Introduction
Gastric cancer, a highly lethal disease, is the most
common malignant neoplasm of the digestive tract and the second
leading cause of cancer death in the world. More than 7000,000
deaths of gastric cancer patients and nearly 1,000,000 new gastric
cancer cases occur globally each year (1). Gastric cancer has an extremely poor
prognosis. The relative 5-year survival rates for gastric cancer
are low in most cuntries at <30% (2). Surgical resection is the only
potentially curative therapy for gastric cancer. Unfortunately,
only a minority of patients with gastric cancer is candidate for
surgical treatment, mostly due to the high proportion of tumor that
is advanced at the time of presentation (2). So, the prognosis of patients with
gastric cancer remains poor. In such cases, it is necessary to
study the molecular mechanisms of effect the gastric cancer
progression, which may be a useful therapeutic for the treatment of
gastric cancer.
As is well-known, TNF-α binds to TNFR1, then
activating both the signaling pathways of live and death (3). RIP1 is recruited to the TNFR1
signaling complex within seconds of TNF-α stimulation, and become
the key molecular of TNF-α-TNFR1 signaling pathway (4). RIP1 was initially described as a
novel molecule with a C-terminal death domain that can interact
with the death domain of one of these receptors, Fas, in a yeast
two-hybrid screen (5). RIP1 plays
an key role during cellular stress caused by inflammation and DNA
damage. Different cell inflammation signals are transmitted by
cellsurface receptors, then cascade of events are initiated by the
activaed RIP1. The most important response is RIP-1 activation of
transcription factor NF-κB (6) and
AP-1 (7), which induces genes
expression and promotes cell survival and differentiation.
Therefore, we concluded that RIP-1 is a crucial
regulator of cell survival and is a pivotal component of the
inflammation-signaling pathway as reported (8). Ting et al (9) shown that the RIP1 gene knockout made
the TNF-α couldnot activate the NF-κB and increased the sensitivity
of apoptosis of TNF-α induced in the Jurkat T cell line. Devin
et al (7) found that TNF-α
activated the ability of P38, ERK, and JUK decreasing
induced apoptosis in the MEF cells with the knockout of the RIP1
gene.
The relationship between RIP1 and the occurrence and
development of tumors are largely unknown. Park et al
(10) reported that patients with
glioblastomas who had increased expression of RIP1 had a
significantly worse prognosis and RIP1 is overexpressed with an
increase in the malignant grade of glioblastoma. A recent study
reported that the K562 and HL60 leukemia cell lines with knockdown
of RIP1 were sensitized by induction of apoptosis (11). Additionally, Liu et al
(12) found that RIP1 is an
oncogenic driver in melanoma. When RIP1 was silenced using siRNA in
A549 and H460 cell lines, the migration and invasion abilities of
these cells lines were reduced (13).
However, the role of RIP-1 regulating the growth and
invasion of gastric cancer cells and detail mechanisms are largely
unclear. Additionally, the precise mechanisms underlying RIP-1
promotion of gastric cancer growth and invasion are not well
understood. Therefore, the aim of this study was to clarify the
role of RIP-1 in gastric cancer. We analyzed the biological
functions and underlying mechanisms of RIP-1 in human gastric
cancer tissue samples and gastric cancer cell lines and addressed
the role of RIP1 using a xenograft mouse model.
Materials and methods
Patient tissues and data collection
This study of human gastric cancer samples was
approved by the Ethics Committee of the First Hospital Affiliated
to Fujian Medical University. Seventy formalin-fixed and
paraffin-embeded gastric cancer tissues, and 19 samples of normal
gastric tissues that were located away from the cancer were used as
contorls. The samples were provied by Pathology Department and
Department of Gastrointestinal Surgery Section II of the First
Hospital Affiliated to Fujian Medical University. The patient group
samples were conserved during the period from 2008 to 2009, and the
patient group consisted of 34 men and 36 women with a median age of
63 years. None of the gastric cancer patients included in this
study had received any preoperative chemotherapy or other therapy
such as radiotherapy. Postoperative chemotherapy was indicated for
patients who had indications of chemotherapy with intravenous
infusion of 5-fluorouracil-based chemotherapy. Follow-up was done
every three months for the first 2 years and annually
thereafter.
Immunohistochemistry and evalution
Immunohistochemistry staining was performed using
the standard immunoperoxidase staning procedure. In brief, serial
4-μm slices were obtained from formalin-fixed and paraffin-embedded
tissues specimens. Then dewaxed in xylene, rehydrated in alcohol,
and incubated with fresh 3% hydrogen peroxide
(H2O2) for 25 min at room temperature.
Sections were antigen-retrieved by boling for 15 min in citrate
buffer (pH 6.0) and followed by a wash step with phosphate buffered
saline (PBS). The non-specific antigen of sections were blocked
with appropriate normal serum in PBS. RIP1 mouse monoclonal
anti-human antibody (1:500, Abcam Biotechnology) was diluted and
placed on the sections overnight in humidified boxes at 4°C. The
sections were then washed with PBS for 6 min followed by an
incubation with an UltraSensitive S-P kit (Maixin-Bio, Fuzhou,
China) according to the manufacturer's instructions. After exposure
to stable 3, 3-diaminobenzidine for 4–6 min, slides were
counterstained with hematoxylin, dehydrated, and mounted. Cells
with a deposition of buffy-colored granules in the sections were
scored as RIP1-positive. The expression of RIP-1 was evaluated
using the Image-Pro Plus 6.0 software (Media Cybernetics, Inc.
USA). Five slices were randomly selected for each group and five
fields were selected for each slice. The expression of RIP-1 in
each group was semiquantitatively analysed using mean optical
density (MOD) = the intergral optical density (IOD) / the positive
area, in 25 fields, respectively.
Cell culture
The gastric cancer cell lines MGC, MKN-74, HGC and
AGS were preserved by the Key Laboratory of the Ministry of
Eduction for Gastrointestinal Cancer, Fujian Medical Universtiy,
Fuzhou, Fujian, China. The MGC, MKN-74 and HGC cell lines were
cultured in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS) (both from Gibco, Carlsbad, CA, USA). AGS was incubated
in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
FBS. All the cells were incubated at 37°C under 95% air and 5%
CO2.
RNA preparation, reverse transcription,
and real-time PCR amplification
Semiquantitative RT-PCR was used to detect the
expression of RIP-1 gene in the gastric cancer cell lines MGC,
MKN-74, HGC and AGS, respectively. Total RNA was isolated from the
cultured cells grown in 6-well plates using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer's
instructions and quantified by UV 260/280 nm to an absorption ratio
of >1.8. Total RNA (2 μg), according to the different
concentrations of sample RNA, were reverse transcribed to cDNA in a
final volume of 20 μl using the AVM First Strand cDNA synthesis kit
(Invitrogen) following the manufacture's instructions.
Additionally, they were used as cDNA template for PCR. The PCR
primers used for amplification are shown in Table I. All PCRs were performed with
Thermo Scientific SYBR Green qPCR kit on an Applied Biosystems
StepOne Real-time PCR System. PCR conditions were: 95°C for 2 min,
95°C for 15 sec, and 60°C for 30 sec for 40 cycles. All gene
transcripts were quantified by the 2−ΔΔCt method.
 | Table IThe gene name and sequence of
primers, and the product length. |
Table I
The gene name and sequence of
primers, and the product length.
| Name | Sequences | Product (bp) |
|---|
| RIP-1 | F:
5′-GTCCTGGTTTGCTCCTTCCC-3′
R: 5′-GTCTCCTTTCCTCCTCTCTGTTG-3′ | 170 |
| VEGF-C | F:
5′-TGTGTGTCCGTCTACAGATGTG-3′,
R: 5′-TCGGCAGGAAGTGTGATTGG-3′ | 165 |
| β-actin | F:
5′-CTGTCTGGCGGCACCACCAT-3′
R: 5′-GCAACTAAGTCATAGTCCGC-3′ | 254 |
Western blot analysis
Western blot analysis and detection of the blotted
product were carried out as described previously (14). The following primary antibodies
were used from the companies as listing, respectively: RIP-1 mouse
monoclonal anti-human antibody (1:1,000), VEGF-C rabbit polyclonal
anti-human antibody (1:1,000), c-jun (AP-1) monoclonal rabbit
anti-human antibody (1:1,000), and p-c-jun (p-AP-1) monoclonal
rabbit anti-human antibody (1:1,000) (all from Abcam
Biotechnology), nuclear factor-κB (NF-κB) (p65) monoclonal mouse
anti-human antibody (1:500), p-NF-κB (p-p65) monoclonal mouse
anti-human antibody (1:500) (both from Cell Signaling Technology,
Danvers, MA, USA), β-actin monoclonal mouse anti-human antibody
(1:1,500) (from Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA).
RIP-1 siRNA plasmid constuction and
transfection
According to the siRNA design principle, four
suitable siRNA target sequences (R-1, R-2, R-3 and R-4), from human
RIP-1 gene GenBank accession no. NM_003804, were synthesized as
follows, respectively: R-1, GCACAAATACGAACTTCAA; R-2,
GGCCAATTCCAAGTCATAT; R-3, TACAACAGAGAGGAGGAAA; and R-4,
TTGTGATAATGACTTCCA. A small hairpin RNA (shRNA) of human RIP-1 in a
phU6 gene transfer vector and the negative control (NC) sequence
encolding an enhanced green fluorescent protein (EGFP) sequence
with the puromycin resistance were constructed by Genechem Co. Ltd.
(Shanghai, China). Additionally, all the plasmids were verified by
DNA sequencing. The AGS and HGC cells were cultured in appropriate
medium supplemented with 10% FBS. When the AGS and HGC cells were
at ~90% confluency, we transfected the plasmids into these cells.
We used Lipofectamine 2000 (Invitrogen) to transfect cells
according to the manufacturer's instructions. We used a microscope
to observe the transfection efficiency by counting the percentage
of cells that were EGFP-positive.
Construction of lentiviral vetors and
infection
In order to create better stable transfection for
later experiments, we constructed lentiviral-mediated siRNA
targeting RIP-1 vector. We constructed lentiviral vectors for human
RIP-1 small hairpin RNA (shRNA) encoding a green fluorescent
protein (GFP). The lentivirus vectors containing RIP-1 shRNA were
constructed by ligating the digested pGCSIL-GFP and the RIP-1 shRNA
PCR product and then verified by DNA sequencing. ShRNA for the
negative controls was preserved in our lab. AGS and HGC cells were
plated in 6-well plates with appropriate medium supplemented with
10% FBS overnight. When these cells were at approximately 35%
confluent, they were used in siRNA virus infection. The
multiplicity of infection of (MOI) = 50 was appropriate quantity of
virus infection and 1 ml complete medium was added to 8 mg/ml
polybrene and mixed with cells and incubated for 12 h at 37°C.
Then, the cells were incubated in fresh complete medium containing
10% FBS for 24 h. Microscope was used to confirm the infection
efficiency by observing the GFP-positive cells.
Cell proliferation assay
Cell proliferation was assessed by MTT assay. Three
groups of cells (blank control group, NC-RNAi-LV group and
R-1-RNAi-LV group) were seeded into 96-well plates at density of
103 cells/well with 100 μl of medium supplemented with
10% FBS. The proliferative activity was determined after 1, 2, 3,
4, and 5 days by the addition of 10 μl of sterile
3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT)
(5 mg/ml; Sigma) to each well. The reaction was terminated after 4
h of incubation at 37°C through the addition of 100 μl of dimethyl
sulfoxide (DMSO; Sigma). The optical density (OD) value was
obtained by measuring absorbance at a wavelength of 450 nm. Each
well test was repeated 6 times.
In vitro cell invasion assay
Invasion assays were performed using Transwell
24-well plates with 8-μm polycarbonate membranes (BD Biosciences).
Briefly, the upper side of the membranes was coated with Matrigel
matrix (20 μg/well) and the membranes were then air-dried for 1 h
of incubation 37°C. The lower side of the membranes was coated with
5 μg fibronectin (BD Biosciences). To test the ability of cell
invasion, blank control group, NC-RNAi-LV group and R-1-RNAi-LV
group gastric cancer cells (2×105) in 200 μl of
RPMI-1640 (DMEM for AGS cells) medium with 2.5% FBS were placed in
the upper chamber. The lower chamber was filled with 700 μl
RPMI-1640 (DMEM for AGS cells) medium with 10% FBS as the
chemoattractant. The invasion chamber was incubated for 8 h at 37°C
and 5% CO2. The cells on the upper surface of the
membrane were removed by gentle scrubbing with a cotton swab.
Membranes were fixed in a stationary liquid of 95% ethanol and 5%
acetic acid for 30 min and stained with crystal violet. The number
of cells on the lower surface of the membrane in 5 random visual
fields (×400) was then counted using a bright field light
microscope. Each assay was repeated in triplicate.
Animal experiments
Male four to six-week-old athymic BALB/c nu/nu mice
were obtained from the Shanghai SLAC Laboratory Animals Co.,
(Shanghai, China) and kept in the Experimental Center of the Fujian
Medical University. All the mice were handled according to the
recommendations of the institutional guidelines and were approved
by the Ethics Committee of the Medical Faculty of the Fujian
Medical University. Blank control group, NC-RNAi-LV group and
R-1-RNAi-LV group were randomly divided into three groups using 15
mice (five mice/group) for the subcutaneous gastric cancer nude
mouse model. Three groups of different gastric cancer cells
(106) in 100 μl serum-free medium were seeded
subcutaneously into the upper flank region of the nude mice. Tumor
growth was monitored and measured in two dimensions (width and
length respectively) every 3 days. The size of tumor growth was
estimated using the formula = width2 (mm2) ×
length (mm)/2 (the width and length are the shortest and longest
diameters of the subcutaneous tumor). After 5 weeks, nude mice were
sacrificed and the tumors were collected and weighed immediately.
Then all the samples were fixed in 10% formalin solution for the
immunohistochemical analysis.
Statistical analysis
The statistical analysis used GraphPad Prism 5
software. Data were analyzed by the one-way ANOVA or Student's
t-test. The data were expressed as the means ± standard deviation
(SD). Survival curves were calculated by the Kaplan-Meier method
and compared by using the log-rank test. A P-value <0.05 was
considered to indicate a statistically significant difference.
Results
Analysis of RIP-1 expression in patients
with gastric cancer by immunohistochemistry staining,
clinicopathologic factors and prognosis
We evaluated the expression of RIP-1 in 70 clinical
samples from gastric cancer patient and 19 normal gastric tissues
distant from the cancer site using immunohistochemistry. In the
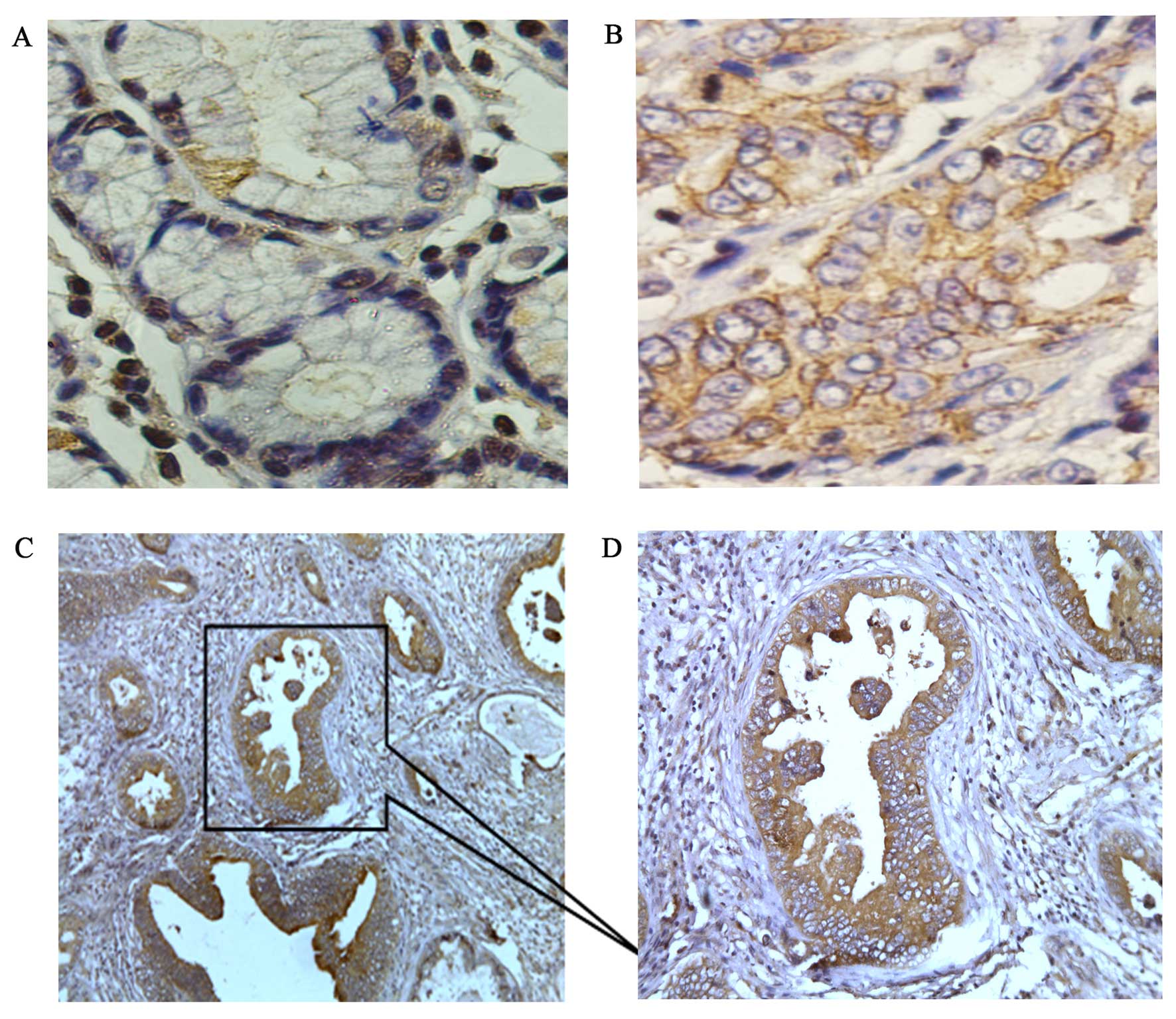
normal gastric tissues, low levels of RIP-1 expression were
detected (Fig. 1A); but, in the
gastric cancer tissues high levels of expression of RIP-1 were
found (Fig. 1B). The MOD of RIP-1
in the normal gastric tissues (0.1478±0.1346) was markedly lower
than gastric cancer tissues (0.3576±0.0961; P<0.001; Table II). Thus, RIP-1 was overexpressed
in the gastric cancer tissues. Importantly, the RIP-1
immunoreactivity was positive at the site of invasion, but little
or no immunoreactivity was detected at the parts of interstitial
substance (Fig. 1C and D). This
suggested that RIP-1 overexpression may promote gastric cancer
invasion.
 | Table IIImmunohistochemistry results of the
expression of RIP-1 in gastric cancer samples, and normal gastric
tissues. |
Table II
Immunohistochemistry results of the
expression of RIP-1 in gastric cancer samples, and normal gastric
tissues.
| Tissues | N | RIP-1
expressiona (MOD ± SD) | P-value |
|---|
| Gastric cancer
tissues | 70 | 0.3576±0.0961 | <0.001 |
| Normal gastric
tissues | 19 | 0.1478±0.1346 |
Table III shows
the relationship between RIP-1 expression and select
clinicopathological parameters. RIP-1 expression did not vary
significantly with age, gender, tumor size and histological grade;
however, there was a significant relationship between RIP-1
overexpression and clinical stage (I–II vs. III–IV; P=0.031), lymph
node metastasis (no vs. positive; P=0.036) and Helicobacter
pylori (negative vs. positive; P=0.0312).
 | Table IIIRelationship between RIP-1 expression
and gastric cancer clinicopathological parameters. |
Table III
Relationship between RIP-1 expression
and gastric cancer clinicopathological parameters.
| Factors | N | RIP-1 expression
(MOD±SD)a | P-value |
|---|
| Age (year) | | | | |
| <60 | 37 | 0.2529±0.0685 | t=0.1018 | 0.7189 |
| ≥60 | 33 | 0.2411±0.0769 | | |
| Gender | | | | |
| Male | 34 | 0.3238±0.0919 | t=0.7067 | 0.3266 |
| Female | 36 | 0.3147±0.0930 | | |
| Tumor size
(cm) | | | | |
| ≤3 | 48 | 0.3675±0.0382 | t=1.138 | 0.5083 |
| >3 | 22 | 0.3957±0.1804 | | |
| Clinical
stageb | | | | |
| I–II | 25 | 0.2176±0.0964 | t=2.665 | 0.0031* |
| III–IV | 45 | 0.3687±0.1512 | | |
| Lymph node
metastasis | | | | |
| No | 21 | 0.2293±0.1432 | t=2.171 | 0.036* |
| Yes | 49 | 0.3385±0.1123 | | |
| Histological
grade | | | | |
| Poorly | 25 | 0.3616±0.1367 | F=0.2786 | 0.1522 |
| Moderately | 19 | 0.2879±0.2812 | | |
| Well | 26 | 0.1914±0.9401 | | |
| Helicobacter
pylori | | | | |
| Negative | 29 | 0.2185±0.1161 | t=2.276 | 0.0312* |
| Positive | 41 | 0.3627±0.0871 | | |
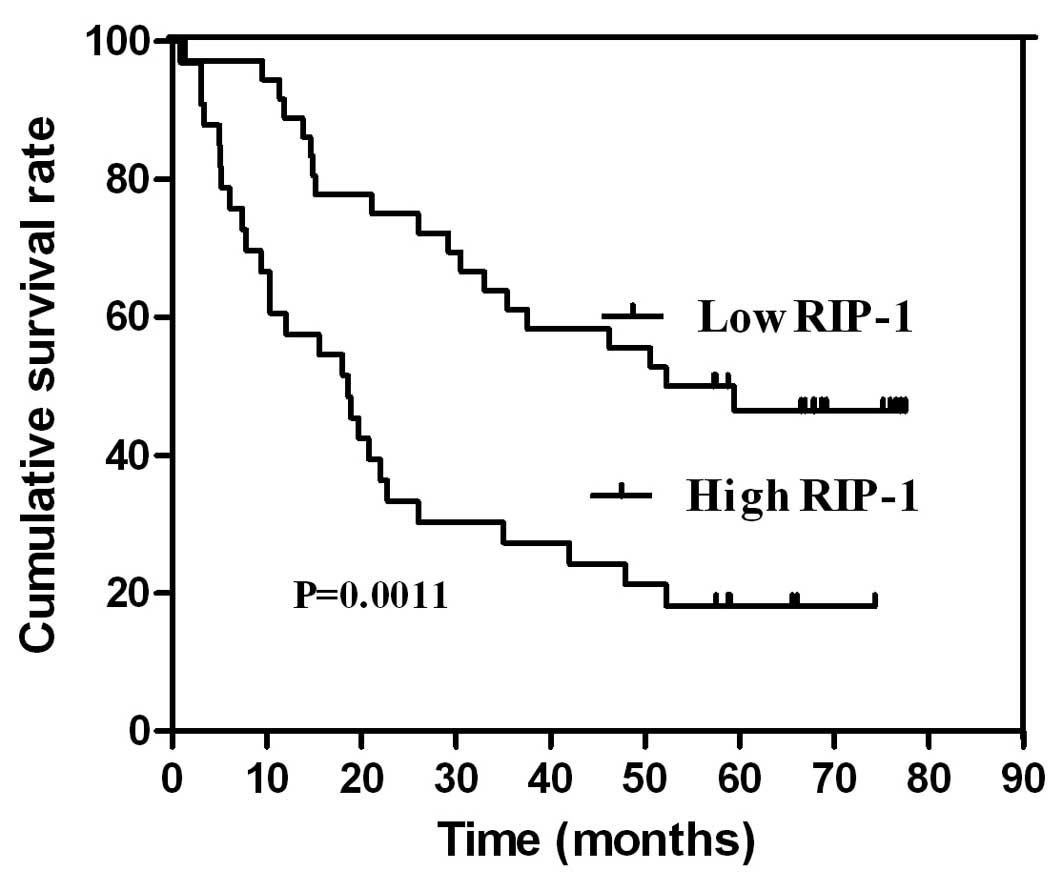
Survival and prognostic analysis
The expression levels of RIP-1 were positively
correlated with the prognosis of gastric cancer patients. The study
samples were split into two groups according to the MOD expression
levels: a low RIP-1 expression levels group (<0.3011, which is
the average MOD values of 70 gastric cancer patients of tissues)
and a high RIP-1 expression levels group (≥0.3011). Additionally,
the patients with high expression of RIP-1 had very discouraging
prognosis; however, the patients with low expression of RIP-1 had a
better prognosis. Thus, the survival and prognosis of the low
expression of RIP-1 group was significantly longer than the high
expression of RIP-1 group (P<0.05) (Fig. 2).
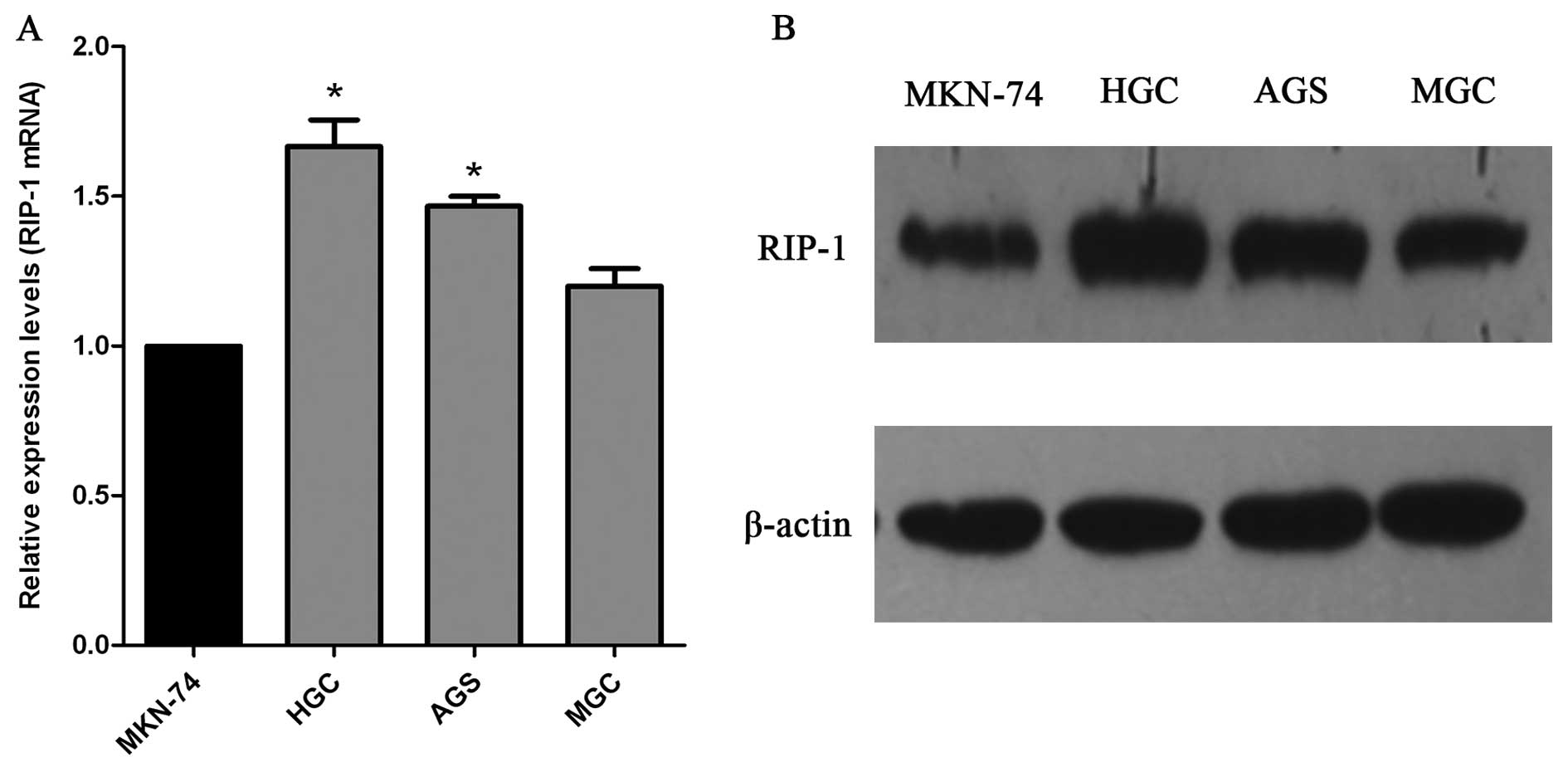
Expression of RIP-1 mRNA and protein in
gastric cancer cell lines
We used qRT-PCR and western blot technology to
detect the expression of RIP-1 in human gastric cancer cell lines
(MGC, MKN-74, HGC, and AGS). RIP-1 mRNA was detected in all four
cell lines by qRT-PCR. In addition, expression of RIP-1 protein in
all four cell lines was confirmed. The expression quantity of RIP-1
mRNA and protein in HGC and AGS cells was higher compared with the
MGC and MKN-74 cells (Fig. 3).
Thus, we used the HGC and AGS cells to further evaluate the role of
RIP-1 in gastric cancer.
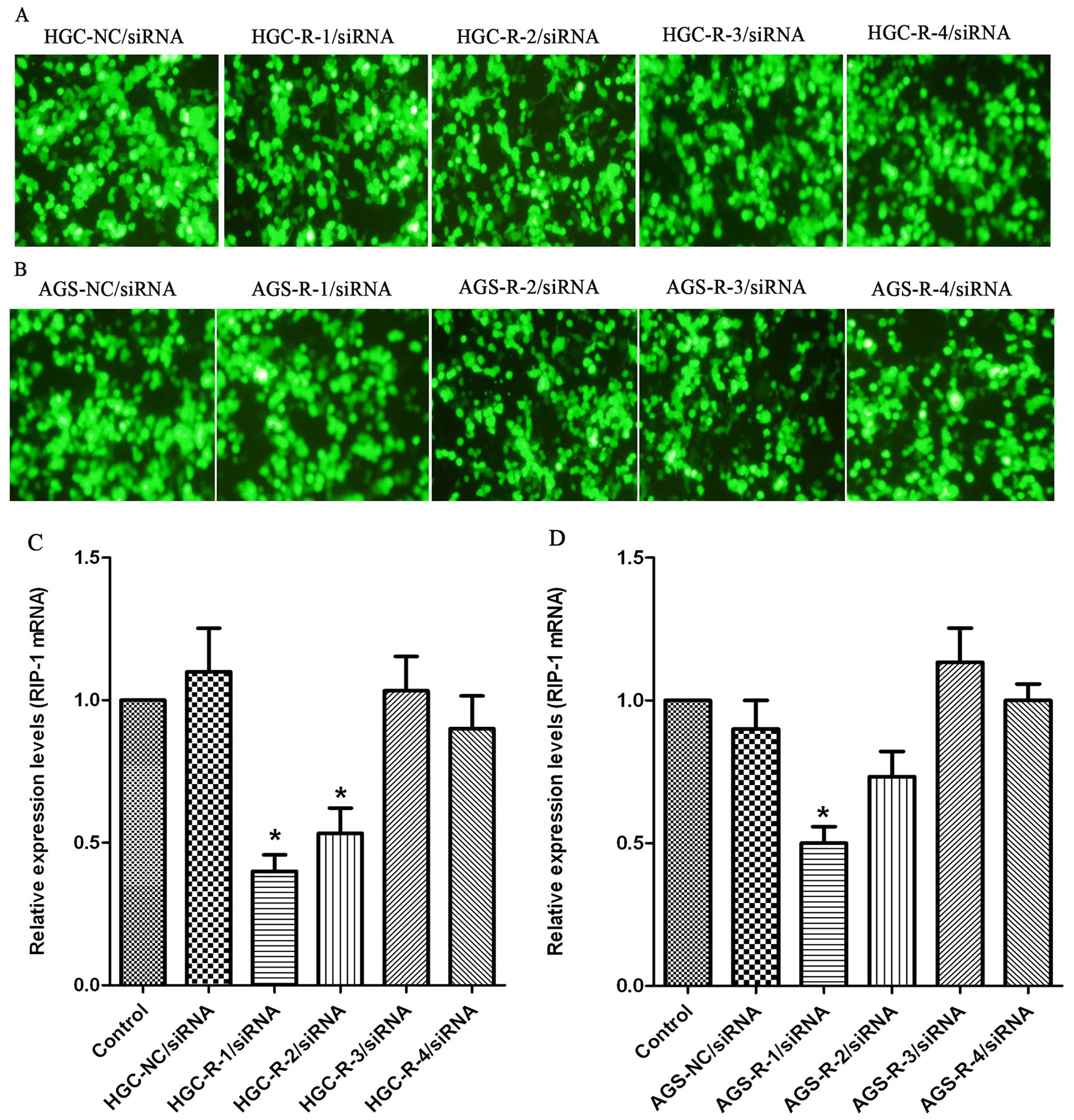
Expression of RIP-1 mRNA after siRNA
transfection
The RNAi effect of silencing RIP-1 gene in the HGC
and AGS cells was assessed. The DNA sequencing results verified
that RIP-1 siRNA plasmid construction was successful. To determine
the level of RIP-1 expression levels 72 h after transfection in HGC
and AGS cells we used qRT-PCR. Expression of the siRNA plasmids
vectors (NC/R-1/R-2/R-3/R-4/siRNA) enhanced green fluorescent
protein (EGFP) 72 h after transfection, as shown by fluorescence
microscopy, in the HGC and AGS cells (Fig. 4A and B). qRT-PCR revealed a
decrease in RIP-1 mRNA in siRNA-transfected cells. The R-1/siRNA
plasmid vector resulted in a higher suppression of the levels of
RIP-1 mRNA expression than other vectors (R-2/siRNA, R-3/siRNA, and
R-4/siRNA), while non-transfected and NC/siRNA vectors had no
effect on the levels of RIP-1 mRNA expression in the HGC and AGS
cells (Fig. 4C and D). Because
semi-quantitative analysis by qRT-PCR showed that there was marked
inhibition between R-1/siRNA and non-transfected siRNA groups in
the HGC and AGS cells, we selected R-1/siRNA sequence to knock down
the RIP-1 in our further studies.
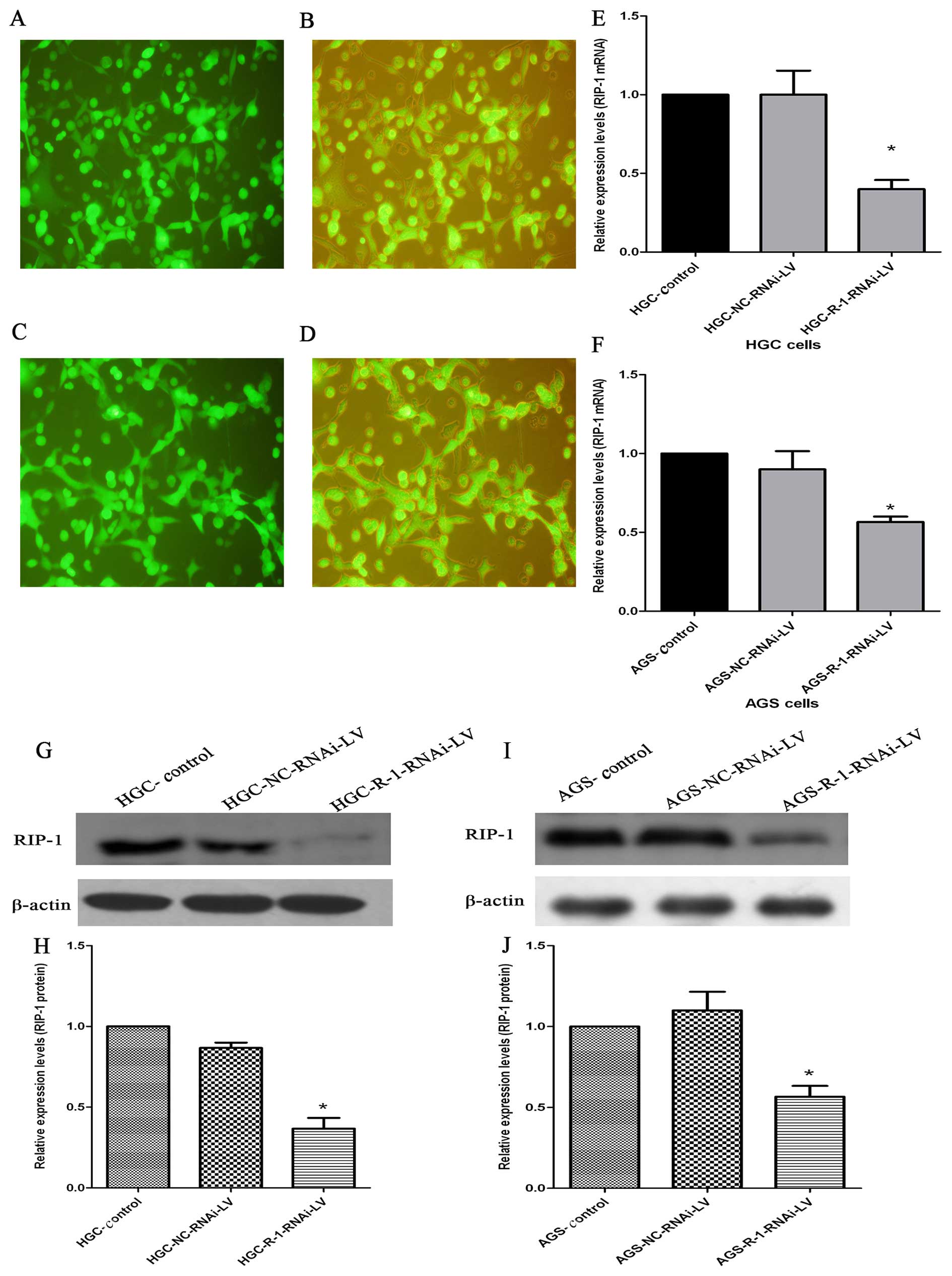
Expression of RIP-1 mRNA and protein
after R-1/siRNA lentiviral vectors infection
We used lentiviral-mediated R-1/siRNA targeting
RIP-1 vector to improve the stable transfection for the next
experiment. Transfection efficiency was quantified by counting the
cells under a fluorescent microscope 72 h after infection, and the
efficiency of HGC and AGS cells were >80% (Fig. 5A–D). The qRT-PCR showed that the
expression of RIP-1 mRNA was significantly inhibited after
infection in HGC and AGS cells. Compared with the control group and
NC-RNAi-LV group, the R-1-RNAi-LV group showed relatively lower
expression of RIP-1 mRNA in HGC and AGS cells (Fig. 5E and F). In accordance with mRNA,
western blotting revealed that RIP-1 protein expression was
suppressed in the R-1-RNAi-LV group compared with the control group
and NC-RNAi-LV group in HGC and AGS cells. There were distinct
densitometric differences between R-1-RNAi-LV group and control
group and NC-RNAi-LV group in HGC and AGS cells (Fig. 5J, H, J and K).
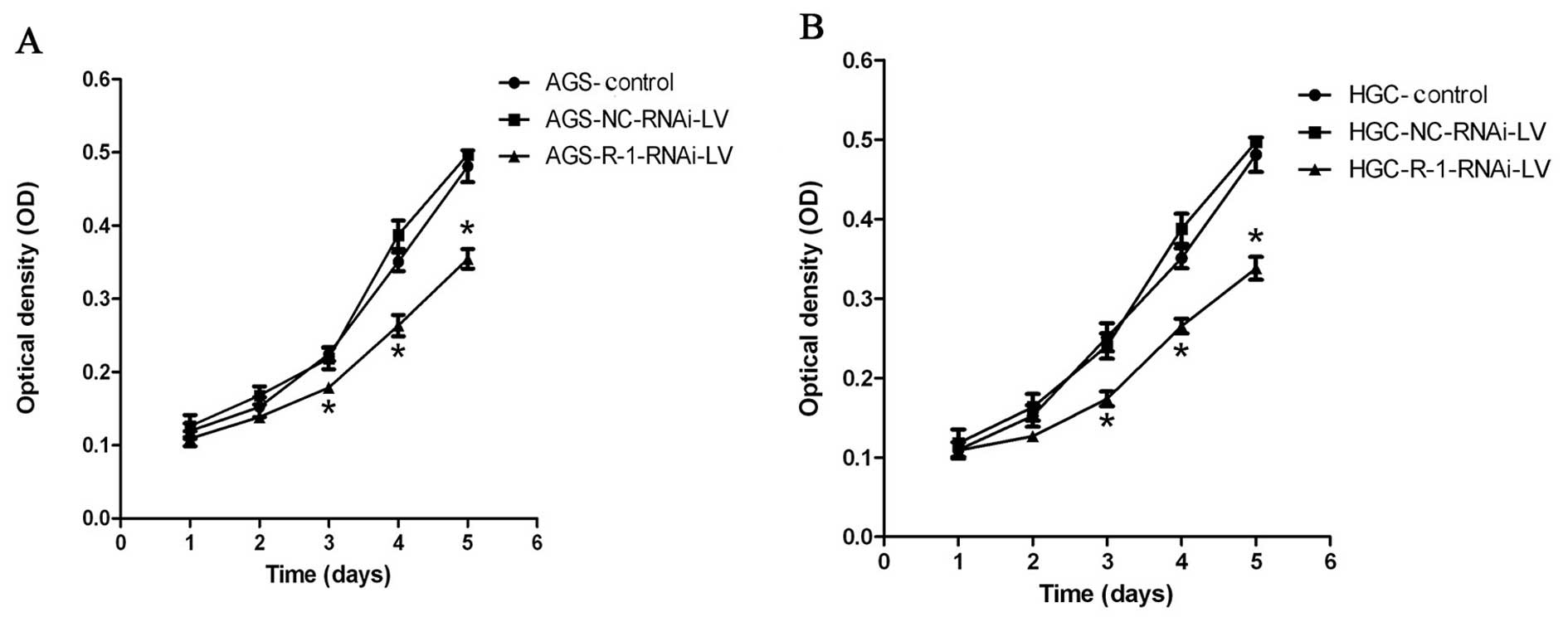
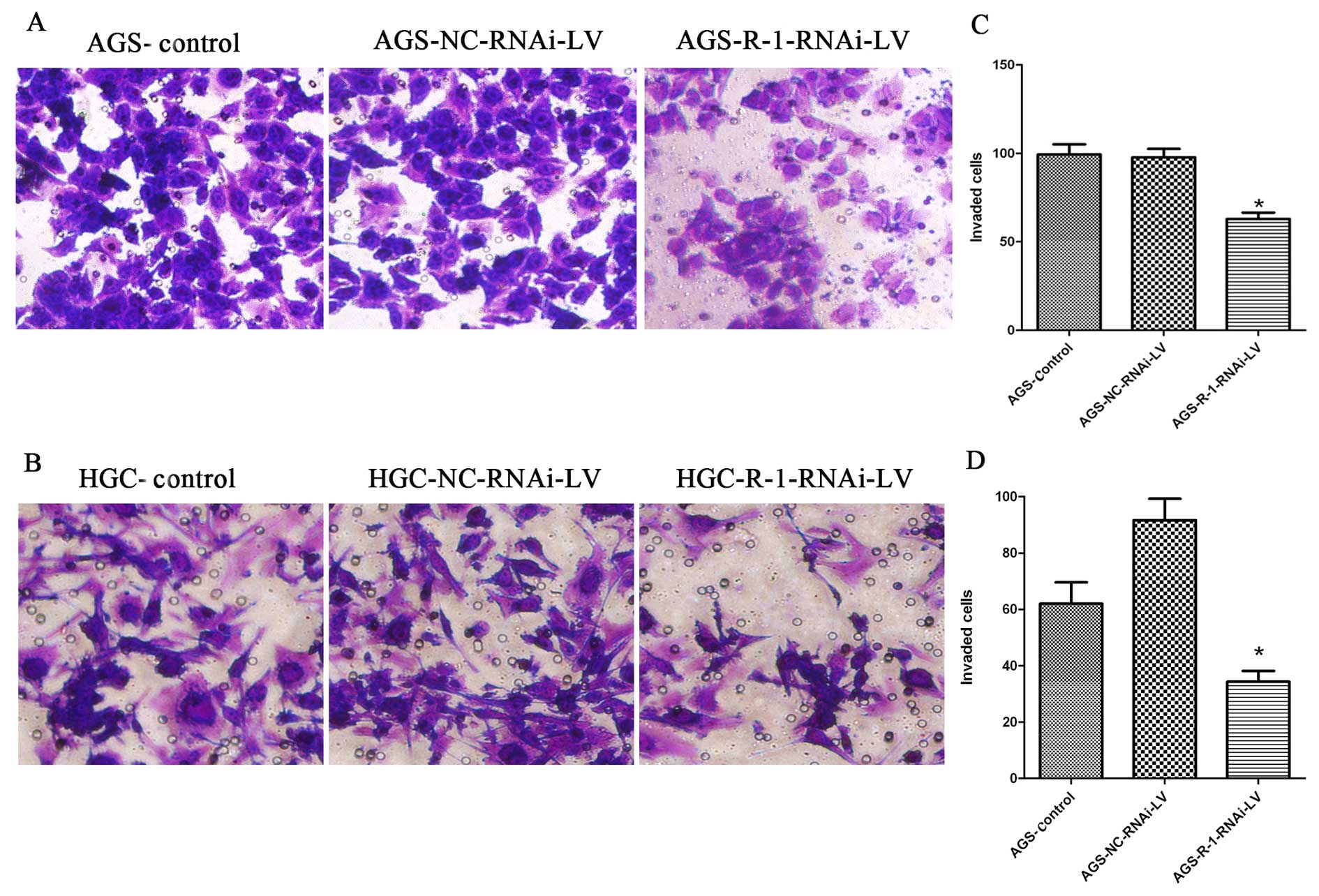
RIP-1 promotes HGC and AGS cell
proliferation and invasion in vitro
To experimentally address the biological function of
RIP-1 in gastric cancer cells, we analyzed the effect of
lentivirus-mediated RIP-1 siRNA on HGC and AGS cell proliferation
and invasion by MTT assay and cell invasion assay, respectively.
MTT experiment results showed that compared to control group and
NC-RNAi-LV group, the cell proliferation of the R-1-RNAi-LV group
was slower in HGC and AGS cells (Fig.
6). The cell viability of R-1-RNAi-LV group was decreased when
compared with control group in both gastric cancer cell lines. The
results from cell in vitro invasion indicated that the
numbers of cells that penetrated the basal membrane from the
R-1-RNAi-LV group was significantly less than the blank control
group, which was similar to the number of cells from the NC-RNAi-LV
group in HGC and AGS cells (Fig.
7).
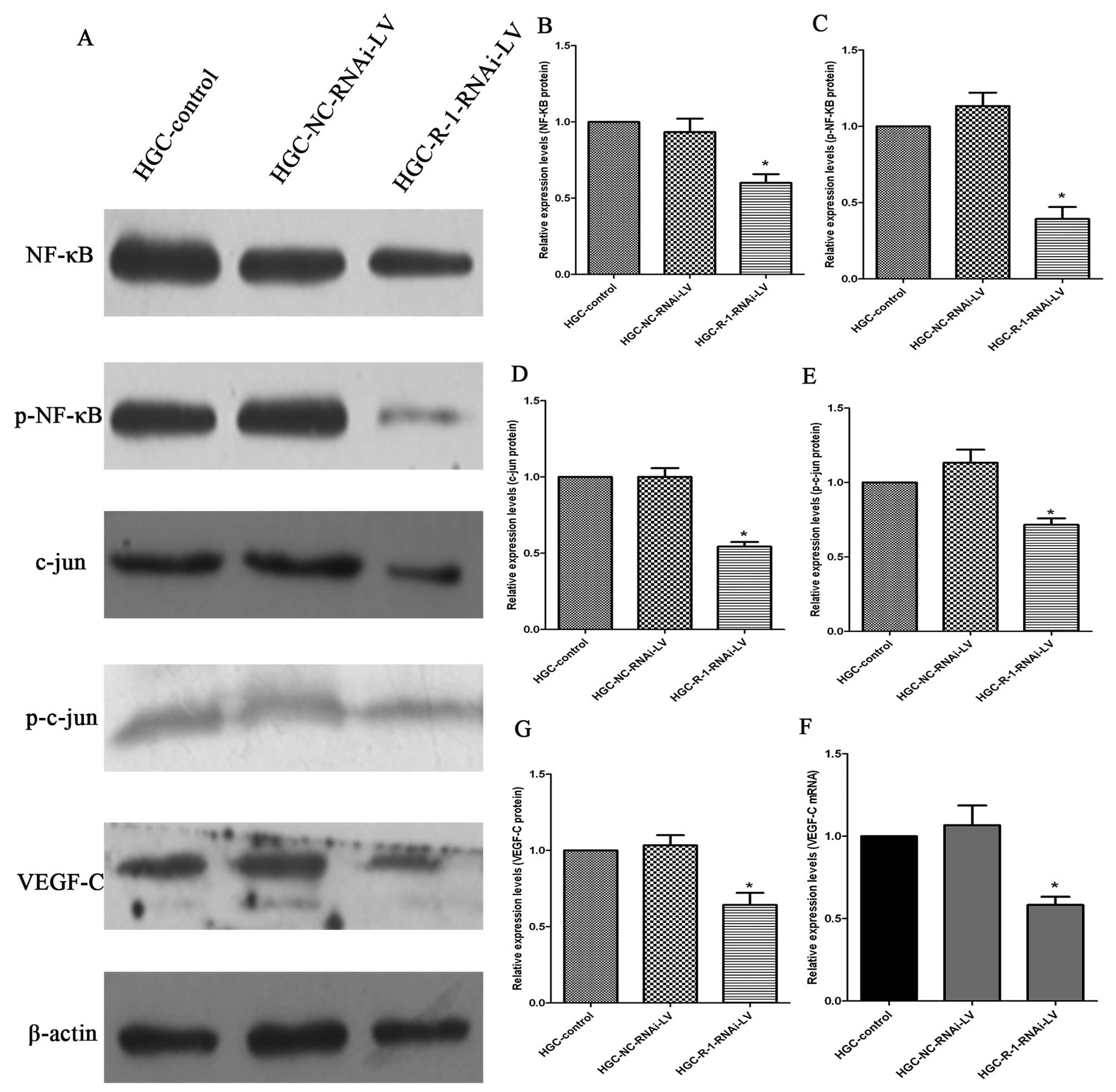
RIP-1 knockdown reduces the activity of
NF-κB (p65) and c-jun (AP-1) and decreases VEGF-C gene
expression
To investigate the mechanisms of RIP-1 silencing
responsible for the decrease in proliferation and invasion, we
assessed the changes in levels of NF-κB (p65), p-NF-κB (p-p65),
c-jun (AP-1), p-c-jun (p-AP-1) and VEGF-C protein and VEGF-C mRNA;
these factors are vital to the growth and invasion of cancer cells.
Western blot analysis revealed that NF-κB (p65), p-NF-κB (p-p65),
c-jun (AP-1), p-c-jun (p-AP-1) and VEGF-C protein levels in the
R-1-RNAi-LV cells decreased compared with the control NC-RNAi-LV
and in HGC cells (Fig. 8). The
changes in VEGF-C mRNA expression in the three groups were the same
as the levels of VEGF-C protein in HGC cells (Fig. 8F).
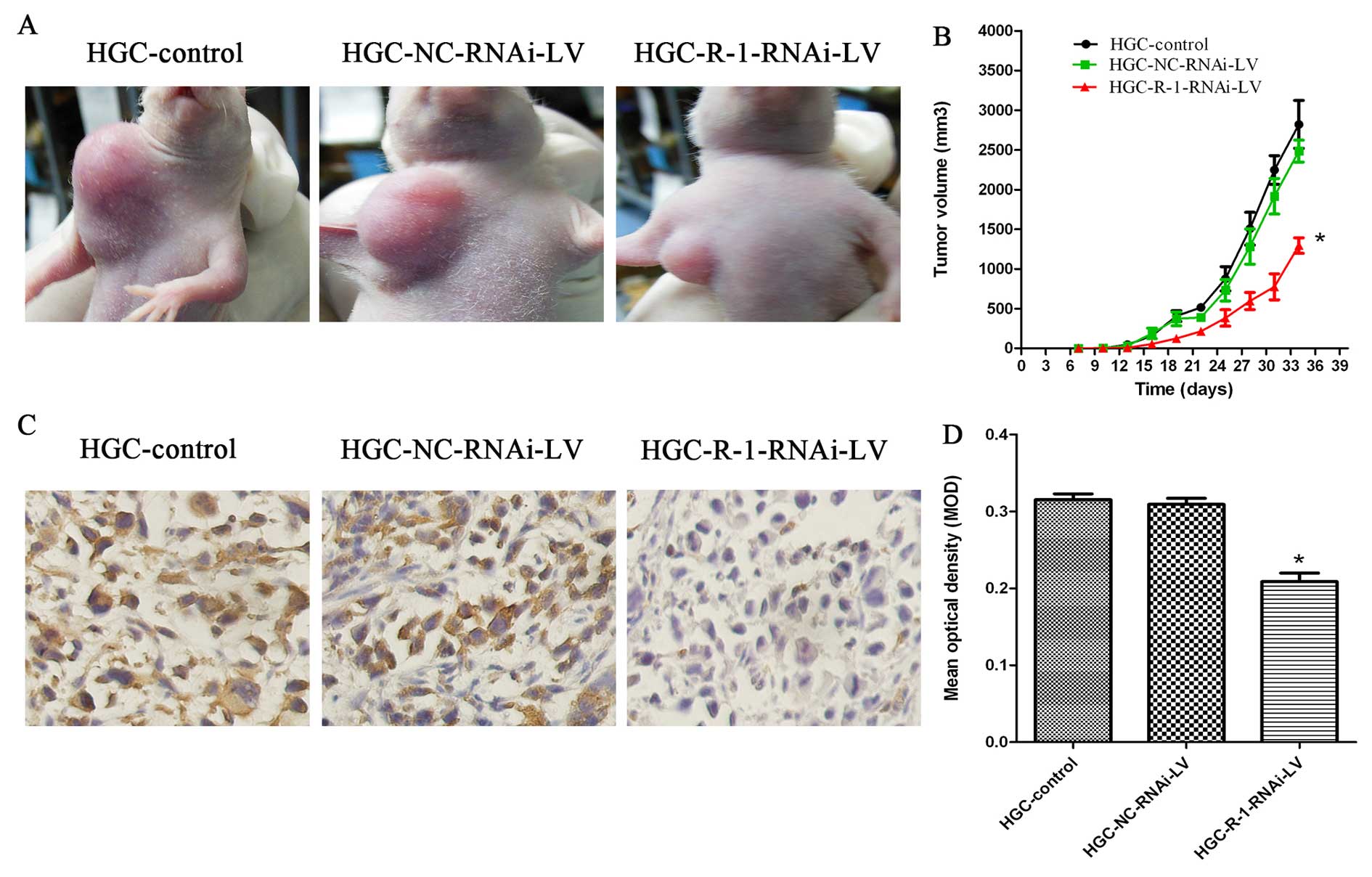
RIP-1 promotes gastric cancer tumor
growth in nude mice with subcutaneous xenograft tumors
To address the impact of RIP-1 expression on gastric
cancer tumor biology in vivo, three groups (HGC-control,
HGC-NC-RNAi-LV, and HGC-R-1-RNAi-LV groups) of HGC cells were
subcutaneously xenografted onto nude mice. Nude mice were all
successfully established in the subcutaneous mouse model. None of
the mice had ascites or metastasis to the liver, lungs, or lymph
nodes at the time of autopsy. At the end of the 5-week experimental
period, the size of the subcutaneous xenograft tumor showed
significant differences between the HGC-R-1-RNAi-LV and
HGC-NC-RNAi-LV groups, which was similar to the HGC-control group.
We found that xenograft growth was markedly slower from the second
week until the day on which the mice were sacrificed (Fig. 9A and B). Because RIP-1 expression
was reduced in the HGC-R-1-RNAi-LV cells, we used
immunohistochemistry to analyze the expression of RIP-1 in the nude
mice. Compared with the HGC-control and HGC-NC-RNAi-LV groups, the
levels of RIP-1 expression were significantly reduced in the
HGC-R-1-RNAi-LV group (Fig. 9C and
D).
Discussion
Aberrant activation of survival signaling pathways
plays a very important role in cancer development, and progression
(15). The activation of NF-κB and
AP-1 in human cancer is common, and the activation promotes growth
and invasion of cancer cells (7,16–19).
RIP-1 is a crucial molecule for NF-κB and AP-1 signaling in
inflammation and cell survival (16–19).
RIP-1 consists of N-terminal kinase domain, an intermediate domain,
a RIP homotypic interaction motif, and a C-terminal death domain
motif (20). RIP-1 is a kinase
with important roles in inflammation, cell survival, and cell
apoptosis (20), mediating both
cell survival and death signaling.
Upon TNFR1 stimulation, RIP-1, along with other
proteins including TRADD, TRAF2, cIAP1, and cIAP2, are recruited to
form the prosurvival complex I (21,22).
Then k63 polyubiquitin and linear ubiquitin chains linked to RIP-1
serve as substrates for binding the TAB/TAK1 complex and NEMO,
leading to activation of NF-κB and AP-1, which plays an important
role in regulating many cellular processes (7,16,23,24).
However, the complicated functions and mechanisms of
RIP-1 regulated cell signaling pathways are largely unknown.
Additionally, many details pertaining to the role of RIP-1 in
cancer cells are unclear. Some recent reports address the functions
of RIP-1 in gioblastomas (10),
leukemia (11), lung cancer
(13), and melanoma (12). Additionally, our study team
reported that RIP-1 promotes the proliferation and invasion via the
signaling pathways of RIP-NF-κB/AP-1-VEGF-C in gallbladder cancer
cells (25). However, the
expressions and roles of RIP-1 in gastric cancer are still unknown.
The specific interactions between RIP-1 and cancers are largely
unclear, so we investigated the biological functions and underlying
mechanisms in vitro and in vivo in gastric
cancer.
In this study, we first demonstrated the roles of
RIP-1 in promoting growth and invasion of human gastric carcinoma.
We used immunohistochemical techniques to evaluate the expression
of RIP-1 in 70 gastric cancer specimens and 19 samples of normal
gastric tissues which were locted distant from the cancer site.
RIP-1 is overexpressed in gastric cancer tissues, and RIP-1 has low
expression in normal tissues. Increased expression of RIP-1 is
common in gastric cancer tissues. This suggested that RIP-1 may be
an important molecule and be of clinical significance in prognosis.
The survival and prognosis of the low expression of RIP-1 group was
significantly longer than the high expression of RIP-1 group. This
agreed with Park et al (10) who reported that RIP-1 is an
independent prognostic factor in glioblastomas. We found that the
RIP-1 immunoreactivity was positive at the site of invasion, but
little or no immunoreactivity was detected at the parts of
interstitial substance and the expression of RIP-1 was markedly
correlated with the clinical stage of gastric cancer patients,
which suggested that RIP-1 may promote gastric cancer invasion and
growth. We also found that RIP-1 expression is significantly
increased in the gastric cancer patients with Helicobacter
pylori infection. The reason may be Helicobacter pylori
infection inducing inflammation and thus increasing the expression
of RIP-1 (26). We also found that
the RIP-1 expression was high in the gastric cancer patients with
lymph node metastasis. Additionally, the VEGF-C is an important
factor in the gastric cancer lymph node metastasis (27,28).
It was reported that VEGF-C could promote tumor cells to invade the
interstitial substance (29).
RNAi technology has served as the most widely used
techique for gene knockdown and provide a method to study gene
expression and function (30). In
addition, siRNA allows us to target any gene with greater
selectivity, stability and efficiency (31) and plays an important role in the
study of gene function in human cancer (32). Thus, we used siRNA to evaluate the
effect of suppressing the RIP-1 gene in gastric cancer in
vitro and in vivo. In this study, we successfully
selected R-1/siRNA as the most effective siRNA to knock down RIP-1
expression after four RIP-1 siRNA plasmid transfections in HGC and
AGS cells. RIP-1 mRNA and protein was inhibited by R-3-RNAi-LV
vector in HGC and AGS cells. In vitro, knockdown of the
RIP-1 expression by siRNA suppressed the proliferation and invasion
of HGC and AGS cells. Our results agree with those of Azijli et
al (13) who reported that
knockdown the RIP-1 gene inhibited the migration and invasion in
A549 and H460 cell lines. Our laboratory showed that the
proliferation and invasion were significantly suppressed in the
gallbladder cancer cells, when silencing the RIP-1 gene expression
in the tumor cells (25). The
results suggest that RIP-1 can affect the biological
characteristics of tumor cells in gastric cancer.
To investigate the mechanisms underlying RIP-1
promotion of growth and invasion of HGC and AGS cells, we used
western blot analysis to evaluate the changes of expression of
NF-κB (p65), p-NF-κB (p-p65), c-jun (AP-1), p-c-jun (p-AP-1) and
VEGF-C protein in HGC cells. These proteins are vital to the
survival of gastric cancer cells and very important in the
inflammation signaling pathways, in which RIP-1 can activate the
activity of NF-κB (p65), p-NF-κB (p-p65), c-jun (AP-1), p-c-jun
(p-AP-1) (7,16–19).
In this study, the expression of NF-κB (p65), p-NF-κB (p-p65),
c-jun (AP-1), p-c-jun (p-AP-1) and VEGF-C protein were decreased in
the HGC-R-1-RNAi-LV group cells compared with HGC-control group and
HGC-NC-RNAi-LV group cells. These results suggested that
RIP-1-NF-κB/AP-1 signaling pathway may regulate the expression of
VEGF-C.
In our murine model gastric cancer, we found that
the mice of HGC-R-1-RNAi-LV group the growth of subcutaneous
xenograft tumors was much slower compared with the subcutaneous
xenograft tumors of mice of HGC-control group and HGC-NC-RNAi-LV
group. In our in vivo model of gastric cancer, we did not
find liver, lung or lymph node metastasis, and ascite had not
developed. This is because the subcutaneous xenograft gastric tumor
model can not provide a suitable microenvironment for tumors
(33). Therefore, we will
establish an orthotopic xenograft model of gastric cancer to futher
study RIP-1 and its influence on the biological behavior of gastric
cancer.
In conclusion, this study demonstrated that the
expression of RIP-1 in the gastric cancer tissues was significantly
higher than the expression in the normal gastric tissues.
Additionally, RIP-1 immunoreactivity was positive at the site of
invasion, but little or no immunoreactivity was detected at the
gastric cancer parts of the interstitial substance. The survival
rate was poor in RIP-1 high expression of patients with gastric
cancer. Futhermore, the RIP-1 expression in the gastric cancer cell
lines were general. Additionally, we provided evidence that
endogenously secreted RIP-1 contributed to the proliferation and
invasion of HGC and AGS cells. We also found that the
RIP-NF-κB/AP-1-VEGF-C signaling pathways have a crucial role in the
regulation of the biological functions of HGC and AGS cells. We
verified the relationship between RIP-1 overexpression and the
growth of gastric cancer using a mouse model. Together, our data
indicate that RIP-1 promote the growth and invasion of gastric
cancer in vitro and in vivo. Additionally, our
results provide evidence that targeting RIP-1 may be useful in the
treatment of gastric cancer.
Acknowledgements
This study was supported by the Key Project of
Science and Technology Reserch Program in Fujian Province (no.
2012B002), the Fujian Provincial Natural Science Foundation (no.
2014J01309), the Backbone Teacher Project of Fujian Medical
University (no. JGG200716), China Non intervention Gastric Cancer
Registration Survey Clinical Research Projects (no. QT-201403), the
Ministry of Health Medicine Science and Technology Development and
Research (no. W2013FZ08) and the National Clinical Key Specialty
Construction Project (General Surgery) of China.
References
|
1
|
Ferlay JSI, Ervik M, et al: GLOBOCAN
2012v1.0, Cancer Incidence and Mortality Worldwide. IARC CancerBase
No. 11 (Internet). International Agency for Research on Cancer;
Lyon: 2013, http://globocan.iarc.fr.
Accessed Apr 25, 2014
|
|
2
|
Carneiro F: Stomach cancer. World Cancer
Report 2014. Steward B and Wild CP: International Agency for
Research on Cancer; Lyon: pp. 383–391. 2014
|
|
3
|
Waters JP, Pober JS and Bradley JR: Tumour
necrosis factor in infectious disease. J Pathol. 230:132–147. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Donnell MA and Ting AT: RIP1 comes back
to life as a cell death regulator in TNFR1 signaling. FEBS J.
278:877–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stanger BZ, Leder P, Lee TH, Kim E and
Seed B: RIP: A novel protein containing a death domain that
interacts with Fas/APO-1 (CD95) in yeast and causes cell death.
Cell. 81:513–523. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hur GM, Lewis J, Yang Q, Lin Y, Nakano H,
Nedospasov S and Liu ZG: The death domain kinase RIP has an
essential role in DNA damage-induced NF-kappa B activation. Genes
Dev. 17:873–882. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Devin A, Lin Y and Liu ZG: The role of the
death-domain kinase RIP in tumour-necrosis-factor-induced
activation of mitogen-activated protein kinases. EMBO Rep.
4:623–627. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bradley JR and Pober JS: Tumor necrosis
factor receptor-associated factors (TRAFs). Oncogene. 20:6482–6491.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ting AT, Pimentel-Muiños FX and Seed B:
RIP mediates tumor necrosis factor receptor 1 activation of
NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J.
15:6189–6196. 1996.PubMed/NCBI
|
|
10
|
Park S, Hatanpaa KJ, Xie Y, Mickey BE,
Madden CJ, Raisanen JM, Ramnarain DB, Xiao G, Saha D, Boothman DA,
et al: The receptor interacting protein 1 inhibits p53 induction
through NF-kappaB activation and confers a worse prognosis in
glioblastoma. Cancer Res. 69:2809–2816. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han W, Xie J, Fang Y, Wang Z and Pan H:
Nec-1 enhances shikonin-induced apoptosis in leukemia cells by
inhibition of RIP-1 and ERK1/2. Int J Mol Sci. 13:7212–7225. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu XY, Lai F, Yan XG, Jiang CC, Guo ST,
Wang CY, Croft A, Tseng HY, Wilmott JS, Scolyer RA, et al: RIP1
kinase is an oncogenic driver in melanoma. Cancer Res.
75:1736–1748. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Azijli K, Yuvaraj S, Peppelenbosch MP,
Würdinger T, Dekker H, Joore J, van Dijk E, Quax WJ, Peters GJ, de
Jong S, et al: Kinome profiling of non-canonical TRAIL signaling
reveals RIP1-Src-STAT3-dependent invasion in resistant non-small
cell lung cancer cells. J Cell Sci. 125:4651–4661. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu G, Du Q, Wang X, Tang N, She F and
Chen Y: TNF-α promotes gallbladder cancer cell growth and invasion
through autocrine mechanisms. Int J Mol Med. 33:1431–1440.
2014.PubMed/NCBI
|
|
15
|
Nikolaou VA, Stratigos AJ, Flaherty KT and
Tsao H: Melanoma: New insights and new therapies. J Invest
Dermatol. 132:854–863. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu ZG, Hsu H, Goeddel DV and Karin M:
Dissection of TNF receptor 1 effector functions: JNK activation is
not linked to apoptosis while NF-kappaB activation prevents cell
death. Cell. 87:565–576. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karin M, Cao Y, Greten FR and Li ZW:
NF-kappaB in cancer: From innocent bystander to major culprit. Nat
Rev Cancer. 2:301–310. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pacifico F and Leonardi A: NF-kappaB in
solid tumors. Biochem Pharmacol. 72:1142–1152. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang CY, Cusack JC Jr, Liu R and Baldwin
AS Jr: Control of inducible chemoresistance: Enhanced anti-tumor
therapy through increased apoptosis by inhibition of NF-kappaB. Nat
Med. 5:412–417. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Festjens N, Vanden Berghe T, Cornelis S
and Vandenabeele P: RIP1, a kinase on the crossroads of a cell's
decision to live or die. Cell Death Differ. 14:400–410. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bertrand MJ, Milutinovic S, Dickson KM, Ho
WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ and
Barker PA: cIAP1 and cIAP2 facilitate cancer cell survival by
functioning as E3 ligases that promote RIP1 ubiquitination. Mol
Cell. 30:689–700. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Imre G, Larisch S and Rajalingam K:
Ripoptosome: A novel IAP-regulated cell death-signalling platform.
J Mol Cell Biol. 3:324–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Niu J, Shi Y, Iwai K and Wu ZH: LUBAC
regulates NF-κB activation upon genotoxic stress by promoting
linear ubiquitination of NEMO. EMBO J. 30:3741–3753. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Blackwell K, Zhang L, Workman LM, Ting AT,
Iwai K and Habelhah H: Two coordinated mechanisms underlie tumor
necrosis factor alpha-induced immediate and delayed IκB kinase
activation. Mol Cell Biol. 33:1901–1915. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu G, Chen X, Wang X, Li X, Du Q, Hong H,
Tang N, She F and Chen Y: Expression of the RIP-1 gene and its role
in growth and invasion of human gallbladder carcinoma. Cell Physiol
Biochem. 34:1152–1165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee TH, Shank J, Cusson N and Kelliher MA:
The kinase activity of Rip1 is not required for tumor necrosis
factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or
for the ubiquitination of Rip1 by Traf2. J Biol Chem.
279:33185–33191. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jüttner S, Wissmann C, Jöns T, Vieth M,
Hertel J, Gretschel S, Schlag PM, Kemmner W and Höcker M: Vascular
endothelial growth factor-D and its receptor VEGFR-3: Two novel
independent prognostic markers in gastric adenocarcinoma. J Clin
Oncol. 24:228–240. 2006. View Article : Google Scholar
|
|
28
|
Choi JH, Oh YH, Park YW, Baik HK, Lee YY
and Kim IS: Correlation of vascular endothelial growth factor-D
expression and VEGFR-3-positive vessel density with lymph node
metastasis in gastric carcinoma. J Korean Med Sci. 23:592–597.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen Y, Jiang L, She F, Tang N, Wang X, Li
X, Han S and Zhu J: Vascular endothelial growth factor-C promotes
the growth and invasion of gallbladder cancer via an autocrine
mechanism. Mol Cell Biochem. 345:77–89. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bantounas I, Phylactou LA and Uney JB: RNA
interference and the use of small interfering RNA to study gene
function in mammalian systems. J Mol Endocrinol. 33:545–557. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kawasaki H and Taira K: Short hairpin type
of dsRNAs that are controlled by tRNA (Val) promoter significantly
induce RNAi-mediated gene silencing in the cytoplasm of human
cells. Nucleic Acids Res. 31:700–707. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Caplen NJ and Mousses S: Short interfering
RNA (siRNA)-mediated RNA interference (RNAi) in human cells. Ann NY
Acad Sci. 1002:56–62. 2003. View Article : Google Scholar
|
|
33
|
Yang B, Tuo S, Tuo CW, Zhang N and Liu QZ:
Establishment of a nude mouse model of highly metastatic gastric
lymphoma constructed with orthotopic transplantation of surgical
specimen. Zhonghua Wei Chang Wai Ke Za Zhi. 13:436–439.
2010.PubMed/NCBI
|