Introduction
Breast cancer is the most frequently diagnosed
cancer and is the leading cause of cancer death among females
worldwide, with an estimated 1.7 million cases and 521,900 deaths
in 2012 (1). Breast cancer alone
accounts for 25% of all cancer cases and 15% of all cancer deaths
among females. If diagnosed at an early stage, breast cancer
patients often completely recover. However, many cases recur and
develop gradual therapeutic resistance. To improve the prognosis of
breast cancer patients, further research is required worldwide to
identify new therapeutic targets.
Stathmin1 (STMN1), also known as oncoprotein 18, is
a cytosolic phosphoprotein and a key regulator of cell division due
to its microtubule depolymerization in a phosphorylation-dependent
manner (2–4). STMN1 interacts with and sequesters
free tubulin leading to microtubule depolymerization in
vitro (5). STMN1 expression is
associated with breast cancer proliferation (6,7).
STMN1 overexpression correlates with low estrogen receptor (ER)
expression, low progesterone receptor (PgR) expression and high
histological grade in human primary breast cancer (6).
Since Perou et al (8) and Sørlie et al (9) performed breast cancer gene expression
profiling using cDNA microarray in 2000, intrinsic subtype
classification based on gene expression profiling has been
attracting attention. According to this classification, an
alternative subtype classification based on the immunohistochemical
analysis of ER, PgR, human epidermal growth factor receptor 2
(HER2) and Ki-67, mainly obtained by common pathological
examination, is used for the clinical strategy (10). Triple-negative breast cancers
(TNBCs) characterized by the absence of ER, PgR and HER2 expression
have relatively poor outcomes and often exhibit treatment
resistance (11). TNBCs are not
eligible for endocrine therapies or anti-HER2-targeted therapies;
this represents a substantial problem as there is no clear
treatment target.
TNBCs are associated with cancer stem cells (CSCs)
in breast cancer (12,13). CSCs are a small cell population
with unique characteristics such as self-renewal and multipotency.
The high CD44/low CD24 breast cancer cell phenotype is associated
with a subpopulation of tumorigenic stem cells (14). Breast cancer cells with increased
aldehyde dehydrogenase 1 (ALDH1) activity have stem cell properties
(15). Breast cancer stem cells
(BCSCs) are associated with therapeutic resistance as well as
growth, diversity and metastasis of breast cancer (16,17).
Therefore, it is hoped that BCSC-targeted therapies will be
developed to overcome therapeutic resistance.
Epithelial-to-mesenchymal transition (EMT) also has an important
role in cancer progression and metastasis. Through EMT, cancer
cells invade the vascular system and metastasize (18,19).
The relationships between CSCs and EMT are still controversial.
High STMN1 expression is associated with poor
prognosis in breast cancer patients (20–22).
Furthermore, taxane sensitivity is low in breast cancer cell lines
with STMN1 overexpression (23).
However, few studies have addressed the relationship between STMN1
and CSCs and EMT, which are attracting attention as a treatment
target in breast cancer, in particular TNBCs.
The present study aimed to determine the clinical
significance of STMN1 and its association with the expression of
CSC markers, EMT markers and several cancer-related markers in
breast cancer. Therefore, we retrospectively investigated the
expression of STMN1 and CSC markers, including CD44/CD24 and ALDH1,
in breast cancer tissue samples using immunohistochemistry to
evaluate whether STMN1 qualifies as a marker of cancer progression
and cancer stem cell type in breast cancer patients. Furthermore,
we evaluated the expression of E-cadherin and epithelial cell
adhesion molecule (EpCAM) as representative epithelial markers and
the expression of vimentin as a representative mesenchymal marker
to determine the association between STMN1 expression and EMT.
Materials and methods
Patients
We retrospectively analyzed tumor specimens from 237
patients with primary breast cancer who underwent primary tumor
excision between January 1999 and October 2010 (180 patients were
randomly selected from patients who underwent surgery between
January 1999 and December 2002, and all patients with TNBC subtype
who underwent surgery between January 2008 and December 2010 were
included) at Breast and Endocrine Surgery of Gunma University
Hospital. The inclusion criteria were as follows: histologically
proven diagnosis of primary breast cancer; potentially curative
operation was performed; and complete pathological records. The
exclusion criteria were as follows: breast cancer with synchronous
multiple cancers; stage IV cancer in preoperative diagnosis; and
lost to the pathological records. The patients included 1 man and
236 women with a median age at surgery of 55 years (range, 28–95
years). Eighty patients had stage I, 101 had stage II and 50 had
stage III breast cancer at the time of the surgery. In addition,
137 (57.8%) patients were negative and 95 (40.1%) patients were
positive for lymph node metastasis, and 132 (55.7%) patients were
ER positive, 99 (41.8%) patients were PgR positive, and 47 (19.8%)
patients had 2+ or 3+ HER2 scores.
Tumor staging was based on the Union for
International Cancer Control TNM classification, seventh edition
(24). The nuclear grades were
defined as the sum of scores for nuclear atypia (1, low-degree
atypia; 2, intermediate-degree atypia; 3, high-degree atypia) and
mitotic count per 10 high-power fields (×40 objective lens; 1, 0–4
mitoses; 2, 5–10 mitoses, 3, ≥11 mitoses). The nuclear grade was 1,
2 and 3 when the sum of scores for nuclear atypia and mitotic
counts were 2–3, 4 and 5–6, respectively (25). This study was in accordance with
the Declaration of Helsinki. The Ethics Committee of Gunma
University approved the study protocol.
Tissue microarray (TMA)
Clinical formalin-fixed paraffin-embedded (FFPE)
samples were stored in the archives of the Clinical Department of
Pathology, Gunma University Hospital. For each patient, one
paraffin block containing representative non-necrotic tumor areas
was selected. Breast cancer tissue cores (2.0-mm diameter per
tumor) were punched out from the representative areas near the
invasive front and transferred into the paired recipient paraffin
block using a tissue array instrument (Beecher Instruments, Silver
Spring, MD, USA).
Immunohistochemistry (IHC)
A 4-µm section was cut from the sample
paraffin blocks. Each section was mounted on a silane-coated glass
slide, deparaffinized, and soaked for 30 min at room temperature in
0.3% H2O2/methanol to block endogenous
peroxidases. The sections were then heated in boiling water and
Immunosaver (Nishin EM, Co., Ltd., Tokyo, Japan) at 98°C for 45
min. Non-specific binding sites were blocked by incubating with
Protein Block serum-free (Dako, Carpinteria, CA, USA) for 30 min. A
mouse monoclonal anti-STMN1 (OP18) antibody (Santa Cruz
Biotechnology, Santa Cruz, CA, USA) was applied at a dilution of
1:100 for 24 h at 4°C. The primary antibody was visualized using
the Histofine Simple Stain PO (M) kit (Nichirei, Tokyo, Japan),
according to the instructions manual. Chromogen
3,3′-diaminobenzidine tetrahydrochloride was applied as a 0.02%
solution containing 0.005% H2O2 in 50 mM
ammonium acetate-citrate acid buffer (pH 6.0). The sections were
lightly counterstained with Mayer's hematoxylin and mounted.
Negative controls were established by omitting the primary
antibody.
Other IHC was performed using the following primary
antibodies: anti-ER (SP1; Ventana Medical Systems, Inc., Tucson,
AZ, USA), anti-PgR (1E2; Ventana Medical Systems), anti-HER2 (4B5;
Ventana Medical Systems), anti-Ki-67 (30–9; Ventana Medical
Systems), anti-epidermal growth factor receptor (EGFR) (31G7;
Nichirei), anti-cytokeratin 5/6 (CK5/6) (D5/16;B4; Dako, Glostrup,
Demark), anti-E-cadherin (36; Ventana Medical Systems), anti-ALDH1
(46/ALDH; BD Biosciences, Franklin Lakes, NJ, USA), anti-CD44
(DF1485; Dako), anti-CD24 (SN3b; Thermo Fisher Scientific, Fremont,
CA, USA), anti-EpCAM (D9S3P; Cell Signaling Technology, Inc.,
Danvers, MA, USA) and anti-vimentin (M725; Dako).
Immunohistochemical evaluation and
subtype classification
The cut-off value for ER and PgR positivity was 1%.
HER2 expression was scored according to the American Society of
Clinical Oncology/College of American Pathologists guidelines (0,
no reactivity or membranous reactivity in <10% of cells; 1+,
faint/barely perceptible membranous reactivity in at least 10% of
cells or reactivity in only part of the cell membrane; 2+, weak to
moderate complete membranous reactivity in at least 10% of tumor
cells; 3+, strong complete membranous reactivity in at least 10% of
tumor cells) (26). The Ki-67
labeling index (LI) was used to calculate the percentage of cells
with high nuclear expression in ~1000 cells/sample (27). The Ki-67 LI assumes a 14% cut-off
value (28). EGFR, CD44 and EpCAM
expression were scored in the same way as HER2 expression; 0 and 1+
scores were considered to be negative, and 2+ and 3+ scores were
considered to be positive. The cut-off values for CK5/6, E-cadherin
and ALDH1 used 10%. If there was even a slightly stained positive
part, the expression of CD24 and vimentin was considered to be
positive.
When the cytoplasm of the cells was stained, the
cells were STMN1-expression positive. In addition, for each case,
we determined a modified Allred score, which is a semi-quantitative
system that takes the proportion of positive cells into
consideration (0, none; 1, 0–1%; 2, 1–10%; 3, 10–33%; 4, 33–66%;
and 5, 66–100%) and staining intensity (0, none; 1, weak; 2,
intermediate; and 3, strong) (29). The proportion score and the
intensity score were then summed to produce total scores of 0 or 2
through 8. A score of 0–3 was defined as low STMN1 expression and a
score of 4–8 was defined as high STMN1 expression.
Based on IHC, we defined the breast cancer subtypes
as follows: luminal A-like (ER+, HER2 0/1+
and Ki-67 low), luminal B-like (ER+, HER2
0/1+ and Ki-67 high), luminal-HER2 (ER+ and
HER2 2+/3+), HER2 (ER− and HER2
2+/3+), and triple negative (ER−
and HER2 0/1+).
Data mining
We used the gene expression-based outcome for breast
cancer online (GOBO) to obtain information on STMN1 expression in
51 breast cancer cell lines (30).
GOBO is an online tool that enables assessment of gene expression
levels in breast cancer specimens and breast cancer cell lines.
We also used an online database Kaplan-Meier (KM)
plotter to validate the association between STMN1 mRNA expression
and overall survival (OS) and post-progression survival in breast
cancer patients (31). The KM
plotter is an entirely independent patient database with
large-scale survival data, which can be stratified by selected
genes and characteristics, such as histology, stage and sex.
Statistical analysis
Statistical analysis was performed using the t-test
for continuous variables and the Chi-square test for categorical
variables. Survival curves were generated according to the
Kaplan-Meier method. Differences between survival curves were
examined using the log-rank test. A result was considered to be
statistically significant when the relevant P-value was <0.05.
All statistical analyses were performed with the IBM SPSS
statistics, version 21.0 (IBM Corp., Armonk, NY, USA).
Results
Immunohistochemical analysis of STMN1
expression in breast cancer
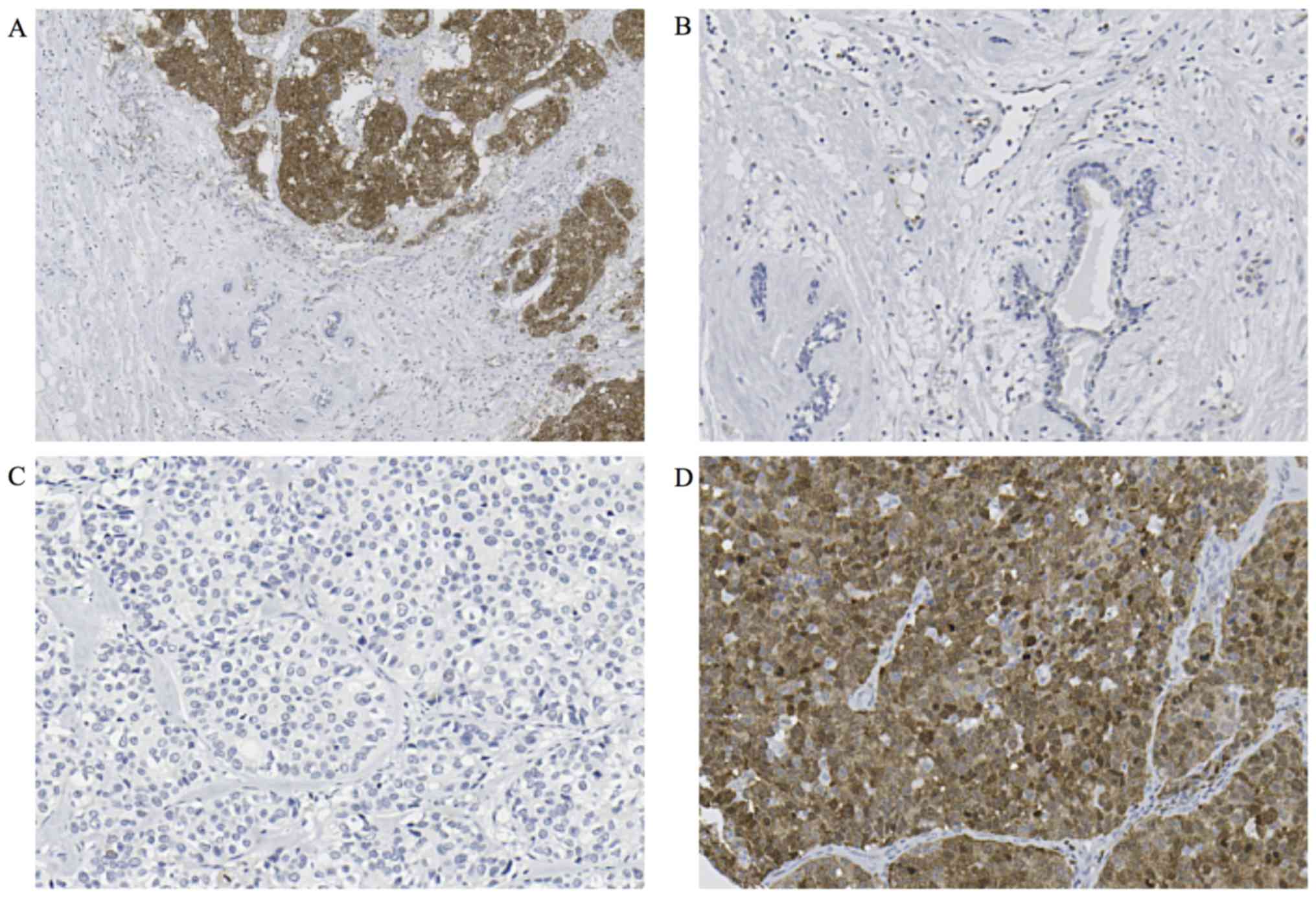
We evaluated STMN1 expression using
immunohistochemistry in 237 breast cancer TMA samples. Cytoplasmic
expression of STMN1 in breast cancer tissue was higher than that in
normal breast tissue (Fig. 1A and
B). In total, 171 (72.2%) breast cancer specimens were assigned
to the low STMN1-expression group (Fig. 1C) and 66 (27.8%) to the high
STMN1-expression group (Fig.
1D).
Association between the expression of
STMN1 and clinicopathological features of breast cancer
The correlations between STMN1 expression in breast
cancer specimens and the clinicopathological characteristics of the
patients are shown in Table I.
Tumor nuclear grade was significantly higher in the
STMN1-overexpression group (P<0.001). For the patients with
tumor assigned to the high STMN1-expression group, there were
significant associations with ER and PgR negativity (P<0.001,
P=0.002). According to IHC-based subtypes, the STMN1 expression
level was significantly higher in the triple-negative subtype
(Table I, P<0.001) (Fig. 2A–D). Moreover, when EGFR positive
or the CK5/6 positive in the triple-negative subtype were defined
as basal-like subtype, the STMN1 expression level was also
significantly higher in the basal-like subtype with EGFR or CK5/6
positivity (Table I, P<0.001)
(Fig. 2A, E and F). We also
examined the association between STMN1 expression and Ki-67 LI.
High STMN1-expressing patients showed significantly higher Ki-67 LI
than low STMN1-expressing patients (Table I, P<0.001) (Fig. 2A and G). There were no correlations
between STMN1 expression and patient age, tumor size, stage, lymph
node metastasis, lymphatic invasion and vascular invasion.
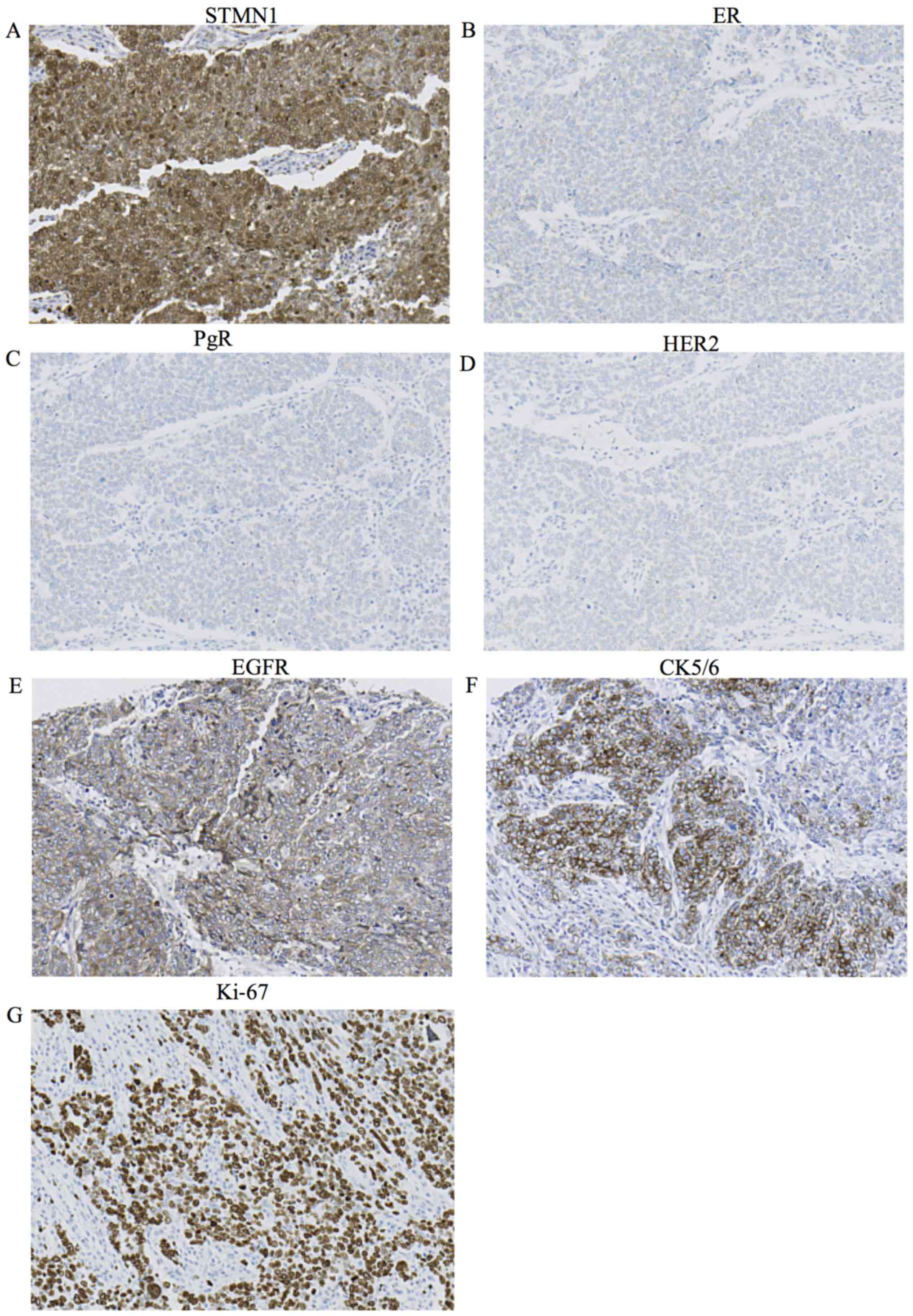 | Figure 2Immunohistochemical analysis of
STMN1, ER, PgR, HER2, EGFR, CK5/6 and Ki-67 expression in the
representative breast cancer tissue from a patient. (A) High STMN1
expression in the breast cancer tissue (magnification, ×200). (B)
Negative ER expression in the breast cancer tissue (magnification,
×200). (C) Negative PgR expression in the breast cancer tissue
(magnification, ×200). (D) Negative HER2 expression in the breast
cancer tissue (magnification, ×200). (E) Positive EGFR expression
in the breast cancer tissue (magnification, ×200). (F) Positive
CK5/6 expression in the breast cancer tissue (magnification, ×200).
(G) High Ki-67 expression in the breast cancer tissue
(magnification, ×200). |
 | Table ICorrelation between the expression of
STMN1 and the clinicopathological characteristics of breast cancer
patients. |
Table I
Correlation between the expression of
STMN1 and the clinicopathological characteristics of breast cancer
patients.
|
Characteristics | STMN1 expression
| P-value |
|---|
| Low expression
(n=171) | High expression
(n=66) |
|---|
| Age (years), mean ±
SE | 56.6±12.11 | 53.8±13.29 | 0.121 |
| Tumor size (cm),
mean ± SE | 2.3±1.51 | 2.6±2.43 | 0.233 |
| Stage | | | 0.7 |
| 0 | 3 | 0 | |
| I | 57 | 23 | |
| II | 71 | 30 | |
| III | 37 | 13 | |
| Unknown | 3 | 0 | |
| Lymph node
metastasis | | | 0.192 |
| Negative | 103 | 34 | |
| Positive | 64 | 31 | |
| Unknown | 4 | 1 | |
| Lymphatic
invasion | | | 0.557 |
| Negative | 56 | 19 | |
| Positive | 115 | 47 | |
| Vascular
invasion | | | 0.66 |
| Negative | 119 | 44 | |
| Positive | 47 | 20 | |
| Unknown | 5 | 2 | |
| Nuclear grade | | | <0.001a |
| NG1 | 25 | 4 | |
| NG2 | 66 | 13 | |
| NG3 | 43 | 47 | |
| Unknown | 37 | 2 | |
| ER | | | <0.001a |
| Negative | 59 | 46 | |
| Positive | 112 | 20 | |
| PgR | | | 0.002a |
| Negative | 89 | 49 | |
| Positive | 82 | 17 | |
| HER2 | | | 0.692 |
| Score 0, 1+ | 136 | 54 | |
| Score 2+, 3+ | 35 | 12 | |
| Ki-67 labeling
index (%), mean ± SE | 12.1±14.09 | 40.6±29.01 | <0.001a |
| Ki-67 | | | <0.001a |
| Low (≤14) | 119 | 17 | |
| High (>14) | 52 | 49 | |
| IHC based
subtypes | | | <0.001a |
| Luminal
A-like | 89 | 13 | |
| Luminal
B-like | 17 | 6 | |
| Luminal-HER2 | 6 | 1 | |
| HER2 | 29 | 11 | |
|
Triple-negative | 30 | 35 | |
| EGFR | | | 0.034a |
| Negative | 160 | 56 | |
| Positive | 11 | 10 | |
| CK5/6 | | | 0.001a |
| Negative | 168 | 58 | |
| Positive | 3 | 8 | |
| Basal-like
typeb | | | <0.001a |
| Basal | 7 | 12 | |
| Non-basal | 164 | 54 | |
| ALDH1 | | | 0.102 |
| Negative | 161 | 58 | |
| Positive | 10 | 8 | |
| CD44 | | | <0.001a |
| Negative | 143 | 34 | |
| Positive | 27 | 32 | |
| Unknown | 1 | 0 | |
| CD24 | | | <0.001a |
| Negative | 18 | 25 | |
| Positive | 153 | 41 | |
| E-cadherin | | | 0.009a |
| Negative | 26 | 2 | |
| Positive | 145 | 64 | |
| EpCAM | | | <0.001a |
| Negative | 122 | 31 | |
| Positive | 48 | 35 | |
| Unknown | 1 | 0 | |
| Vimentin | | | <0.001a |
| Negative | 153 | 35 | |
| Positive | 16 | 30 | |
| Unknown | 2 | 1 | |
We examined the association between STMN1 expression
and immunohistochemical staining of existing BCSC markers ALDH1,
CD44 and CD24. High STMN1 expression had a strong association with
high CD44/low CD24 expression and a tendency with high ALDH1
expression related to the BCSC phenotypes (Table I, P<0.001, P<0.001) (Fig. 3A–D). We also examined the
association between the expression on STMN1 and that of epithelial
markers such as E-cadherin and EpCAM. High STMN1 expression was
associated with high E-cadherin expression and high EpCAM
expression (Table I, P=0.009,
P<0.001) (Fig. 3A, E and F).
Furthermore, we examined the association between STMN1 expression
and vimentin expression. High STMN1 expression was associated with
high vimentin expression (Table I,
P<0.001) (Fig. 3A and G).
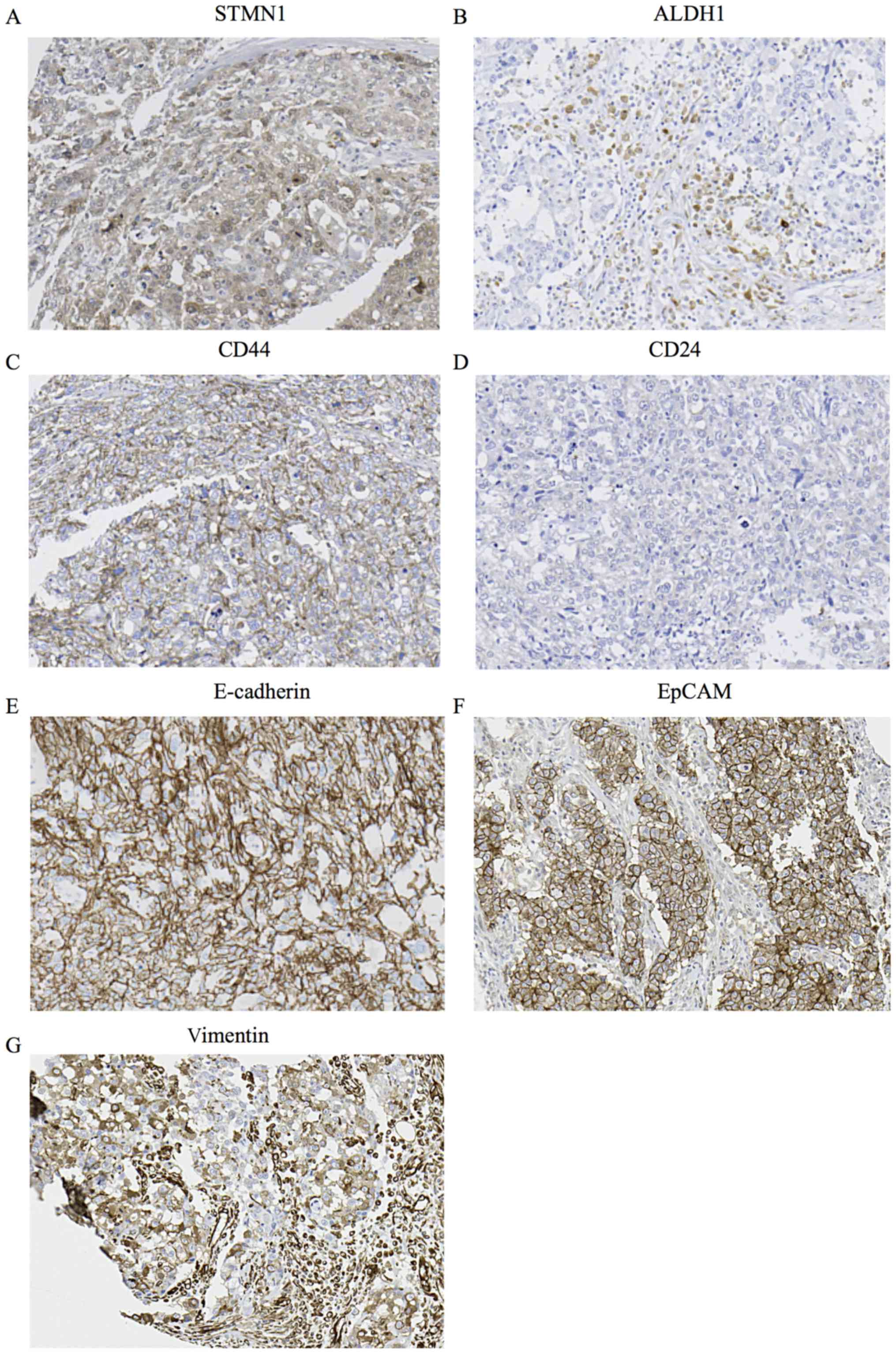 | Figure 3Immunohistochemical analysis of
STMN1, ALDH1, CD44, CD24, E-cadherin, EpCAM and vimentin expression
in a representative breast cancer tissue from a patient. (A) High
STMN1 expression in the breast cancer tissue (magnification, ×200).
(B) High ALDH1 expression in the breast cancer tissue
(magnification, ×200). (C) High CD44 expression in the breast
cancer tissue (magnification, ×200). (D) Low CD24 expression in the
breast cancer tissue (magnification, x200). (E) High E-cadherin
expression in the breast cancer tissue (magnification, ×200). (F)
High EpCAM expression in the breast cancer tissue (magnification,
×200). (G) High vimentin expression in the breast cancer tissue
(magnification, ×200). |
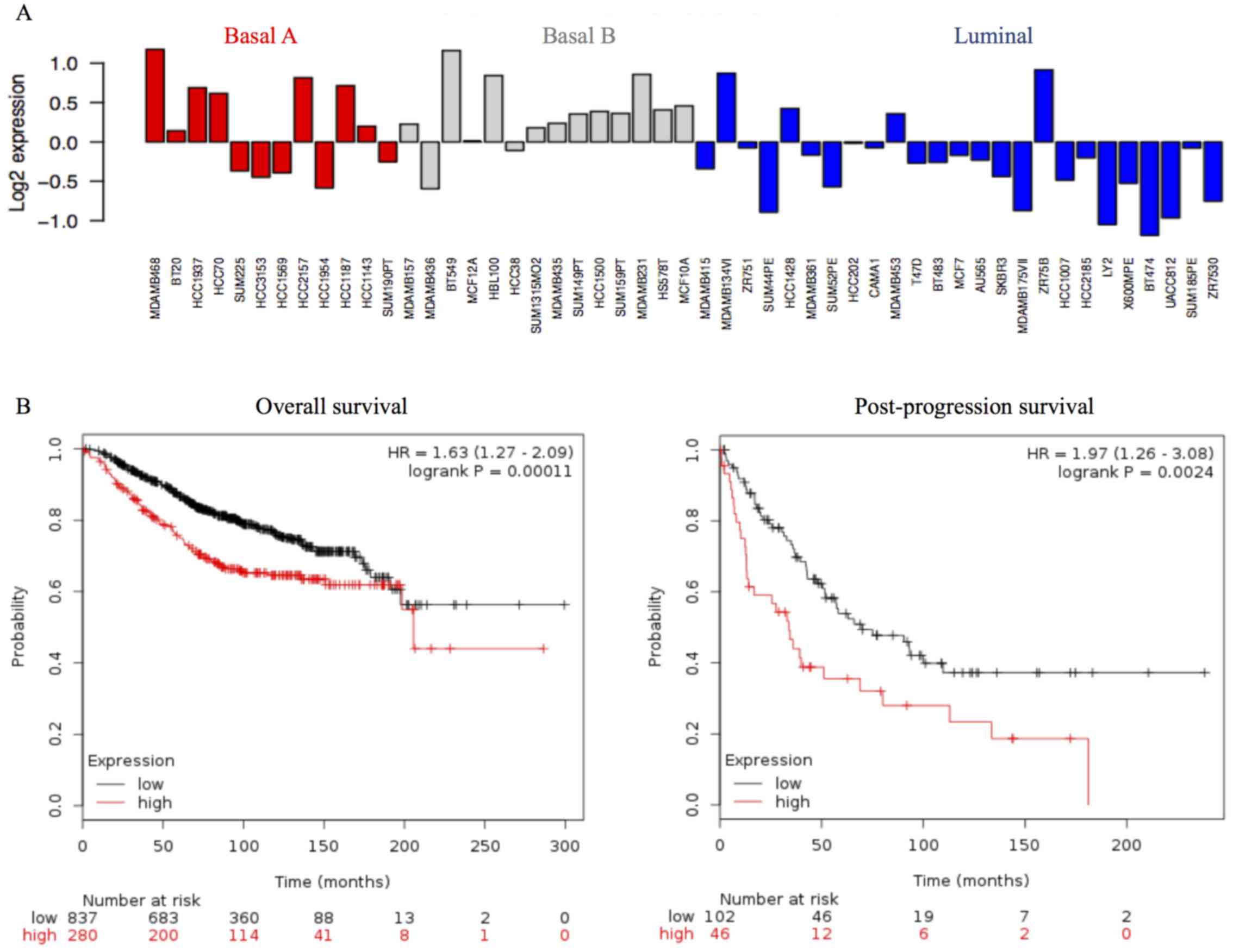
We investigated STMN1 mRNA expression levels in the
51 breast cancer cell lines using the public breast cancer database
GOBO. STMN1 mRNA expression was higher in the basal A and basal B
subgroups than in the luminal subgroups (Fig. 4A).
Association between the expression of
STMN1 and clinicopathological features of TNBCs
The correlations between the expression of STMN1 in
TNBC specimens and the clinicopathological features of the patients
are shown in Table II. Tumor
nuclear grade and Ki-67 LI were significantly higher in the
STMN1-overexpression group in the TNBC subtype (Table II, P=0.007, P<0.001).
Furthermore, high STMN1 expression had a strong association with
high CD44/low CD24 expression in the TNBC subtype (Table II, P=0.035, P=0.035).
 | Table IICorrelation between the expression of
STMN1 and the clinicopathological characteristics of TNBCs. |
Table II
Correlation between the expression of
STMN1 and the clinicopathological characteristics of TNBCs.
|
Characteristics | STMN1 expression
| P-value |
|---|
| Low expression
(n=30) | High expression
(n=35) |
|---|
| Age (years), mean ±
SE | 61.4±10.89 | 55.5±14.17 | 0.067 |
| Tumor size (cm),
mean ± SE | 2.3±1.12 | 2.7±2.88 | 0.448 |
| Stage | | | 0.352 |
| 0 | 0 | 0 | |
| I | 11 | 12 | |
| II | 10 | 17 | |
| III | 9 | 6 | |
| Lymph node
metastasis | | | 0.124 |
| Negative | 19 | 15 | |
| Positive | 11 | 19 | |
| Unknown | 0 | 1 | |
| Lymphatic
invasion | | | 0.332 |
| Negative | 12 | 10 | |
| Positive | 18 | 25 | |
| Vascular
invasion | | | 0.354 |
| Negative | 20 | 19 | |
| Positive | 9 | 14 | |
| Unknown | 1 | 2 | |
| Nuclear grade | | | 0.007a |
| NG1 | 4 | 0 | |
| NG2 | 5 | 2 | |
| NG3 | 15 | 32 | |
| Unknown | 6 | 1 | |
| Ki-67 labeling
index (%), mean ± SE | 19.4±23.79 | 55.0±27.20 | <0.001a |
| Ki-67 | | | <0.001a |
| Low (≤14) | 17 | 3 | |
| High (>14) | 13 | 32 | |
| EGFR | | | 0.964 |
| Negative | 23 | 27 | |
| Positive | 7 | 8 | |
| CK5/6 | | | 0.27 |
| Negative | 28 | 29 | |
| Positive | 2 | 6 | |
| Basal-like
typeb | | | 0.333 |
| Basal | 7 | 12 | |
| Non-basal | 23 | 23 | |
| ALDH1 | | | 0.455 |
| Negative | 25 | 32 | |
| Positive | 5 | 3 | |
| CD44 | | | 0.035a |
| Negative | 19 | 13 | |
| Positive | 11 | 22 | |
| CD24 | | | 0.035a |
| Negative | 11 | 22 | |
| Positive | 19 | 13 | |
| E-cadherin | | | 0.087 |
| Negative | 5 | 1 | |
| Positive | 25 | 34 | |
| EpCAM | | | 0.077 |
| Negative | 15 | 10 | |
| Positive | 15 | 25 | |
| Vimentin | | | 0.001a |
| Negative | 21 | 10 | |
| Positive | 9 | 25 | |
Prognostic significance of STMN1
expression in breast cancer patients
In our breast cancer cohort, RFS and OS in relation
to STMN1 expression were not significant (data not shown). However,
the survival time in breast cancer patients with high STMN1
expression was slightly worse than those with low STMN1 expression.
The median follow-up period of OS was 110 months.
To examine the prognostic significance of STMN1 in a
large cohort of breast cancer patients, we examined the correlation
between STMN1 mRNA expression and prognosis using the public
database KM plotter. High STMN1 mRNA expression correlated with
poor OS in 1117 breast cancer patients [Fig. 4B, left panel, hazard ratio (HR),
1.63, 95% confidence interval (CI), 1.27–2.09; P<0.001] and poor
post-progression survival in 148 breast cancer patients (Fig. 4B, right panel; HR, 1.97, 95% CI,
1.26–3.08; P=0.0024).
Discussion
In the present study, we determined that high levels
of STMN1 expression are associated with nuclear grade progression,
TNBC phenotype and Ki-67 expression in patients with breast cancer.
Moreover, we demonstrated that STMN1 expression was related to
CSC-marker expression, such as high CD44/low CD24 expression and
ALDH1.
STMN1 favors microtubule depolymerization by binding
to tubulin heterodimers (5).
Taxanes are microtubule-stabilizing agents commonly used in
chemotherapy for treating breast cancer (32). STMN1 overexpression decreases
microtubule polymerization and the breast cancer cell bond for
paclitaxel weakens, leading to therapeutic resistance (23). The effect of preoperative
chemotherapy containing docetaxel was low in the
STMN1-overexpression group (33).
In the present study, using the KM plotter, it was suggested that
post-progression survival was significantly worse and the response
to treatment after recurrence was lower in the STMN1-overexpression
group. Furthermore, silencing STMN1 induces microtubule
polymerization and sensitizes STMN1-overexpressing breast cancer
cells to antimicrotubule agents (34). Taxol and anti-STMN1 therapy have a
synergistic anticancer effect on a leukemic cell line (35). In the future, therapeutic
resistance to taxanes may be overcome by developing STMN1-targeted
treatments.
CSCs are a small cell population with unique
characteristics, such as self-renewal and multipotency, and show
aggressive phenotypes and therapeutic resistance by various
mechanisms (e.g., ABC transporter, ALDH activity, DNA repair and
reactive oxygen species scavenging) (36,37).
Therefore, CSCs are resistant to many cancer treatments and cause
new recurrence and metastasis by their aggressive phenotypes.
Therefore, as CSCs are closely associated with cancer progression
and metastasis, CSC-targeted therapy development may exterminate a
cancer. EMT also has an important role in cancer progression and
metastasis. Through EMT, cancer cells lose cell adhesion, gain
invasive ability and cause vascular invasion and metastases
(18,19). Although some studies have indicated
a close association between CSCs and EMT state acquisition
(38), others have suggested that
EMT and CSC states are independent (39,40).
EMT induction in human mammary epithelial cells by transcription
factor expression, such as TGF-β or snail, results in mesenchymal
trait acquisition and stem-cell marker expression (38). In contrast, Biddle et al
(39) and Liu et al
(40) suggested the presence of
EMT CSCs and non-EMT CSCs. Non-EMT CSCs, similar to normal
epithelial stem cells, have the ability of self-renewal and cell
proliferation. EMT CSCs can migrate and are characterized by
transient expression of EMT-associated genes, which can be reversed
by MET, and therefore, enable secondary tumor formation at a
metastatic site. Non-EMT CSCs and EMT CSCs can switch their
epithelial or mesenchymal traits to reconstitute the cellular
heterogeneity, which is characteristic of CSCs. There are a few
reports that have described an association between STMN1, CSCs and
EMT. Siva1 suppresses EMT and metastasis of tumor cells by
inhibiting STMN1 and stabilizing microtubules and an association
was suggested between STMN1 and EMT CSC (41). In this study, we demonstrated that
high STMN1 expression had a strong association with high CD44/low
CD24 expression and suggested an association between STMN1
expression and CSCs. We also demonstrated that the expression of
STMN1 expression correlated with that of E-cadherin and EpCAM,
which are epithelial markers, and vimentin, which is a mesenchymal
marker. In other words, it was difficult to distinguish EMT CSCs
and non-EMT CSCs by STMN1 expression in this study. However, these
two states can switch their epithelial or mesenchymal traits, and
the presence of cells that co-express epithelial and mesenchymal
markers has been suggested (42).
Furthermore, a study by Abell et al (43) showed that CSCs may represent a
population of cells in an intermediate state of EMT. These cells
express low-to-moderate levels of E-cadherin, and simultaneously,
they exhibit mesenchymal features. STMN1 may be a marker detecting
such an intermediated phenotype harboring both of EMT and
non-EMT.
Because there is no indication for TNBCs in
endocrine therapy or HER2 inhibitors, novel molecular-targeted
therapies against TNBCs are crucially needed. TNBCs have loss of
PTEN more frequently, and the PI3K pathway is strongly activated in
these tumors (44–46). PTEN loss correlates with STMN1
expression, and STMN1 expression becomes a good marker of the PI3K
pathway activation (20). In this
study, the STMN1 expression level was significantly higher in the
TNBCs. Assessment of STMN1 expression may be a clinically useful
test for the stratification of patients for anti-PI3K pathway
therapy and for monitoring therapeutic efficacy.
As described above, it is hoped that STMN1 becomes a
good therapeutic target in refractory breast cancer and recurrent
breast cancer. However, there are several limitations to this
study. First, due to the small number of patients, there was not a
significant difference between STMN1 expression and prognosis.
Second, there were many older patients in whom the treatment
regimen differed from present regimens. Therefore, in the future,
large cohort prospective validation studies are needed. However,
for TNBCs with STMN1 overexpression in this study, preoperative
chemotherapy is often currently recommended. Therefore, the
evaluation of needle biopsy tissues is required to assess STMN1
expression in treatment-free tissue. Because STMN1 expression has
relatively little heterogeneity in the tissues, we were able to
show a significant association between STMN1 expression and CSCs by
evaluating TMAs. It is expected that large cohort prospective
studies using needle biopsy tissues before treatment will be
conducted in the future to examine the significance of STMN1 as a
predictive marker for therapeutic effect and as a prognostic
marker.
In conclusion, we found that high STMN1 expression
could be a powerful marker of cancer cell proliferation, TNBC
phenotypes and cancer stem cells in breast cancer patients.
Abbreviations:
|
ER
|
estrogen receptor
|
|
PgR
|
progesterone receptor
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
IHC
|
immunohistochemistry
|
|
EGFR
|
epidermal growth factor receptor
|
|
ALDH1
|
aldehyde dehydrogenase 1
|
|
STMN1
|
stathmin1
|
|
GOBO
|
gene expression-based outcome for
breast cancer online
|
|
KM
|
Kaplan-Meier
|
|
BCSCs
|
breast cancer stem cells
|
|
TMA
|
tissue microarray
|
|
TNBC
|
triple-negative breast cancer
|
|
CSC
|
cancer stem cells
|
|
LI
|
labeling index
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
RFS
|
recurrence-free survival
|
|
OS
|
overall survival
|
|
CI
|
confidence interval
|
|
HR
|
hazard ratio
|
Acknowledgments
The present study was supported by Grants-in-Aid for
Scientific Research from the Japan Society for the Promotion of
Science (grant no. 26461939). This study was also supported in part
by Uehara Zaidan; the Medical Research Encouragement Prize of The
Japan Medical Association; the Promotion Plan for the Platform of
Human Resource Development for Cancer and New
Paradigms-Establishing Centers for Fostering Medical Researchers of
the Future programs by the Ministry of Education, Culture, Sports,
Science and Technology of Japan; and the Gunma University
Initiative for Advanced Research.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Belmont LD and Mitchison TJ:
Identification of a protein that interacts with tubulin dimers and
increases the catastrophe rate of microtubules. Cell. 84:623–631.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Curmi PA, Gavet O, Charbaut E, Ozon S,
Lachkar-Colmerauer S, Manceau V, Siavoshian S, Maucuer A and Sobel
A: Stathmin and its phosphoprotein family: General properties,
biochemical and functional interaction with tubulin. Cell Struct
Funct. 24:345–357. 1999. View Article : Google Scholar
|
|
4
|
Cassimeris L: The oncoprotein 18/stathmin
family of microtubule destabilizers. Curr Opin Cell Biol. 14:18–24.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Curmi PA, Andersen SS, Lachkar S, Gavet O,
Karsenti E, Knossow M and Sobel A: The stathmin/tubulin interaction
in vitro. J Biol Chem. 272:25029–25036. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Curmi PA, Noguès C, Lachkar S, Carelle N,
Gonthier MP, Sobel A, Lidereau R and Bièche I: Overexpression of
stathmin in breast carcinomas points out to highly proliferative
tumours. Br J Cancer. 82:142–150. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arnedos M, Drury S, Afentakis M, A'Hern R,
Hills M, Salter J, Smith IE, Reis-Filho JS and Dowsett M: Biomarker
changes associated with the development of resistance to aromatase
inhibitors (AIs) in estrogen receptor-positive breast cancer. Ann
Oncol. 25:605–610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goldhirsch A, Winer EP, Coates AS, Gelber
RD, Piccart-Gebhart M, Thürlimann B, Senn HJ, Albain KS, André F,
Bergh J, et al Panel members: Personalizing the treatment of women
with early breast cancer: Highlights of the St Gallen International
Expert Consensus on the Primary Therapy of Early Breast Cancer
2013. Ann Oncol. 24:2206–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giatromanolaki A, Sivridis E, Fiska A and
Koukourakis MI: The CD44+/CD24− phenotype
relates to 'triple-negative' state and unfavorable prognosis in
breast cancer patients. Med Oncol. 28:745–752. 2011. View Article : Google Scholar
|
|
13
|
Idowu MO, Kmieciak M, Dumur C, Burton RS,
Grimes MM, Powers CN and Manjili MH:
CD44+/CD24−/low cancer stem/progenitor cells
are more abundant in triple-negative invasive breast carcinoma
phenotype and are associated with poor outcome. Hum Pathol.
43:364–373. 2012. View Article : Google Scholar
|
|
14
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ginestier C, Hur MH, Charafe-Jauffret E,
Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG,
Liu S, et al: ALDH1 is a marker of normal and malignant human
mammary stem cells and a predictor of poor clinical outcome. Cell
Stem Cell. 1:555–567. 2007. View Article : Google Scholar
|
|
16
|
Shafee N, Smith CR, Wei S, Kim Y, Mills
GB, Hortobagyi GN, Stanbridge EJ and Lee EY: Cancer stem cells
contribute to cisplatin resistance in Brca1/p53-mediated mouse
mammary tumors. Cancer Res. 68:3243–3250. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
To K, Fotovati A, Reipas KM, Law JH, Hu K,
Wang J, Astanehe A, Davies AH, Lee L, Stratford AL, et al: Y-box
binding protein-1 induces the expression of CD44 and CD49f leading
to enhanced self-renewal, mammosphere growth, and drug resistance.
Cancer Res. 70:2840–2851. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guarino M, Rubino B and Ballabio G: The
role of epithelial-mesenchymal transition in cancer pathology.
Pathology. 39:305–318. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saal LH, Johansson P, Holm K,
Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA,
Malmström P, Memeo L, et al: Poor prognosis in carcinoma is
associated with a gene expression signature of aberrant PTEN tumor
suppressor pathway activity. Proc Natl Acad Sci USA. 104:7564–7569.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Golouh R, Cufer T, Sadikov A, Nussdorfer
P, Usher PA, Brünner N, Schmitt M, Lesche R, Maier S, Timmermans M,
et al: The prognostic value of Stathmin-1, S100A2, and SYK proteins
in ER-positive primary breast cancer patients treated with adjuvant
tamoxifen monotherapy: An immunohistochemical study. Breast Cancer
Res Treat. 110:317–326. 2008. View Article : Google Scholar
|
|
22
|
Baquero MT, Hanna JA, Neumeister V, Cheng
H, Molinaro AM, Harris LN and Rimm DL: Stathmin expression and its
relationship to microtubule-associated protein tau and outcome in
breast cancer. Cancer. 118:4660–4669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alli E, Bash-Babula J, Yang JM and Hait
WN: Effect of stathmin on the sensitivity to antimicrotubule drugs
in human breast cancer. Cancer Res. 62:6864–6869. 2002.PubMed/NCBI
|
|
24
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumours. 7th edition.
Wiley-Blackwell; 2009
|
|
25
|
Tsuda H, Akiyama F, Kurosumi M, Sakamoto G
and Watanabe T; Japan National Surgical Adjuvant Study of Breast
Cancer(NSAS-BC) Pathology Section: Establishment of histological
criteria for high-risk node-negative breast carcinoma for a
multi-institutional randomized clinical trial of adjuvant therapy.
Jpn J Clin Oncol. 28:486–491. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al American Society of Clinical Oncology; College
of American Pathologists: Recommendations for human epidermal
growth factor receptor 2 testing in breast cancer: American Society
of Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dowsett M, Nielsen TO, A'Hern R, Bartlett
J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, et
al International Ki-67 in Breast Cancer Working Group: Assessment
of Ki-67 in breast cancer: Recommendations from the International
Ki-67 in Breast Cancer working group. J Natl Cancer Inst.
103:1656–1664. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheang MC, Chia SK, Voduc D, Gao D, Leung
S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, et al: Ki67
index, HER2 status, and prognosis of patients with luminal B breast
cancer. J Natl Cancer Inst. 101:736–750. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Allred DC, Harvey JM, Berardo M and Clark
GM: Prognostic and predictive factors in breast cancer by
immunohistochemical analysis. Mod Pathol. 11:155–168.
1998.PubMed/NCBI
|
|
30
|
Ringnér M, Fredlund E, Häkkinen J, Borg Å
and Staaf J; GOBO: Gene expression-based outcome for breast cancer
online. PLoS One. 6:e179112011. View Article : Google Scholar
|
|
31
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar
|
|
32
|
McGrogan BT, Gilmartin B, Carney DN and
McCann A: Taxanes, microtubules and chemoresistant breast cancer.
Biochim Biophys Acta. 1785:96–132. 2008.
|
|
33
|
Meng XL, Su D, Wang L, Gao Y, Hu YJ, Yang
HJ and Xie SN: Low expression of stathmin in tumor predicts high
response to neoadjuvant chemotherapy with docetaxel-containing
regimens in locally advanced breast cancer. Genet Test Mol
Biomarkers. 16:689–694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Alli E, Yang JM, Ford JM and Hait WN:
Reversal of stathmin-mediated resistance to paclitaxel and
vinblastine in human breast carcinoma cells. Mol Pharmacol.
71:1233–1240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iancu C, Mistry SJ, Arkin S and Atweh GF:
Taxol and anti-stathmin therapy: A synergistic combination that
targets the mitotic spindle. Cancer Res. 60:3537–3541.
2000.PubMed/NCBI
|
|
36
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao J: Cancer stem cells and
chemoresistance: The smartest survives the raid. Pharmacol Ther.
160:145–158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Biddle A, Liang X, Gammon L, Fazil B,
Harper LJ, Emich H, Costea DE and Mackenzie IC: Cancer stem cells
in squamous cell carcinoma switch between two distinct phenotypes
that are preferentially migratory or proliferative. Cancer Res.
71:5317–5326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu
Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al:
Breast cancer stem cells transition between epithelial and
mesenchymal states reflective of their normal counterparts. Stem
Cell Rep. 2:78–91. 2013. View Article : Google Scholar
|
|
41
|
Li N, Jiang P, Du W, Wu Z, Li C, Qiao M,
Yang X and Wu M: Siva1 suppresses epithelial-mesenchymal transition
and metastasis of tumor cells by inhibiting stathmin and
stabilizing microtubules. Proc Natl Acad Sci USA. 108:12851–12856.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu M, Bardia A, Wittner BS, Stott SL, Smas
ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al:
Circulating breast tumor cells exhibit dynamic changes in
epithelial and mesenchymal composition. Science. 339:580–584. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abell AN and Johnson GL: Implications of
mesenchymal cells in cancer stem cell populations: Relevance to
EMT. Curr Pathobiol Rep. 2:21–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Depowski PL, Rosenthal SI and Ross JS:
Loss of expression of the PTEN gene protein product is associated
with poor outcome in breast cancer. Mod Pathol. 14:672–676. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Saal LH, Holm K, Maurer M, Memeo L, Su T,
Wang X, Yu JS, Malmström PO, Mansukhani M, Enoksson J, et al:
PIK3CA mutations correlate with hormone receptors, node metastasis,
and ERBB2, and are mutually exclusive with PTEN loss in human
breast carcinoma. Cancer Res. 65:2554–2559. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Perren A, Weng LP, Boag AH, Ziebold U,
Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, Mutter GL,
et al: Immunohistochemical evidence of loss of PTEN expression in
primary ductal adenocarcinomas of the breast. Am J Pathol.
155:1253–1260. 1999. View Article : Google Scholar : PubMed/NCBI
|