Introduction
Neuroblastoma is the most common solid tumor in
children less than 5 years old, and is derived from primordial
neural crest cells that eventually reside in the sympathetic
nervous system (1,2). Amplification of a 130 kb core genomic
DNA region containing the MYCN oncogene, the MYCN
antisense NCYM gene and the long non-coding RNA
lncUSMycN gene, occurs in more than a quarter of human
neuroblastoma tissues (3). More
than 70% of patients with neuroblastoma associated with the
amplification of the 130 kb core genomic DNA region die of the
disease in spite of current intensive multimodal therapies
(4,5).
The MYCN oncogene encodes the transcription
factor N-Myc which plays important roles in cell proliferation and
differentiation during embryonic development (6), and induces neuroblastoma initiation
and progression by regulating target gene expression (7).
Long non-coding RNAs are transcripts longer than 200
nucleotides without protein-coding potential, and can be divided
into five different classes according to their gene position
relevant to neighbouring protein-coding genes: sense, antisense,
bidirectional, intronic and intergenic (8,9).
Long non-coding RNAs have recently emerged as important regulators
of gene transcription, malignant transformation, tumor initiation,
progression and metastasis (10,11).
For example, lincRNA-p21 is activated by the tumor suppressor p53,
represses the transcription of the genes that interfere with
apoptosis, and plays a critical role in p53-mediated apoptosis
(12). The long non-coding RNA
NORAD is induced after DNA damage, and maintains genomic stability
by sequestering PUMILIO proteins (13). We have recently shown that the long
non-coding RNA lncUSMycN increases N-Myc mRNA expression by
interacting with the RNA-binding protein NonO (14).
While N-Myc has been extensively studied, the
function of the antisense NCYM gene did not attract research
until recently. NCYM has been shown to stabilize N-Myc protein by
inhibiting the activity of GSK3β, a kinase that promotes N-Myc
protein degradation (15), and to
promote calpain-mediated Myc-nick production in human neuroblastoma
cells (16). In addition, NCYM
plays an essential role in neuroblastoma cell metastasis (15). In this study, we examined how
NCYM gene expression was regulated, whether NCYM regulated
the expression of its neighbouring protein-coding gene MYCN,
and whether a high level of NCYM expression in human neuroblastoma
tissues was a marker for poor patient survival.
Materials and methods
Cell culture
Neuroblastoma BE(2)-C cells were cultured in
Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%
fetal calf serum. Kelly cells were cultured in Roswell Park
Memorial Institute (RPMI)-1640 medium supplemented with 10% fetal
calf serum and 1% L-glutamine. All cell lines were maintained in a
humidified incubator at 37°C and 5% CO2 in air. The
identity of all cell lines was authenticated by small tandem repeat
profiling conducted at the Garvan Institute or Cellbank
Australia.
siRNA transient transfection
Tumor cells were transfected with siRNAs using
Lipofectamine 2000 reagent according to the manufacturer’s
instructions (Invitrogen, Carlsbad, CA, USA). Pre-designed control
siRNA and siRNA specifically targeting NCYM or N-Myc were purchased
from Qiagen (Qiagen, Hamburg, Germany), and the lncUSMycN siRNAs
were custom synthesized by Dharmacon (Lafayette, CO, USA) (14,17).
Quantitative real-time RT-PCR
NCYM and N-Myc expression was examined by
quantitative real-time RT-PCR with Power SYBR Green Master Mix
(Life Technologies, Grand Island, NY, USA) as a fluorescent dye on
ABI-7900 (Applied Biosystems, Grand Island, NY, USA) according to
the manufacturer’s instructions and as we described previously
(18,19). The expression of genes of interest
was determined after normalisation according the expression level
of the housekeeping gene β-actin in the total RNA samples.
Immunoblotting
Cells were lysed, protein was extracted and
separated by gel electrophoresis. After western transfer, membranes
were probed with an anti-N-Myc (1:2,000) (Santa Cruz Biotech, Santa
Cruz, CA, USA) antibody followed by a horseradish
peroxidase-conjugated anti-mouse (1:10,000) antibody (Santa Cruz
Biotech). Protein bands were visualized with SuperSignal (Pierce,
Rockford, IL, USA). Finally, the membranes were re-probed with an
anti-actin antibody (1:10,000; Sigma, St. Louis, MO, USA) as
loading controls.
Cell proliferation assays
Cell proliferation was examined with Alamar blue
assays as we described previously (20). Briefly, cells were plated into
96-well plates and transfected with various siRNAs. Ninety-one
hours later, cells were incubated with Alamar blue (Invitrogen) for
five hours, and plates were read on a micro-plate reader at 570/595
nm. Results were calculated according to the readings (optical
density absorbance units) and expressed as percentage changes in
cell numbers.
RNA immunoprecipitation assays
RNA immunoprecipitation assays were performed using
Magna RIP kit from Merck Millipore according to the manufacturer’s
instructions (Merck Millipore, Billerica, MA, USA), with 5 μg of
control IgG or anti-NonO antibody for immunoprecipitation and
primers targeting NCYM or the negative control U1 RNA for RT-PCR
(14).
Patient tumor sample analyses
NCYM, N-Myc, NonO and lncUSMycN expression in
neuroblastoma tissues was analysed in 88 (Versteeg dataset) and 476
(Kocak dataset) human neuroblastoma samples in the publicly
available gene expression databases (http://r2.amc.nl).
Clinical information for the 88 patients in the Versteeg dataset
was directly downloaded from http://r2.amc.nl,
and clinical information for the 476 patients in the Kocak dataset
was extracted from the authors’ reports (21,22).
Correlation between NCYM expression and N-Myc, NonO
and lncUSMycN expression in human neuroblastoma tissues was
examined with Pearson’s correlation. For patient prognosis studies,
overall survival was defined as the time from diagnosis until
death, or otherwise as the time until last contact. The patient
cohort was dichotomized into two groups (low versus high NCYM
expression). Survival analyses were performed according to the
method of Kaplan and Meier and comparisons of survival curves were
made using two-sided log-rank tests (23).
Statistical analyses
All data for statistical analysis were calculated as
mean ± standard error. Differences were analyzed for significance
using two-sided unpaired t-test for two groups, or multiple
comparison one-way analysis of variance (ANOVA) for more than two
groups. A probability value of ≤0.05 was considered
significant.
Results
The long non-coding RNA lncUSMycN
upregulates NCYM expression by inducing its gene transcription
The lncUSMycN, NCYM and MYCN genes are
within the 130 kb core genomic DNA region amplified in a quarter of
human neuroblastoma tissues (3).
As long non-coding RNAs are well-known to upregulate the expression
of neighboring genes in cis (24), we investigated whether lncUSMycN
modulated NCYM gene expression. BE(2)-C and Kelly
neuroblastoma cells were transfected with control siRNA or two
independent siRNAs targeting different regions of lncUSMycN. The
lncUSMycN siRNAs had been previously validated to knock down
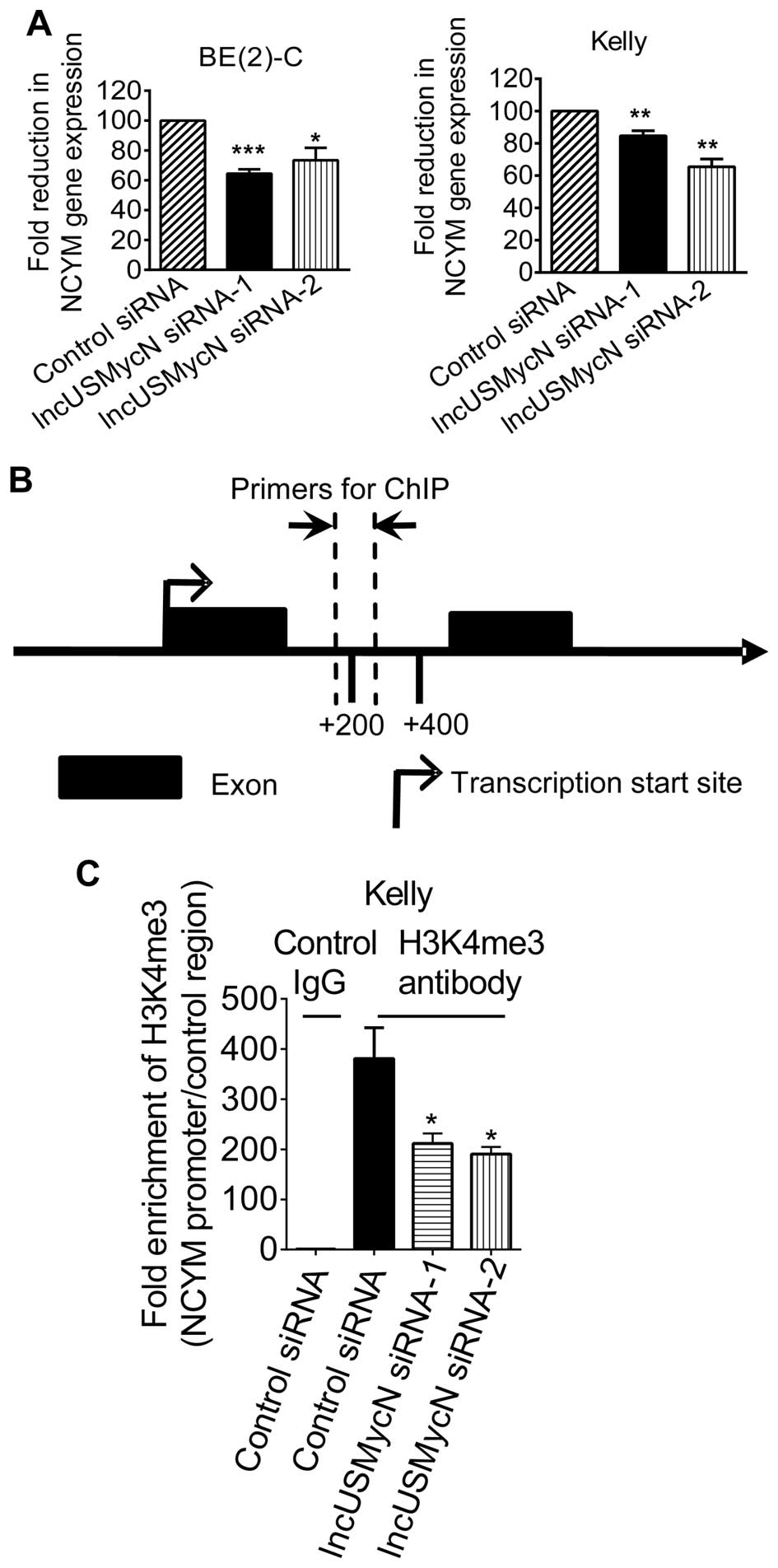
lncUS-MycN RNA expression in BE(2)-C and Kelly cells (14). As shown in Fig. 1A, transfection with lncUSMycN
siRNAs significantly reduced NCYM RNA expression in the MYCN
oncogene-amplified BE(2)-C and Kelly neuroblastoma cells.
To understand whether lncUSMycN upregulated NCYM
expression through activating NCYM gene transcription, we performed
chromatin immunoprecipitation (ChIP) assays with a control antibody
(Ab) or an antibody against trimethylated histone H3 lysine 4
(H3K4me3), a marker for active gene transcription. As shown in
Fig. 1B and C, the ChIP assays
showed that knocking down lncUSMycN expression with siRNAs reduced
histone H3K4 trimethylation at the NCYM gene promoter
(intron 1). Taken together, the data suggest that lncUSMycN
upregulates NCYM expression by enhancing its gene
transcription.
NCYM upregulates N-Myc expression and
induces neuroblastoma cell proliferation
As the NCYM gene is antisense to the
MYCN oncogene, we examined whether NCYM regulated N-Myc
expression. BE(2)-C and Kelly cells were transfected with two
independent siRNAs targeting different regions of NYCM RNA or a
control siRNA for 48 h. As shown in Fig. 2A, transfection with NCYM siRNAs
efficiently reduced NCYM expression. RT-PCR analysis showed that
knocking down NCYM expression with siRNAs reduced N-Myc mRNA
expression (Fig. 2B), and
immunoblot analysis showed that knocking down NCYM expression also
reduced N-Myc protein expression (Fig.
2C). Consistent with the RT-PCR data, the reduction in N-Myc
protein expression by NCYM siRNA-1 was weaker than NCYM siRNA-2,
indicating variation in the efficacy of NCYM siRNA-1 and NCYM
siRNA-2.
As N-Myc is well-known to induce neuroblastoma cell
proliferation, we performed Alamar blue cell proliferation assays
in BE(2)-C and Kelly cells after transfection with control siRNA,
NCYM siRNA-1 or NCYM siRNA-2 for 48 h. Alamar blue assays showed
that knocking down NCYM expression with siRNAs reduced the number
of neuroblastoma cells (Fig. 2D).
Taken together, these data suggest that NCYM upregulates N-Myc
expression, leading to neuroblastoma cell proliferation.
NCYM RNA binds to the RNA-binding protein
NonO
We have previously shown that the RNA-binding
protein NonO upregulates N-Myc mRNA expression (14). Next we performed RNA
immunoprecipitation assays with an anti-NonO antibody or control
IgG, followed by RT-PCR analysis of NCYM RNA. As shown in Fig. 3, the anti-NonO antibody, compared
with the control IgG, efficiently immunoprecipitated NCYM RNA,
compared with the negative control U1 RNA. The data suggest that
NCYM RNA forms a complex with NonO protein, leading to N-Myc mRNA
upregulation.
BET bromodomain inhibitors reduce NCYM
gene expression
The BET bromodomain inhibitors JQ1 and I-BET151 have
been shown to reduce oncogene expression (25,26).
We next examined whether the BET bromodomain inhibitors modulated
NCYM expression. BE(2)-C and Kelly cells were treated with vehicle
control or the BET bromodomain inhibitor JQ1 or I-BET151. RT-PCR
analysis showed that treatment with JQ1 (Fig. 4A) or I-BET151 (Fig. 4B) reduced NCYM RNA expression in
both BE(2)-C and Kelly cells. The data suggest that NCYM gene
expression is regulated by BET bromodomain proteins, and that BET
bromodomain inhibitors can be employed to suppress NCYM
expression.
NCYM expression in neuroblastoma tissues
positively correlates with N-Myc, NonO and lncUSMycN expression and
poor patient prognoses
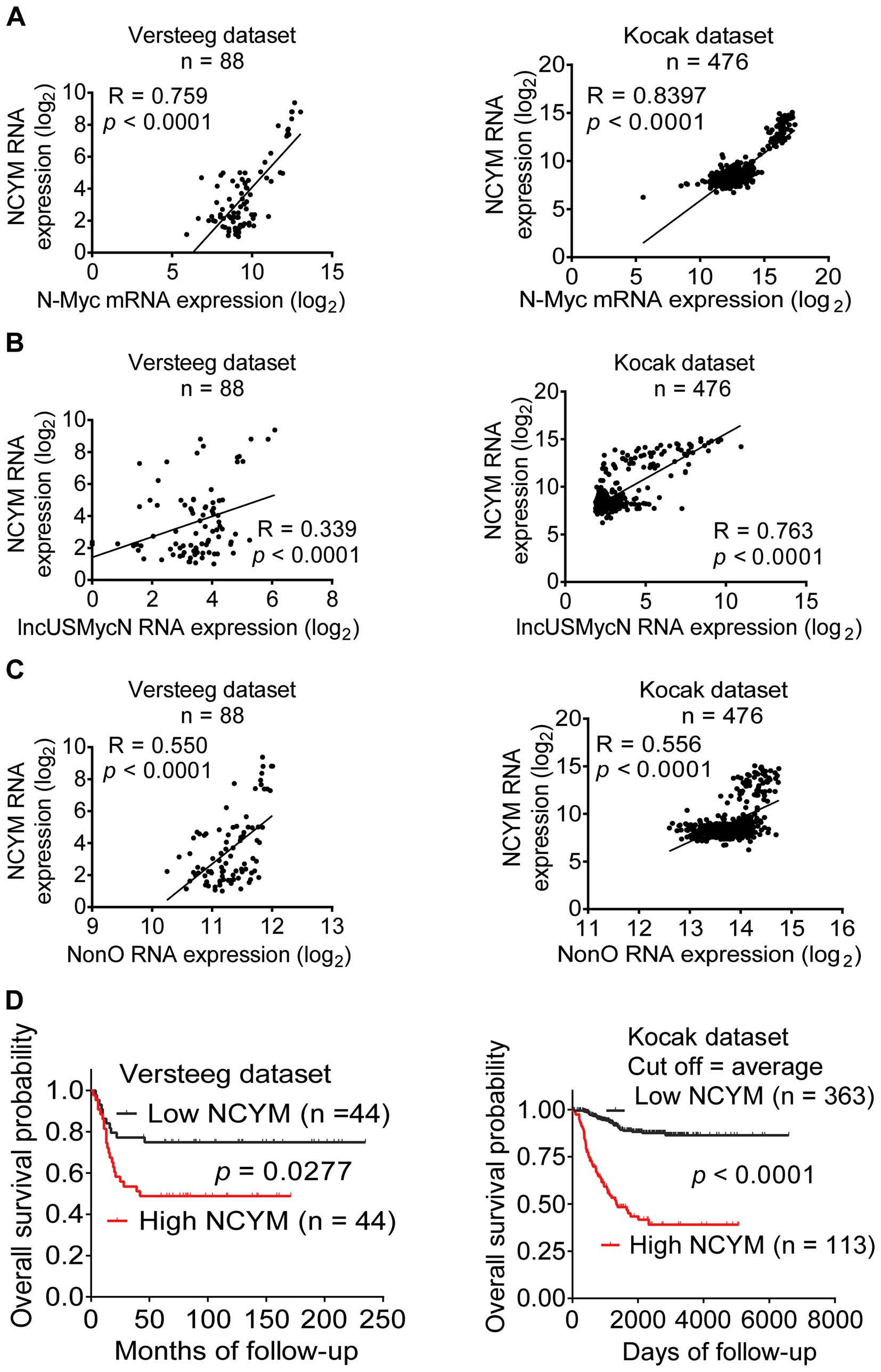
We finally examined correlation of NCYM expression
in human neuroblastoma tissues with N-Myc, NonO and lncUSMycN
expression and patient prognosis, using the publicly available
(http://r2.amc.nl) Versteeg and Kocak (21,22)
microarray gene expression datasets. Probes for NCYM, N-Myc, NonO
and lncUSMycN in the Affymetrix HG-U133_Plus_2 microarray in the
Versteeg dataset were 207028_at, 209757_s_at, 200057_s_at and
244438_at, respectively. Probes for NCYM, N-Myc, NonO and lncUSMycN
in the Agilent Technologies oligonucleotide microarrays in the
Kocak dataset were UKv4_A_23_P67952, UKv4_A_24_ P94402,
UKv4_A_24_P151727 and UKv4_A_24_P486173, respectively. As shown in
Fig. 5A–C, NCYM RNA expression was
positively correlated with N-Myc, NonO and lncUS-MycN in the
cohorts of 88 and 476 neuroblastoma patients, respectively.
Kaplan-Meier analysis showed that high levels of NCYM expression in
neuroblastoma tissues correlated with poor patient prognoses
(Fig. 5D). Taken together, the
data suggest that high levels of NCYM expression in human
neuroblastoma tissues positively correlate with N-Myc, NonO and
lncUSMycN expression and predict poor patient survival.
Discussion
The MYCN antisense gene NCYM, like the
MYCN and the long non-coding RNA lncUSMYCN genes, is
amplified in approximately a quarter of human neuroblastoma tissues
(15). Long non-coding RNAs are
now well-known to exert biological effects through both
transcriptional and post-transcriptional mechanisms and to regulate
gene transcription both in cis and in trans (24,27).
We have recently reported that the long non-coding RNA lncUSMycN
upregulates N-Myc mRNA expression by binding to the RNA-binding
protein NonO, but does not directly regulate MYCN gene
transcription since knocking down lncUSMycN expression does not
have an effect on histone H3K4 trimethylation, a marker for active
gene transcription, at the MYCN gene promoter (14). In this study, we found that
lncUSMycN upregulates NCYM expression in human neuroblastoma cells,
and that knocking down lncUSMycN expression reduces histone H3K4
trimethylation at the NCYM gene promoter. The data suggest
that lncUS-MycN upregulates NCYM expression by activating its gene
transcription.
This study demonstrates that NCYM upregulates N-Myc
mRNA and protein expression, and that NCYM RNA binds to the
RNA-binding protein NonO. As our previous study shows that NonO
protein binds to N-Myc mRNA and post-transcriptionally upregulates
N-Myc mRNA expression (14), we
propose that NCYM RNA upregulates N-Myc mRNA expression by binding
to NonO protein. In addition, we have confirmed that knocking down
NCYM expression leads to neuroblastoma cell growth inhibition.
Taken together, the data suggest that NCYM RNA overexpression
upregulates N-Myc mRNA expression by binding to NonO protein,
leading to neuroblastoma cell proliferation.
This study shows that high levels of NCYM expression
in two independent cohorts of human neuroblastoma tissues
positively correlate with high levels of N-Myc, NonO and lncUSMycN
expression. Importantly, Kaplan-Meier analysis shows that high
levels of NCYM expression in neuroblastoma tissues correlate with
poor prognosis in the two cohorts of 88 and 476 patients. These
data therefore demonstrate that high levels of NCYM expression in
human neuroblastoma tissues can be employed as a marker for poor
prognosis and a therapeutic target.
Histone lysine acetylation plays a pivotal role in
regulating chromatin dynamics and gene transcription. The BET
bromodomain is a conserved structural unit in chromatin-associated
proteins and histone acetyltranferases, known to recognize
acetyl-lysine residues on proteins (28). BET bromodomain inhibitors have been
shown to exert anticancer effects against neuroblastoma by blocking
BET bromodomain protein binding to the MYCN gene intron 1, a
region shared by the MYCN and the NCYM genes, and
consequently reducing N-Myc expression (17,26).
In this study, we found that suppression of BET bromodomain
proteins by the BET bromodomain inhibitors JQ1 and I-BET151
considerably reduces NCYM expression, and that knocking down NCYM
expression with siRNAs considerably reduces neuroblastoma cell
proliferation. Our data suggest that BET bromodomain inhibitors
reduce N-Myc expression directly by repressing MYCN gene
transcription and indirectly by repressing NCYM gene
transcription, and exert anticancer effects against neuroblastoma
by simultaneously blocking the expression of N-Myc and NCYM.
In conclusion, NCYM, MYCN and
lncUSMycN genes are co-amplified in human neuroblastoma. Our
data demonstrate that the long non-coding RNA lncUSMycN upregulates
NCYM expression by activating its gene transcription, and that NCYM
RNA upregulates N-Myc expression by binding to the RNA-binding
protein NonO. High levels of NCYM expression in human neuroblastoma
tissues correlate with high levels of N-Myc, NonO and lncUSMycN
expression and poor patient survival, and treatment with BET
bromodomain inhibitors reduces NCYM expression. Our findings
therefore provide further evidence for the application of BET
bromodomain inhibitors for the therapy of neuroblastoma
characterized by MYCN/NCYM gene locus amplification.
Acknowledgements
This study was supported by National Health and
Medical Research Council (NHMRC) Australia grants. T.L. is a
recipient of an Australian Research Council Future Fellowship.
Children’s Cancer Institute Australia is affiliated with UNSW
Australia and Sydney Children’s Hospital.
References
|
1
|
Colon NC and Chung DH: Neuroblastoma. Adv
Pediatr. 58:297–311. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matthay KK, George RE and Yu AL: Promising
therapeutic targets in neuroblastoma. Clin Cancer Res.
18:2740–2753. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reiter JL and Brodeur GM: High-resolution
mapping of a 130-kb core region of the MYCN amplicon in
neuroblastomas. Genomics. 32:97–103. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maris JM, Hogarty MD, Bagatell R and Cohn
SL: Neuroblastoma. Lancet. 369:2106–2120. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Matthay KK, Villablanca JG, Seeger RC,
Stram DO, Harris RE, Ramsay NK, Swift P, Shimada H, Black CT,
Brodeur GM, et al; Children’s Cancer Group. Treatment of high-risk
neuroblastoma with intensive chemotherapy, radiotherapy, autologous
bone marrow transplantation, and 13-cis-retinoic acid. N Engl J
Med. 341:1165–1173. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Knoepfler PS, Cheng PF and Eisenman RN:
N-myc is essential during neurogenesis for the rapid expansion of
progenitor cell populations and the inhibition of neuronal
differentiation. Genes Dev. 16:2699–2712. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang M and Weiss WA: Neuroblastoma and
MYCN. Cold Spring Harb Perspect Med. 3:a0144152013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO proteins. Cell.
164:69–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu PY, Erriquez D, Marshall GM, Tee AE,
Polly P, Wong M, Liu B, Bell JL, Zhang XD, Milazzo G, et al:
Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc
expression and neuroblastoma progression. J Natl Cancer Inst.
106:dju1132014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suenaga Y, Islam SM, Alagu J, Kaneko Y,
Kato M, Tanaka Y, Kawana H, Hossain S, Matsumoto D, Yamamoto M, et
al: NCYM, a Cis-antisense gene of MYCN, encodes a de novo evolved
protein that inhibits GSK3β resulting in the stabilization of MYCN
in human neuroblastomas. PLoS Genet. 10:e10039962014. View Article : Google Scholar
|
|
16
|
Shoji W, Suenaga Y, Kaneko Y, Islam SM,
Alagu J, Yokoi S, Nio M and Nakagawara A: NCYM promotes
calpain-mediated Myc-nick production in human MYCN-amplified
neuroblastoma cells. Biochem Biophys Res Commun. 461:501–506. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shahbazi J, Liu PY, Atmadibrata B, Bradner
JE, Marshall GM, Lock RB and Liu T: The bromodomain inhibitor JQ1
and the histone deacetylase inhibitor panobinostat synergistically
reduce N-Myc expression and induce anticancer effects. Clin Cancer
Res. 22:2534–2544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu PY, Xu N, Malyukova A, Scarlett CJ,
Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK, et al: The
histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death
Differ. 20:503–514. 2013. View Article : Google Scholar :
|
|
19
|
Tee AE, Liu B, Song R, Li J, Pasquier E,
Cheung BB, Jiang C, Marshall GM, Haber M, Norris MD, et al: The
long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by
up-regulating pro-angiogenic gene expression. Oncotarget.
7:8663–8675. 2016.PubMed/NCBI
|
|
20
|
Liu T, Liu PY, Tee AEL, Haber M, Norris
MD, Gleave ME and Marshall GM: Over-expression of clusterin is a
resistance factor to the anti-cancer effect of histone deacetylase
inhibitors. Eur J Cancer. 45:1846–1854. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kocak H, Ackermann S, Hero B, Kahlert Y,
Oberthuer A, Juraeva D, Roels F, Theissen J, Westermann F, Deubzer
H, et al: Hox-C9 activates the intrinsic pathway of apoptosis and
is associated with spontaneous regression in neuroblastoma. Cell
Death Dis. 4:e5862013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oberthuer A, Juraeva D, Li L, Kahlert Y,
Westermann F, Eils R, Berthold F, Shi L, Wolfinger RD, Fischer M,
et al: Comparison of performance of one-color and two-color
gene-expression analyses in predicting clinical endpoints of
neuroblastoma patients. Pharmacogenomics J. 10:258–266. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kaplan EL and Meier P: Nonparametric
estimation from incomplete observations. Breakthroughs in
Statistics. Kotz S and Johnson N: Springer; New York, NY: pp.
319–337. 1992, View Article : Google Scholar
|
|
24
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wyce A, Ganji G, Smitheman KN, Chung CW,
Korenchuk S, Bai Y, Barbash O, Le B, Craggs PD, McCabe MT, et al:
BET inhibition silences expression of MYCN and BCL2 and induces
cytotoxicity in neuroblastoma tumor models. PLoS One. 8:e729672013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Puissant A, Frumm SM, Alexe G, Bassil CF,
Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P,
et al: Targeting MYCN in neuroblastoma by BET bromodomain
inhibition. Cancer Discov. 3:308–323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sanchez R and Zhou MM: The role of human
bromodomains in chromatin biology and gene transcription. Curr Opin
Drug Discov Devel. 12:659–665. 2009.PubMed/NCBI
|