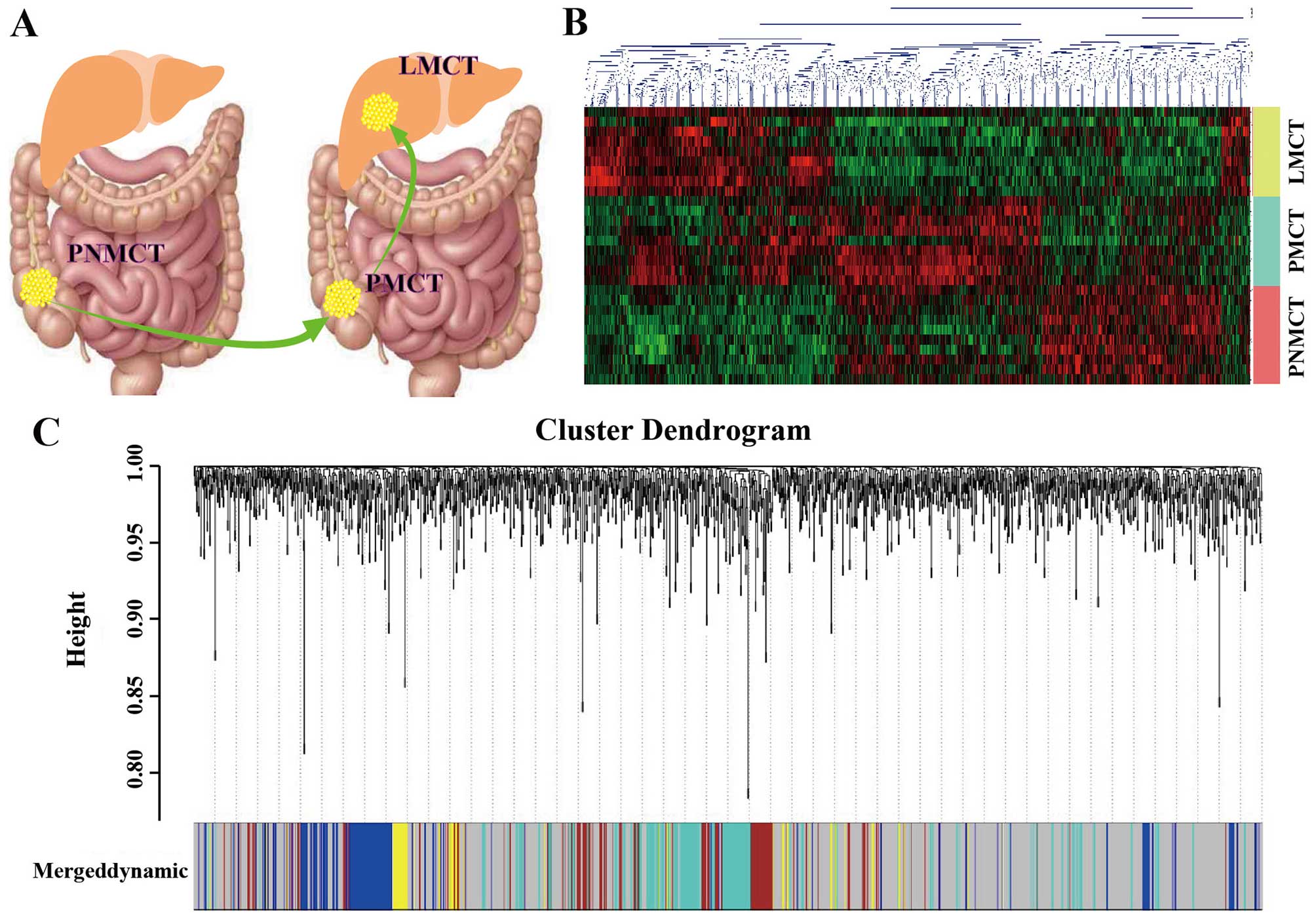
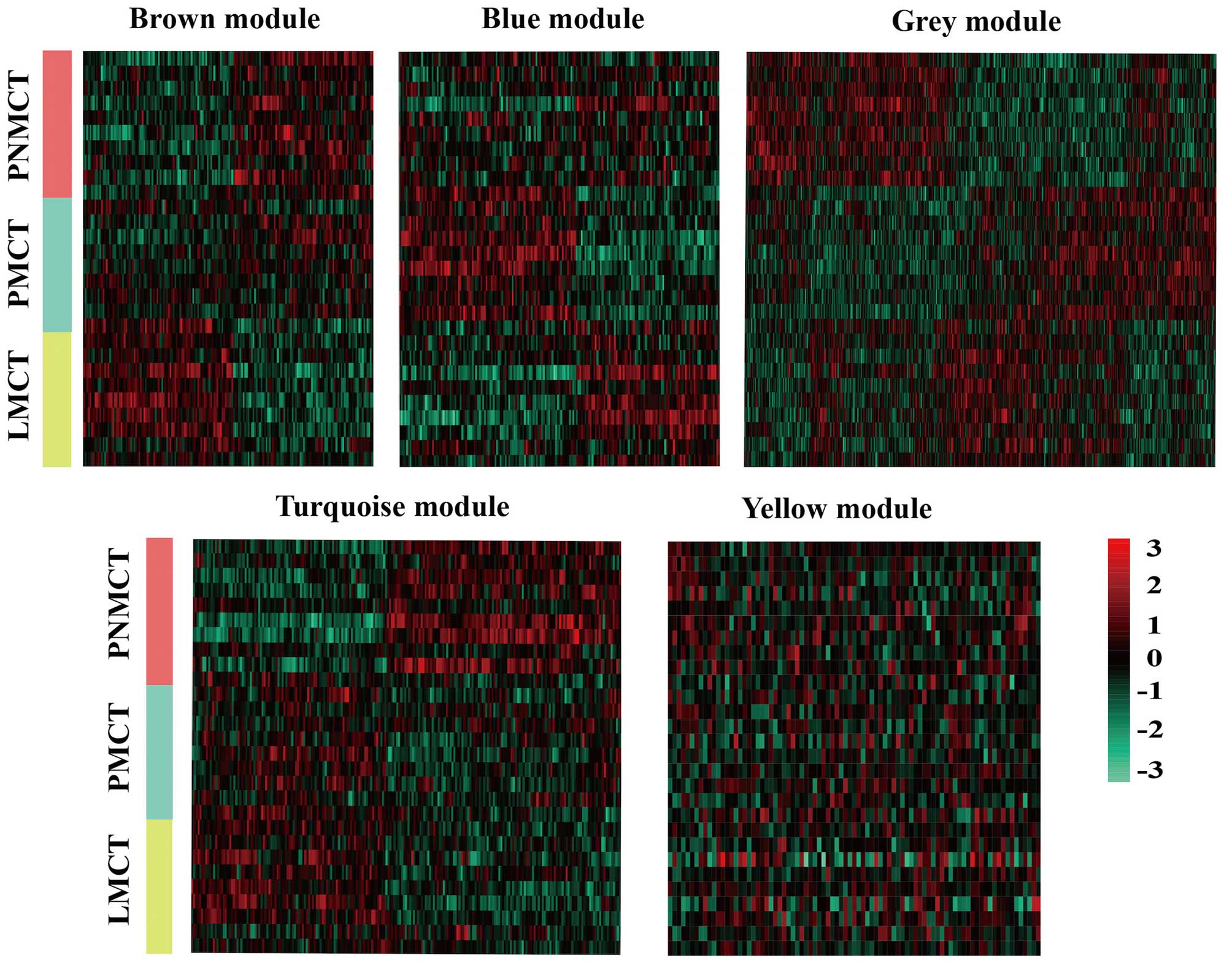
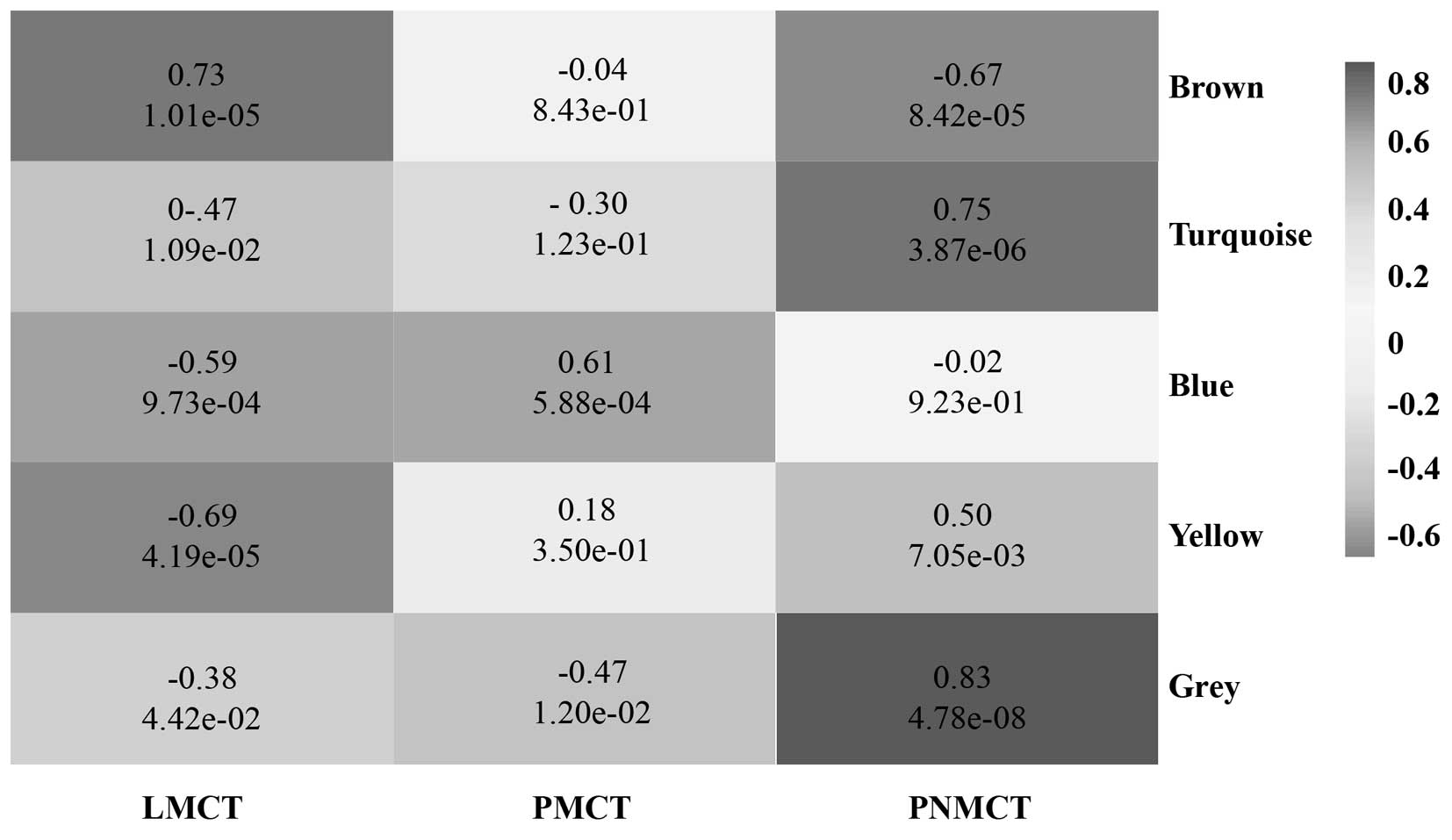
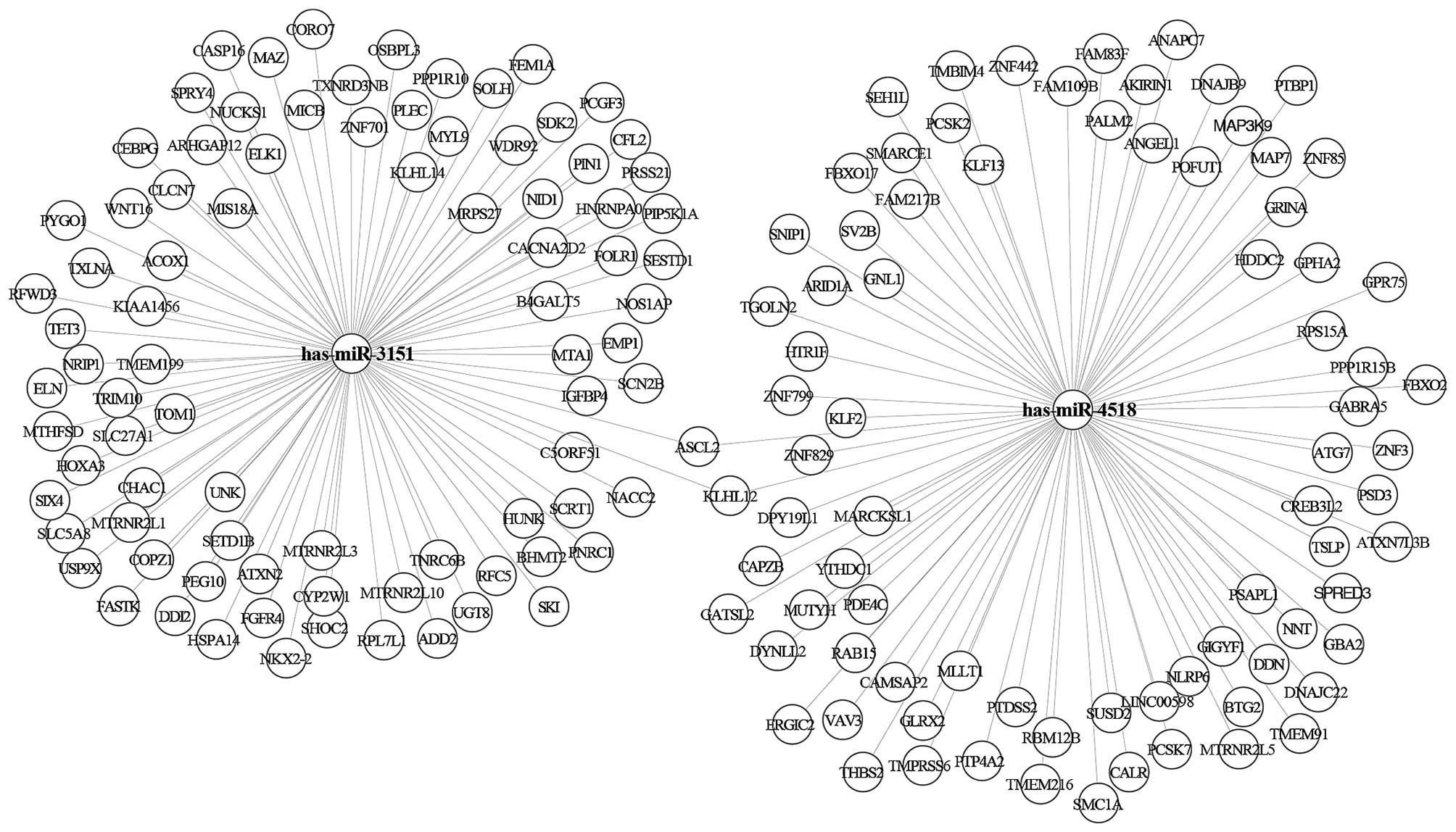
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wiseman M: The second World Cancer
Research Fund/American Institute for Cancer Research expert report.
Food, nutrition, physical activity, and the prevention of cancer: A
global perspective. Proc Nutr Soc. 67:253–256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
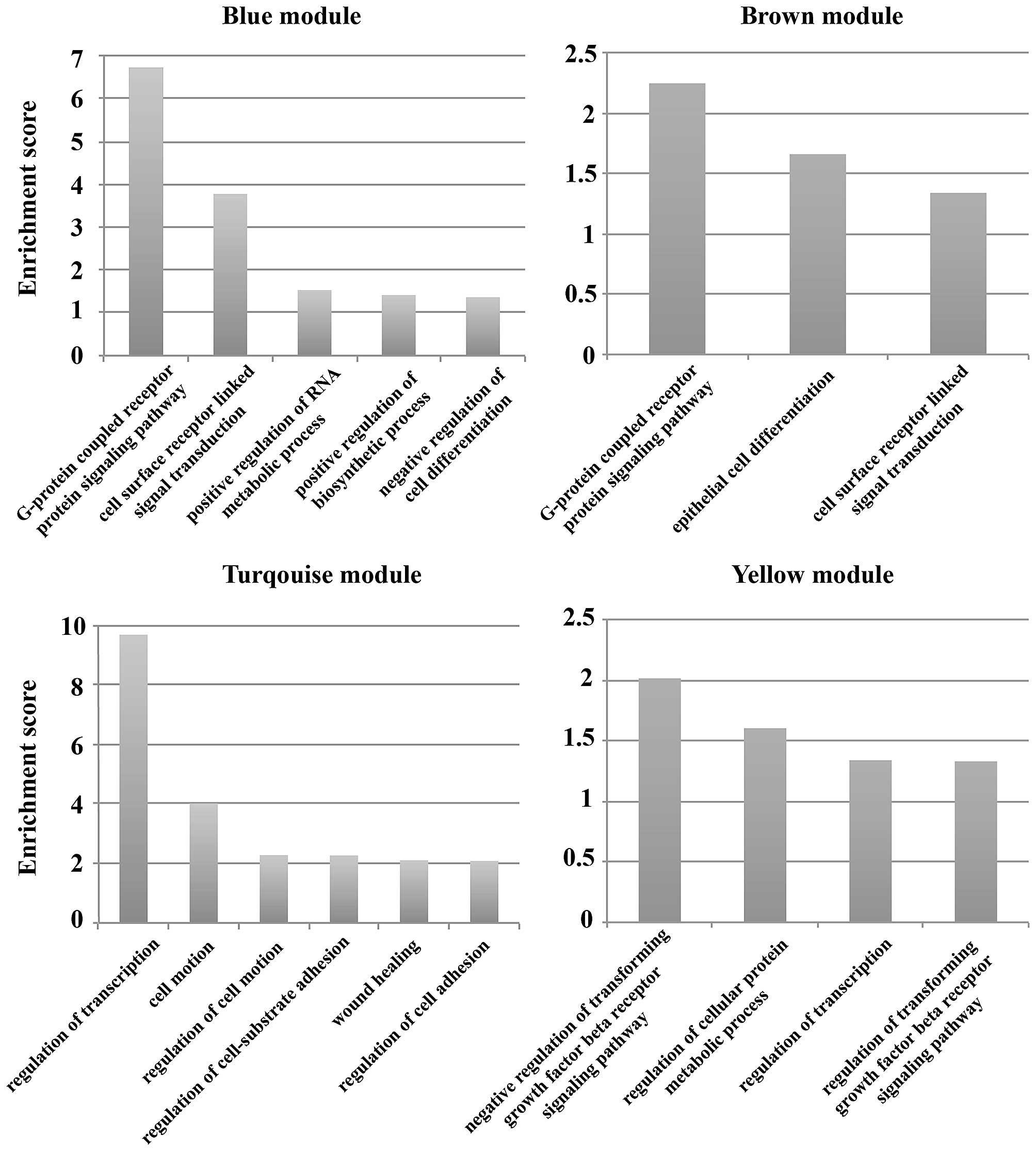
3
|
Pasetto LM, Jirillo A, Iadicicco G, Rossi
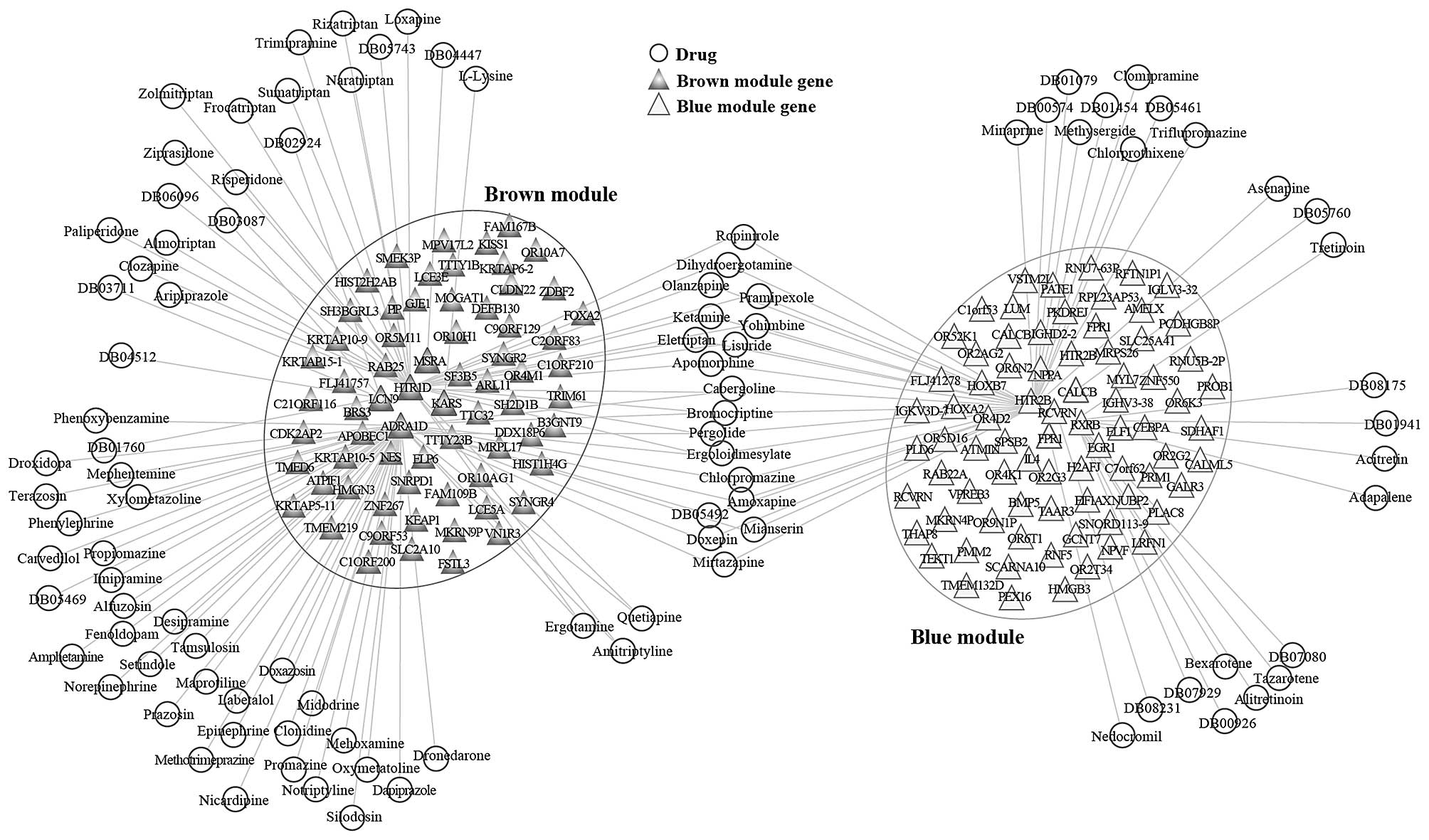
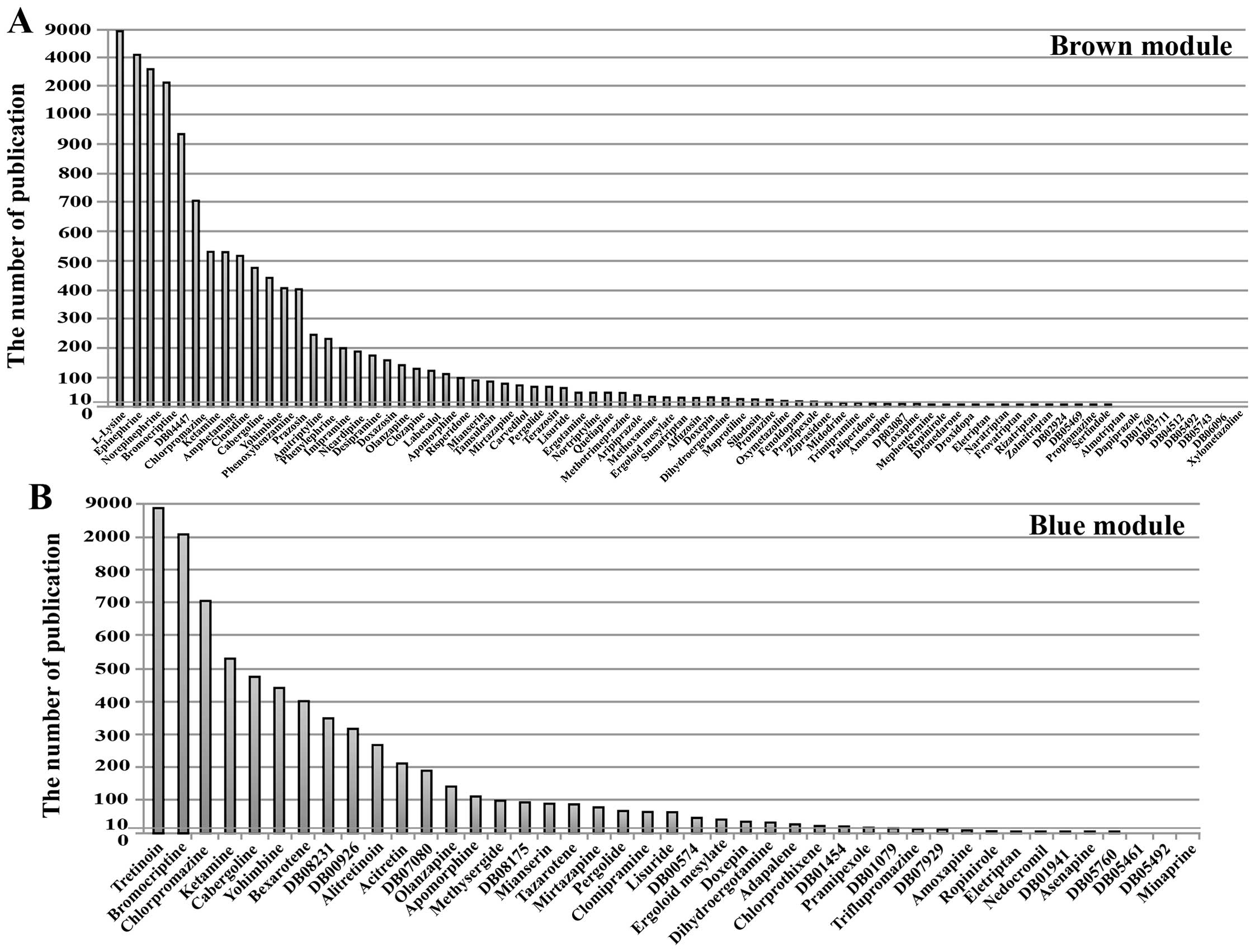
E, Paris MK and Monfardini S: FOLFOX versus FOLFIRI: A comparison
of regimens in the treatment of colorectal cancer metastases.
Anticancer Res. 25B:563–576. 2005.
|
|
4
|
Zhu D, Ren L and Xu J: Interpretation of
guidelines for the diagnosis and comprehensive treatment of
colorectal cancer liver metastases in China (v2013). Zhonghua Wei
Chang Wai Ke Za Zhi. 17:525–529. 2014.(In Chinese). PubMed/NCBI
|
|
5
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Akgül Ö, Çetinkaya E, Ersöz Ş and Tez M:
Role of surgery in colorectal cancer liver metastases. World J
Gastroenterol. 20:6113–6122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Postriganova N, Kazaryan AM, Rosok BI,
Fretland A, Barkhatov L and Edwin B: Margin status after
laparoscopic resection of colorectal liver metastases: does a
narrow resection margin have an influence on survival and local
recurrence? HPB (Oxford). 16:822–829. 2014. View Article : Google Scholar
|
|
8
|
Pan JG, Liu M and Zhou X: Relationship
between lower urinary tract symptoms and metabolic syndrome in a
Chinese male population. J Endocrinol Invest. 37:339–344. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Simpson AL, Leal JN, Pugalenthi A, Allen
PJ, DeMatteo RP, Fong Y, Gönen M, Jarnagin WR, Kingham TP, Miga MI,
et al: Chemotherapy-induced splenic volume increase is
independently associated with major complications after hepatic
resection for metastatic colorectal cancer. J Am Coll Surg.
220:271–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimada Y: Chemotherapy and
molecular-targeted treatment for unresectable hepatic metastases: A
Japanese perspective. J Hepatobiliary Pancreat Sci. 19:515–522.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang PI and Marcotte EM: It’s the machine
that matters: Predicting gene function and phenotype from protein
networks. J Proteomics. 73:2277–2289. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hedges SB: The origin and evolution of
model organisms. Nat Rev Genet. 3:838–849. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amrine KC, Blanco-Ulate B and Cantu D:
Discovery of core biotic stress responsive genes in Arabidopsis by
weighted gene co-expression network analysis. PLoS One.
10:e01187312015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Law V, Knox C, Djoumbou Y, Jewison T, Guo
AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al:
DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids
Res. 42(D1): D1091–D1097. 2014. View Article : Google Scholar :
|
|
15
|
Song WM and Zhang B: Multiscale Embedded
Gene Co-expression Network Analysis. PLOS Comput Biol.
11:e10045742015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang X, Jin R, Qu G, Wang X, Li Z, Yuan Z,
Zhao C, Siwko S, Shi T, Wang P, et al: GPR116, an adhesion
G-protein-coupled receptor, promotes breast cancer metastasis via
the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res. 73:6206–6218.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan M, Li H, Zhu M, Zhao F, Zhang L, Chen
T, Jiang G, Xie H, Cui Y, Yao M, et al: G protein-coupled receptor
87 (GPR87) promotes the growth and metastasis of CD133(+) cancer
stem-like cells in hepatocellular carcinoma. PLoS One.
8:e610562013. View Article : Google Scholar
|
|
18
|
Kim KB, Yi JS, Nguyen N, Lee JH, Kwon YC,
Ahn BY, Cho H, Kim YK, Yoo HJ, Lee JS, et al: Cell-surface receptor
for complement component C1q (gC1qR) is a key regulator for
lamellipodia formation and cancer metastasis. J Biol Chem.
286:23093–23101. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo QH, Yang HJ and Wang SD: Olanzapine
inhibits the proliferation and induces the differentiation of
glioma stem-like cells through modulating the Wnt signaling pathway
in vitro. Eur Rev Med Pharmacol Sci. 19:2406–2415. 2015.PubMed/NCBI
|
|
20
|
Lee WY, Lee WT, Cheng CH, Chen KC, Chou
CM, Chung CH, Sun MS, Cheng HW, Ho MN and Lin CW: Repositioning
antipsychotic chlorpromazine for treating colorectal cancer by
inhibiting sirtuin 1. Oncotarget. 6:27580–27595. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grisoli F, Vincentelli F, Foa J, Lavail G
and Salamon G: Effect of bromocriptine on brain metastasis in
breast cancer. Lancet. 2:745–746. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lissoni P, Vaghi M, Villa S, Bodraska A,
Cerizza L, Tancini G and Gardani GS: Antiprolactinemic approach in
the treatment of metastatic breast cancer: A phase II study of
polyneuroendocrine therapy with LHRH-analogue, tamoxifen and the
long-acting antiprolactinemic drug cabergoline. Anticancer Res.
23B:733–736. 2003.
|
|
23
|
O‘Dwyer PJ, Ravikumar TS, McCabe DP and
Steele G Jr: Effect of 13-cis-retinoic acid on tumor prevention,
tumor growth, and metastasis in experimental colon cancer. J Surg
Res. 43:550–557. 1987. View Article : Google Scholar
|
|
24
|
Adachi Y, Itoh F, Yamamoto H, Iku S,
Matsuno K, Arimura Y and Imai K: Retinoic acids reduce matrilysin
(matrix metalloproteinase 7) and inhibit tumor cell invasion in
human colon cancer. Tumour Biol. 22:247–253. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sarrouilhe D, Clarhaut J, Defamie N and
Mesnil M: Serotonin and cancer: What is the link? Curr Mol Med.
15:62–77. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lamm V, Hara H, Mammen A, Dhaliwal D and
Cooper DK: Corneal blindness and xenotransplantation.
Xenotransplantation. 21:99–114. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morelli MB, Amantini C, Nabissi M,
Liberati S, Cardinali C, Farfariello V, Tomassoni D, Quaglia W,
Piergentili A, Bonifazi A, et al: Cross-talk between
alpha1D-adrenoceptors and transient receptor potential vanilloid
type 1 triggers prostate cancer cell proliferation. BMC Cancer.
14:9212014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Yang Y, Chen L and Zhang J:
Expression analysis of genes and pathways associated with liver
metastases of the uveal melanoma. BMC Med Genet. 15:292014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soll C, Jang JH, Riener MO, Moritz W, Wild
PJ, Graf R and Clavien PA: Serotonin promotes tumor growth in human
hepatocellular cancer. Hepatology. 51:1244–1254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu RZ, Graham K, Glubrecht DD, Lai R,
Mackey JR and Godbout R: A fatty acid-binding protein 7/RXRβ
pathway enhances survival and proliferation in triple-negative
breast cancer. J Pathol. 228:310–321. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng CH, Jiang YZ, Tai AS, Liu CB, Peng
SC, Liao CT, Yen TC and Hsieh WP: Causal inference of gene
regulation with subnetwork assembly from genetical genomics data.
Nucleic Acids Res. 42:2803–2819. 2014. View Article : Google Scholar :
|
|
32
|
Are C, Simms N, Rajput A and Brattain M:
The role of transforming growth factor-beta in suppression of
hepatic metastasis from colon cancer. HPB (Oxford). 12:498–506.
2010. View Article : Google Scholar
|
|
33
|
Chai J, Wang S, Han D, Dong W, Xie C and
Guo H: MicroRNA-455 inhibits proliferation and invasion of
colorectal cancer by targeting RAF proto-oncogene
serine/threonine-protein kinase. Tumour Biol. 36:1313–1321. 2015.
View Article : Google Scholar
|
|
34
|
Wu K, He Y, Li G and Peng J: Expression
and proliferative regulation of miR-204 related to mitochondrial
transcription factor A in colon cancer. Zhonghua Wei Chang Wai Ke
Za Zhi. 18:1041–1046. 2015.(In Chinese). PubMed/NCBI
|
|
35
|
Zhang H, Li W, Nan F, Ren F, Wang H, Xu Y
and Zhang F: MicroRNA expression profile of colon cancer stem-like
cells in HT29 adenocarcinoma cell line. Biochem Biophys Res Commun.
404:273–278. 2011. View Article : Google Scholar
|