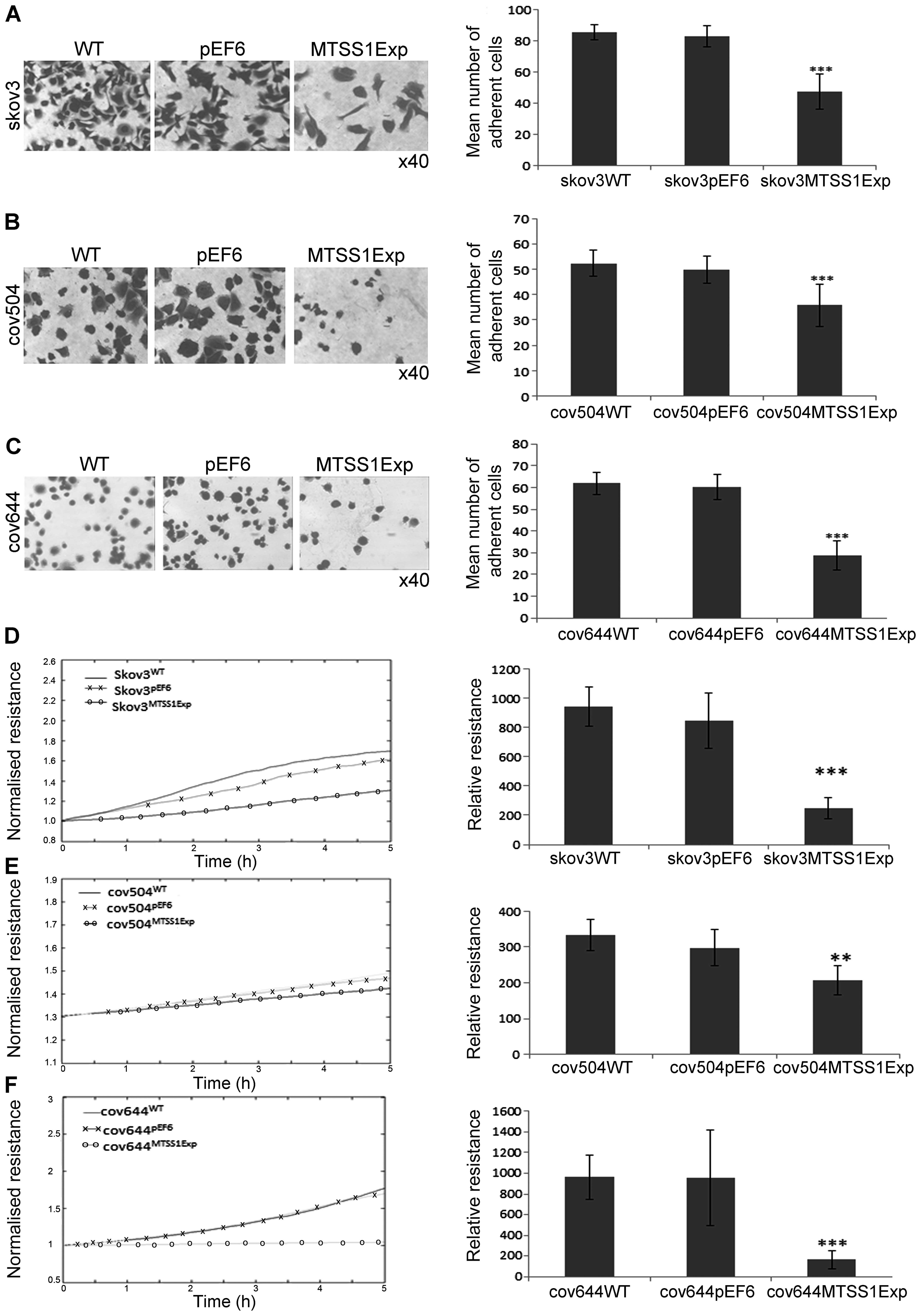
The effect of MTSS1 on the ability of EOC cells to
adhere to Matrigel matrix was examined (Fig. 4A–C). Over-expression of MTSS1
protein caused a significant (P<0.001) inhibitory effect of ~40%
on cell-matrix adhesion in the SKOV3 cells compared to both WT and
pEF6 controls (Fig. 4A). Compared
with COV504WT and COV504pEF6, the number of
COV504MTSS1Exp cells that adhered was also significantly
reduced (P<0.001) by ~40% (Fig.
4B). In COV644 cells, MTSS1 overexpression significantly
reduced (P<0.001) Matrigel adhesion by 50% compared to controls.
The ECIS system was also used to confirm the inhibitory effect of
enhanced expression of MTSS1 on SKOV3, COV504 and COV644 cell
adhesion. This was measured by change in resistance formed over the
growth surface as cells attached from 0 to 5 h (Fig. 4D–E). Compared with the appropriate
WT and pEF6 controls, the resistance was significantly reduced in
SKOV3MTSS1Exp, COV504MTSS1Exp and
COV644MTSS1Exp cells confirming that high expression of
MTSS1 in ovarian cells reduced adhesive capability.
Effect of MTSS1 overexpression on the
invasion of EOC cells
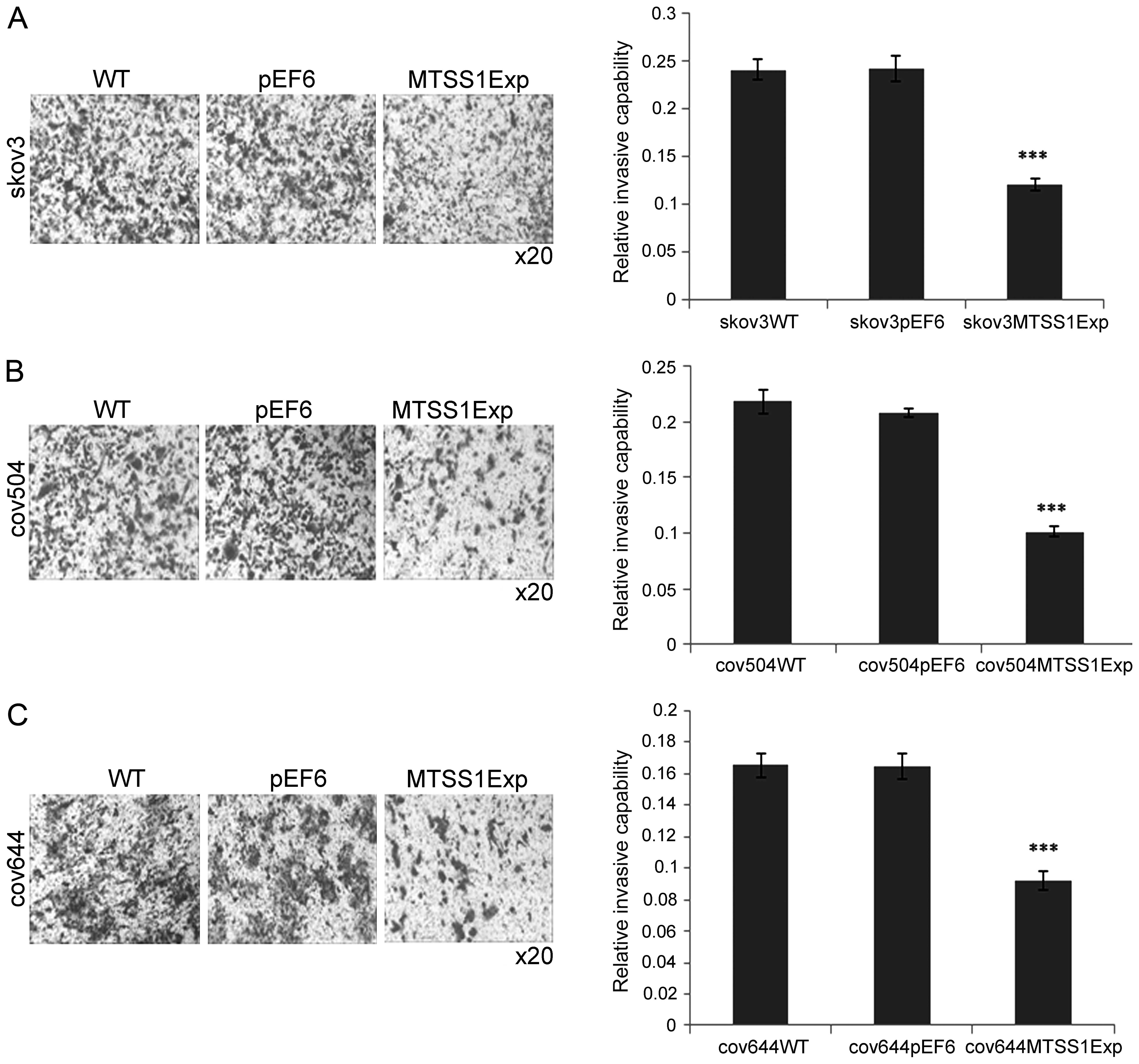
The potential biological relevance of increased
MTSS1 expression was further investigated using in vitro
invasion assays over the artificial basement membrane, Matrigel.
The wild-type ovarian cell lines all had invasive capability. In a
typical experiment, ~320 SKVO3 and COV644 cells invaded per
membrane. COV504 cells had a slightly lower (not significant)
invasive capability with 230 cells/inset (data not shown).
Increased expression of MTSS1 in all of these cell lines caused a
50% reduction (P<0.001) in basal invasion compared to the pEF6
controls (Fig. 5).
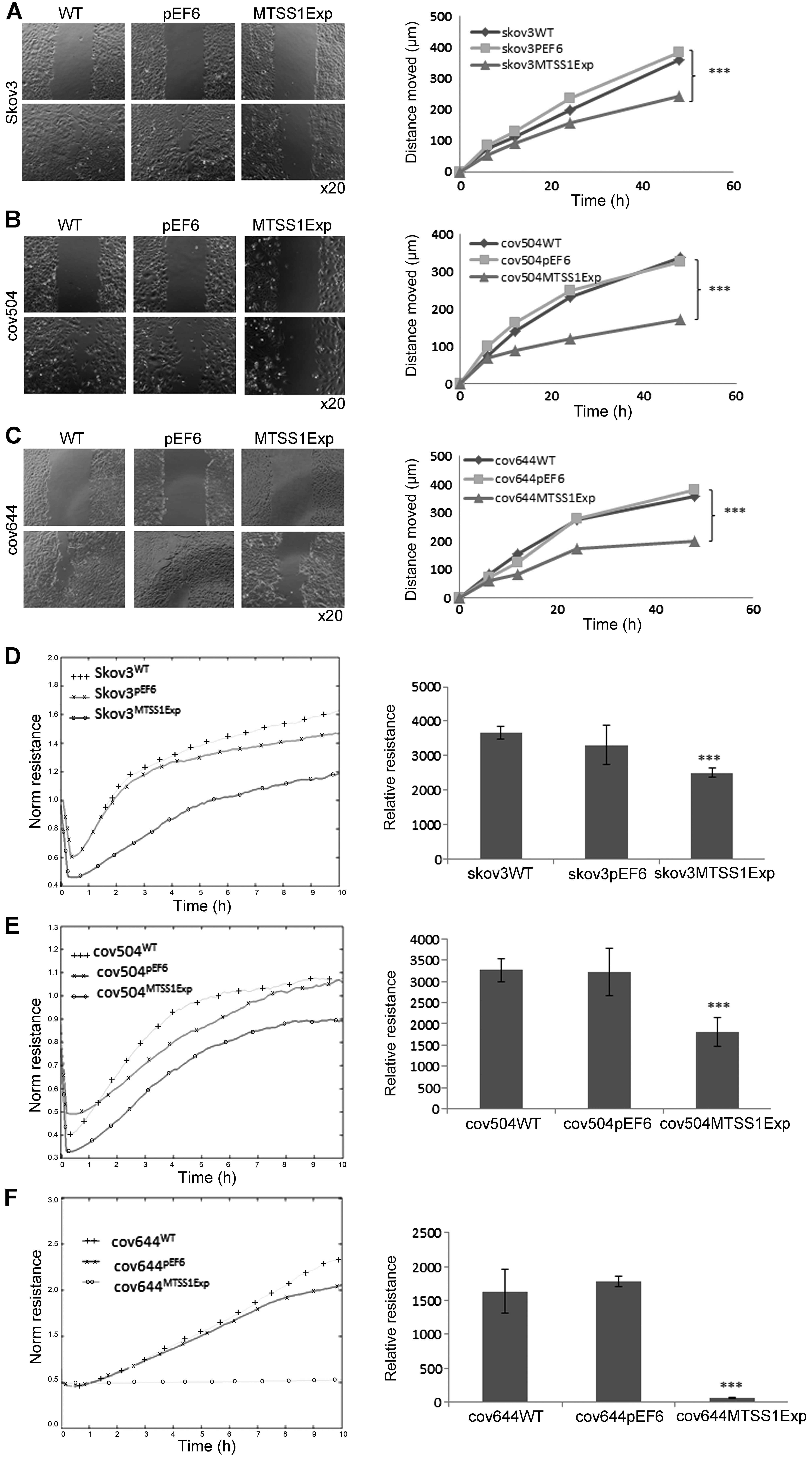
Effect of MTSS1 on wounding/migration of
EOC cells
A cellular wounding assay was used to compare the
ability of wild-type and MTSS1 overexpressing cells to migrate.
SKVO3WT cells migrated slightly faster than COV504 and
COV644 cells, but increased expression of MTSS1 in all three cell
lines caused a marked (up to 50%) reduction in migration capability
(P<0.001).
The ability of increased MTSS1 expression to reduce
ovarian cell motility was also confirmed by measuring the ability
of cell lines to recover from an electrical wound generated using
the ECIS system (Fig. 6D–F).
Measurements taken 10 h post-wound also showed that the migration
capacity of SKVO3MTSS1Exp, COV504MTSS1Exp and
COV644MTSS1Exp were markedly reduced (P<0.001) in
comparison with wild-type and pEF6 control cells (Fig. 6D–F).
Discussion
Over the past decade, prognosis for patients with
EOC has improved little, with the cancer recurring in 70–80% of the
patients who ultimately succumb to the disease (20). There are a number of genetic and
epigenetic changes that lead to transformation of ovarian
epithelial cells into tumor cells (21). To improve the prognosis, assessment
and treatment of EOC patients, it is crucial that we identify the
key molecular regulators of tumorigenesis and understand the key
molecular pathways involved. Although many of these tumorigenesis
mechanisms remain largely unknown, over-expression of certain
oncoproteins (22) or
downregulation of tumor suppressor proteins (23) has been demonstrated to play
important roles in the process of tumor growth and metastasis.
Downregulation of the potential tumor suppressor
MTSS1, has been observed in a number of human cancer types and
complete loss of MTSS1 can be associated with poorly differentiated
metastatic tumors and with poor survival rates (9). However, the available data are
controversial and whether or not MTSS1 serves as a metastasis
suppressor has not been clearly defined. To date, the role of MTSS1
in ovarian cancer remains largely unknown. To the best of our
knowledge, the present study is the first to have examined the
staining pattern of MTSS1 in human EOC tissues and to test the
impact of MTSS1 on the growth, adhesion, invasion and migration of
EOC cells by genetically manipulating the expression of MTSS1.
The present study determined whether there was a
relationship between MTSS1 protein expression and the
aggressiveness of clinical ovarian cancer. Conclusions were limited
by the relatively small sample size used, but immunohistochemical
analysis clearly demonstrated that the samples we examined
expressed a range of MTSS1 protein. We detected relatively high
levels of MTSS1 in non-cancerous ovarian tissue and in some
metastatic and non-metastatic tumors. However, typically the MTSS1
staining in samples from tumors that had metastasised was weaker
than that detected in non-metastatic cancers. We also detected a
higher percentage (40%) of completely MTSS1 negative metastatic
compared to non-metastatic (20%) samples. Due to the relatively low
number of clinical samples included in our study, the results did
not enable statistical analysis. It has been previously reported
that MTSS1 can be lost in aggressive tumors in a number of tumor
types including bladder (5),
prostate (6), kidney (7) and gastric (8) cancer. However, as seen in
hepatocellular carcinoma (10) and
currently in our ovarian tumor samples, MTSS1 can also frequently
be overexpressed in primary tumors. It has been suggested that in
primary tumor formation, high MTSS1 expression may initially be
selectively beneficial by increasing plasma membrane EGFR
expression, resulting in increased EGF signalling, Erk1/2
activation and cancer proliferation and survival (25). As cell density increases with tumor
growth, MTSS1 may switch to the inhibitory tumor suppressor role
for which it is better known; when it inhibits EGFR and Akt
signalling and cells retain the epithelial-like morphology
consistent with an anti-metastatic role (25). In some tumor types, including
breast and oesophageal cancer, MTSS1 may be considered a suitable
biomarker for tumor progression, with high MTSS1 being associated
with favourable prognosis and reduced MTSS1 a poorer outcome
(9,26). How MTSS1 can be overexpressed in
early tumor formation and lost in later metastatic stages remains
unclear, although methylation has been considered as one mechanism
used to silence the gene expression (28). Our clinical results show that
although there is a trend for reduced MTSS1 expression in
metastatic tumors, MTSS1 expression is variable between individuals
which may reflect a range of differences in ovarian cancer etiology
or disease stages when samples were taken. Conclusive results can
only be obtained if a larger cohort is used in the study.
A relatively small number of in vitro studies
have been previously reported in which the function of MTSS1 is
characterized. Our studies have shown that 3 different ovarian
tumor cell lines, SKVO3, COV504 and COV644, which expressed a
reasonable (but not high) invasive capability, all expressed a
similar low level of MTSS1 mRNA and protein. Cellular function
tests further demonstrated that the presence of MTSS1 was related
to the inhibition of the ovarian cancer cell aggressiveness.
Previous studies with a panel of human cell lines have shown MTSS1
is differentially expressed with an inverse correlation between
cell differentiation, invasive capability and MTSS1 expression
(9,25,26).
These results were not without exceptions as some cell lines, like
the breast cancer cell ZR75.1, considered non-invasive, were
negative for MTSS1 expression suggesting other factors in addition
to MTSS1 are involved in regulating invasion (9). We have demonstrated here that MTSS1
overexpression resulted in a dramatic reduction in ovarian cancer
cell line growth, adhesion, invasion and migration, in comparison
with control cells. The inhibitory effect of MTSS1 on ovarian
cancer cell growth is in agreement with the findings in kidney,
bladder, oesophageal, breast and prostate cell lines (7,9,26,27,29).
Although the precise molecular mechanisms by which MTSS1 inhibits
tumor growth remains unknown, at high cell density MTSS1 has been
shown to dampen EFG signalling which inhibits proliferation in
epithelial-like layers (25).
MTSS1 is a member of a growing family of cytoskeletal components
that associate with transcription factors to affect nuclear
signaling. MTTS1 is known to behave as a sonic hedgehog (Shh)
responsive gene and may use this pathway to modulate responses in
cancer growth and development (30,31).
MTSS1 overexpression resulting in dramatic
inhibition of tumor cell adhesion and migration and invasion over
Matrigel has also been previously reported in the ovarian cell line
SKVO3 (14) as well as in breast
and prostate cell lines (9,27).
There are, however, differences between cancer types, because
overexpression of MTSS1 in bladder cancer cell lines has been
reported to inhibit growth and adhesion but have no effect on
invasion or migration (29). In
contrast, in breast cancer, increased MTSS1 expression inhibits
invasion but has no effect of cell adhesion (9). Cytoskeletal rearrangement is a
critical event in cell motility. MTSS1 is known to act as a
cytoskeletal scaffold protein where it regulates plasma membrane
dynamics and actin filaments in a complex fashion. The detailed
mechanism of the MTSS1 effect on cell motility remains to be
further defined. MTSS1 protein appears to control the formation of
lamellipodia, membrane ruffles and filopodia and enhance changes in
cell shape (32). In contrast,
MTSS1 also promotes cell-cell junction assembly through recruiting
small GTPase and actin, which drives junction maintenance.
Therefore, loss of MTSS1 in cancers may lead to the loss of
junction stability, which ultimately promotes EMT and metastasis
(24). Furthermore, MTSS1 is known
to negatively regulate the epidermal growth factor signaling to
suppress metastasis (25). Further
studies are required to reveal the exact molecular mechanisms and
signaling pathways through which MTSS1 modulates cancer cell
migration and invasion.
In summary, the present study shows that the
overexpression of the MTSS1 protein can suppress the aggressiveness
of human ovarian cancer. Our work further suggests that preventing
MTSS1 degradation, or partially restoring MTSS1 expression, could
be a possible novel strategy to treat aggressive ovarian cancer
growth and metastasis.
Acknowledgements
The authors are grateful to Cancer Research Wales
and the Albert Hung Foundation for their support and funding of the
present study.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee YG, Macoska JA, Korenchuk S and Pienta
KJ: MIM, a potential metastasis suppressor gene in bladder cancer.
Neoplasia. 4:291–294. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nixdorf S, Grimm MO, Loberg R, Marreiros
A, Russell PJ, Pienta KJ and Jackson P: Expression and regulation
of MIM (Missing In Metastasis), a novel putative metastasis
suppressor gene, and MIM-B, in bladder cancer cell lines. Cancer
Lett. 215:209–220. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Loberg RD, Neeley CK, Adam-Day LL, Fridman
Y, St John LN, Nixdorf S, Jackson P, Kalikin LM and Pienta KJ:
Differential expression analysis of MIM (MTSS1) splice variants and
a functional role of MIM in prostate cancer cell biology. Int J
Oncol. 26:1699–1705. 2005.PubMed/NCBI
|
|
7
|
Du P, Ye L, Li H, Yang Y and Jiang WG: The
tumour suppressive role of metastasis suppressor-1, MTSS1, in human
kidney cancer, a possible connection with the SHH pathway. J Exp
Ther Oncol. 10:91–99. 2012.
|
|
8
|
Liu K, Wang G, Ding H, Chen Y, Yu G and
Wang J: Downregulation of metastasis suppressor 1 (MTSS1) is
associated with nodal metastasis and poor outcome in Chinese
patients with gastric cancer. BMC Cancer. 10:4282010. View Article : Google Scholar
|
|
9
|
Parr C and Jiang WG: Metastasis suppressor
1 (MTSS1) demonstrates prognostic value and anti-metastatic
properties in breast cancer. Eur J Cancer. 45:1673–1683. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma S, Guan XY, Lee TK and Chan KW:
Clinicopathological significance of missing in metastasis B
expression in hepato-cellular carcinoma. Hum Pathol. 38:1201–1206.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamagishi A, Masuda M, Ohki T, Onishi H
and Mochizuki N: A novel actin bundling/filopodium-forming domain
conserved in insulin receptor tyrosine kinase substrate p53 and
missing in metastasis protein. J Biol Chem. 279:14929–14936. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mattila PK, Salminen M, Yamashiro T and
Lappalainen P: Mouse MIM, a tissue-specific regulator of
cytoskeletal dynamics, interacts with ATP-actin monomers through
its C-terminal WH2 domain. J Biol Chem. 278:8452–8459. 2003.
View Article : Google Scholar
|
|
13
|
Woodings JA, Sharp SJ and Machesky LM:
MIM-B, a putative metastasis suppressor protein, binds to actin and
to protein tyrosine phosphatase delta. Biochem J. 371:463–471.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Liu J, Segura MF, Shao C, Lee P,
Gong Y, Hernando E and Wei JJ: MiR182 overexpression in
tumorigenesis of high-grade ovarian papillary serous carcinoma. J
Pathol. 228:204–215. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang WG, Davies G, Martin TA, Parr C,
Watkins G, Mason MD and Mansel RE: Expression of membrane type-1
matrix metal-loproteinase, MT1-MMP in human breast cancer and its
impact on invasiveness of breast cancer cells. Int J Mol Med.
17:583–590. 2006.PubMed/NCBI
|
|
16
|
Jiang WG, Davies G, Martin TA, Parr C,
Watkins G, Mason MD, Mokbel K and Mansel RE: Targeting matrilysin
and its impact on tumor growth in vivo: The potential implications
in breast cancer therapy. Clin Cancer Res. 11:6012–6019. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang WG, Hiscox S, Singhrao SK, Nakamura
T, Puntis MC and Hallett MB: Inhibition of HGF/SF-induced membrane
ruffling and cell motility by transient elevation of cytosolic free
Ca2+. Exp Cell Res. 220:424–433. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang WG, Hiscox S, Hallett MB, Horrobin
DF, Scott C and Puntis MCA: Inhibition of invasion and motility of
human colon cancer cells by gamma linolenic acid. Br J Cancer.
71:744–752. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang WG, Martin TA, Lewis-Russell JM,
Douglas-Jones A, Ye L and Mansel RE: Eplin-alpha expression in
human breast cancer, the impact on cellular migration and clinical
outcome. Mol Cancer. 7:71–80. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Onda T and Yoshikawa H: Neoadjuvant
chemotherapy for advanced ovarian cancer: Overview of outcomes and
unanswered questions. Expert Rev Anticancer Ther. 11:1053–1067.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kurman RJ, Visvanathan K, Roden R, Wu TC
and Shih IeM: Early detection and treatment of ovarian cancer:
Shifting from early stage to minimal volume of disease based on a
new model of carcinogenesis. Am J Obstet Gynecol. 198:351–356.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang JL, Ow KT, Russell PJ, Ham JM and
Crowe PJ: Higher expression of oncoproteins c-myc, c-erb B-2/neu,
PCNA, and p53 in metastasizing colorectal cancer than in
nonmetastasizing tumors. Ann Surg Oncol. 3:574–579. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Goncharuk VN, del-Rosario A, Kren L, Anwar
S, Sheehan CE, Carlson JA and Ross JS: Co-downregulation of PTEN,
KAI-1, and nm23-H1 tumor/metastasis suppressor proteins in
non-small cell lung cancer. Ann Diagn Pathol. 8:6–16. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mustafa N, Martin TA and Jiang WG:
Metastasis tumour suppressor-1 and the aggressiveness of prostate
cancer cells. Exp Ther Med. 2:157–162. 2011.PubMed/NCBI
|
|
25
|
Dawson JC, Timpson P, Kalna G and Machesky
LM: Mtss1 regulates epidermal growth factor signaling in head and
neck squamous carcinoma cells. Oncogene. 31:1781–1793. 2012.
View Article : Google Scholar
|
|
26
|
Xie F, Ye L, Chen J, Wu N, Zhang Z, Yang
Y, Zhang L and Jiang WG: The impact of metastasis suppressor-1,
MTSS1, on oesophageal squamous cell carcinoma and its clinical
significance. J Transl Med. 9:952011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dawson JC, Bruche S, Spence HJ, Braga VM
and Machesky LM: Mtss1 promotes cell-cell junction assembly and
stability through the small GTPase Rac1. PLoS One. 7:e311412012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu
X, Zhao Z, Kong KL, Dong S, Song Y, Chan THM and Guan XY: MTSS1, a
novel target of DNA methyltransferase 3B, functions as a tumor
suppressor in hepatocellular carcinoma. Oncogene. 31:2298–2308.
2012. View Article : Google Scholar
|
|
29
|
Du P, Ye L, Ruge F, Yang Y and Jiang WG:
Metastasis suppressor-1, MTSS1, acts as a putative tumour
suppressor in human bladder cancer. Anticancer Res. 31:3205–3212.
2011.PubMed/NCBI
|
|
30
|
Callahan CA, Ofstad T, Horng L, Wang JK,
Zhen HH, Coulombe PA and Oro AE: MIM/BEG4, a Sonic
hedgehog-responsive gene that potentiates Gli-dependent
transcription. Genes Dev. 18:2724–2729. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie F, Ye L, Ta M, Zhang L and Jiang WG:
MTSS1: A multi-functional protein and its role in cancer invasion
and metastasis. Front Biosci (Schol Ed). 3:621–631. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhong J, Shaik S, Wan L, Tron AE, Wang Z,
Sun L, Inuzuka H and Wei W: SCF β-TRCP targets MTSS1 for
ubiquitination-mediated destruction to regulate cancer cell
proliferation and migration. Oncotarget. 4:2339–2353. 2013.
View Article : Google Scholar : PubMed/NCBI
|