Introduction
Ovarian cancer is the most lethal gynecologic
malignancy. Approximately 70% of cases are diagnosed at an advanced
stage with metastases, and the 5-year survival of patients with
advanced stage ovarian cancer is <30% (1). The high rate of lethality from
ovarian cancer is mainly due to the early progression of the
diseases and lack of effective therapies for advanced ovarian
cancers (2). Ovarian cancer
displays highly molecular and genetic heterogeneity and complexity,
although most cases of ovarian cancer are treated in a similar
manner. A comprehensive understanding of molecular basis in ovarian
cancer development and progression will help to identify better
therapeutic strategies for this disease. Whole genome profiling of
ovarian cancer has identified many dysregulated genes which are
implicated in cell proliferation, invasion, motility, chromosomal
instability, and gene silencing (3–5).
However, the biological functions of these genes in ovarian cancer
have not been fully studied.
IQ motif-containing GTPase activating protein 2
(IQGAP2) belongs to the IQGAP family which contains a conventional
Ras GTPase-activating protein (RasGAP) domain and an IQ motif
(6,7). The human IQGAP gene family consists
of 3 members, IQGAP1, IQGAP2, and IQGAP3. IQGAP1 is the best
characterized and is considered to be a scaffolding protein
integrating diverse signaling pathways involved in cell
proliferation, adhesion and migration (8,9).
Although IQGAP2 and IQGAP3 proteins exhibit a high level of
sequence homology with IQGAP1, but their biological roles are
poorly defined. Cumulative evidence from clinical specimens suggest
IQGAP2 may be a tumor suppressor. Loss of IQGAP2 has been observed
in gastric cancer, hepatocellular carcinoma and advanced prostate
cancer (10–12), while the status of IQGAP2 in
ovarian cancer remain unclear.
In the present study, by analysis of the
differential epigenetic features between normal ovary and ovarian
cancer, we identified that IQGAP2 was significantly downregulated
in ovarian cancer due to elevated DNA methylation and was
associated with patient prognosis. Biological function studies
indicated that loss of IQGAP2 induced ovarian cancer cell
epithelial-mesenchymal transition (EMT) and thus promoted cell
migration and invasion. Further mechanism dissection demonstrated
that loss of IQGAP2 potentiated Wnt/β-catenin signaling rather than
activating the Ras in ovarian cancer cells.
Materials and methods
Human ovarian cancer specimen
Five frozen human serous ovarian cancer and matched
normal ovary specimens were obtained from patients who underwent
surgical resection with the approval of the Institutional Review
Board (IRB) of the First Hospital of Medical College of Xi'an
Jiaotong University. Whole genomic DNA and total proteins were
extracted for gene methylation and protein expression assays.
Bioinformatic analysis
HumanMethylation450 BeadChip arrays-based
genome-scale DNA methylation data (Batch 9 and 40, update to
February 4, 2014), RNA-Seq-based gene expression data for IQGAP2,
and the clinical annotation data of serous ovarian cancer samples
from The Cancer Genome Atlas (TCGA) were all retrieved through the
CGDS server of the cBioportal hosted by the Memorial
Sloan-Kettering Cancer Center. Gene microarray data for IQGAP2 of
normal ovarian tissues and different subtypes of ovarian cancer
tissues were retrieved from the GEO datasets (GSE26712, GSE6008).
The X-tile which is a bioinformatics tool for biomarker assessment
and outcome-based cut-point optimization was used to generate an
optimal cut-off point to dichotomize IQGAP2 mRNA level as ‘High’
and ‘Low’ using a Monte Carlo P-value <0.05.
Cell culture
Human ovarian cancer cell SKOV3 and SW626 were
maintained in RPMI-1640 (Gibco, San Diego, CA, USA) medium
supplemented with 10% fetal bovine serum (FBS), CAOV3 was
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco)
supplemented with FBS. All cells were cultured in a humidified
incubator at 37°C with 5% CO2.
Methylation specific PCR (MS-PCR)
Genomic DNA from normal and ovarian cancer tissues
was extracted using DNAse kit (Qiagen, Hilden, Germany), 2 μg of
genomic DNA was denatured by NaOH and modified by sodium bisulfite,
and the modified DNA was purified using a Wizard DNA clean-up
system (Promega, Madison, WI, USA). Bisulfite-treated DNA was then
amplified as described (10).
Briefly, PCR was performed by using methylation specific or
unmethylation specific primers. Primers used for methylated IQGAP2
were: 5′-GGAGTGGGTCGTAGATTTTCGGGC-3′ (sense) and
5′-CTACCCTCGCTAACCAAACTCGCG-3′ (antisense), and primers used for
unmethylated IQGAP2 were: 5′-AGGGAGTGGGTTGTAGATTTTTGGGT-3′ (sense)
and 5′-CAACTACCCTCACTAACCAAACTCACA-3′ (antisense). The PCR reaction
was performed for 35 cycles of 95°C for 30 sec, 58°C for 30 sec and
72°C for 30 sec.
Reverse transcriptional (RT) real-time
PCR
Cells were treated with control (dimethyl sulfoxide,
DMSO) or the indicated dose of methylation inhibitor
5-aza-2′-deoxycytidine (5-Aza; Sigma, St. Louis, MO, USA) for 7
days. Total RNA was extracted and 1 μg RNA was reverse transcribed
with a cDNA synthesis kit (Invitrogen). Real-time PCR analysis was
set up with SYBR Green qPCR Supermix kit (Invitrogen) and carried
out in the iCycler thermal cycler. The relative level of mRNA
expression of each gene was determined by normalizing with β-actin.
The primers used are: for β-actin, 5′-TACCACAGGCATTGTGATGG-3′
(forward) and 5′-TTTGATGTCACGCACGATTT-3′ (reverse); for IQGAP2,
5′-TCAAGATTGGACTGCTGGTG-3′ (forward) and 5′-AGG
TTTGGTCTGGAGGAGGT-3′ (reverse).
Western blotting
Western blotting was performed as described
(13). Briefly, cell lysates were
harvested, equivalent amount of proteins were separated by 10%
sodium dodecyl sulfate polyacrylamide gel electrophoresis and
transferred to nitrocellulose membranes. Membranes were blocked
with 5% skim milk, incubated with anti-IQGAP2 antibody (Santa Cruz,
1:500) overnight at 4°C and followed by incubation with horseradish
peroxidase conjugated secondary antibodies for 1 hour (h) at room
temperature. Signals were then detected by chemiluminescence
(Pierce, Rockford, IL, USA).
GST pull-down active Ras detection
Cells transiently transfected with different dose of
IQGAP2 cDNA were then serum-starved for 12 h. Active Ras (Raf1
binding Ras) was determined by the glutathione S-transferase
(GST)-Raf1-RBD pull-down assay (Thermo Scientific, Waltham, MA,
USA) followed by western blotting with Ras antibody. Briefly, cells
were lysed in 500 μl of lysis/binding buffer (25 mM Tris HCl, pH
7.2, 150 mM NaCl, 5 mM MgCl2, 1% NP-40 and 5% glycerol)
for 5 min. Insoluble cellular debris was removed by centrifugation
at 14,000 rpm for 15 min at 4°C. Cell lysate (250 μg) was incubated
with 80 μg of GST-Raf1-RBD protein fused to glutathione agarose
resin at 4°C for 60 min. Cell lysate incubated with GTPγS or GDP
were set up as positive or negative control. After the incubation,
samples were washed three times, resuspended in 30 μl of SDS sample
buffer and heated for 10 min. Active Ras was detected by western
blotting probed with Ras antibody.
Dual-luciferase reporter assay
β-catenin reporter gene luciferase assay was
performed as described previously (14). Cells seeded in 24-well plates were
transfected with 1 μg β-catenin firefly responsive luciferase
constructs TOP and 2 ng Renilla luciferase construct as internal
control. Forty-eight hours after transfection, TOP luciferase
activity was measured using dual luciferase assay kit according to
the manufacturer's protocol (Promega).
Immunofluorescence (IF)
The cells were fixed with 4% paraformaldehyde for 20
min, permeabilized with 0.1% Triton X-100 and blocked with 3%
bovine serum albumin for 1 h. Cells were then incubated with
β-catenin primary antibody (Cell Signaling Technology, 1:200)
overnight at 4°C followed by TRITC labeled conjugates (Sigma;
1:500). Cells were counterstaining with
4,6-diamidino-2-phenylindole and fluorescence was visualized by
fluorescence microscopy (Olympus Optical Co., Japan).
Transfection
Cells (2×105) at 60% confluence were
seeded in a 6-well plate prior to the transfection, IQGAP2
overexpressing vector (pcDNA3-IQGAP2) and vector control (VC) were
transfected into cells by Xfect™ Transfection reagent (Clontech,
Palo Alto, CA, USA), 48 h after the transfection, G418 was used to
select stable clones. IQGAP2 siRNAs were from RiboBio (Guangzhou,
China) and transfected by X-tremeGENE siRNA transfection reagent
(Roche Diagnostics, Mannheim, Germany) according to the
manufacturer's instructions.
Transwell invasion assay
Matrigel-coated Transwell was used to examine the
invasive ability of cells. Briefly, 5×104 cells in 100
μl of medium containing 0.5% FBS were seeded into the upper
chamber, and 1 ml of medium containing 20% FBS was added to the
lower chamber. Cells in Transwell were then cultured at 37°C in 5%
CO2 for 48 h. Cells on the lower surface of the membrane
were stained with 0.5% crystal violent (Sigma) and photographed and
counted.
Statistical analysis
The Kaplan-Meier method was used to analyze patient
survival, and the log-rank test was used to assess the differences
between groups. All the data from in vitro assay are
presented as the mean ± SEM from three independent experiments and
the differences between two groups were compared by the Student's
t-test. All statistical analyses were performed using GraphPad
Prism 6.0 and SPSS16.0 software.
Results
IQGAP2 is hypermethylated in ovarian
cancer
DNA methylation is an epigenetic mark which can be
associated with transcriptional inactivity, it is essential for
normal development (15) and has
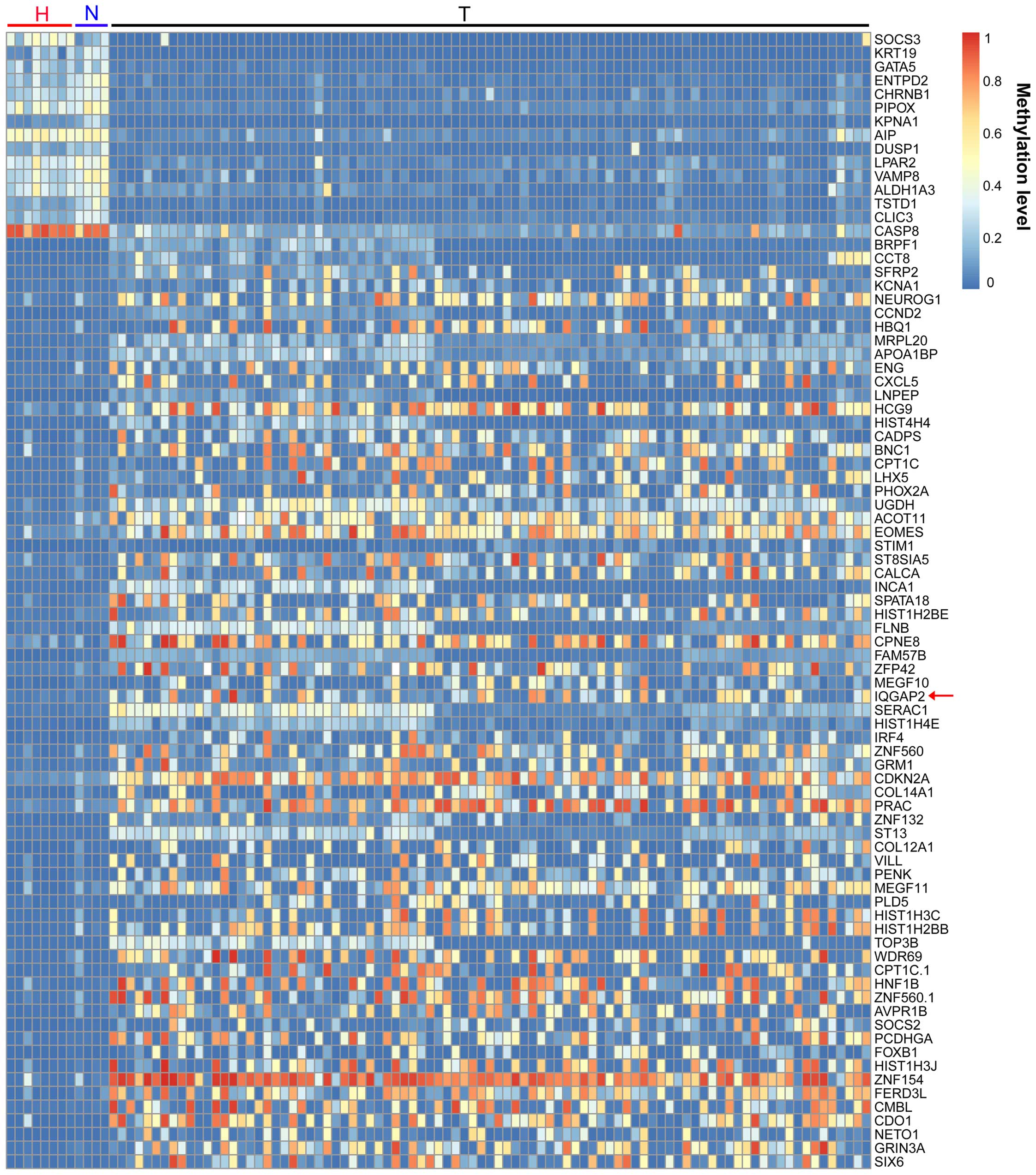
been implicated in many pathologies including cancer (16). We firstly analyzed the genome-scale
DNA methylation profiles of 8 healthy ovary tissues, 89 ovarian
cancers and the corresponding 4 normal ovary tissues from TCGA
(17) to identify the altered gene
methylation in ovarian cancers (Fig.
1). Within those differential genes, IQGAP2, a Ras
GTPase-activating protein coding gene, was significantly
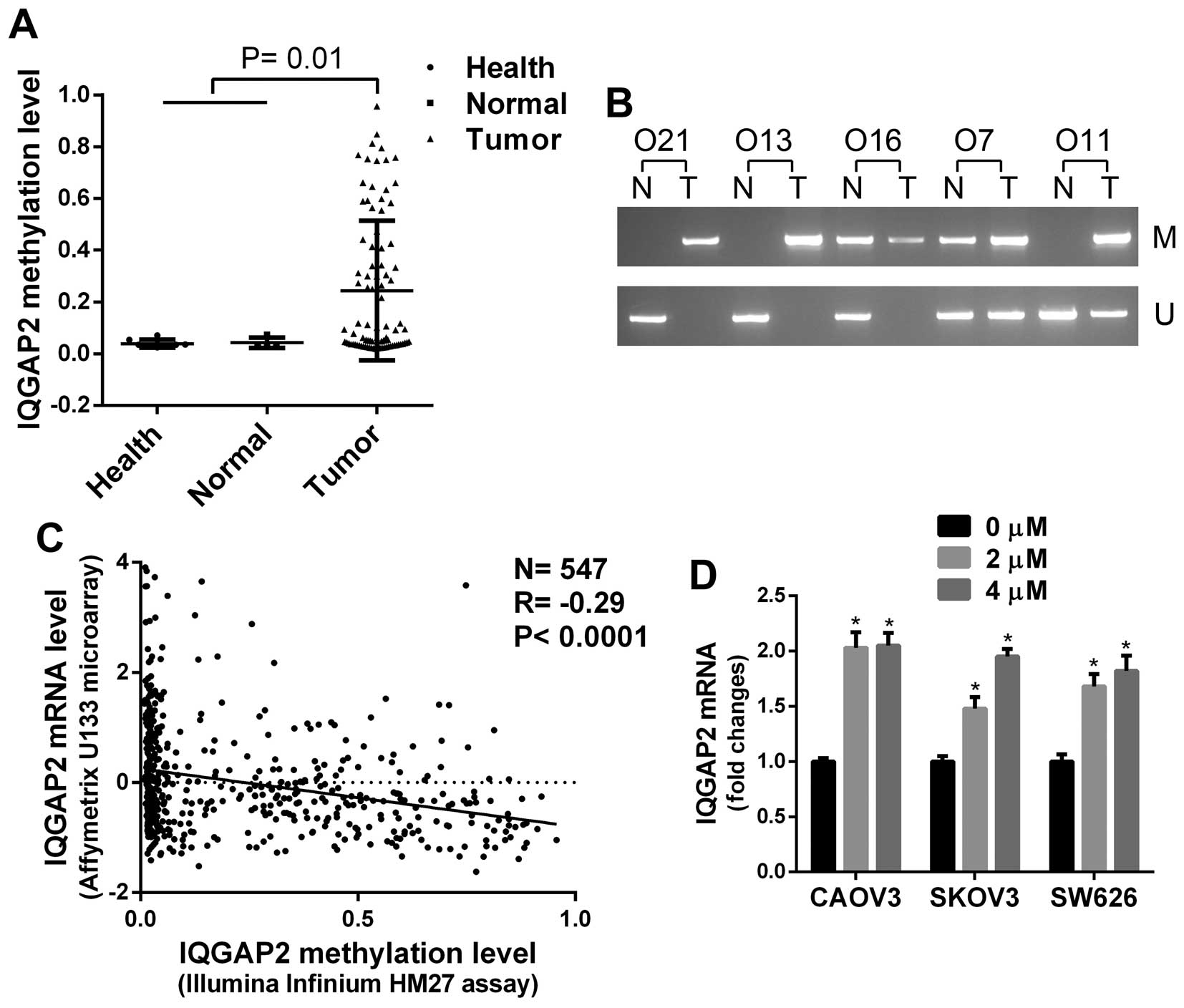
hypermethylated in ovarian cancers (P<0.05, Figs. 1 and 2A). To verify the data from TCGA, the
methylation status of IQGAP2 in 5 serous ovarian cancers and the
corresponding normal ovary tissues from an independent cohort was
examined by MS-PCR. Four of 5 ovarian cancers exhibited increased
methylation of IQGAP2 compared to normal tissues (Fig. 2B).
To determine whether aberrant methylation results in
inactivation of IQGAP2 in ovarian cancer, the association between
IQGAP2 DNA methylation and mRNA expression in 547 serous ovarian
cancers from TCGA was analyzed, and a reverse correlation was found
(P<0.001, Fig. 2C).
Additionally, ovarian cancer cells were treated with a DNA
methylation inhibitor, 5-Aza, and the expression of IQGAP2 in cells
were significantly induced (Fig.
2D). Taken together, these data indicate that IQGAP2 is
hypermethylated in ovarian cancer.
IQGAP2 is downregulated in ovarian cancer
and associated with a poor survival of patient
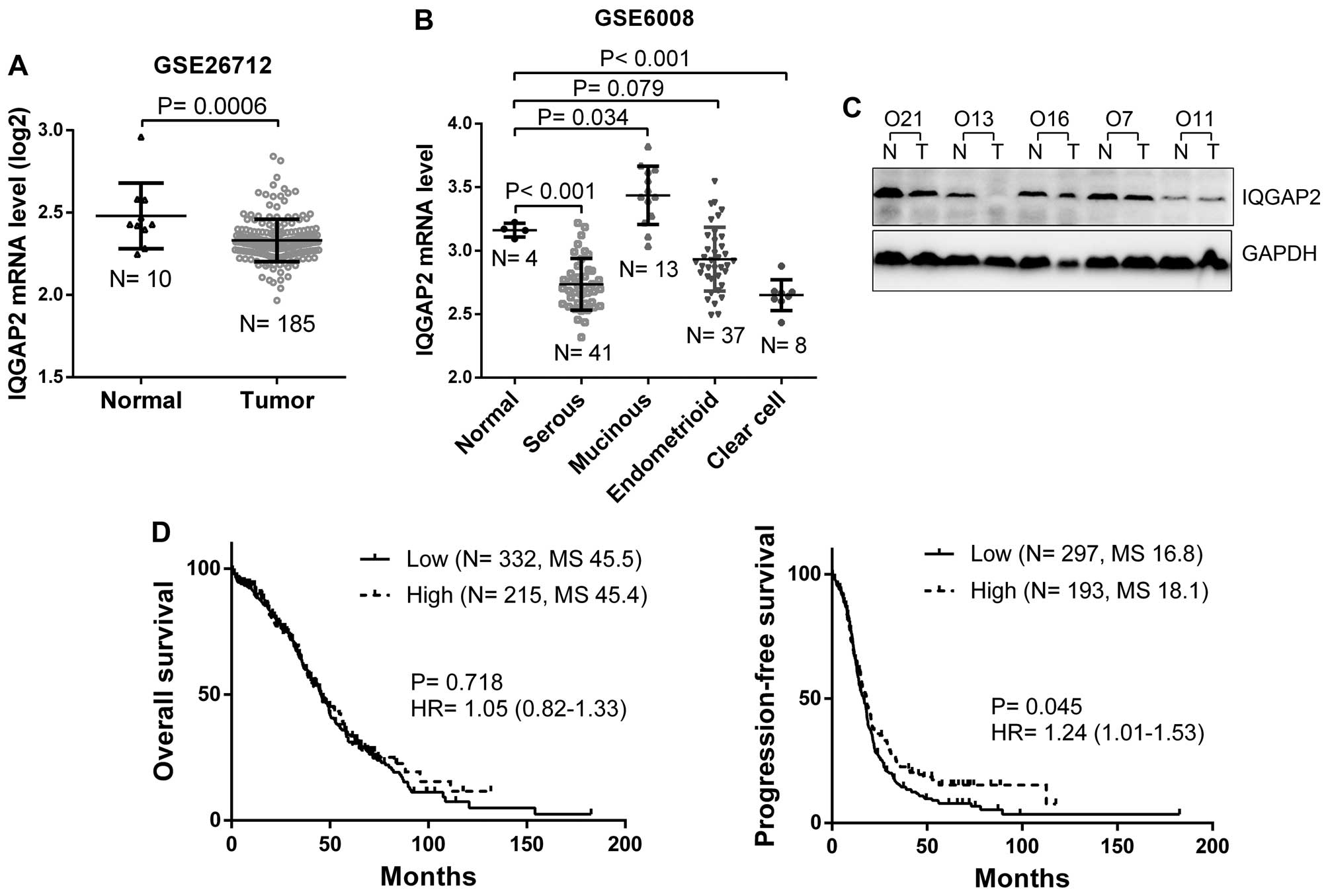
We further analyzed the expression of IQGAP2 in 10
normal ovary and 185 ovarian cancer tissues from GEO database and
found IQGAP2 mRNA was significantly decreased in ovarian cancers
(P=0.0006, Fig. 3A). When subtypes
were stratified, serous and clear cell ovarian cancers showed
decreased IQGAP2 mRNA expression (P<0.05), while there was no
significant difference in IQGAP2 level between endometrioid and
normal tissues (P=0.079, Fig. 3B).
In addition, the expression of IQGAP2 protein was examined in those
5 serous ovarian cancers, 2 of them showed significantly decreased
IQGAP2 expression compared to the corresponding normal ovary
tissues (Fig. 3C). Furthermore, to
determine whether IQGAP2 expression is a prognostic factor for
ovarian cancer, Kaplan-Meier (log-rank test) survival assay was
performed to determine the association between IQGAP2 level and
patient survival from TCGA. The data indicated that although IQGAP2
was not correlated with patient overall survival (P=0.718),
decreased IQGAP2 mRNA was correlated with a worse progression-free
survival of ovarian cancer patients (P=0.045, Fig. 3D).
IQGAP2 negatively regulates the
invasiveness of ovarian cancer
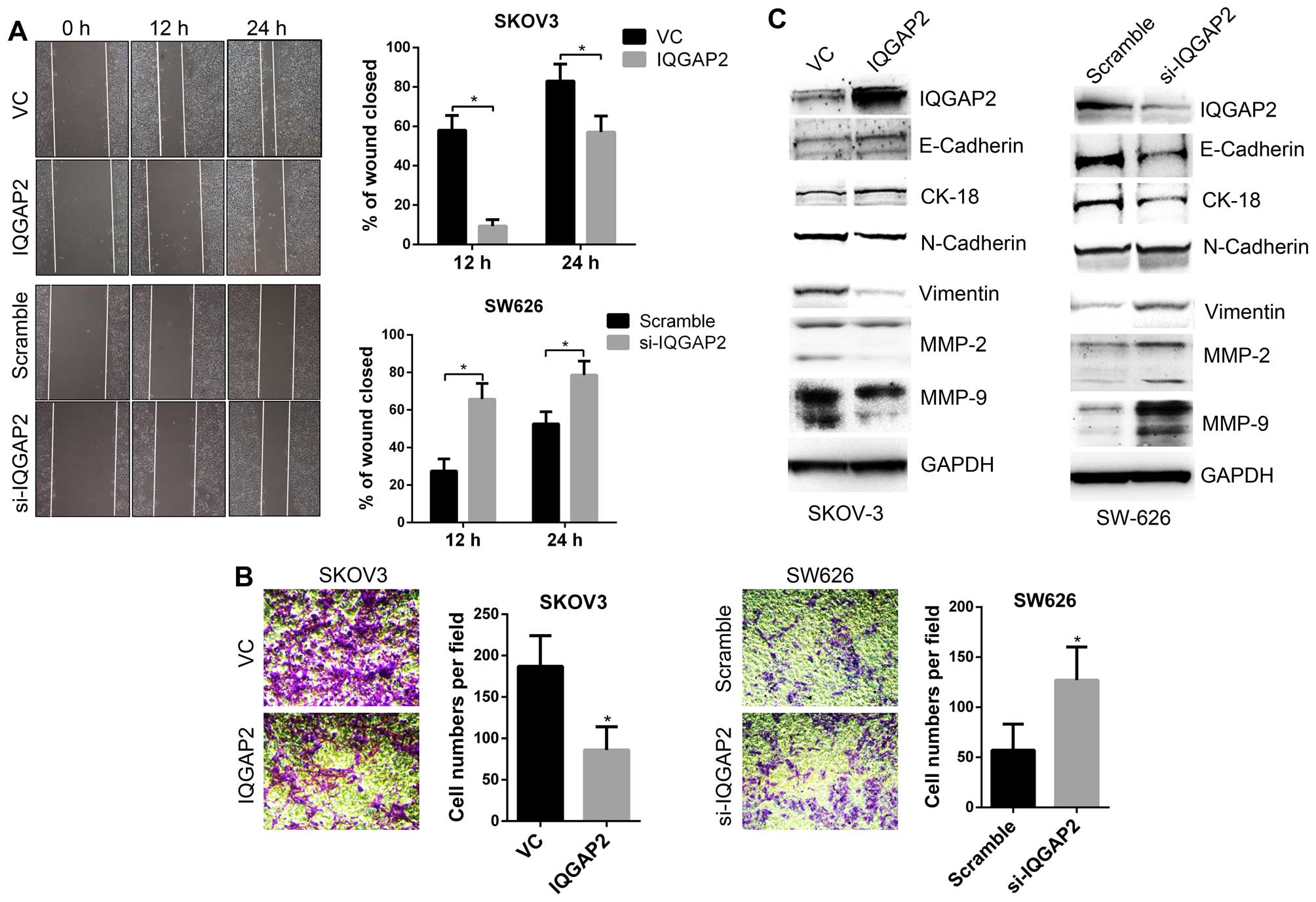
To determine the biological function of IQGAP2 in
ovarian cancer, stable IQGAP2-overexpressing (IQGAP2) and vector
control (VC) cells from SKOV3, and IQGAP2 silencing (si-IQGAP2) and
control (Scramble) cells from SW626 were established. In
vitro cell migration and invasion assay demonstrated that
overexpression of IQGAP2 suppressed while loss of IQGAP2 promoted
the migration and invasion of ovarian cancer cells (Fig. 4A and B). EMT is a process thought
to initiate metastasis by enhancing the motility of tumor cells
(18). We examined the expression
of EMT markers in IQGAP2-overexpressing or IQGAP2-silenced cells,
and the results showed that IQGAP2 upregulated epithelial markers,
such as E-cadherin and CK-18, and downregulated mesenchymal makers,
such as vimentin, MMP-2 and MMP-9 (Fig. 4C). These data indicate IQGAP2 as a
suppressor for the invasiveness of ovarian cancer cells.
IQGAP2 suppresses cell invasiveness via
inhibition of Wnt/β-catenin signaling
To investigate the mechanism of IQGAP2 in the
regulation of cell invasiveness, we firstly determined the effect
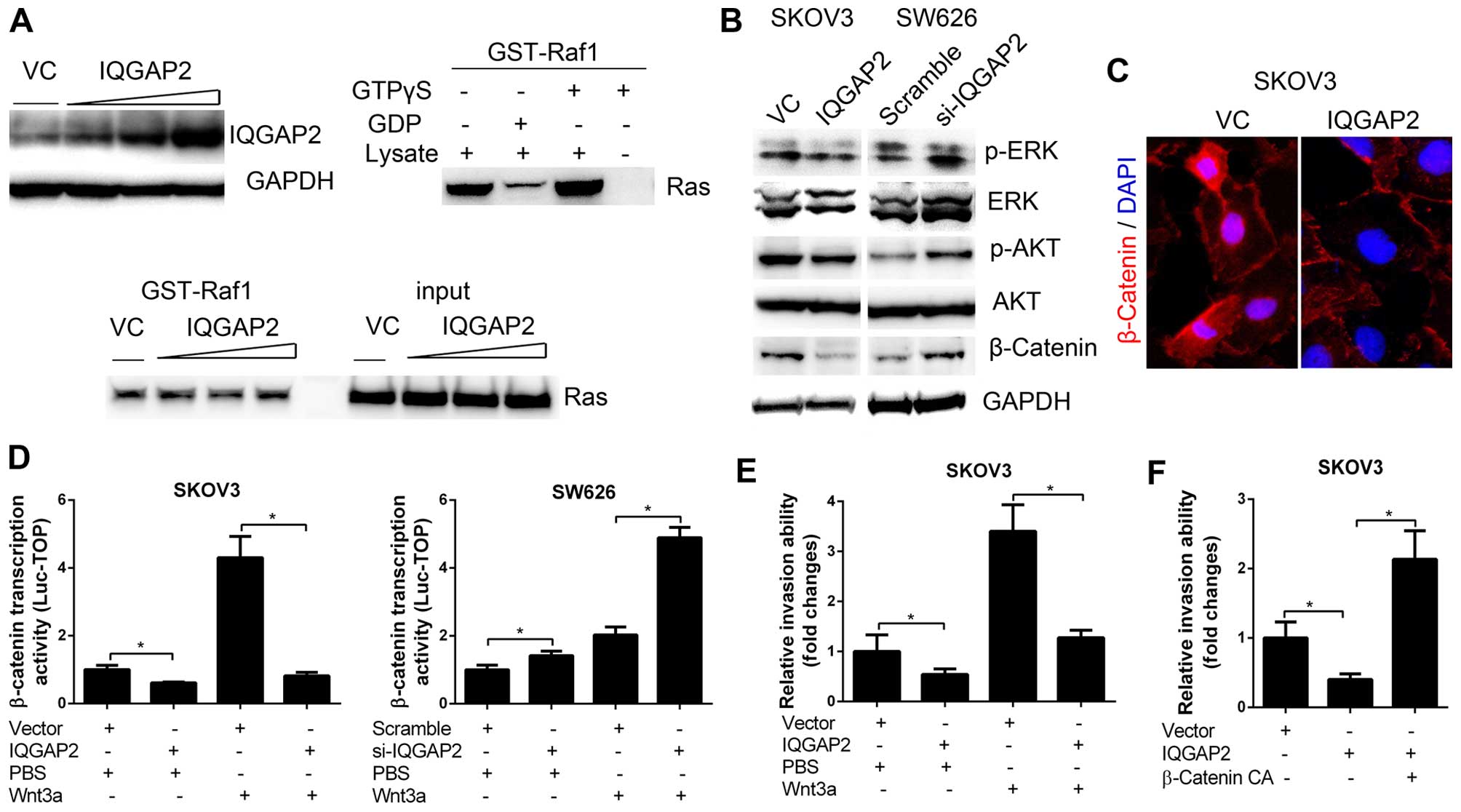
of IQGAP2 on the activity of Ras in ovarian cancer cells. SKOV3
cells were transfected with different doses of IQGAP2 cDNA and GST
pull-down active Ras assay demonstrated that the activity of Ras in
cells were not affected by IQGAP2 (Fig. 5A). We then determined the
activities of ERK, AKT and β-catenin signaling which have been
reported to be regulated by IQGAP2 in other cancers (11,12),
and found that IQGAP2 significantly suppressed β-catenin protein
expression as well as its nuclear translocation (Fig. 5B and C). Notably, IQGAP2 was also
able to inhibit Wnt3a-induced transcriptional activity of β-catenin
(Fig. 5D), which indicated that
IQGAP2 was a suppressor of Wnt/β-catenin signaling. We further
found that overexpression of IQGAP2 could ablate Wnt3a-induced cell
invasion (Fig. 5E). To determine
whether the inactivation of β-catenin is a critical for
IQGAP2-regulated cell invasion, IQGAP2-overexpressing SKOV3 cells
were transfected with a constitutively activated β-catenin
(β-catenin CA), and the results showed that β-catenin CA
significantly rescued the invasiveness of cells (Fig. 5F). Collectively, these data
indicate that IQGAP2 suppresses ovarian cancer cell invasiveness
via the inhibition of Wnt/β-catenin signaling.
Discussion
While the molecular basis for ovarian cancer remains
poorly defined, studies suggest many gene alterations due to
mutation, copy number variation or epigenetic abnormalities may
contribute to this disease. The well-known oncogenes and tumor
suppressors in ovarian cancer include TP53, PIK3C, BRCA1 and
BRCA2 (19,20). In particular, TP53 is the
most common mutated gene occurring in >70% of the advanced
cases. More recently, along with the improvement of whole genomic
analysis tools, other genes have also been identified, such as
EMSY, PTEN, RAD51C and CTTNB1 (21). DNA methylation is associated with
gene transcription silencing, dysregulated gene methylation has
been implicated in almost all types of cancer (16). We compared gene methylation level
between healthy/normal ovaries and ovarian cancers, and found that
many critical proto-oncogenes and antioncogenes such as BRAF1,
SIX6, FOXB1, SFRP2, CDKN2A and CASP8 are dysregulated by
DNA methylation in ovarian cancers. Functions of these genes in
ovarian cancer need to be further defined. Among those altered
genes, IQGAP2 is hypermethylated and its expression is decreased in
ovarian cancers. Indeed, loss of heterozygosity of IQGAP2 is
reported in ovarian cancer (22).
Hypermethylation of IQGAP2 is also observed in gastric cancer
(10). Notably, we found decreased
IQGAP2 level is associated with a poor survival of patient,
indicating IQGAP2 as a new prognostic marker for ovarian
cancer.
The Ras pathway is one of the most commonly
deregulated pathways in human cancer. Hyperactivation of Ras
pathway can be caused by mutations in Ras genes or loss of RasGAP
proteins (23). There are 14
predicted RasGAP genes in the human genome and they all contain a
RasGAP domain but share little similarity in other regions. Except
for a RasGAP domain, the IQGAP subfamily also contain an
actin-binding calponin homology domain (CH), a polyproline-binding
domain (WW) and four IQ calmodulin-binding motifs (IQ) (24). IQGAP1 functions as a scaffolding
protein that is required for RAS-driven tumorigenesis and
metastasis, many studies indicate a positive role for IQGAP1 in
cancer. It modulates MAPK signaling pathway to regulate cell
proliferation and differentiation and interacts with β-catenin and
E-cadherin to regulate intercellular adhesion and migration
(25,26). Although IQGAP2 shares similarity in
sequencing with IQGAP1, it exhibits opposite roles and functions as
a tumor suppressor. Deficiency of IQGAP2 leads to the development
of hepatocellular carcinoma (27).
IQGAP2 is abnormally methylated in gastric cancers and is
significantly associated with tumor invasion and a poor prognosis
(10). IQGAP2 is lost in advanced
prostate cancers and inhibits EMT and attenuates serum induced AKT
activation (12). We found IQGAP2
is downregulated in ovarian cancers and suppresses cell migration
and invasion. Mechanistically, loss of IQGAP2 is able to activate
ERK, AKT and Wnt/β-catenin signaling pathways, but it fails to
affect the activity of Ras. This suggests that IQGAP2 does not
exhibit RasGAP activity in ovarian cancer cells. In particular,
Wnt/β-catenin seems to be the critical mediator for
IQGAP2-regulated migration and invasion in ovarian cancer cells. In
hepatocytes, IQGAP2 interacts with β-catenin and anchors β-catenin
at the submembrane region along with E-cadherin, it also, as a part
of the β-catenin destruction complex, consists of GSK3β kinase,
Axin and adenomatous polyposis coli (APC) (28). We demonstrated that IQGAP2 promotes
β-catenin cytosol localization and thus suppresses its
transcriptional activity.
Many patients with ovarian cancer exhibit early
extension of tumors to the outside of ovaries at the time of
diagnosis. EMT is a process characterized by the gain of
mesenchymal markers (e.g., N-cadherin, vimentin) and the loss of
epithelial markers (e.g., E-cadherin, cytokeratin), as well as
altered morphological features associated with this process
inducing increased motility and invasion of cancer cells (29). EMT occurs during ovarian cancer
progression, however, the underlying mechanisms are not well
established. The Wnt/β-catenin signaling pathway has been reported
to induce EMT in ovarian cancer and other tumor cells (30,31).
In the present study, we demonstrate a novel role of IQGAP2 in
suppressing ovarian cancer EMT through regulating Wnt/β-catenin
signaling, providing a new biomarker and potential therapeutic
strategy for ovarian cancer.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Berek JS, Crum C and Friedlander M: Cancer
of the ovary, fallopian tube, and peritoneum. Int J Gynaecol
Obstet. 119(Suppl 2): S118–S129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Quitadamo A, Tian L, Hall B and Shi X: An
integrated network of microRNA and gene expression in ovarian
cancer. BMC Bioinformatics. 16(Suppl 5): S52015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ying H, Lv J, Ying T, Jin S, Shao J, Wang
L, Xu H, Yuan B and Yang Q: Retraction note: Gene-gene interaction
network analysis of ovarian cancer using TCGA data. J Ovarian Res.
8:182015. View Article : Google Scholar :
|
|
5
|
Shah RH, Scott SN, Brannon AR, Levine DA,
Lin O and Berger MF: Comprehensive mutation profiling by
next-generation sequencing of effusion fluids from patients with
high-grade serous ovarian carcinoma. Cancer Cytopathol.
123:289–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Briggs MW and Sacks DB: IQGAP proteins are
integral components of cytoskeletal regulation. EMBO Rep.
4:571–574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maertens O and Cichowski K: An expanding
role for RAS GTPase activating proteins (RAS GAPs) in cancer. Adv
Biol Regul. 55:1–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L,
Dikalov SI, Chen YE, Tojo T, Fukai T, Fujimoto M, Patrushev NA,
Wang N, et al: IQGAP1, a novel vascular endothelial growth factor
receptor binding protein, is involved in reactive oxygen species -
dependent endothelial migration and proliferation. Circ Res.
95:276–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Katata T, Irie K, Fukuhara A, Kawakatsu T,
Yamada A, Shimizu K and Takai Y: Involvement of nectin in the
localization of IQGAP1 at the cell-cell adhesion sites through the
actin cytoskeleton in Madin-Darby canine kidney cells. Oncogene.
22:2097–2109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin SH, Akiyama Y, Fukamachi H, Yanagihara
K, Akashi T and Yuasa Y: IQGAP2 inactivation through aberrant
promoter methylation and promotion of invasion in gastric cancer
cells. Int J Cancer. 122:1040–1046. 2008. View Article : Google Scholar
|
|
11
|
White CD, Khurana H, Gnatenko DV, Li Z,
Odze RD, Sacks DB and Schmidt VA: IQGAP1 and IQGAP2 are
reciprocally altered in hepatocellular carcinoma. BMC
Gastroenterol. 10:1252010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie Y, Yan J, Cutz JC, Rybak AP, He L, Wei
F, Kapoor A, Schmidt VA, Tao L and Tang D: IQGAP2, A candidate
tumour suppressor of prostate tumorigenesis. Biochim Biophys Acta.
1822:875–884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou J, Deng Z, Chen Y, Gao Y, Wu D, Zhu
G, Li L, Song W, Wang X, Wu K, et al: Overexpression of FABP7
promotes cell growth and predicts poor prognosis of clear cell
renal cell carcinoma. Urol Oncol. 33:113.e9–113.e17. 2015.
View Article : Google Scholar
|
|
14
|
Zhou J, Wu K, Gao D, Zhu G, Wu D, Wang X,
Chen Y, Du Y, Song W, Ma Z, et al: Reciprocal regulation of
hypoxia-inducible factor 2α and GLI1 expression associated with the
radioresistance of renal cell carcinoma. Int J Radiat Oncol Biol
Phys. 90:942–951. 2014. View Article : Google Scholar
|
|
15
|
Reik W: Stability and flexibility of
epigenetic gene regulation in mammalian development. Nature.
447:425–432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sowter HM and Ashworth A: BRCA1 and BRCA2
as ovarian cancer susceptibility genes. Carcinogenesis.
26:1651–1656. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kohler MF, Kerns BJ, Humphrey PA, Marks
JR, Bast RC Jr and Berchuck A: Mutation and overexpression of p53
in early-stage epithelial ovarian cancer. Obstet Gynecol.
81:643–650. 1993.PubMed/NCBI
|
|
21
|
Bell D, Berchuck A, Birrer M, Chien J,
Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P, et al; Cancer
Genome Atlas Research Network. Integrated genomic analyses of
ovarian carcinoma. Nature. 474:609–615. 2011. View Article : Google Scholar
|
|
22
|
Gorringe KL, Ramakrishna M, Williams LH,
Sridhar A, Boyle SE, Bearfoot JL, Li J, Anglesio MS and Campbell
IG: Are there any more ovarian tumor suppressor genes? A new
perspective using ultra high-resolution copy number and loss of
heterozygosity analysis. Genes Chromosomes Cancer. 48:931–942.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Karnoub AE and Weinberg RA: Ras oncogenes:
Split personalities. Nat Rev Mol Cell Biol. 9:517–531. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bernards A and Settleman J: GAP control:
Regulating the regulators of small GTPases. Trends Cell Biol.
14:377–385. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fukata M, Watanabe T, Noritake J, Nakagawa
M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F and Kaibuchi
K: Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170.
Cell. 109:873–885. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Roy M, Li Z and Sacks DB: IQGAP1 is a
scaffold for mitogen-activated protein kinase signaling. Mol Cell
Biol. 25:7940–7952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schmidt VA, Chiariello CS, Capilla E,
Miller F and Bahou WF: Development of hepatocellular carcinoma in
Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol.
28:1489–1502. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schmidt VA: Watch the GAP: Emerging roles
for IQ motif-containing GTPase-activating proteins IQGAPs in
hepatocellular carcinoma. Int J Hepatol. 2012:9586732012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshioka S, King ML, Ran S, Okuda H,
MacLean JA II, McAsey ME, Sugino N, Brard L, Watabe K and Hayashi
K: WNT7A regulates tumor growth and progression in ovarian cancer
through the WNT/β-catenin pathway. Mol Cancer Res. 10:469–482.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu Y, Ginther C, Kim J, Mosher N, Chung S,
Slamon D and Vadgama JV: Expression of Wnt3 activates Wnt/β-catenin
pathway and promotes EMT-like phenotype in trastuzumab-resistant
HER2-overexpressing breast cancer cells. Mol Cancer Res.
10:1597–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|