Introduction
Breast cancer is one of the most commonly diagnosed
cancers and the leading cause of cancer-related death in females
worldwide (1). The factors that
increase risk include: late age of first birth, alcohol
consumption, and postmenopausal hormone therapy or oral
contraceptives (2–4). Breast cancer can be classified into
different molecular subtypes based on the hormone responsive
surface receptors (5). Luminal A
[estrogen receptor (ER)+, progesterone receptor
(PR)+/−, human epidermal growth factor receptor 2
(HER2)]−, luminal B (ER+, PR+/−,
HER2+), HER2 (ER−, PR−,
HER2+), and basal (ER−, PR−,
HER2−). Luminal A tumors tend to have a good prognosis,
with high survival rates and fairly low recurrence rates (6,7); in
addition, ≤15% of luminal A tumors have p53 mutations. Luminal B
tumors are larger, HER2-positive, and ≤30% have p53 mutations.
Luminal A tumors grow very slowly compared with luminal B tumors
(8). Basal tumors (triple-negative
breast cancers - TNBC) represent ~20% of breast cancers (8–10)
and they tend to occur in younger women and in African American
women (8,11). Approximately 20% of breast cancers
are HER2-positive due to over-production of the HER2 protein. This
type tends to be aggressive and fast-growing (12–15)
and ≤75% of cases have p53 mutations; in addition, this type has a
poor prognosis and is prone to frequent recurrence and metastasis
(6,8).
There are four different types of HER receptors
(HER1, HER2, HER3, HER4) (16).
The HER gene is located on chromosome 17. Activation of the
HER2/neu oncogene leads to the production of HER2 (17). Overexpression of the HER2/neu gene
is associated with breast, ovarian, and several other types of
cancer with high transition probability (18). The binding of specific ligands
leads to the activation of the tyrosine kinase activity of HER2 and
cell survival, which in turn leads to the uncontrolled growth of
cancer cells. The STAT proteins are expressed in all types of
breast cancer cells and tissues (19) and have pivotal roles in apoptosis,
differentiation, and proliferation (20). The Jak/STAT pathways are activated
by various factors and cytokines, leading to the activation of Jak
tyrosine kinase followed by tyrosine phosphorylation of the
receptors. The role of the HER2 and STAT3 signaling network has
been confirmed in breast cancer cells (21), and HER2 expression can be modulated
through the STATs.
Methylsulfonylmethane (MSM) is a simple organic
sulfur-containing compound and a stable, odorless, colorless,
non-toxic, crystalline product (22). MSM is found in foods, including
fruits, vegetables, and grains (23–25).
It is known for its effects on allergies, skin diseases, and
arthritis (26,27). In addition to these medicinal
properties, MSM showed growth promoting activities by enhancing the
differentiation of mesenchymal stem cells (28). In addition, MSM suppressed tumor
growth and progression (29,30).
We reported that MSM suppresses breast cancer growth through
modulating STAT3 and STAT5b pathways (29). In addition, MSM is active against
HER2-positive breast cancers; although, the molecular mechanism
behind this activity is unclear. The expression levels of HER2 are
associated with a number of factors. In the present study, we
examined MSM in breast cancer cell lines and hypothesized that this
compound inhibits HER2 gene expression through STAT5b in SK-BR3
cells.
Materials and methods
Antibodies and reagents
The following were purchased from the indicated
sources: penicillin-streptomycin solution and fetal bovine serum
(FBS) from Hyclone (South Logan, UT, USA); RPMI-1640 from Sigma
Chemical Co. (St. Louis, MO, USA); trypsin-EDTA (0.05%) from
Gibco-BRL (Grand Island, NY, USA); STAT5b, HER2 antibodies, and
secondary antibody (goat anti-mouse and rabbit IgG-horseradish
peroxidase) from Santa Cruz Biotechnology (Santa Cruz, CA, USA);
phosphorylated STAT5 from Upstate Biotechnology (Lake Placid, NY,
USA); β-actin from Sigma Chemical Co.; the enhanced
chemiluminescence (ECL) detection kit from Amersham Pharmacia
Biotech (Piscataway, NJ, USA); Restore™ Western Blot Stripping
Buffer and NE-PER kit from Pierce (Rockford, IL, USA); the
electrophoretic mobility shift assay (EMSA) kit, oligonucleotide
probes (STAT5b), luciferase assay substrates, and reporter lysis
buffer from Promega Corp. (Madison, WI, USA); FuGENE6 transfection
reagent from Roche (Basel, Switzerland); RNeasy mini kit and
Qiaprep spin miniprep kit from Qiagen (Hilden, Germany); the RT-PCR
Premix kit and VEGF, IGF-1R, 18s primer for RT-PCR were synthesized
by Bioneer (Dajeon, Korea); imprint chromatin immunoprecipitation
assay kit from Sigma Chemical Co.; and MSM from Fluka/Sigma Co.
(St. Louis, MO, USA).
Cell culture and treatment
The human breast adenocarcinoma cell lines, SK-BR3
and MCF-7, were maintained in RPMI-1640 medium containing 10% FBS
and 100 U/ml penicillin and streptomycin at 37°C in 5%
CO2. The cells were placed in airtight chambers (Nu
Aire, Plymouth, MN, USA). At the beginning of each experiment, the
cells were resuspended in the medium at a density of
2.5×105 cells/ml. Cells were treated with 300 mM
MSM.
Cell proliferation inhibition
Cell viability was assayed by measuring blue
formazan that was metabolized from
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)
by mitochondrial dehydrogenase, which is only active in live cells.
The cells were resuspended in the medium one day before drug
treatment, at a density of 3×103 cells per well in
96-well culture plates. Liquid medium was replaced with fresh
medium containing dimethyl sulfoxide (DMSO) as a control (vehicle).
Cells were incubated with various concentrations of MSM. Then, MTT
(5 mg/ml) was added to each well and incubated for 4 h at 37°C. The
formazan product formed was dissolved by adding 200 μl DMSO to each
well, and the absorbance was measured at 550 nm on an Ultra
Multifunctional Microplate Reader (Tecan, Durham, NC, USA). All
measurements were performed in triplicate, and were repeated at
least three times.
Western blotting
The SK-BR3 and MCF-7 cell lines were treated with
MSM. Whole cells were lysed on ice with radioimmunoprecipitation
(RIPA) lysis buffer, containing phosphatase and protease
inhibitors. Cells were disrupted by aspiration through a 23-gauge
needle, and centrifuged at 15,000 rpm for 10 min at 4°C to remove
cellular debris. Protein concentrations were measured using the
Bradford method. Equal amounts of proteins were resolved with
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred onto nitrocellulose membranes. The blots
were blocked for 1 h with 5% skim milk. Membranes were probed
overnight at 4°C with a primary antibody followed by horseradish
peroxidase-conjugated secondary antibodies. Detection was performed
using the ECL Plus detection kit and a LAS-4000 imaging device
(Fujifilm, Japan).
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was extracted using the RNeasy Mini kit
(Qiagen) and quantified spectrometrically at 260 nm. Then, RT-PCR
analysis for HER2 and 18s RNA was performed. The cDNA was
synthesized from total RNA by RT at 42°C for 1 h and 95°C for 5 min
using first strand cDNA synthesis kits (Bioneer, Korea). The cDNA
was used in PCR with the following primers: HER2 sense,
5′-TGCGGCTCGTACACAGGGACTT-3′ and HER2 antisense,
5′-TGCGGAGAATTCAGACACCAACT-3′, with a 420-bp amplified HER2 mRNA
fragment; 18s sense, 5′-CGGCTACCACATCCAAGGAA-3′ and 18s antisense,
5′-CCGGCGTCCCTCTTAATC-3′, with a 489-bp amplified 18s mRNA
fragment. The PCR conditions consisted of denaturation for 1 min at
95°C, annealing for 1 min at 58°C, and extension for 1 min at 72°C.
The PCR products were analyzed on a 1% agarose gel stained with
ethidium bromide.
Electrophoretic mobility shift assay
(EMSA)
The DNA binding activity of STAT5b was assessed
using EMSA, in which labeled double-stranded DNA was used as a DNA
probe to bind active STAT5b proteins in nuclear extracts. Nuclear
protein extracts were prepared with a nuclear extract kit
(Panomics, AY2002). The EMSA experiment was performed by incubating
a biotin-labeled transcription factor-STAT5b probe with treated and
untreated nuclear extracts. Proteins were resolved on a
non-denaturing 6% polyacrylamide gel (Bio-Rad, Korea). The proteins
in the gel were transferred to a nylon membrane and detected using
streptavidin-horseradish peroxidase and a chemiluminescent
substrate.
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assay was performed using the Imprint
chromatin immunoprecipitation kit (Sigma) according to the
manufacturer's protocol. Briefly, SK-BR3 cells were fixed with 1%
formaldehyde and quenched with 1.25 M glycine. After washing with
PBS, the cells were suspended in nuclei preparation buffer and
shearing buffer and sonicated under optimized conditions. This
sheared DNA was then centrifuged and the cleared supernatant used
for protein/DNA immunoprecipitation. The clarified supernatant was
diluted with buffer (1:1 ratio) and 5 μl of the diluted samples was
removed as an internal control. The diluted supernatant was
incubated with antibody (STAT5b) in pre-coated wells for 90 min.
The negative and positive controls were normal goat IgG and
anti-RNA polymerase II, respectively. The unbound DNA was washed
off with IP wash buffer and the bound DNA was collected by cross
link reversal using DNA release buffer containing proteinase K. The
released DNA and DNA from the internal control were purified with
the GenElute Binding Column G. The DNA was then quantified using
conventional PCR.
Expression vectors, transfection, and the
luciferase reporter assay
Cells were co-transfected with various combinations
of the following constructs: wild-STAT5b (pMX/STAT5b; kindly
provided by Dr Koichi Ikuta, Kyoto University, Japan), constructed
as previously described (38), and
the HER2 reporter construct containing 5.6 kb of the HER2 promoter
region. Transfected cells were washed with ice-cold PBS and lysed.
Lysates were used directly to measure luciferase activity. The
luciferase activity of each sample was determined by measuring
luminescence for 10 sec on a Lumat LB 9507 luminometer (EG&G
Berthold, TN, USA). The experiments were performed in triplicate,
and similar results were obtained from at least three independent
experiments.
Small interference RNA (siRNA)
analysis
SK-BR3 cells (1×105) were cultured on
6-well plates and grown to 50% confluence. The cells were then
transfected with On-Target plus SMARTpool siRNA targeting STAT5b or
On-Target plus non-targeting siRNA (Dharmacon, Chicago, IL, USA)
using FuGENE6 (Roche, IN, USA), according to the manufacturer's
instructions. Following transfection with this mixture for 48 h,
invasion assays were conducted without adding drugs for an
additional 24 h. Different areas were captured and the cells were
counted.
Results
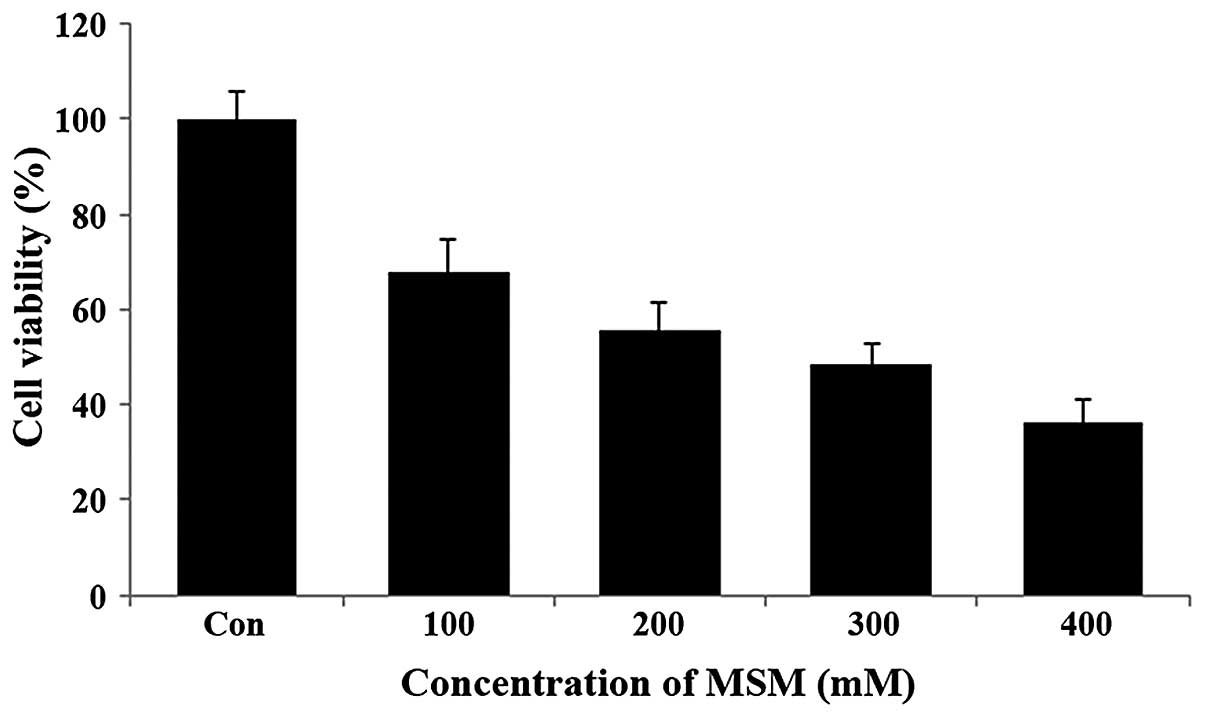
MSM inhibits SK-BR3 cell
proliferation
The effect of MSM on the viability of the human
breast cancer cell line SK-BR3 was examined using the MTT assay.
The SK-BR3 cells were exposed to increasing concentrations of MSM
(100, 200, 300 and 400 mM) for a period of 24 h. Following this,
the metabolically viable cells were quantified based on the amount
of formazan crystals formed. Treatment with MSM substantially
decreased the viability of SK-BR3 cells in a dose-dependent manner.
It was observed that 100 mM MSM inhibited SK-BR3 cell growth by
32%, 200 mM MSM by 45%, and 300 mM MSM by 51% (Fig. 1). Therefore, the 300-mM
concentration of MSM is considered the IC50 dosage and
was used in the further experiments.
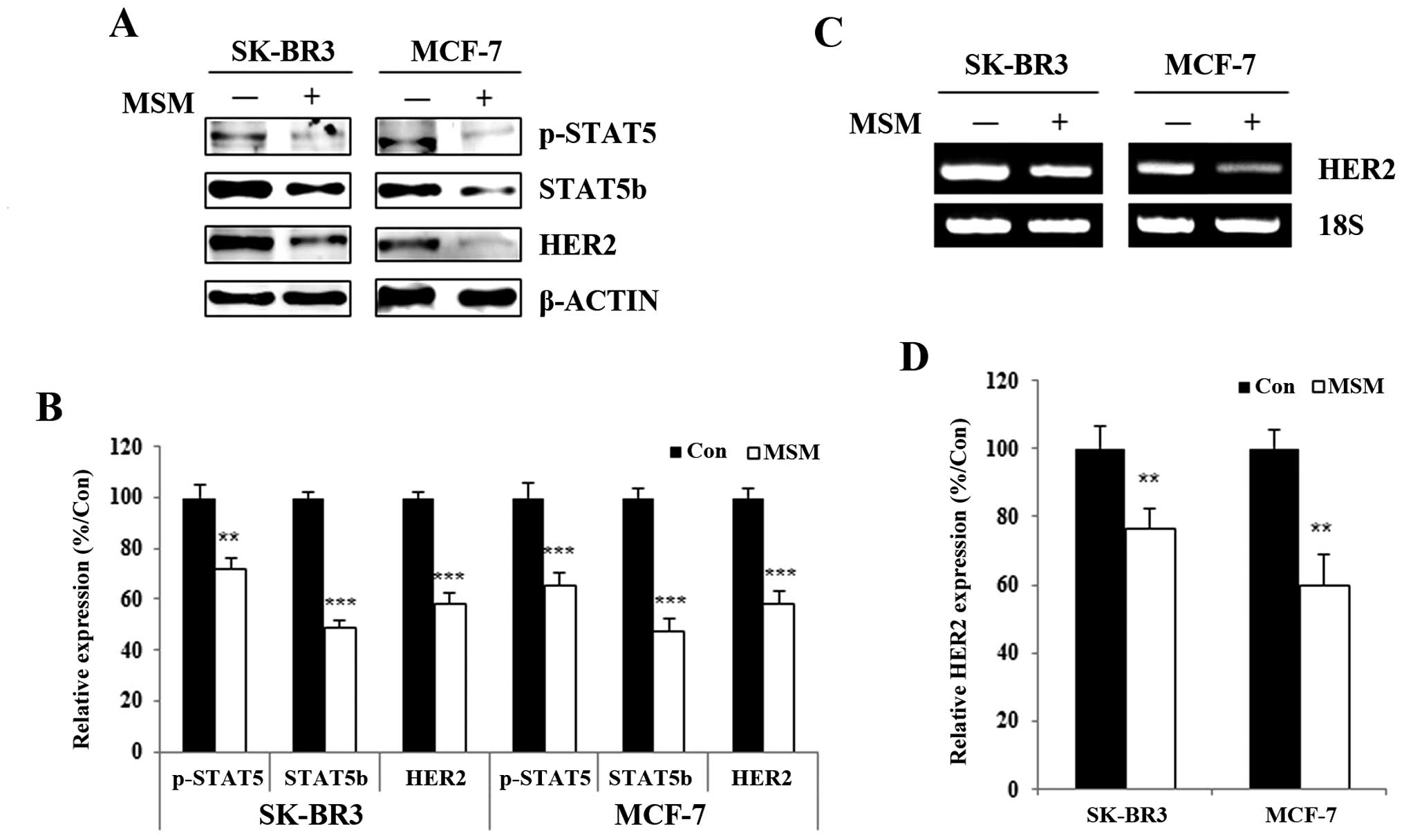
MSM suppressed the expression, as well as
phosphorylation, of STAT5b and HER2 in breast cancer cells
To study the effect of MSM on STAT5b and HER2,
SK-BR3 and MCF-7 cells were treated with 300 mM MSM for 24 h. Whole
cell lysates were prepared in 1X RIPA lysis buffer containing 1X
protease and 1X phosphatase inhibitor. Western blotting of the
whole cell lysates prepared from MSM-treated and non-treated cells
showed a decrease in the expression and phosphorylation of STAT5 in
MSM-treated cells. Concurrently, the expression of HER2 also
decreased in both SK-BR3 and MCF-7 cells (Fig. 2A). Compared with the control group,
the MSM-treated group showed a 30% decrease in STAT5
phosphorylation (Fig. 2B); while,
STAT5b and HER2 expression levels were inhibited ~50 and 40%,
respectively (Fig. 2B).
MSM suppresses the transcription
activation functions of STAT5b
We analyzed the expression of the STAT5b target
gene, HER2. The RT-PCR analysis showed a decrease in the
transcription of HER2 mRNA in MSM-treated cells (Fig. 2C) when amplifying HER2 with
gene-specific primers and using 18S RNA as a loading control.
Treated cells had ~25 and 40% inhibition of HER2 expression when
compared with non-treated control SK-BR3 and MCF-7 cells,
respectively (Fig. 2D). The
ability of MSM to suppress HER2 expression was confirmed at the
translational level.
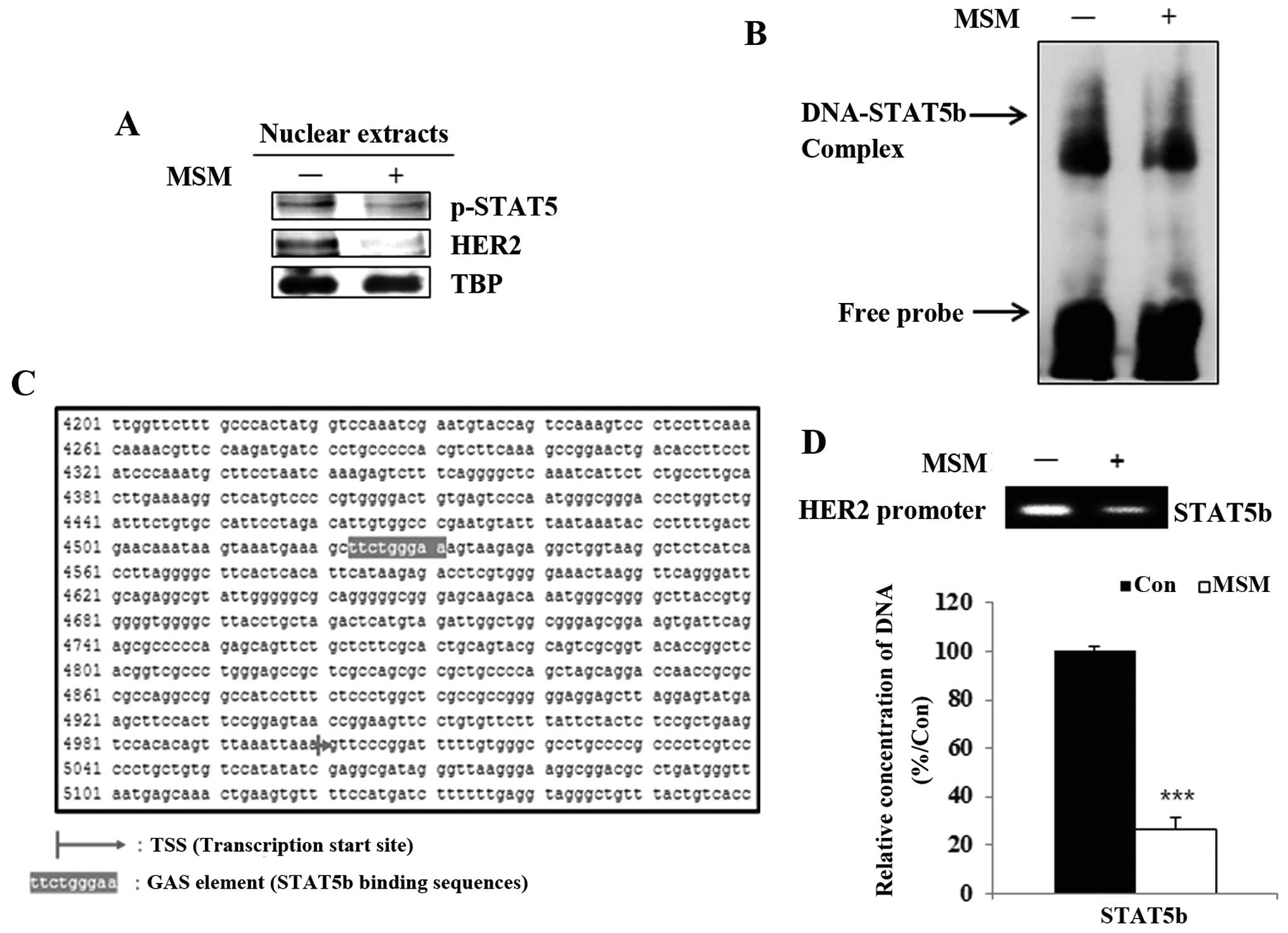
MSM inhibits the binding of STAT5b to the
HER2 gene promoter
A previous study showed STAT5 was a transcription
factor for HER2. We then analyzed the nuclear level expressions of
STAT5b and HER2 after MSM treatment. The nuclear extracts of the
MSM-treated groups showed decreased levels of p-STAT5 and HER2
(Fig. 3A). Results from the
electrophoretic mobility shift assay indicated that MSM suppressed
the STAT5b-DNA binding activity in SK-BR3 cells (Fig. 3B). Therefore, it was necessary to
determine the DNA-binding site of STAT5b, which we found
corresponds to the interaction of STAT5b with the GAS element
(TTCagcGAA) of the HER2 gene (Fig.
3C). There results proved that MSM inhibited the binding of
p-STAT5 to the HER2 gene promoter site.
The DNA binding of STAT5b is inhibited by
MSM
To perform transcriptional functions, phosphorylated
STAT5b should translocate to the nucleus from the cytosol. Thus,
these nuclear translocations were analyzed using the ChIP assay. We
found that MSM treatment led to a decrease in the STAT5b binding to
the HER2 promoter in SK-BR3 cells. The quantitative analysis by
qPCR showed that MSM treatment inhibited binding of STAT5b to the
HER2 promoter region in SK-BR3 cells (Fig. 3D). There was significant
downregulation of DNA binding of STAT5b in MSM-treated cells. These
results indicate MSM plays an important role in the suppression of
binding.
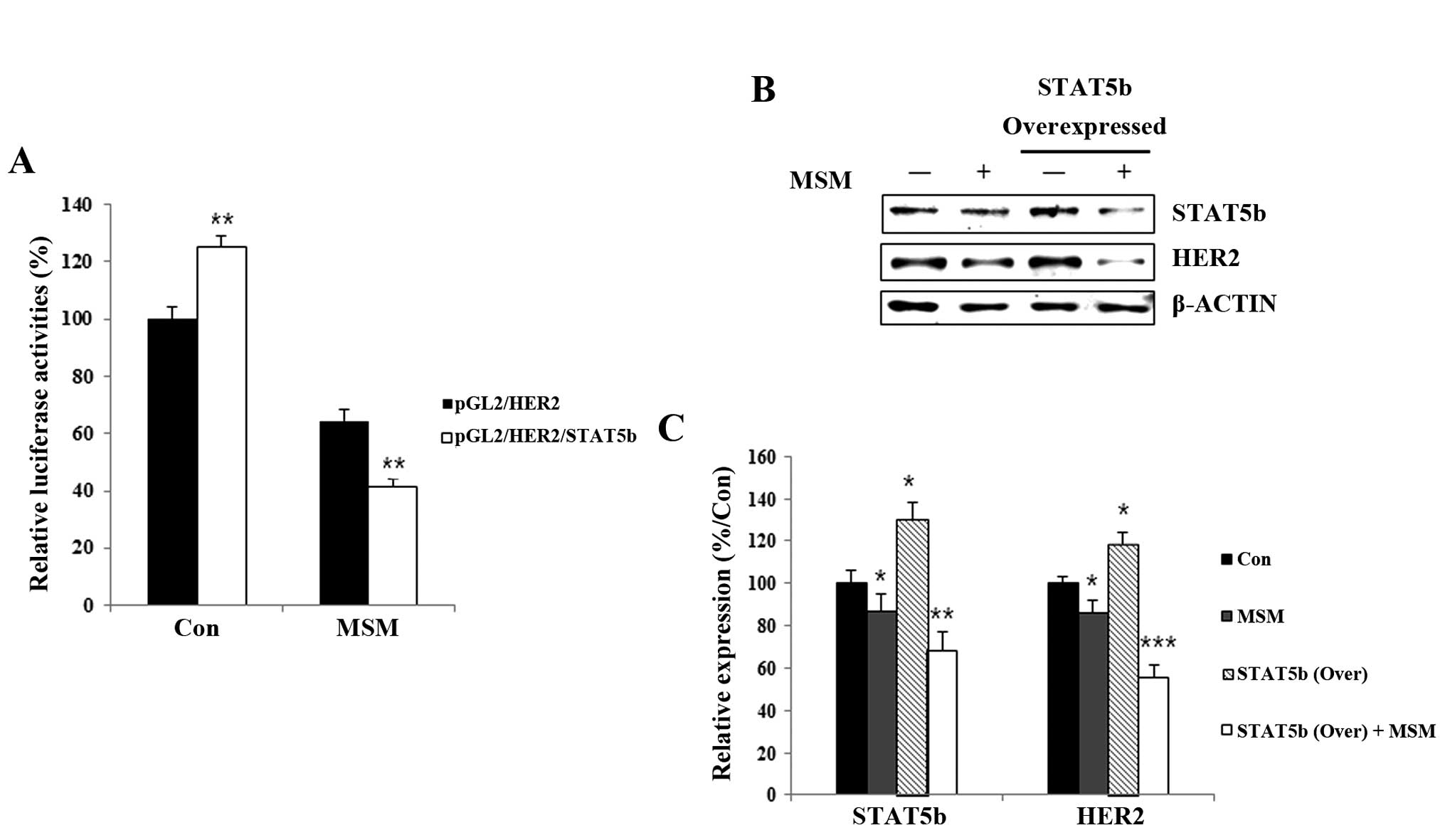
MSM suppresses transcriptional activity
between STAT5b and HER2 genes in SK-BR3 cells
The transcription activation functions of STAT5b
after MSM treatment were studied using a luciferase assay. After
treatment with MSM for 24 h, relative luciferase activity was
decreased significantly for STAT5b/HER2 (Fig. 4A, ***P<0.001). This
finding confirms the critical role of MSM in inhibiting the
transcription promoter activities of STAT5b.
MSM inhibits STAT5b and HER2 expression
at the translational and transcriptional levels
To analyze the correlation between STAT5b and HER2,
we overexpressed the STAT5b protein in MCF-7 cells. After
transfection with STAT5b for overexpression, we analyzed the
expression of STAT5b and HER2 using western blotting (Fig. 4B). Treatment with MSM led to ~15%
inhibition of STAT5b and HER2 compared with the control in MCF-7
cells. In cells overexpressing STAT5b, the MSM treatment led to
~60% inhibition of STAT5b and HER2 compared with non-transfected
MSM-treated MCF-7 cells (Fig. 4C).
Expression levels of STAT5b and HER2 after MSM treatment showed
similar expression patterns in cell lysates, demonstrating the
ability of MSM to suppress STAT5b and HER2 expression.
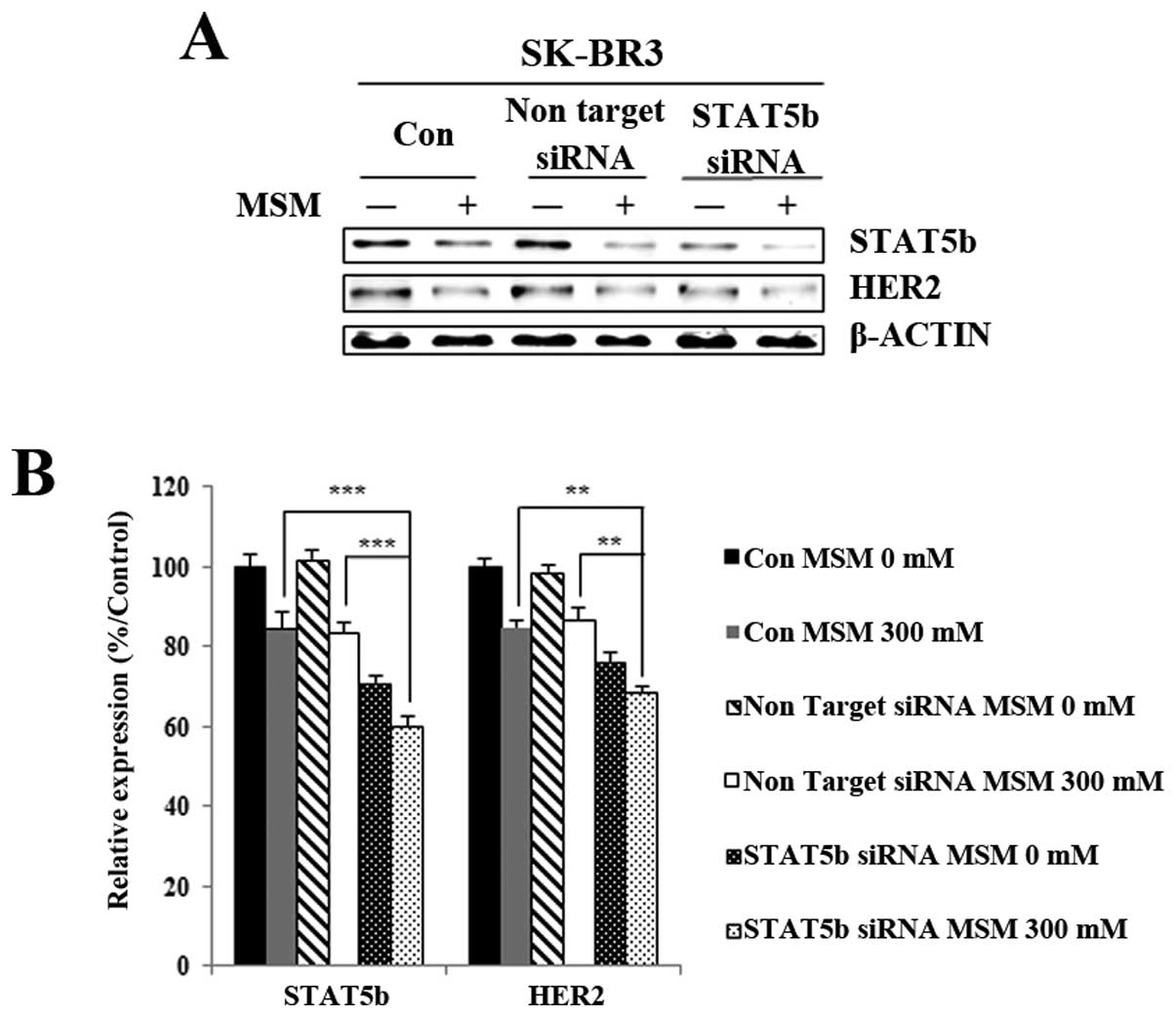
MSM inhibits HER2 expression in a
STAT5b-dependent manner
To explore whether STAT5b is involved in HER2
expression, we used the siRNA strategy. Before MSM treatment,
STAT5b in SK-BR3 cells was knocked down with a specific STAT5b
siRNA. Interestingly, the results showed a similar pattern for
STAT5b and HER2, with the knockdown of STAT5b leading to decreased
HER2 protein expression. After MSM treatment, the HER2 expression
was less in cells targeted with siSTAT5b than in cells treated with
non-targeting siRNA (Fig. 5A). The
relative expression of proteins, with respect to actin, gave a
clear view of the effect of regulating STAT5b-related HER2
expression with MSM (Fig. 5B).
From there results, we concluded that STAT5b plays an essential
role in HER2 activation.
Discussion
Overexpression of HER2 is reported in ~25% of breast
cancer cases and is associated with an unfavorable prognosis
(31). When a breast cell
expresses abnormally high levels of HER2, it drives breast cancer
growth and metastasis. Increased receptor activation and signaling
contributes to a more aggressive tumor biology with prominent
metastasis to the visceral and central nervous systems, recurrence,
and mortality (32,33). Hence, immense research has been
carried out worldwide to inhibit HER2 and the HER2 subtype of
breast cancer. Multiple approaches have been developed and examined
to suppress ligand binding to the receptor or inhibit receptor
activation and thereby block the HER2 signaling cascade (16,17,34,35).
Recently, we reported that the natural compound MSM suppresses HER2
(29). The result draws attention
to solve the various problems caused by the over- expression of
Her2.
In the present study, we examined the anticancer
effects of MSM on the HER2 subtype of breast cancer. The
proliferation inhibition analysis showed the suitable concentration
of MSM was 300 mM, and this was used for further analysis (Fig. 1). The role of STAT as a
transcription factor for regulating HER2 expression was
demonstrated in previous studies. Both STAT3 and STAT5b have some
role in modulating the expression of HER2 (28,29,36).
The ability of MSM to modulate the expression, as well as
phosphorylation, of STAT5b was studied using western blotting. The
results were consistent with our hypothesis that MSM inhibits HER2
gene expression through STAT5b. The findings were also similar in a
luminal A subtype of breast cancer with a basal level expression of
HER2 (5), demonstrated the
significance of using MSM in multiple breast cancer subtypes
(Fig. 2A and B).
The STAT5b signaling is dependent on its ability to
translocate to the nucleus and bind to the nuclear response element
(19). The western blot analysis
of nuclear extracts showed a decline in the phosphorylated STAT5
level, indicating that the nuclear translocation was also affected
by MSM treatment (Fig. 3A).
Moreover, transcriptional level expression studies of HER2 also
showed a prominent inhibition by MSM treatment (Fig. 2C and D). We hypothesized that
STAT5b-transcription factor (STAT5b-TF) binds to the promoter
region of the HER2 (ERBB2) gene and inhibits the transcription of
HER2. In support of our hypothesis, EMSA data showed inhibition in
the DNA binding activities of STAT5b-TF after MSM treatment
(Fig. 3B). Sequence analysis
showed a GAS element in the Her2 gene promoter. The DNA binding was
re-confirmed by ChIP analysis (Fig.
3D). Similar to the EMSA data, the MSM treatment drastically
inhibited the STAT5b-TF/DNA binding.
The promoter regulatory functions of STAT5b-TF were
studied with a luciferase assay. The result confirmed that MSM
decreased the STAT5b-TF/DNA binding together with the promoter
activities (Fig. 4A). To confirm
our hypothesis, we used luminal A type MCF-7 cells, which do not
overexpress HER2 (37). The MCF-7
cells were induced for STAT5b overexpression and analyzed for HER2
expression levels (Fig. 4B and C).
Consistent with our hypothesis, STAT5b over-expression led to the
overexpression of HER2 and the results were reversed with MSM
treatment. In addition, the STAT5b knockdown studies confirmed the
role of STAT5b in MSM- mediated downregulation of HER2 (Fig. 5A and B).
In conclusion, we confirmed that MSM has the ability
to regulate the expression, as well as phosphorylation, of STAT5b.
This, in turn, inhibits the STAT5b-TF functions and the expression
of HER2 in this subtype of breast cancer. Hence, MSM should be
evaluated as a trial drug for targeting HER2-positive breast
cancers.
Acknowledgements
This study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF); Ministry of Education (2013R1A1A2057942).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hulka BS and Moorman PG: Breast cancer:
Hormones and other risk factors. Maturitas. 38:103–113; discussion
113–116. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baan R, Straif K, Grosse Y, Secretan B, El
Ghissassi F, Bouvard V, Altieri A and Cogliano V: WHO International
Agency for Research on Cancer Monograph Working Group:
Carcinogenicity of alcoholic beverages. Lancet Oncol. 8:292–293.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Key J, Hodgson S, Omar RZ, Jensen TK,
Thompson SG, Boobis AR, Davies DS and Elliott P: Meta-analysis of
studies of alcohol and breast cancer with consideration of the
methodological issues. Cancer Causes Control. 17:759–770. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bertucci F, Finetti P and Birnbaum D:
Basal breast cancer: A complex and deadly molecular subtype. Curr
Mol Med. 12:96–110. 2012. View Article : Google Scholar :
|
|
6
|
Lumachi F, Brunello A, Maruzzo M, Basso U
and Basso SM: Treatment of estrogen receptor-positive breast
cancer. Curr Med Chem. 20:596–604. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arvold ND, Taghian AG, Niemierko A, Abi
Raad RF, Sreedhara M, Nguyen PL, Bellon JR, Wong JS, Smith BL and
Harris JR: Age, breast cancer subtype approximation, and local
recurrence after breast-conserving therapy. J Clin Oncol.
29:3885–3891. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carey LA, Perou CM, Livasy CA, Dressler
LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S,
et al: Race, breast cancer subtypes, and survival in the Carolina
Breast Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Potemski P, Kusinska R, Watala C,
Pluciennik E, Bednarek AK and Kordek R: Prognostic relevance of
basal cytokeratin expression in operable breast cancer. Oncology.
69:478–485. 2005. View Article : Google Scholar
|
|
10
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lund MJ, Butler EN, Hair BY, Ward KC,
Andrews JH, Oprea-Ilies G, Bayakly AR, O'Regan RM, Vertino PM and
Eley JW: Age/race differences in HER2 testing and in incidence
rates for breast cancer triple subtypes: A population-based study
and first report. Cancer. 116:2549–2559. 2010.PubMed/NCBI
|
|
12
|
Mayer IA: Treatment of HER2-positive
metastatic breast cancer following initial progression. Clin Breast
Cancer. 9(Suppl 2): S50–S57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Slamon DJ, Clark GM, Wong SG, Levin WJ,
Ullrich A and McGuire WL: Human breast cancer: Correlation of
relapse and survival with amplification of the HER-2/neu oncogene.
Science. 235:177–182. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Slamon DJ, Godolphin W, Jones LA, Holt JA,
Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al:
Studies of the HER-2/neu proto-oncogene in human breast and ovarian
cancer. Science. 244:707–712. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Murphy CG and Fornier M: HER2-positive
breast cancer: Beyond trastuzumab. Oncology (Williston Park).
24:410–415. 2010.
|
|
17
|
Zhou Z and Hick DG: HER2 Amplification or
Overexpression in Upper GI Tract and Breast Cancer with Clinical
Diagnosis and Treatment. Siregar Y: INTECH Open Access Publisher;
2013
|
|
18
|
Xing X, Wang SC, Xia W, Zou Y, Shao R,
Kwong KY, Yu Z, Zhang S, Miller S, Huang L, et al: The Ets protein
PEA3 suppresses HER-2/neu overexpression and inhibits
tumorigenesis. Nat Med. 6:189–195. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Furth PA: STAT signaling in different
breast cancer sub-types. Mol Cell Endocrinol. 382:612–615. 2014.
View Article : Google Scholar
|
|
20
|
Bowman T, Garcia R, Turkson J and Jove R:
STATs in oncogenesis. Oncogene. 19:2474–2488. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duru N, Fan M, Candas D, Menaa C, Liu HC,
Nantajit D, Wen Y, Xiao K, Eldridge A, Chromy BA, et al:
HER2-associated radioresistance of breast cancer stem cells
isolated from HER2-negative breast cancer cells. Clin Cancer Res.
18:6634–6647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
William EJ: MSM reviewed. J Equine Vet
Sci. 7:1987.
|
|
23
|
Silva Ferreira AC, Rodrigues P, Hogg T and
Guedes De Pinho P: Influence of some technological parameters on
the formation of dimethyl sulfide, 2-mercaptoethanol, methionol,
and dimethyl sulfone in port wines. J Agric Food Chem. 51:727–732.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rose SE, Chalk JB, Galloway GJ and
Doddrell DM: Detection of dimethyl sulfone in the human brain by in
vivo proton magnetic resonance spectroscopy. Magn Reson Imaging.
18:95–98. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Caron JM, Bannon M, Rosshirt L, Luis J,
Monteagudo L, Caron JM and Sternstein GM: Methyl sulfone induces
loss of metastatic properties and reemergence of normal phenotypes
in a metastatic cloudman S-91 (M3) murine melanoma cell line. PLoS
One. 5:e117882010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barrager E, Veltmann JR Jr, Schauss AG and
Schiller RN: A multicentered, open-label trial on the safety and
efficacy of methylsulfonylmethane in the treatment of seasonal
allergic rhinitis. J Altern Complement Med. 8:167–173. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim LS, Axelrod LJ, Howard P, Buratovich N
and Waters RF: Efficacy of methylsulfonylmethane (MSM) in
osteoarthritis pain of the knee: A pilot clinical trial.
Osteoarthritis Cartilage. 14:286–294. 2006. View Article : Google Scholar
|
|
28
|
Joung YH, Lim EJ, Darvin P, Chung SC, Jang
JW, Do Park K, Lee HK, Kim HS, Park T and Yang YM: MSM enhances GH
signaling via the Jak2/STAT5b pathway in osteoblast-like cells and
osteoblast differentiation through the activation of STAT5b in
MSCs. PLoS One. 7:e474772012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lim EJ, Hong DY, Park JH, Joung YH, Darvin
P, Kim SY, Na YM, Hwang TS, Ye SK, Moon ES, et al:
Methylsulfonylmethane suppresses breast cancer growth by
down-regulating STAT3 and STAT5b pathways. PLoS One. 7:e333612012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim JH, Shin HJ, Ha HL, Park YH, Kwon TH,
Jung MR, Moon HB, Cho ES, Son HY and Yu DY: Methylsulfonylmethane
suppresses hepatic tumor development through activation of
apoptosis. World J Hepatol. 6:98–106. 2014.PubMed/NCBI
|
|
31
|
Ross JS: Update on HER2 testing for breast
and upper gastrointestinal tract cancers. Biomarkers Med.
5:307–318. 2011. View Article : Google Scholar
|
|
32
|
Winstanley J, Cooke T, Murray GD,
Platt-Higgins A, George WD, Holt S, Myskov M, Spedding A,
Barraclough BR and Rudland PS: The long term prognostic
significance of c-erbB-2 in primary breast cancer. Br J Cancer.
63:447–450. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hicks DG, Yoder BJ, Short S, Tarr S,
Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, et
al: Loss of breast cancer metastasis suppressor 1 protein
expression predicts reduced disease-free survival in subsets of
breast cancer patients. Clin Cancer Res. 12:6702–6708. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gajria D and Chandarlapaty S:
HER2-amplified breast cancer: Mechanisms of trastuzumab resistance
and novel targeted therapies. Expert Rev Anticancer Ther.
11:263–275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Emde A, Köstler WJ and Yarden Y:
Association of Radiotherapy and Oncology of the Mediterranean arEa
(AROME): Therapeutic strategies and mechanisms of tumorigenesis of
HER2-over-expressing breast cancer. Crit Rev Oncol Hematol.
84(Suppl 1): e49–e57. 2012. View Article : Google Scholar
|
|
36
|
Chung SS, Giehl N, Wu Y and Vadgama JV:
STAT3 activation in HER2-overexpressing breast cancer promotes
epithelial-mesenchymal transition and cancer stem cell traits. Int
J Oncol. 44:403–411. 2014.
|
|
37
|
Subik K, Lee JF, Baxter L, Strzepek T,
Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG, et
al: The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and
AR by immunohistochemical analysis in breast cancer cell lines.
Breast Cancer (Auckl). 4:35–41. 2010.
|
|
38
|
Joung YH, Lim EJ, Lee MY, Park JH, Ye SK,
Park EU, Kim SY, Zhang Z, Lee KJ, Park DK, et al: Hypoxia activates
the cyclin D1 promoter via the Jak2/STAT5b pathway in breast cancer
cells. Exp Mol Med. 37:353–364. 2005. View Article : Google Scholar : PubMed/NCBI
|