Introduction
Lung cancer is one of the leading threats to human
health globally. It is associated with high morbidity and mortality
and a poor survival rate (1).
Non-small cell lung cancer (NSCLC) accounts for approximately
85–90% of all lung cancers and compared with other types is
relatively insensitive to treatment (2). Numerous studies have investigated the
prevention and treatment of NSCLC; however, despite the gain in
knowledge it remains a largely intractable disease (3). One promising treatment is targeted at
the specific molecules borne by patients with mutations in
oncogenes such as epidermal growth factor receptor (EGFR),
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) and
anaplastic lymphoma kinase (ALK) (4). However, not all patients benefit and
many exhibit tolerance. Hence, a search for novel molecular
mechanisms involved in the etiology of NSCLC and its progression is
required, with the ultimate aim of developing targeted therapy.
In recent years, proteomic studies demonstrated that
more than 2000 proteins involved in diverse cellular processes
could be acetylated, indicating that lysine acetylation is a
promising target for cancer treatment (5–7).
Deacetylases such as sirtuin-3 (SIRT3), a primary mitochondrial
deacetylase, mediate a wide range of cellular processes involving
acetylation (8). SIRT3 is involved
in many diseases including diabetes, myocardial injury and cancer
(9–12). However, SIRT3 functions as either
an oncogene or an anti-oncogene in different cancers; the
distinction is associated with complex biologic networks of
signaling pathways depending upon the different genetic backgrounds
in different cancer types (12–16).
To date, there are still only few data available on
the relationship between SIRT3 and NSCLC. In the present study, we
investigated the relationship between SIRT3 and NSCLC in both
clinical samples and cell lines to determine whether SIRT3
correlates with malignancy and examined the underlying
mechanism.
Materials and methods
Patients and tissue specimens
NSCLC specimens accompanied by detailed clinical and
pathological data were collected from 70 NSCLC patients who were
diagnosed with NSCLC between April 2009 and January 2013 and
underwent surgical resection at the Department of Thoracic Surgery,
Tangdu Hospital, the Fourth Military Medical University (Xi'an,
China). Cancerous tissue and adjacent normal lung tissue was
obtained from each individual after resection of the tumor. All
tissue specimens (n=140) were snap-frozen in liquid nitrogen
immediately after the collection for subsequent analysis. The
cohort comprised 61 (87.1%) males and 9 (12.9%) females. The
average age was 60 years (range, 31–79 years). Mean survival time
was 21.1 months. Tumor stage was defined according to the
tumor-node-metastasis (TNM) classification of the American Joint
Committee on Cancer (AJCC)/Union for International Cancer Control
(UICC), the main tumor-staging system used in clinical practice and
research.
Cell culture
NSCLC cell lines H520 and SW900 were purchased from
the Cell Bank of the Chinese Academy of Sciences (Shanghai, China).
Cells were cultured in RPMI-1640 medium supplemented with 10% fetal
bovine serum (FBS), 100 mg/l penicillin, and 100 mg/l streptomycin
at 37°C in a humidified 5% CO2 atmosphere.
SIRT3 siRNA transfection
Briefly, SW900 and H520 cells were transfected with
SIRT3 siRNA and control siRNA using Lipofectamine 2000 according to
the manufacturer's instructions. SIRT3 siRNA1:
5′-CCAGCAUGAAAUACAUUUATT-3′, SIRT3 siRNA2:
5′-GCCCGACAUUGUGUUCUUUTT-3′; negative control siRNA:
5′-UUCUCCGAACGUGUCACGUTT-3′.
Plasmid construction and cell
transfection
SIRT3 full-length cDNA was synthesized from total
RNA extracted from H520 cells using RT-PCR. Primers were designed
as follows: forward, 5′-CCGCTCGAGATGGCGTTCTGGGGTTGG-3′ and reverse,
5′-CGCGGATCCCTATTTGTCTGGTCCATCAAGC-3′.
The cDNA was then cloned into pcDNA3.1 (−)
expression vector (Invitrogen, Carlsbad, CA, USA). The empty
pcDNA3.1 (−) was used as a control. H520 cells were transfected
with pcDNA3.1 (−)-SIRT3 or control empty pcDNA3.1 (−) using
Lipofectamine 2000 according to the manufacturer's
instructions.
Tissue microarray construction
The 140 specimens were fixed in 4% paraformaldehyde
for 24 h, dehydrated in a series of graded ethanol concentrations
and then embedded in paraffin. According to hematoxylin and
eosin-stained sections, representative areas were selected from
which to construct a tissue microarray (TMA) using a
tissue-arraying instrument (Quant Center Pannoramic MIDI). Each TMA
contained 70 cancerous tissues and 70 corresponding adjacent normal
tissues.
Immunohistochemistry
Paraformaldehyde-fixed and paraffin-embedded NSCLC
TMA sections (4 µm thick) were dewaxed in xylene and graded
alcohols, hydrated and washed in phosphate-buffered saline (PBS).
After pretreatment in a microwave oven, endogenous peroxidase was
inhibited with 3% hydrogen peroxide for 25 min. After blocking with
3% bovine serum albumin (BSA), slides were then incubated with the
primary antibody (anti-SIRT3, sc-99143; anti-Ki-67, #9449 Cell
Signaling Technology; anti-Phospho-Akt Ser473, #4060; Cell
Signaling Technology) overnight in a moist chamber at 4°C, washed
in PBS and incubated with the appropriate horseradish peroxidase
(HRP)-labeled goat anti-rabbit/mouse antibody. Slides were
developed with liquid 3, ′3-diaminobenzidine tetrahydrochloride
(DAB) + Substrate Chromogen System (Dako) and counterstained with
hematoxylin.
Immunofluorescence
Paraformaldehyde-fixed cells were incubated with
primary antibody (SIRT3, sc-99143; Akt, 60203–1-Ig; Proteintech,
Chicago, IL, USA) at 4°C overnight in a moist chamber, washed in
PBS and incubated with Alexa Fluor 488 and CY3-conjugated secondary
antibodies and 4′,6-diamidino-2-phenylindole (DAPI) was used to
stain the nucleus.
Immunohistochemistry evaluation
Semi-quantitative immunohistochemistry (IHC)
detection was used to determine the protein levels, according to
the intensity of staining and the percentage of positive cells. For
antigens other than Ki-67, an H-score was assigned, derived from H
= Σ(pi*i), in which 'pi' represents the percentage of positive
cells and 'i' represents the intensity (17). For Ki-67, the index was defined as
the percentage of Ki-67-positive cells among all the cells of the
TMA section. Slides were scanned and digitalized using a Pannoramic
MIDI (3DHISTECH, Ltd., Budapest, Hungary) and analyzed using a
Pannoramic Viewer v. 1.15.3 and NuclearQuant application for PV
v.2.0.0.46136, both manufactured by 3DHISTECH (18).
Selection of cut-off score
The cut-off value used to differentiate SIRT3
expression into high and low levels was defined using a receiver
operating characteristic (ROC) curve using the 0.1 criterion
(19). The value closest to the
point with both maximum sensitivity and specificity [i.e., the
point (0.0, 0.1) on the curve] was selected as the cut-off value,
leading to the largest number of tumors correctly classified as
having or not having the clinical outcome (20,21).
High SIRT3 expressing samples were those with scores above the
cut-off value, while the low samples had scores below or equal to
the value.
Quantitative real-time PCR
Total RNA was extracted from the frozen tissue
samples using RNAiso reagent (Takara Bio, Tokyo, Japan) according
to the manufacturer's instructions, and then reverse transcribed to
cDNA using PrimeScript RT Master Mix (Takara Bio). Levels of SIRT3
and β-actin were examined by real-time PCR (qPCR) using SYBR Premix
Ex Taq II (Tli RNaseH Plus; Takara Bio) with a CFX96 Real-Time PCR
detection system (Bio-Rad Laboratories, Hercules, CA, USA). Primers
were designed as follows: SIRT3, forward, 5′-GCATCCCTGCCTCAAAGC-3′
and reverse, 5′-GTCAGCCCGAATGTCCTC-3′; β-actin, forward,
5′-GATCATTGCTCCTCCTGAGC-3′ and reverse, 5′-ACTCCTGCTTGCTGATCCAC-3′.
Conditions were as follows: one cycle of 95°C for 3 min, followed
by 45 amplification cycles at 95°C for 10 sec, and annealing and
elongation at 55°C for 30 sec. The relative expression of SIRT3 was
normalized to endogenous β-actin using the comparative threshold
cycle (2−ΔCT) method. Similarly, the relative expression
of SIRT3 was normalized to the control groups using the comparative
threshold cycle (2−ΔΔCT) method.
Western blotting
Cell lysates were prepared from the frozen tissue
samples using radio immunoprecipitation (RIPA) lysis buffer
combined with a phosphatase and protease inhibitor cocktail.
Lysates were boiled with sodium dodecyl sulfate (SDS) loading
buffer and then separated by SDS-polyacrylamide gel electrophoresis
(PAGE). Proteins were transferred to a nitrocellulose membrane,
which was later incubated with a primary specific antibody
(anti-SIRT3, #2627; Cell Signaling Technology; anti-pan Akt, #4691;
Cell Signaling Technology; anti-Phospho-Akt Ser473, #4060; Cell
Signaling Technology; anti-β-actin: #3700; Cell Signaling
Technology), all purchased in 5% non-fat milk, followed by
HRP-conjugated anti-rabbit/mouse secondary antibody. ECL detection
reagent (Merck Millipore, Darmstadt, Germany) was used to
demonstrate the protein bands.
Immunoprecipitation
Primary antibody was added to magnetic beads diluted
in PBS with Tween-20, and then incubated with rotation at room
temperature to allow crosslinking. After centrifugation, the
supernatant was removed and the beads-antibody complex was washed
with the aid of a magnet. Cell samples extracted through NP40 cell
lysis buffer with phosphatase and protease inhibitor cocktail were
added to the beads-antibody complex to immunoprecipitate the target
antigen. Finally, after removing the supernatant, and washing the
beads-antibody-antigen complex, SDS loading buffer was added for
examination on a gel. Conformation-specific mouse anti-rabbit
HRP-conjugated anti-rabbit/mouse secondary antibody (#5127; Cell
Signaling Technology) was used to avoid IgG subunit
interference.
Statistical analysis
Statistical analyses were performed using SPSS 16.0
software (SPSS, Inc., Chicago, IL, USA). ROC curve analysis was
applied to define the cut-off value for high expression of SIRT3
using the 0.1 criterion, and the area under the curve (AUC) was
measured. A Wilcoxon matched paired test, paired t-test, one-way
ANOVA analysis and SNK-q test were used to compare the SIRT3
expression pattern according to numeric data types. Pearson
correlation analysis was used to estimate the relationship between
SIRT3 expression and other proteins. A χ2 test was
performed to analyze the correlation between SIRT3 expression and
clinicopathological parameters. Overall survival (OS) was defined
as the time from initial diagnosis to death and assessed using the
Kaplan-Meier method and compared by the log-rank test. P=0.05
(two-tailed) was considered statistically significant.
Results
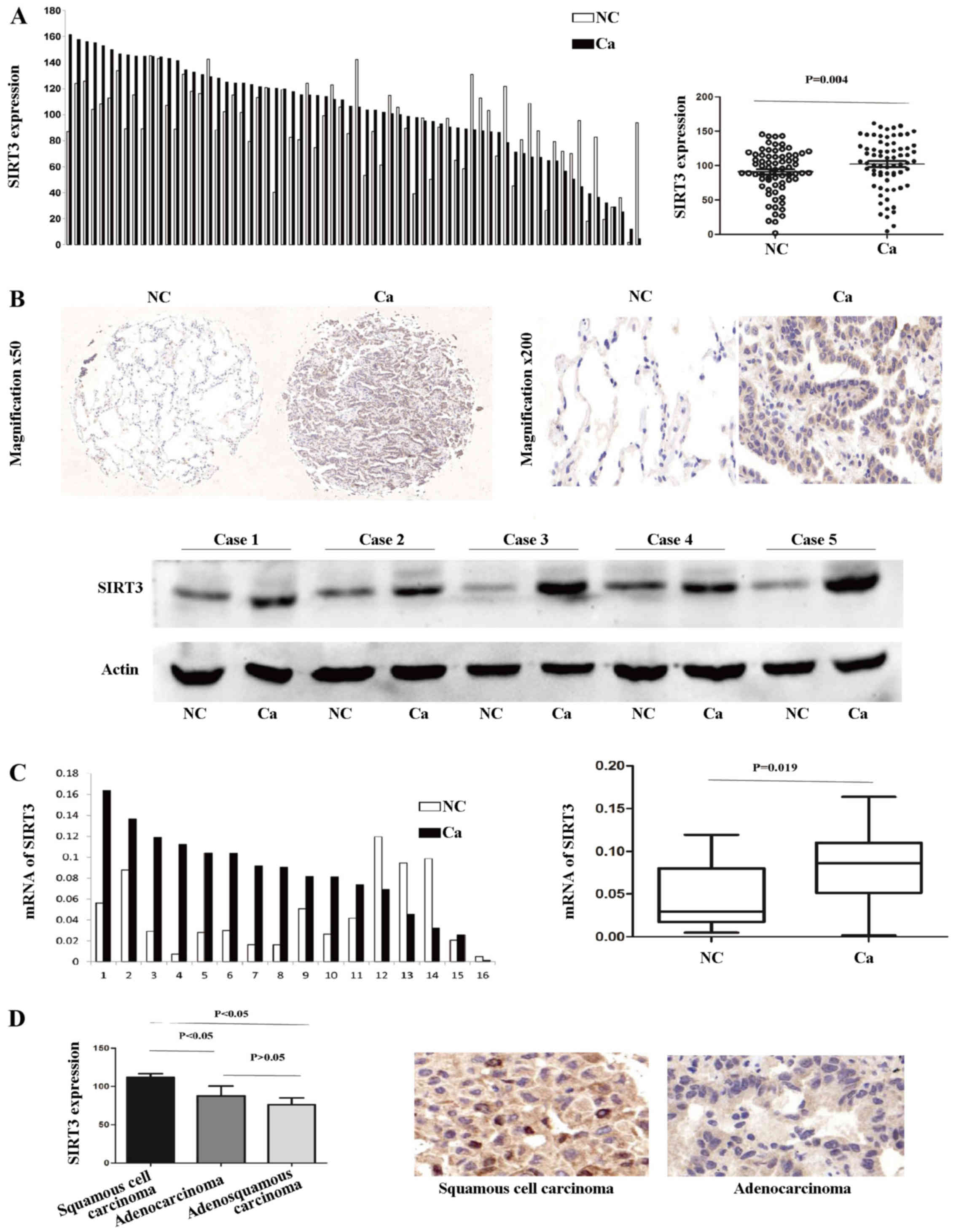
The expression of SIRT3 is upregulated in
human NSCLC tissue
To investigate the expression pattern of SIRT3 in
NSCLC, we first examined the protein level in 70 paired clinical
samples of NSCLC and adjacent normal lung tissue by IHC. According
to the results of the TMA-based IHC, SIRT3 was noticeably
upregulated in the cancerous tissue, compared with the normal
tissues. A paired t-test showed that there was a significant
difference between the cancerous and normal sample groups (P=0.004;
Fig. 1A). A representative pair of
IHC-stained tissues and five paired western blotting results
demonstrating the aberrant upregulated SIRT3 expression in NSCLC
are shown in Fig. 1B. Next, we
investigated SIRT3 mRNA expression levels in 16 paired tissue
samples. SIRT3 mRNA levels were markedly higher in the cancerous,
compared with the normal lung, tissue group (Wilcoxon matched
paired test, P=0.019; Fig. 1C). In
addition, SIRT3 was more upregulated in squamous cell carcinoma
type NSCLC (Fig. 1D).
Taken together, these data demonstrate that SIRT3
expression is upregulated in human NSCLC cancerous tissue compared
with normal lung tissue, and to an even greater extent in squamous
cell carcinoma.
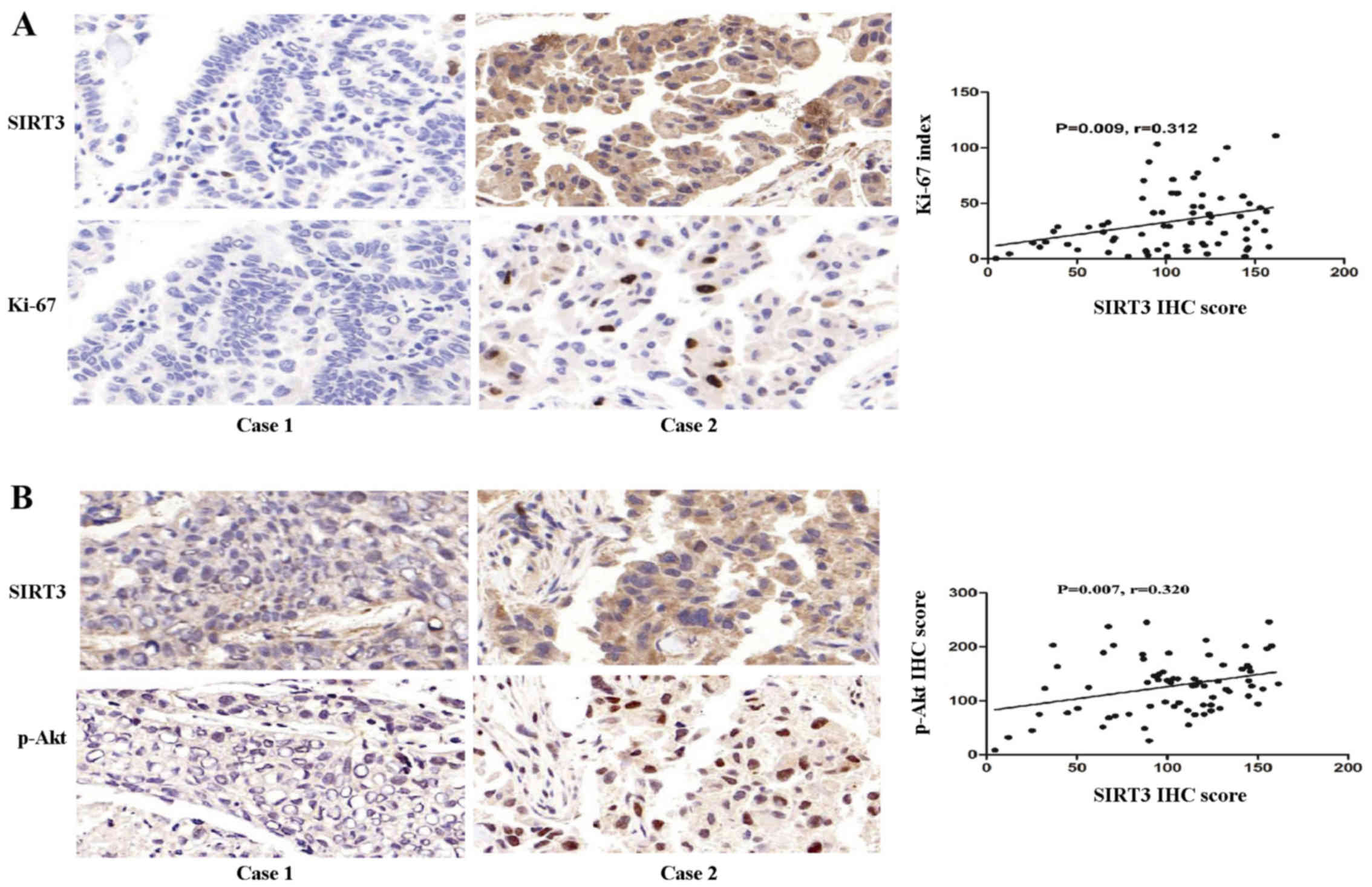
SIRT3 expression positively correlates
with the malignant biomarker Ki-67 and oncogene p-Akt in NSCLC
Next, we investigated the inter-relationship between
SIRT3 expression and Ki-67, a biomarker of malignant proliferation
and the oncogene p-Akt, in NSCLC. The results showed that SIRT3
expression was positively correlated with Ki-67 expression
(P=0.009, coefficient of correlation (r)=0.312; Fig. 2A). SIRT3 expression was also
positively correlated with p-Akt (ser473) (P=0.007, r=0.320;
Fig. 2B).
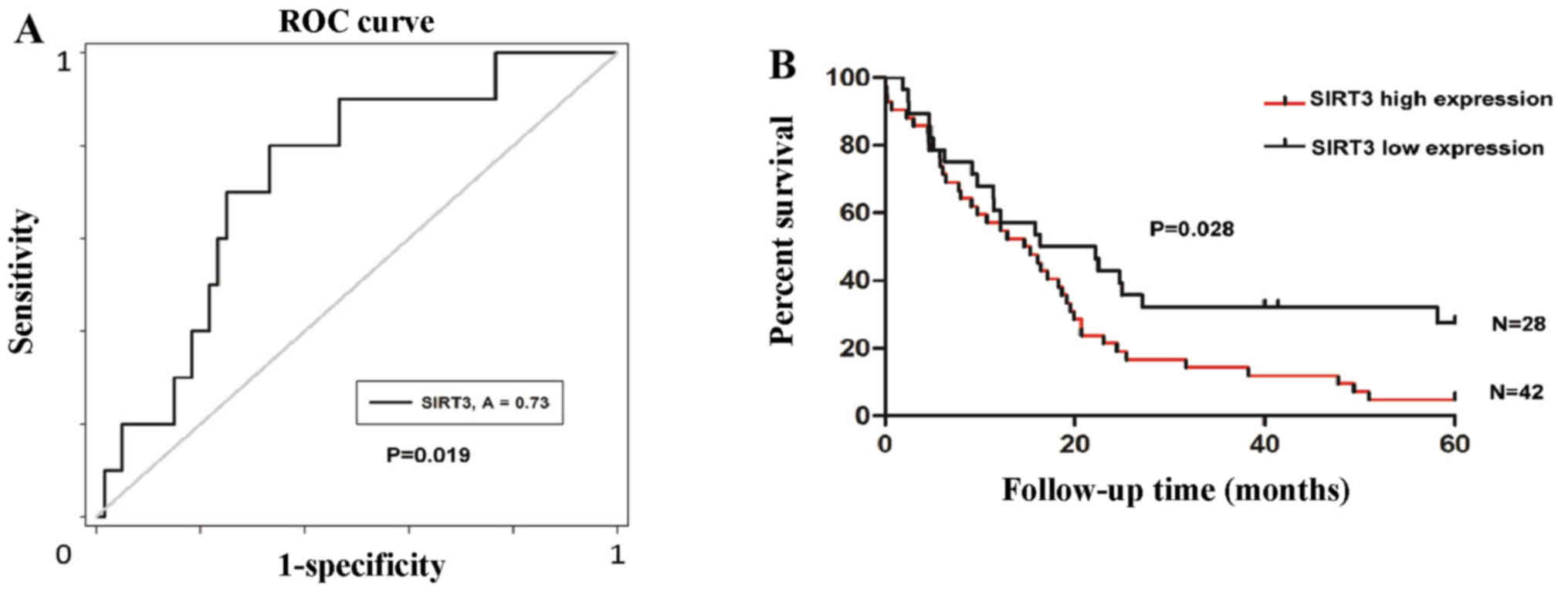
High SIRT3 expression indicates a shorter
OS
To determine the significance of SIRT3 expression on
prognosis in NSCLC patients, we defined an optimal cut-off value
for high or low SIRT3 expression using ROC curve analysis. The
results demonstrated that divided cut-off value for survival status
has the shortest distance from the curve to the point (0.0, 1.0)
(P=0.019; Fig. 3A). We then
carried out a retrospective analysis of the outcomes of the 70
study patients. The median survival time was 14.97 months for the
patients with high SIRT3 expression, while it was 19.29 months for
patients with a low level of SIRT3. Kaplan-Meier survival analyses
showed that patients with high SIRT3 expression had a shorter OS
duration (P=0.028; Fig. 3B).
No correlation was found between the
SIRT3 expression and the clinicopathological variables of NSCLC
except cancer types
To estimate the clinical significance of SIRT3
expression in NSCLC, we analyzed the association between the SIRT3
expression and the clinicopathological characteristics.
Correlations were observed between the SIRT3 expression and the
pathological type (P= 0.001). There were no statistical connections
between the SIRT3 expression and the other clinicopathological
parameters, such as age, gender, tumor location, clinical stage and
therapeutic method (P>0.05; Table
I).
 | Table ICorrelation between SIRT3 expression
and clinical variables of the NSCLC cases. |
Table I
Correlation between SIRT3 expression
and clinical variables of the NSCLC cases.
| Variables | Cases
(n=70) | SIRT3 expression
| P-value
(χ2 test) |
|---|
High
(n=42) | Low
(n=28) |
|---|
| Age (years) | | | | 0.052 |
| >60 | 37 | 18 | 19 | |
| ≤60 | 33 | 24 | 9 | |
| Gender | | | | 0.732 |
| Male | 61 | 36 | 25 | |
| Female | 9 | 6 | 3 | |
| Tumor location | | | | 1 |
| Left lung | 30 | 18 | 12 | |
| Right lung | 40 | 24 | 16 | |
| Tumor size
(cm) | | | | 0.215 |
| >5 | 44 | 29 | 15 | |
| ≤5 | 26 | 13 | 13 | |
| Lymphatic
invasion | | | | |
| Yes | 44 | 24 | 20 | 0.313 |
| No | 26 | 18 | 8 | |
| Tumor stage | | | | |
| T1/T2 | 27 | 13 | 14 | 0.136 |
| T3/T4 | 43 | 29 | 14 | |
| Clinical stage | | | | 0.467 |
| Stage II | 29 | 19 | 10 | |
| Stage III | 41 | 23 | 18 | |
| Pathology type | | | | 0.001 |
|
Adenocarcinoma | 13 | 5 | 8 | |
| Squamous
carcinoma | 47 | 35 | 12 | |
| Adenosquamous
carcinoma | 10 | 2 | 8 | |
| Therapeutic
method | | | | 0.609 |
| Surgery +
chemotherapy | 47 | 27 | 20 | |
| Surgery | 23 | 15 | 8 | |
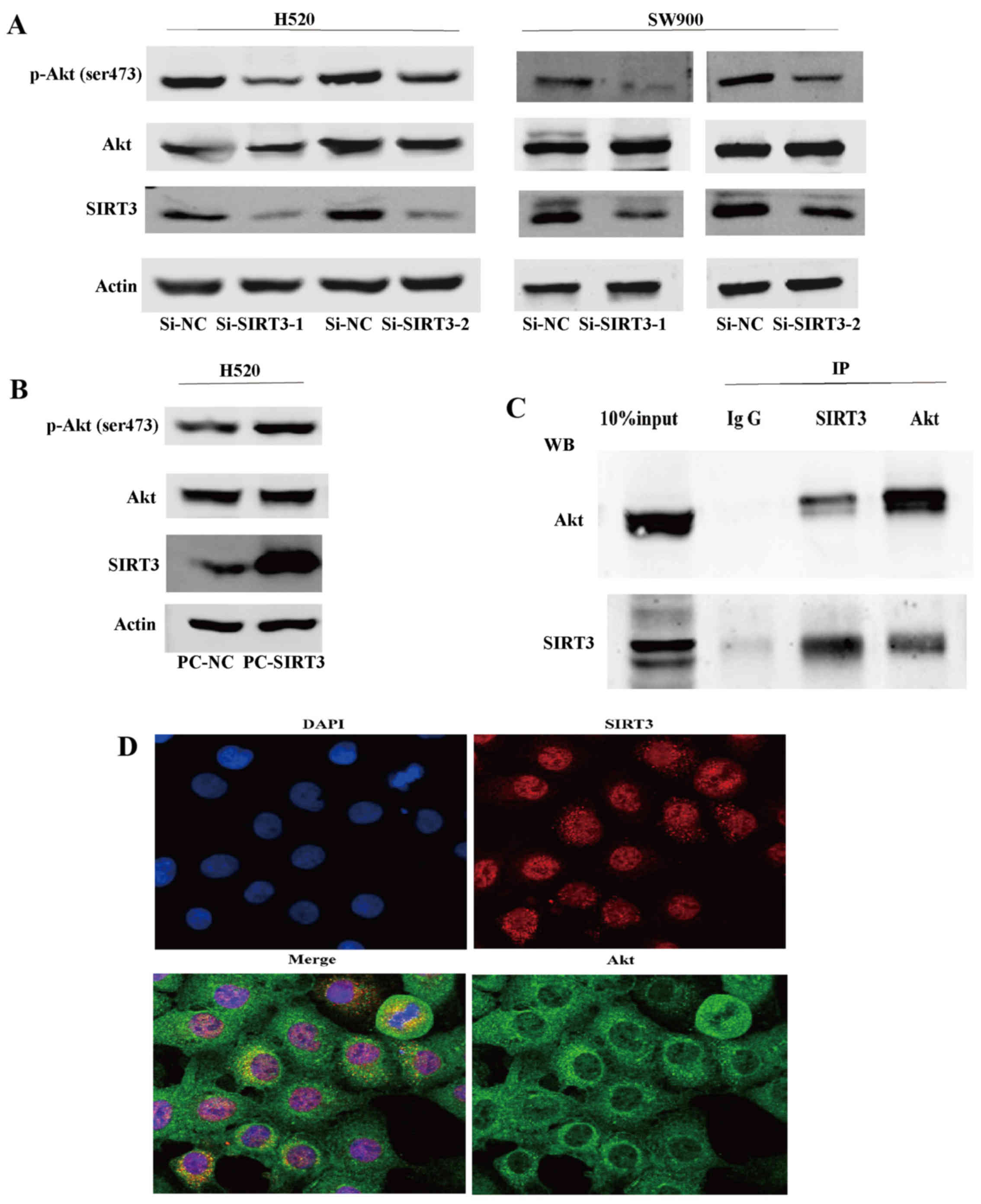
SIRT3 promotes the activation of the Akt
signaling pathway in NSCLC
To explore the mechanism underlying the involvement
of SIRT3 in NSCLC, we first generated two NSCLC cell lines (H520
and SW900) with downregulated SIRT3 using RNA interference. We
found that, in both SIRT3 downregulated cell lines, expression of
p-Akt (ser473), a surrogate marker of Akt activity, was decreased,
while expression of total Akt showed no significant difference
(Fig. 4A). We then generated SIRT3
overexpressing H520 cells using the pcDNA3.1 expression vector and
found that SIRT3 overexpression caused the activation of Akt
(Fig. 4B). Lastly, we demonstrated
that SIRT3 could co-precipitate and was co-located with Akt and
vice versa in endogenous H520 cell state, indicating that SIRT3 may
mediate activation of Akt by their binding or function in a protein
complex (Fig. 4C and D). These
results indicate that SIRT3 promotes activation of Akt signaling
pathways in NSCLC.
Discussion
We propose that SIRT3 has oncogenic activity in
NSCLC, and that the underlying molecular mechanism is associated
with activation of Akt signaling pathways. The present study showed
the oncogene function of SIRT3 in NSCLC, and it suggests it is most
strongly associated with squamous cell carcinoma. We demonstrated
that SIRT3 can activate the Akt signaling pathway and that this
interaction may involve post-transcriptional acetylation
modification of SIRT3. However, the study was limited, as currently
we cannot account for the possible involvement of SIRT3 in other
signaling networks that initiate and promote the progress of NSCLC.
Further systematic research will be necessary to elucidate the
specific pathologic activity of SIRT3.
In recent years, numerous studies have indicated
ambiguity in the function of SIRT3 in different cancer types
(13–15). For example, in breast cancer, SIRT3
suppresses proliferation and anaerobic glycolysis, and increases
apoptosis via the depletion of reactive oxygen species (ROS)
(22,23). Furthermore, in some cancers SIRT3
appears to act as an anti-oncogene by delaying the degradation of
p53 (24,25). By contrast, in some cancer types,
such as esophageal cancer and oral squamous cell carcinoma, SIRT3
is considered to promote carcinogenesis (26,27).
However, with respect to NSCLC, according to the small amount of
research available in the literature, there is no consensus on
SIRT3 function, and while it has apparent clinical significance,
data are insufficient to explain the underlying molecular
mechanisms (25,28). In addition, regarding the
relationship between Akt and SIRT3, previous reports propose that
SIRT3 could inhibit the activation of Akt by preventing
ROS-mediated Ras-PI3K-Akt activation (29–31).
However, as well as ROS-mediated activation of Akt, other recent
research points to post-translational modifications like
acetylation being another significant means of regulating this
central node of activity, where acetylation modification of Akt or
its critical activator PDK1 could enhance their membrane transport
and interaction with each other (32,33).
This breakthrough in finding a mediation mechanism for oncogenes
prompted us to explore the deacetylation function of SIRT3 upon
critical proteins in malignant transformation.
In the present study, we demonstrated that SIRT3
expression was significantly increased in cancerous tissues
compared with normal lung tissues at both the transcription and
protein level, which suggested that SIRT3 had a significant
possibility of being involved in promoting the malignancy of NSCLC.
Our finding that SIRT3 expression was positively correlated with
established malignant biomarkers of NSCLC, Ki-67 (a specific
cellular biomarker of proliferation) (34) and p-Akt (representing the
activation status of oncogene Akt), strongly suggests that SIRT3 is
also positively correlated with the malignancy of NSCLC.
Furthermore, we showed that patients with high expression of SIRT3
have a poor prognosis, indicating that SIRT3 has prognostic
clinical significance. In addition, we also found that the only
clinicopathological variable with which high SIRT3 expression
significantly correlated was pathologic type, namely, squamous cell
carcinoma. This finding is consistent with previous studies that
demonstrated that SIRT3 can play conflicting roles in cancer
because there are distinct types. Akt signaling pathways are
classical oncogenic pathways in numerous cancers via diverse
mechanisms (35,36); hence the finding that SIRT3 was
involved in the pathway further implicates it in the malignancy of
NSCLC. We showed that overexpression of SIRT3 enhanced the
activation of Akt, and our co-precipitation and co-location studies
suggested that SIRT3 was deacetylating Akt, in a similar role to
that known for other SIRTs that deacetylate and activate Akt or an
upstream mediator like PDK1 (31,37).
These results provided clues to understand the
mechanisms underlying the malignant function of SIRT3 in NSCLC and
simultaneously disclosed a novel relationship between SIRT3 and
Akt, namely, that SIRT3 could function as an activator upstream of
the Akt signaling pathway but not via the blocking of ROS-mediated
Ras-PI3K-Akt activation (29–31).
How SIRT3 regulates Akt now requires further investigation; it
could be via lysine deacetylation of Akt itself or of numerous
known substrates of SIRT3, containing some classical or novel
regulators of the Akt signaling pathway.
The present study also revealed clues to help
interpret the conflicting role of SIRT3 in cancer by comparing the
genetic background of squamous cell with other types of carcinoma.
Previous studies show that, in squamous cell carcinomas, such as
esophageal cancer and oral squamous cell carcinoma, SIRT3 behaves
as an oncogene but, in cancers composed mainly of adenocarcinoma,
like breast cancer, SIRT3 acts as an anti-oncogene. However,
ascribing these distinctions is likely to be oversimplifying as the
role of SIRT3 in other cancers, like melanoma and renal cell
cancer, is contradictory (38,39).
We surmise that this phenomenon can be attributed to different
types of 'oncogene addiction' at play in different cancers
depending upon the molecular pathways involved (40,41),
thereby allowing SIRT3 to play variable roles. Unraveling this
complex interplay is likely to provide novel cancer therapeutic
targets and remains an important scientific challenge.
In summary, our studies indicate that SIRT3 could be
applied as a novel target for investigating NSCLC, helping us to
gain a better understanding of the initiation and progression of
NSCLC. We now plan to investigate SIRT3 function in NSCLC in more
specific and comprehensive ways and to further elucidate the
relationship between SIRT3 and NSCLC.
Acknowledgments
We would like to thank all the members of the
Department of Thoracic Surgery, Tangdu Hospital, Fourth Military
Medical University, Xi'an, China and of the State Key Laboratory of
Cancer Biology, Department of Biochemistry and Molecular Biology,
Fourth Military Medical University, Xi'an, China for providing the
clinically annotated samples and fine experimental conditions. The
present study was supported by funds from the Special Clinical
Research Fund from the Wu JiePing Medical Foundation
(320.6750.15095), and the Natural Science and Technology Foundation
Research Key Project of Shaanxi Province (2015JZ 024), China.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choudhary C, Kumar C, Gnad F, Nielsen ML,
Rehman M, Walther TC, Olsen JV and Mann M: Lysine acetylation
targets protein complexes and co-regulates major cellular
functions. Science. 325:834–840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim SC, Sprung R, Chen Y, Xu Y, Ball H,
Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al: Substrate and
functional diversity of lysine acetylation revealed by a proteomics
survey. Mol Cell. 23:607–618. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guan KL and Xiong Y: Regulation of
intermediary metabolism by protein acetylation. Trends Biochem Sci.
36:108–116. 2011. View Article : Google Scholar :
|
|
8
|
Ahn BH, Kim HS, Song S, Lee IH, Liu J,
Vassilopoulos A, Deng CX and Finkel T: A role for the mitochondrial
deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad
Sci USA. 105:14447–14452. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jing E, Emanuelli B, Hirschey MD, Boucher
J, Lee KY, Lombard D, Verdin EM and Kahn CR: Sirtuin-3 (Sirt3)
regulates skeletal muscle metabolism and insulin signaling via
altered mitochondrial oxidation and reactive oxygen species
production. Proc Natl Acad Sci USA. 108:14608–14613. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sundaresan NR, Samant SA, Pillai VB,
Rajamohan SB and Gupta MP: SIRT3 is a stress-responsive deacetylase
in cardiomyocytes that protects cells from stress-mediated cell
death by deacetylation of Ku70. Mol Cell Biol. 28:6384–6401. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kumar S and Lombard DB: Mitochondrial
sirtuins and their relationships with metabolic disease and cancer.
Antioxid Redox Signal. 22:1060–1077. 2015. View Article : Google Scholar :
|
|
12
|
Haigis MC, Deng CX, Finley LWS, Kim HS and
Gius D: SIRT3 is a mitochondrial tumor suppressor: A scientific
tale that connects aberrant cellular ROS, the Warburg effect, and
carcinogenesis. Cancer Res. 72:2468–2472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Finley LWS and Haigis MC: Metabolic
regulation by SIRT3: Implications for tumorigenesis. Trends Mol
Med. 18:516–523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alhazzazi TY, Kamarajan P, Verdin E and
Kapila YL: Sirtuin-3 (SIRT3) and the hallmarks of cancer. Genes
Cancer. 4:164–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Y, Fu LL, Wen X, Wang XY, Liu J,
Cheng Y and Huang J: Sirtuin-3 (SIRT3), a therapeutic target with
oncogenic and tumor-suppressive function in cancer. Cell Death Dis.
5:e10472014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiong Y, Wang M, Zhao J, Han Y and Jia L:
Sirtuin 3: A Janus face in cancer (Review). Int J Oncol.
49:2227–2235. 2016.PubMed/NCBI
|
|
17
|
Budwit-Novotny DA, McCarty KS, Cox EB,
Soper JT, Mutch DG, Creasman WT, Flowers JL and McCarty KS Jr:
Immunohistochemical analyses of estrogen receptor in endometrial
adenocarcinoma using a monoclonal antibody. Cancer Res.
46:5419–5425. 1986.PubMed/NCBI
|
|
18
|
Shin SJ, Roh J, Cha HJ, Choi YD, Kim JM,
Min SK, Kim JE, Eom DW, Lee H, Kim HJ, et al: TCL1 expression
predicts overall survival in patients with mantle cell lymphoma.
Eur J Haematol. 95:583–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
DeLong ER, DeLong DM and Clarke-Pearson
DL: Comparing the areas under two or more correlated receiver
operating characteristic curves: A nonparametric approach.
Biometrics. 44:837–845. 1988. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cai MY, Zhang B, He WP, Yang GF, Rao HL,
Rao ZY, Wu QL, Guan XY, Kung HF, Zeng YX, et al: Decreased
expression of Pinx1 protein is correlated with tumor development
and is a new independent poor prognostic factor in ovarian
carcinoma. Cancer Sci. 101:1543–1549. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zlobec I, Steele R, Terracciano L, Jass JR
and Lugli A: Selecting immunohistochemical cut-off scores for novel
biomarkers of progression and survival in colorectal cancer. J Clin
Pathol. 60:1112–1116. 2007. View Article : Google Scholar
|
|
22
|
Bell EL, Emerling BM, Ricoult SJH and
Guarente L: SirT3 suppresses hypoxia inducible factor 1α and tumor
growth by inhibiting mitochondrial ROS production. Oncogene.
30:2986–2996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei L, Zhou Y, Qiao C, Ni T, Li Z, You Q,
Guo Q and Lu N: Oroxylin A inhibits glycolysis-dependent
proliferation of human breast cancer via promoting SIRT3-mediated
SOD2 transcription and HIF1α destabilization. Cell Death Dis.
6:e17142015. View Article : Google Scholar
|
|
24
|
Zhao K, Zhou Y, Qiao C, Ni T, Li Z, Wang
X, Guo Q, Lu N and Wei L: Oroxylin A promotes PTEN-mediated
negative regulation of MDM2 transcription via SIRT3-mediated
deacetylation to stabilize p53 and inhibit glycolysis in wt-p53
cancer cells. J Hematol Oncol. 8:412015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao K, Jiang J, Wang W, Cao S, Zhu L,
Zeng H, Ouyang R, Zhou R and Chen P: Sirt3 is a tumor suppressor in
lung adenocarcinoma cells. Oncol Rep. 30:1323–1328. 2013.PubMed/NCBI
|
|
26
|
Zhao Y, Yang H, Wang X, Zhang R, Wang C
and Guo Z: Sirtuin-3 (SIRT3) expression is associated with overall
survival in esophageal cancer. Ann Diagn Pathol. 17:483–485. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Alhazzazi TY, Kamarajan P, Joo N, Huang
JY, Verdin E, D'Silva NJ and Kapila YL: Sirtuin-3 (SIRT3), a novel
potential therapeutic target for oral cancer. Cancer.
117:1670–1678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li H, Feng Z, Wu W, Li J, Zhang J and Xia
T: SIRT3 regulates cell proliferation and apoptosis related to
energy metabolism in non-small cell lung cancer cells through
deacetylation of NMNAT2. Int J Oncol. 43:1420–1430. 2013.PubMed/NCBI
|
|
29
|
Zhang YY and Zhou LM: Sirt3 inhibits
hepatocellular carcinoma cell growth through reducing Mdm2-mediated
p53 degradation. Biochem Biophys Res Commun. 423:26–31. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Quan Y, Wang N, Chen Q, Xu J, Cheng W, Di
M, Xia W and Gao WQ: SIRT3 inhibits prostate cancer by
destabilizing oncoprotein c-MYC through regulation of the PI3K/Akt
pathway. Oncotarget. 6:26494–26507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pillai VB, Sundaresan NR and Gupta MP:
Regulation of Akt signaling by sirtuins: Its implication in cardiac
hypertrophy and aging. Circ Res. 114:368–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chan CH, Jo U, Kohrman A, Rezaeian AH,
Chou PC, Logothetis C and Lin HK: Posttranslational regulation of
Akt in human cancer. Cell Biosci. 4:592014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Risso G, Blaustein M, Pozzi B, Mammi P and
Srebrow A: Akt/PKB: One kinase, many modifications. Biochem J.
468:203–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Scholzen T and Gerdes J: The Ki-67
protein: From the known and the unknown. J Cell Physiol.
182:311–322. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bauer TM, Patel MR and Infante JR:
Targeting PI3 kinase in cancer. Pharmacol Ther. 146:53–60. 2015.
View Article : Google Scholar
|
|
36
|
Testa JR and Bellacosa A: AKT plays a
central role in tumorigenesis. Proc Natl Acad Sci USA.
98:10983–10985. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sundaresan NR, Pillai VB, Wolfgeher D,
Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta
M and Gupta MP: The deacetylase SIRT1 promotes membrane
localization and activation of Akt and PDK1 during tumorigenesis
and cardiac hypertrophy. Sci Signal. 4:ra462011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
George J, Nihal M, Singh CK, Zhong W, Liu
X and Ahmad N: Pro-proliferative function of mitochondrial sirtuin
deacetylase SIRT3 in human melanoma. J Invest Dermatol.
136:809–818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Choi J, Koh E, Lee YS, Lee HW, Kang HG,
Yoon YE, Han WK, Choi KH and Kim KS: Mitochondrial Sirt3 supports
cell proliferation by regulating glutamine-dependent oxidation in
renal cell carcinoma. Biochem Biophys Res Commun. 474:547–553.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Weinstein IB: Cancer. Addiction to
oncogenes - the Achilles heal of cancer. Science. 297:63–64. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fisher GH, Wellen SL, Klimstra D,
Lenczowski JM, Tichelaar JW, Lizak MJ, Whitsett JA, Koretsky A and
Varmus HE: Induction and apoptotic regression of lung
adenocarcinomas by regulation of a K-Ras transgene in the presence
and absence of tumor suppressor genes. Genes Dev. 15:3249–3262.
2001. View Article : Google Scholar : PubMed/NCBI
|