Introduction
Based on GLOBOCAN 2012, gastric cancer (GC) is the
fifth most frequently diagnosed cancer and the third leading cause
of death from cancer worldwide (1). A recent study shows that an estimated
679,100 new GC cases and 498,000 deaths occurred in China (2). Except for a few regions where
screening is performed widely, GC is often presented in the
advanced stages in most parts of world. Surgery remains the primary
treatment for the patients with GC. Non-surgical treatments such as
chemotherapy radiotherapy and targeted therapies also play a key
role in prolonging patient life (3).
Radiation therapy can be an important part of
treatment for GC. Several randomized trials had assessed the
clinical effect of radiation therapy (preoperative, postoperative
or palliative) as treatment for GC (4–7).
Yet, these studies have provided neither uniform nor positive
results. In order to reduce radiation toxicity as much as possible,
intensity-modulated and 3-D conformal radiotherapy had been
developed (8). In the present
study with a different direction, we considered if there is a set
of genes that could predict the radiosensitivity of patients. This
would allow these patients to obtain the maximum benefit from
radiotherapy. The identification of molecular markers is a useful
tool for clinical management in GC patients, assisting in
diagnosis, in evaluation of response to treatment, and in
development of novel therapeutic modalities. Furthermore, an
identification of radiosensitive or non-radiosensitive signatures
for GC would be beneficial in guiding radiotherapy in clinical
practice.
We obtained the RNA sequence data for GC from The
Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/). An internal cross
validation was developed using the cross-validated adaptive
signature design. This design combined the gene signature
development and the validation test into a single data set, as
introduced by Freidlin et al (9), Freidlin and Simon (10) and Tang et al (11). Using this novel idea of combination
as a guide, we extended the approach to the proportional hazard
model and developed a radiosensitive gene signature for predicting
radiosensitive patients with GC.
Material and methods
Study samples
All data including clinical information and RNAseq
expression were downloaded from The Cancer Genome Atlas (TCGA,
http://cancergenome.nih.gov/, update at
March 2016). First, we combined the clinical information, including
survival time, radiotherapy, chemotherapy, and other information
from clinical files downloaded from TCGA. Clinical data are
available for 445 patients. Then, we filtered the patients with
missing survival information and obtained the data for 418
patients. After removing duplicated patients from raw data with 452
samples and 20,532 genes, expression data were selected including
418 patients and 20,502 genes with clearly identified gene names.
We merged the clinical and the expression data and obtain 393
patients for further analysis. Thereafter, we filtered the genes
with a maximum expression value <10 as they showed almost no
expression. Genes with proportion of zero expression >75% were
also removed. We calculated the variance of expression for each
gene and kept the genes with variance >20% quartile. Then we
standardized the expression data after filtering the patients with
missing radiotherapy information, obtaining data for 371 patients
with 16,125 genes expression profiles for the final analysis.
Lastly, we entered the missing values in clinical data by multiple
imputations using the R package mice. The cleaned clinical data are
summarized in Table I.
 | Table IPatients clinical characteristics and
results of univariate and multivariate Cox regression analysis. |
Table I
Patients clinical characteristics and
results of univariate and multivariate Cox regression analysis.
|
Characteristics | No. | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | p-values | HR (95% CI) | p-values |
|---|
| Sex | | | | | |
| Female | 130 | | | | |
| Male | 241 | 1.243
(0.881–1.752) | 0.2140 | 1.227
(0.824–1.826) | 0.3126 |
| Age (median, 65,
range, 30–90) | | | | | |
| <60 | 115 | | | | |
| ≥60 | 253 | 1.407
(0.978–2.024) | 0.0652 | 1.492
(0.956–2.327) | 0.0778 |
| (Unknown) | 3 | | | | |
| Race | | | | | |
| White | 239 | | | | |
| Non-white | 97 | 0.897
(0.602–1.336) | 0.5940 | 1.145
(0.719–1.821) | 0.5674 |
| (Unknown) | 35 | | | | |
| Histologic
diagnosis | | | | | |
| PT+TT | 82 | | | | |
| SRT+DT+MT | 96 | 0.877
(0.543–1.417) | 0.5930 | 0.991
(0.518–1.895) | 0.9776 |
| NOS | 193 | 1.172
(0.778–1.765) | 0.4460 | 1.205
(0.708–2.048) | 0.4918 |
| Tumor grade | | | | | |
| G1 | 9 | | | | |
| G2 | 132 | 1.104
(0.343–3.548) | 0.8680 | 1.266
(0.286–5.594) | 0.7559 |
| G3 | 221 | 1.514
(0.479–4.780) | 0.4800 | 1.643
(0.385–7.008) | 0.5025 |
| (Unknown) | 9 | | | | |
| Tumor stage | | | | | |
| I | 49 | | | | |
| II | 122 | 1.586
(0.806–3.117) | 0.1810 | 1.193
(0.400–3.550) | 0.7518 |
| III | 156 | 2.448
(1.297–4.622) | 0.0057 | 1.511
(0.364–6.268) | 0.5697 |
| IV | 29 | 3.636
(1.729–7.646) | 0.0007 | 4.878
(0.842–28.247) | 0.0769 |
| (Unknown) | 15 | | | | |
| T stage | | | | | |
| 1 | 18 | | | | |
| 2 | 81 | 6.124
(0.831–45.100) | 0.0752 | 3.977
(0.505–31.311) | 0.1898 |
| 3 | 167 | 9.082
(1.261–65.380) | 0.0285 | 3.518
(0.395–31.323) | 2.59e-01 |
| 4 | 101 | 9.911
(1.365–71.960) | 0.0233 | 3.296
(0.353–30.706) | 0.2951 |
| (Unknown) | 4 | | | | |
| N stage | | | | | |
| 1 | 112 | | | | |
| 2 | 105 | 1.693
(1.063–2.697) | 0.0266 | 1.745
(0.867–3.512) | 0.1186 |
| 3 | 68 | 1.640
(0.975–2.758) | 0.0621 | 1.370
(0.568–3.303) | 4.83e-01 |
| 4 | 75 | 2.652
(1.654–4.250) | 5.07E-05 | 2.110
(0.864–5.149) | 0.1009 |
| (Unknown) | 11 | | | | |
| M stage | | | | | |
| 1 | 339 | | | | |
| 2 | 18 | 1.831
(0.988–3.392) | 0.0545 | 0.267
(0.066–1.074) | 0.0629 |
| 3 | 14 | 1.637
(0.764–3.505) | 0.2048 | 1.935
(0.781–4.797) | 0.1537 |
| Target therapy | | | | | |
| No | 196 | | | | |
| Yes | 171 | 0.672
(0.483–0.932) | 0.0175 | 0.750
(0.475–1.185) | 0.2179 |
| (Unknown) | 4 | | | | |
| Radiotherapy | | | | | |
| No | 295 | | | | |
| Yes | 76 | 0.405
(0.253–0.646) | 0.0002 | 0.556
(0.307–1.004) | 0.0516 |
| Status | | | | | |
| Dead | 151 | | | | |
| Censor | 220 | | | | |
| Survival time
(month) | | | | | |
| Median (95%
CI) | | 30.86
(25.6–57.4) | | | |
| 5-Year survival
rate (%) | | 36.60
(28.0–47.8) | | | |
Methods
Gene signature development
In the present study, the radiosensitive patients
were defined as a group of patients who had better survival if they
received radiotherapy. To develop the patient radioactive sensitive
signature for predicting radio-sensitive patients, we used the
following modeling assumption: there is a subset of S
predictive/sensitive genes that significantly interact with
radiotherapy. The survival benefit of radiotherapy is associated
with these predictive genes through the Cox proportional hazards
model with the following equation:
h(t/X)=h0(t)exp(rλ+x1b1+x2b2+…+xSbS+rx1i1+rx2i2+…+rxSiS)
where h0(t) is the baseline hazard
function; λ is the effect of radiotherapy; r is an
indicator for radiotherapy with '1' indicating radiotherapy and '0'
otherwise; b1 to bS are the
main effects for these S sensitive genes; and
i1 to iS are
radiotherapy-expression interaction effects that reflect the degree
by which the effect of radiotherapy on survival is influenced by
the expression levels of sensitive genes.
If the main effects and radiotherapy-expression
interaction effects are negative, patients who overexpress the
sensitive genes will have a higher survival probability under
radiotherapy as compared with non-radiotherapy. We assume that a
fraction of the patient population overexpress some, but not
necessarily all, of the sensitive genes. The total hazard ratio
(HR) would tend to be less than a preset threshold value (such as
<1). Then, these patients who have a relative high probability
of survival are called radiosensitive patients.
Cross-validation procedure
Freidlin et al (9), and Freidlin and Simon (10) developed a novel cross-validated
adaptive signature design to identify sensitive patients in
clinical trial for binary outcome. Following their framework, we
extended and modified this approach to a proportional hazards model
(11) and applied it to develop a
radiosensitive gene signature for the present data. A K-fold
cross-validated procedure for gene signature development is
described by the following three-step procedure.
Step 1. Training step. The data were randomly split
into K parts with the same sample size (usually
K=10). Then, K-1 parts were used as training data to
fit the model and to predict the radiosensitive patients in the
left-out part (validation data). In the training data for each gene
j, Cox proportional hazards model was fit using the
following equation:
h(t/X)=h0(t)exp(rλ+xjbj+rxjij).
Then, the p-values for ij were used to rank and
select the genes.
Step 2. Prediction step. The top significant
g genes was used to build a gene signature, and to calculate
an index called nominal HR (nHR) using the equation
for patients in the validation data (k-th part). Here,
λ was the value averaged over the estimates from g
single gene models. Patients in the validation set who had nHR
lower than a specified threshold R were classified as
radiosensitive patients.
Step 3. Validation step. The above two procedures in
steps 1 and 2 were cycled through and validated on each of the
K pieces in turn. Each study patient only appears once in
one of the validation data. After the cross validation, each
patient is classified as either radiosensitive or not
radiosensitive. For radiosensitive patients, a log-rank test was
then performed to test the survival difference between radiotherapy
and non-radiotherapy groups at a specified significance level, such
as 0.05. A significant result indicated that radiotherapy is
beneficial for predicted radiosensitive patients. Then the gene
signature was considered potentially effective and the prediction
of radiosensitive patients was accurate.
In the above procedure, there are two key tuning
parameters g and R in the prediction step. The
optimal values of the tuning parameters g and R are
usually not known in advance. Therefore, all the possible
combinations for g and R were tried and tested. We
used a nested inner loop of K-fold cross-validation approach
on the training data to select the best tuning parameter values
without affecting statistical validity of the procedure. A similar
procedure could be found in Freidlin et al (9) and Freidlin and Simon (10). More details with a work flow plot
can also be found in the supplementary files in our previous study
(11).
The 10-fold cross validation in the above procedure
was recommended which permitted the maximization of the portion of
study patients contributing to the development of the diagnostic
signature and the minimization of prediction error (12). Beyond 10-fold cross validation,
leave-one-out cross-validation (LOOCV) is often mentioned in
internal validation. It is known that LOOCV could provide similar
and stable results, compared with 10-fold cross-validation.
However, LOOCV can be very time-consuming to implement (12).
Results
Survival analysis on clinical
information
Table I summarizes
the results of the clinical information. The median survival is
30.86 months with the 95% CI (25.6–57.4). The 5-year survival rate
is 36.60% (28.0–47.8%). Univariate analysis shows that several
clinical factors, including radiotherapy, are significant factors
for overall survival. However, multivariate survival analysis shows
that radiotherapy did not show significance for the overall
survival, with an HR of 0.556(0.307–1.004) and p-value of
0.0516.
Development of radiosensitive gene
signature
Following the proposed three-step procedure, we
analyzed the present data to get the tuning parameters by 10-fold
cross-validation. Then, the gene signature was developed for
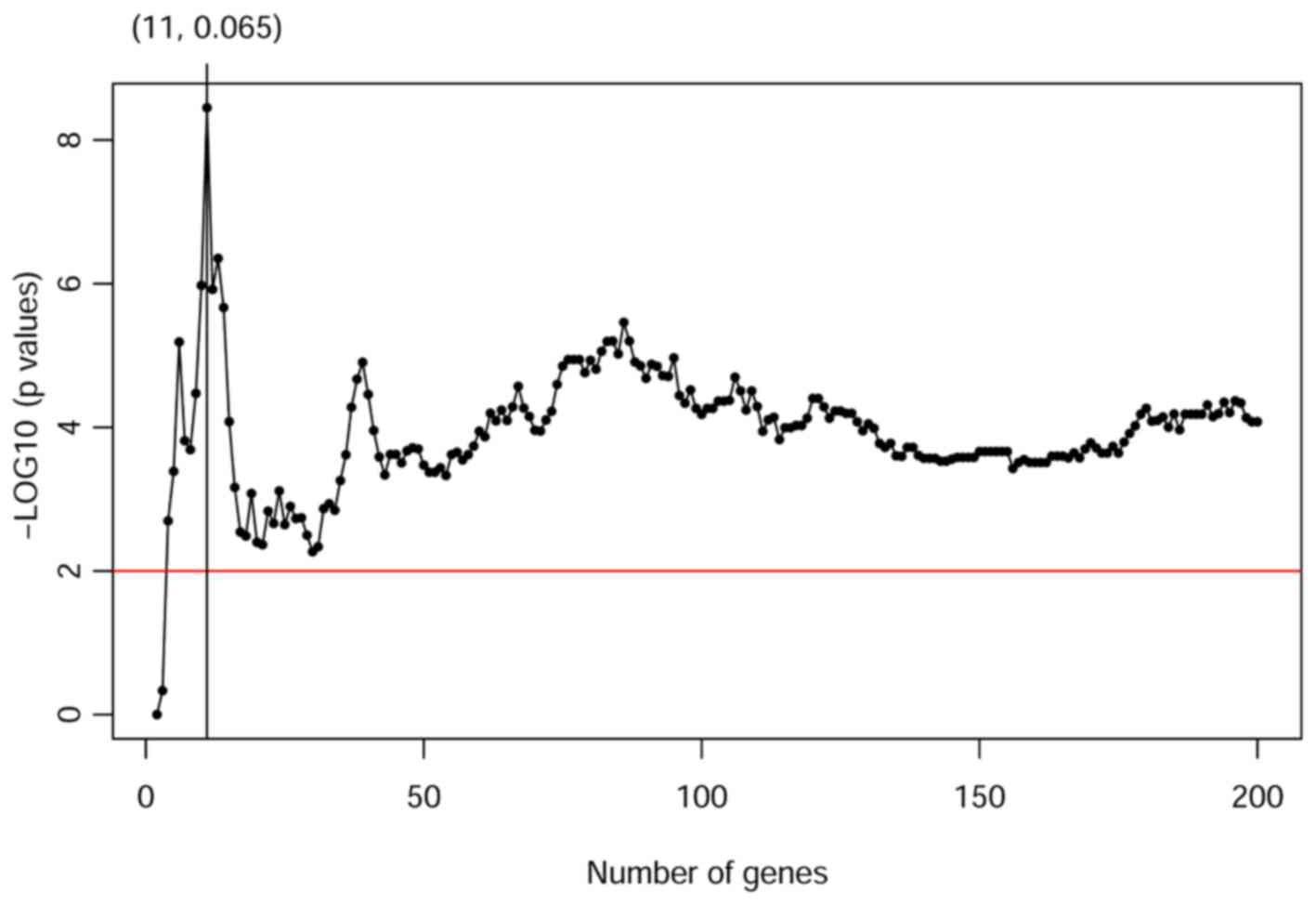
predicting radiosensitivity. Fig.
1 shows the corresponding p-values profiled by log-rank test
among radiotherapy and non-radiotherapy groups for predicted
sensitive patients. Our result shows that gene signatures including
the top 11 significant genes can provide a powerful prediction with
the smallest p-value as 3.810E-09. The smallest p-value appears
when the tuning parameters g and R are 11 and 0.065,
respectively. Table II summarized
the 11 genes included in the radiosensitive gene signature and
their interaction effects with radiotherapy.
 | Table IIThe 11 genes included in the
radiosensitive gene signature and their interaction effects with
radiotherapy. |
Table II
The 11 genes included in the
radiosensitive gene signature and their interaction effects with
radiotherapy.
| Gene names | Main effects of
genes (SE) | p-values | Main effects of
radiotherapy (SE) | p-values | Interaction effects
(SE) | p-values |
|---|
| 1.
GLMN | −1.0481
(0.2760) | 0.0001 | 0.1221
(0.0925) | 0.1870 | −1.3360
(0.3227) | 0.0000 |
| 2.
C14orf135 | −0.8550
(0.2435) | 0.0004 | 0.4067
(0.0934) | 0.0000 | −1.0661
(0.2604) | 0.0000 |
| 3.
KIAA0586 | −0.9099
(0.2475) | 0.0002 | 0.2797
(0.0895) | 0.0018 | −0.8933
(0.2252) | 0.0001 |
| 4.
SFRS5 | −1.0882
(0.2695) | 0.0001 | 0.1512
(0.0876) | 0.0844 | −0.9563
(0.2443) | 0.0001 |
| 5.
C9orf16 | −0.9900
(0.2570) | 0.0001 | −0.2145
(0.0856) | 0.0122 | 1.0739
(0.2831) | 0.0001 |
| 6.
SPSB2 | −1.1388
(0.2847) | 0.0001 | −0.1390
(0.0893) | 0.1197 | 1.0097
(0.2749) | 0.0002 |
| 7.
MUDENG | −1.0455
(0.2687) | 0.0001 | 0.0463
(0.0922) | 0.6156 | −0.8751
(0.2414) | 0.0003 |
| 8.
DNAL4 | −0.8938
(0.2480) | 0.0003 | −0.1486
(0.0859) | 0.0836 | 0.9083
(0.2510) | 0.0003 |
| 9.
ACD | −0.9373
(0.2521) | 0.0002 | −0.1245
(0.0866) | 0.1506 | 1.0289
(0.2852) | 0.0003 |
| 10. TRMT5 | −1.0208
(0.2639) | 0.0001 | 0.2305
(0.0895) | 0.0100 | −0.8575
(0.2378) | 0.0003 |
| 11. ZMYM6 | −0.8313
(0.2392) | 0.0005 | 0.1353
(0.0879) | 0.1235 | −0.7539
(0.2199) | 0.0006 |
Validation of radiosensitive gene
signature
Following the standard validation procedure we
proposed herein, 189 patients were predicted as radiosensitive
patients, while the other 182 patients considered
non-radiosensitive. We compared the survival of radiosensitive
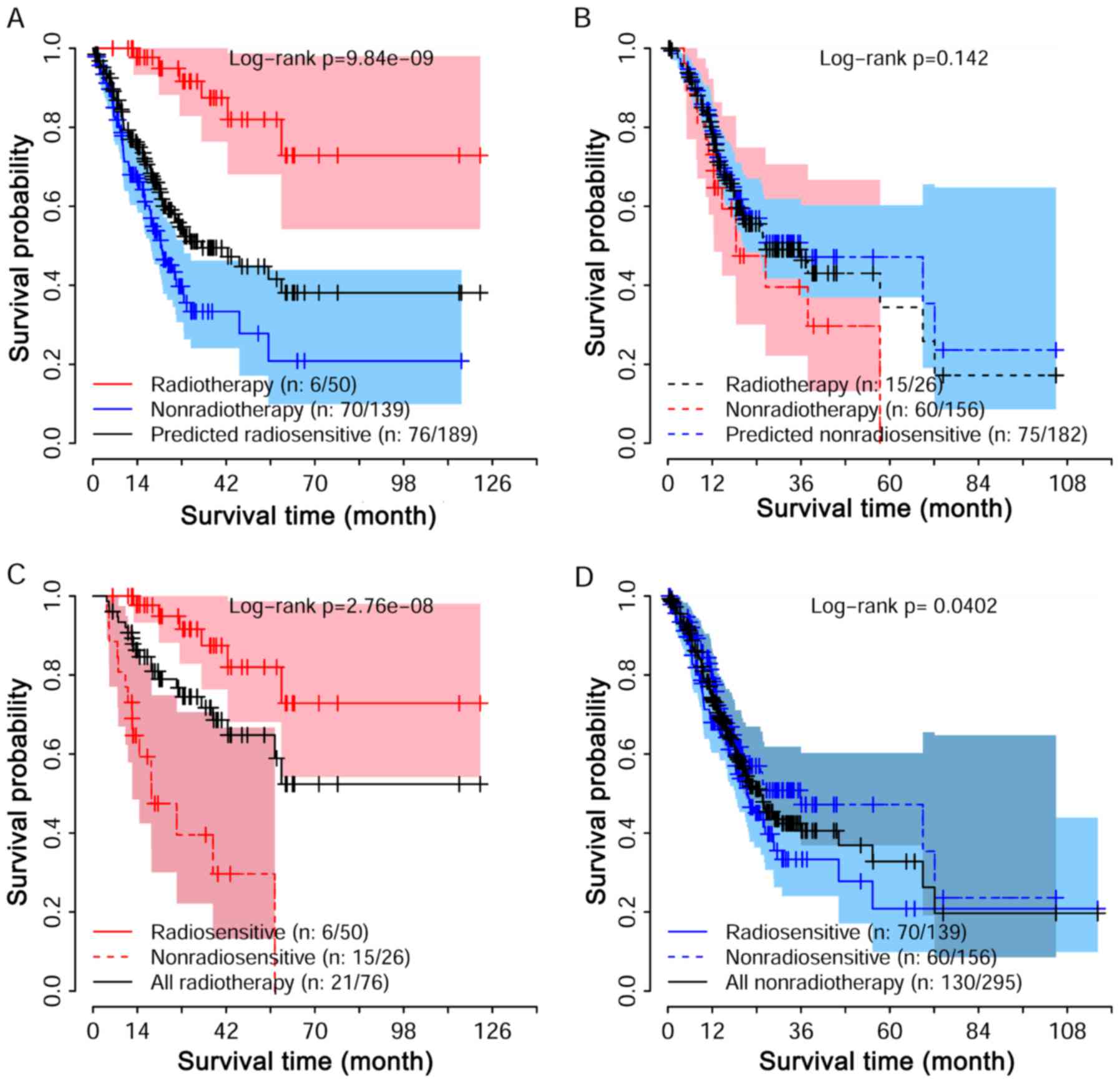
patients who received radiotherapy and non-radiothreapy. Fig. 2A shows the survival curve for these
predicted radiosensitivity patients. The significant difference
with p-value 9.84e-09 suggested that the predicted radiosensitive
patients were reasonable, as they strongly benefited from
radiotherapy. Fig. 2B shows the
comparison for non-radiosensitive patients under radiotherapy and
non-radiotherapy. No obvious difference was detected between the
two groups, suggesting that the benefit of radiotherapy on these
non-radiosensitive patients may not be as expected. We further
compared the survival among radiosensitive and non-radiosensitive
patients when they were all under radiotherapy treatment as shown
in Fig. 2C. As expected, a strong
positive effect of radiotherapy on radiosensitive patients was
observed. Taken together, the predicted radiosensitive and
non-radiosensitive patients were reasonable results. The
radiosensitive gene signature is predictive for both radiosensitive
and non-radiosensitive in radiotherapy. Fig. 1D shows that predicted
radiosensitive patients had a poorer survival than the predicted
non-radiosensitive patients when they were all under
non-radiotherapy.
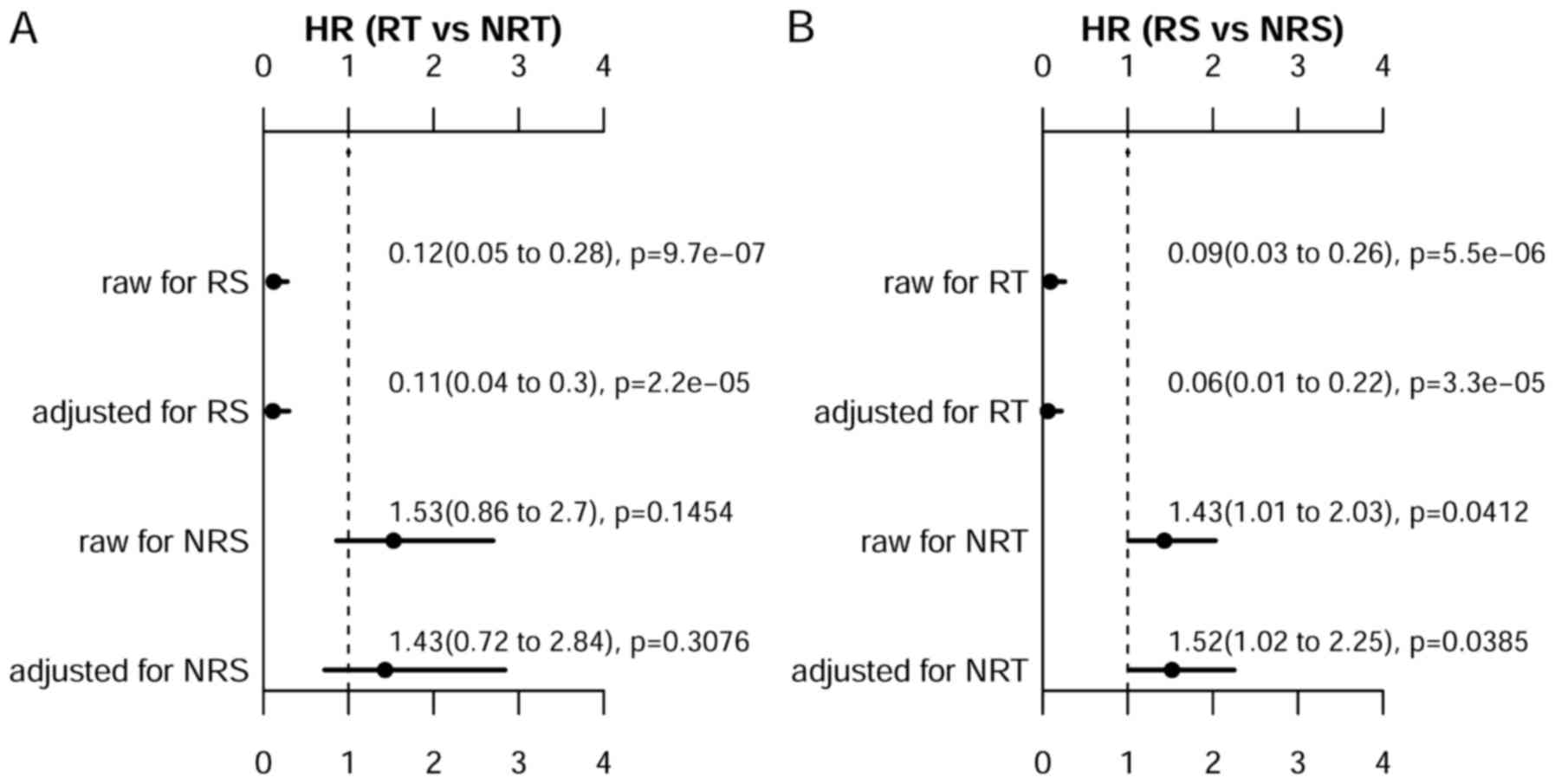
We further performed univariate and multivariate
analyses using the cox proportional hazards regression analysis to
assess prognostic benefit amount of radiotherapy. Fig. 3A demonstrates that radiotherapy was
strongly associated with the improved survival for
non-radiosensitive patients, with adjusted HR of 0.11 (0.04–0.30).
For non-radiosensitive patients, radiotherapy may not improve the
overall survival, with adjusted HR of 1.43 (0.72–2.84). We also
compared the survival between predicted radiosensitive patients and
non-radiosensitive, when both of these patients received
radiotherapy as shown in Fig. 3B.
It is clearly shown that there is a significant survival benefit
for predicted radiosensitive patients as compared with
non-radiosensitive patients, with adjusted HR of 0.06 (0.01–0.22).
These results suggest that the prediction on radiosensitive patient
was accurate, and the positive radiotherapy effect on predicted
radiosensitive patients was effectively validated as expected.
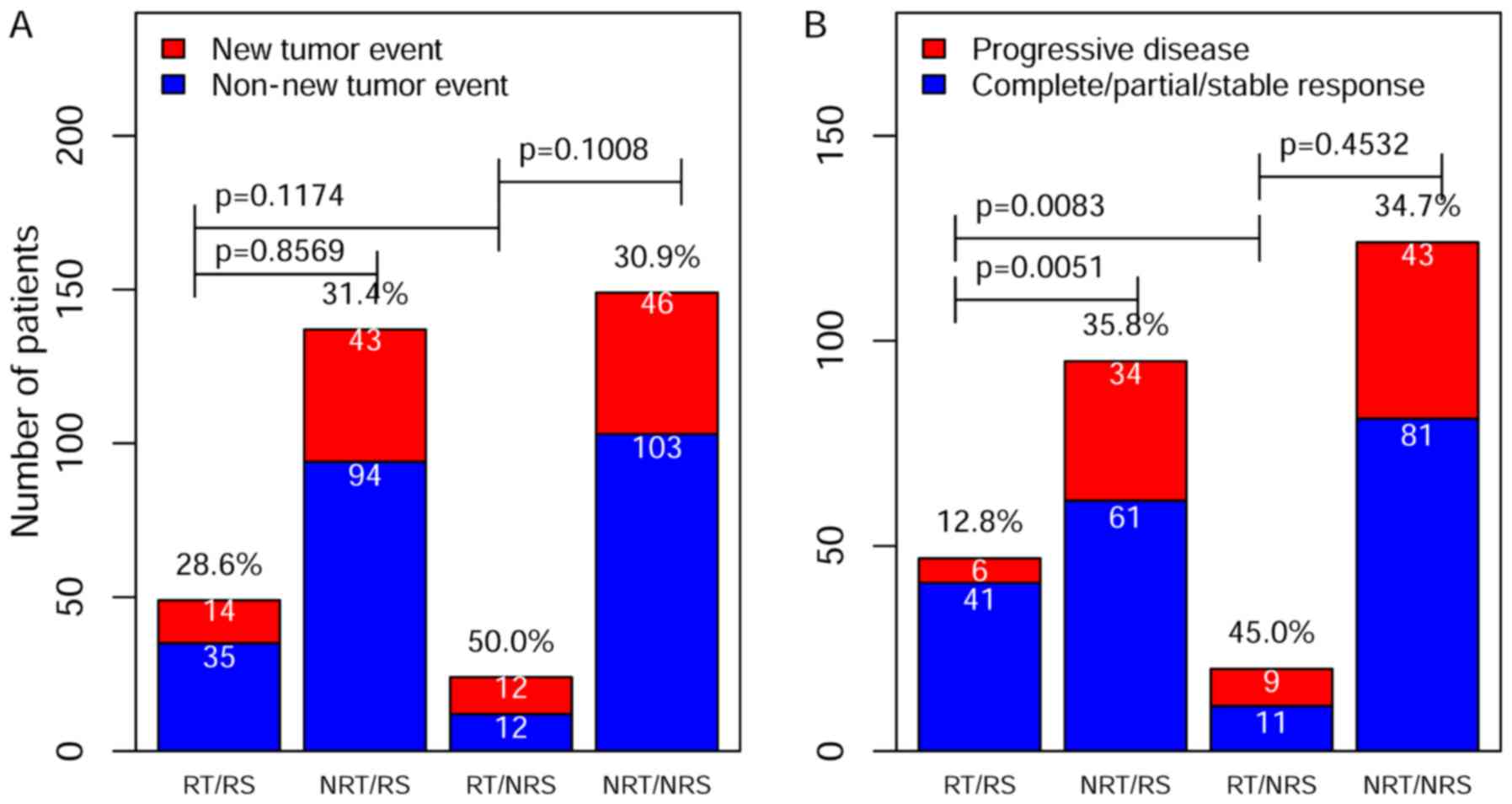
Association among radiotherapy and
clinical assessments after adjuvant treatments
To further validate the predicted radiosensitive and
non-radiosensitive patients, we performed an association study
between radiotherapy and two clinical assessment indexes: the new
tumor event and the progressive disease. The new tumor event is the
important clinical index for prognostic outcome. According to TCGA,
the new tumor event was defined as metastatic recurrent and new
primary tumor after initial treatment. The results of Chi-square
analyses are summarized in Fig. 4A
for the new tumor event and in Fig.
4B for the progressive disease. Although the results did not
suggest a significant difference in a lower rate of the new tumor
event among different groups, the rate of progressive disease were
significantly lower for predicted radiosensitive patients who
received radiotherapy (Fig. 4B).
These results were consistent with survival analysis and further
validated our predicted sensitivity results.
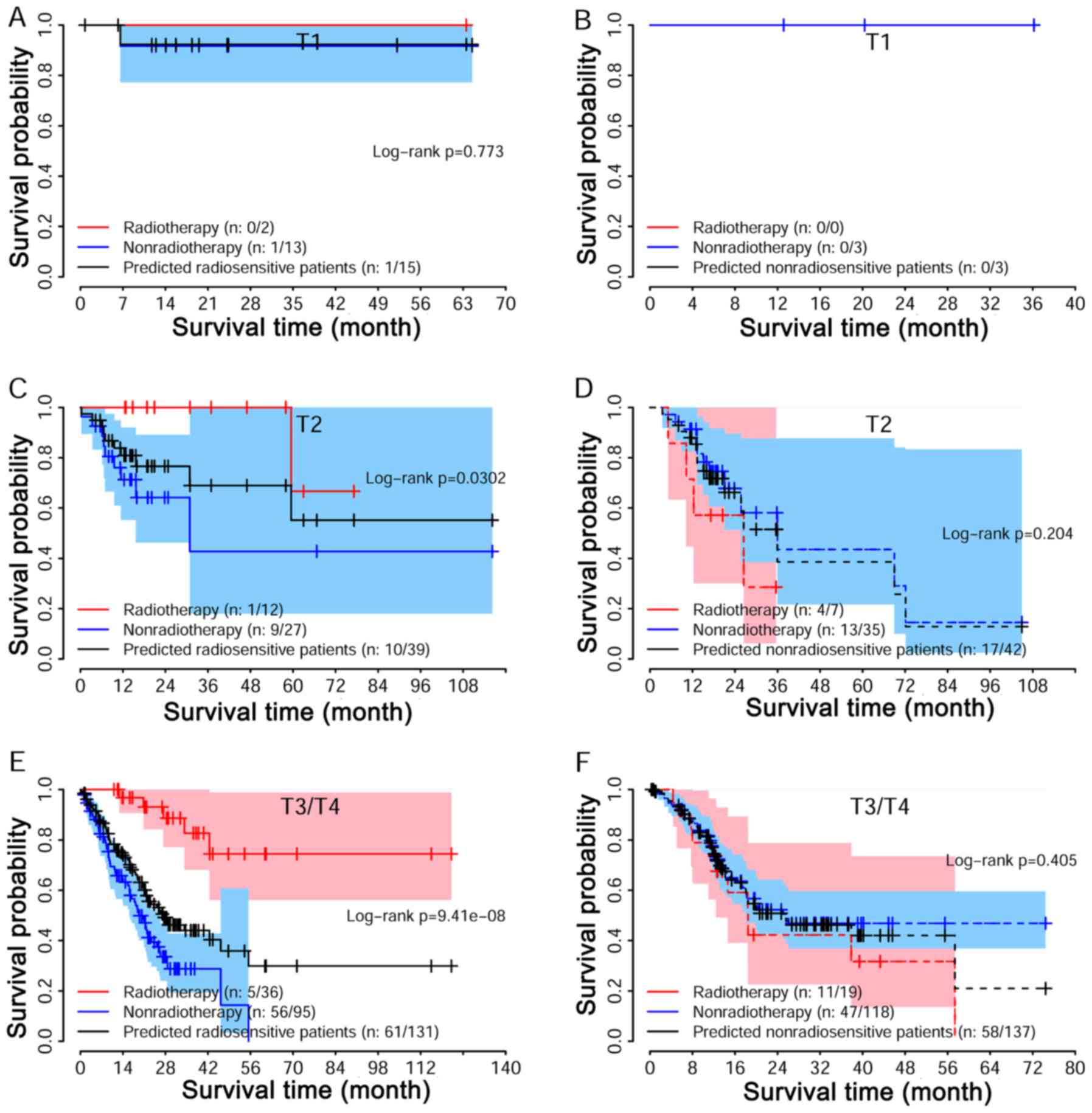
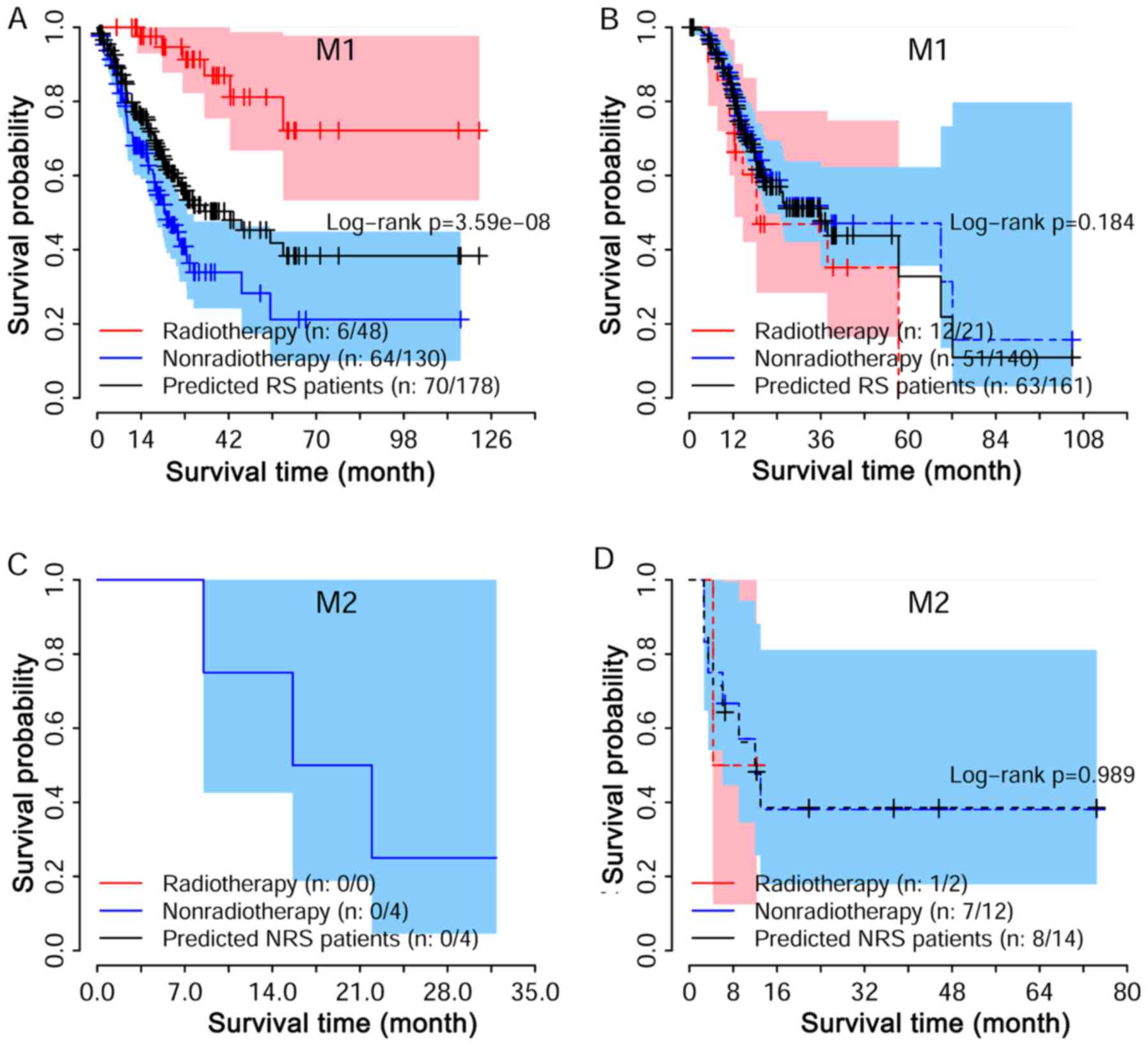
Associations among radiosensitivity and
clinical factors
To find the association between predicted
sensitivity and clinical factors, we performed univariate and
multivariable logistic analyses. Table III summarizes the results. The
univariate and multivariable analyses suggested that only T and M
stages had a significant association with predicted
radiosensitivity. We performed a strata analysis for T and M
stages. Log-rank tests suggested that the predicted radio-sensitive
patients in the radiotherapy group had better survival as compared
with non-radiotherapy group, independent of stages (Fig. 5A, C and E). For non-radiosensitive
patients, radiotherapy may not be a factor that is beneficial
(Fig. 5B, D and F). A similar
result was also reached for different M stage (Fig. 6).
 | Table IIIAssociation study among predicted
radiosensitivity and clinical factors. |
Table III
Association study among predicted
radiosensitivity and clinical factors.
| Characteristic | Sensitive
patients | Non-sensitive
patients | Univariate analysis
p-values | Multivariate
analysis p-values |
|---|
| Sex | | | 0.4173 | 0.4727 |
| Female | 62 | 68 | | |
| Male | 127 | 114 | | |
| Age (years) | 0.5014 | 0.4672 | | |
| <60 | 55 | 60 | | |
| ≥60 | 131 | 122 | | |
| Race | | | 0.5472 | 0.9313 |
| White | 113 | 126 | | |
| Non-white | 50 | 47 | | |
| Histologic | | | 0.0583 | |
| diagnosis | | | | |
| PT+TT | 51 | 31 | | |
| SRT+DT+MT | 45 | 51 | | 0.0643 |
| NOS | 90 | 100 | | 0.0560 |
| Tumor grade | | | | 0.3653 |
| G1 | 5 | 4 | | |
| G2 | 74 | 58 | | 0.8374 |
| G3 | 107 | 114 | | 0.7570 |
| Tumor stage | | | 0.08427 | |
| I | 30 | 19 | | |
| II | 52 | 70 | | 0.1186 |
| III | 85 | 71 | | 0.1295 |
| IV | 13 | 16 | | 0.4059 |
| T stage | | | | 0.0001 |
| 1 | 15 | 3 | | |
| 2 | 39 | 42 | | 0.0457 |
| 3 | 68 | 99 | | 0.1004 |
| 4 | 63 | 38 | | 0.9298 |
| N stage | | | | 0.7032 |
| 1 | 52 | 60 | | |
| 2 | 55 | 50 | | 0.1103 |
| 3 | 34 | 34 | | 0.2484 |
| 4 | 41 | 34 | | 0.0939 |
| M stage | | | | 0.0427 |
| 1 | 178 | 161 | | |
| 2 | 4 | 14 | | 0.0075 |
| 3 | 7 | 7 | | 0.4680 |
| Target therapy | | | 0.0595 | 0.0562 |
| No | 91 | 105 | | |
| Yes | 97 | 74 | | |
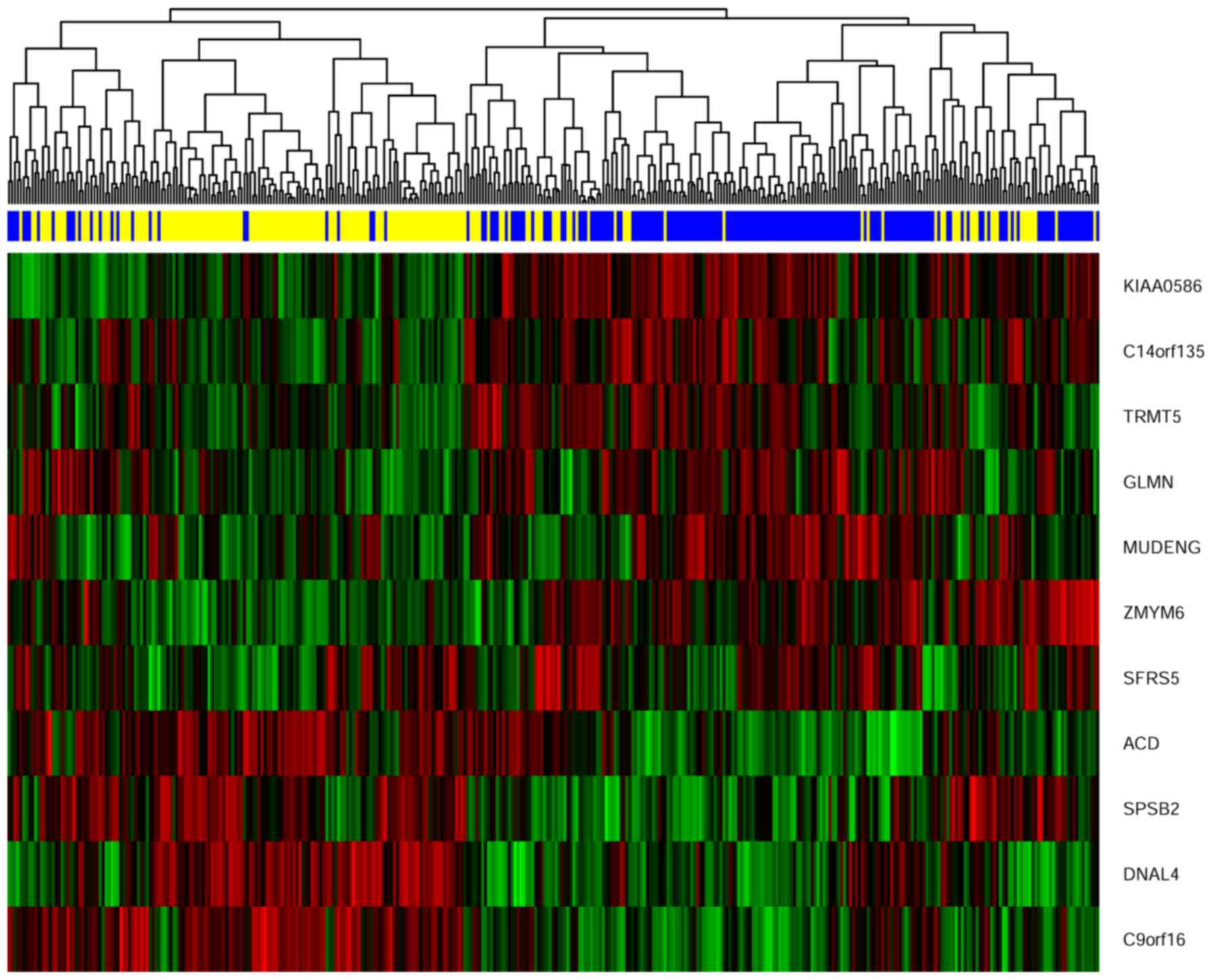
Gene signature and cluster analysis
We extracted the expression pattern of these 11
genes to perform a hierarchical cluster analysis by using R
packages pheatmap. As shown in Fig.
7. All patients were classified into two groups according to
the hierarchical cluster analysis. The blue bar below the
dendrogram denotes the predicted radiosensitive patients, while the
yellow bar denotes the predicted non-radiosensitive patients.
Interestingly, most of the predicted radiosensitive and
non-radiosensitive patients were well matched with the result of
the hierarchical cluster based upon the selected gene signature.
The dendrogram shows 162 out of all 189 predicted radiosensitive
patients located on the right branch and 128 out of all 182
predicted non-radiosensitive patients located the left branch. This
result further validated the predictions of radiosensitive patients
and suggested that the radiosensitive gene signature we developed
is predictive and reasonable.
Discussion
GC is a radioresponsive cancer. Radiotherapy plays
an important role in the treatment of locally advanced GC in
preoperative, intraoperative, postoperative or palliative settings
(5,7,13–18).
The clinical trial conducted by Zhang et al suggested that
preoperative radiotherapy improves resection rates and survival
(4). But radiotherapy as a single
modality treatment cannot improve survival in patients with locally
unresectable GC (5). There are
several trials that have evaluated radiotherapy in a combination
modality treatment for GC. For instance, in 2001, the pivotal
intergroup trial SWOG 9008/INT-0116 has established a standard
postoperative chemoradiation therapy (CRT) in the management of GC
(6). Their results suggested that
postoperative CRT should be considered for all patients at high
risk for recurrence of GC and who have undergone curative resection
with D0 or D1 lymph node dissection. However, D2 lymph node
dissection, which is considered a standard lymph node dissection
for advanced GC, was not widely performed at the time. Therefore,
the recently completed phase III trial, the ARTIST trial, evaluated
the role of postoperative CRT in patients who received curative
resection with D2 lymph node dissection. Regrettably, their results
showed that postoperative CRT did not significantly reduce
recurrence (7).
Radiotherapy still has controversial effects on
treatment for GC. In theory, radiotherapy is a double-edged sword,
which not only can kill tumors but also can cause damage to
surrounding normal tissue. In fact, 41% of patients in INT-0116 had
grade 3 or 4 toxic effects, with 17% stopped treatment because of
toxic effect (6). Therefore, in
order to solve this problem, most studies have focused on how to
more accurately carry out radiotherapy to reduce the damage of
normal tissue (8,19–21).
In this study, we questioned whether we could find a way to
identify the patients who would be more sensitive to radiotherapy.
These patients could then receive the maximum therapeutic effect
with minimal toxicity. As advocated by the principles of precision
medicine, the treatments should be targeted on the need of
individual patients on the basis of genetic characteristics
(22). With continued progress in
identifying biomarkers of radiotherapy response, the role of
radiotherapy in GC treatments will likely become better defined.
Therefore, we developed a radiosensitive gene signature for GC
patients.
External validation is the best way to develop gene
signatures, especially the gene signature development based on
high-dimensional gene expression data (23–25).
In this study, a nested inner loop 10-fold cross validation was
used to find the radiosensitive gene signature and radiosensitive
patients. The 10-fold cross validation maximized the portion of
patients in the study contributing to the development of a gene
signature and minimized prediction error (12). In addition, it also maximized the
size of the sensitive patient subset used to validate the signature
error (12). Moreover, the
proposed cross-validation procedure could evaluate the major
clinical outcomes, including side effects of radiotherapy on
radiosensitive patients. Except for the 10-fold cross validation, a
split-sample method and LOOCV are frequently used for internal
validation. For computationally burdensome analyses, 10-fold CV may
be preferable to LOOCV (12). For
current nested inner loop cross validation procedure, LOOCV may
lengthen the time it takes to complete the analysis. Usually, the
results provided by the 10-fold cross validation is very similar to
LOOCV (12).
In this study, we developed a new index, nominal HR,
in a three-step procedure to identify GC patients that would be
more sensitive by radiotherapy. Using the new index makes a clear
separation of the patients, which makes it easy to identify
radiosensitive patients. A radiosensitive patient is predicted if
the estimated odds ratio (for binary outcome) appears below a
specified threshold (R) for at least g of the
significant genes (9,10). Our model using a product clearly
estimates the sensitivity amount of each patient, which improves
the ability to predict. The proposed model evaluated the whole
genome expression data. The 11 genes were found to be the
radiosensitive gene signature for GC patients.
Our results showed that the predicted radiosensitive
patients who received radiotherapy had significantly better
survival than both the radiosensitive patients without radiotherapy
and non-radiosensitive patients who received radiotherapy (Fig. 2A and C). However, radiotherapy did
not improve the survival of predicted non-radiosensitive patients
(Fig. 2B). After adjusting for
other clinical factors, a multivariate analysis suggested that
radiotherapy was an independent factor of benefit on the predicted
radiosensitive patients (Fig. 3).
The reduced rate of the new tumor event and progressive disease
were observed for predicted radiosensitive patients who received
radiotherapy, which further provided strong positive evidence for
our prediction (Fig. 4). Although
the clinical stage was strongly associated with the predicted
radiosensitivity, the survival of the predicted radiosensitive
patients who received radiotherapy was significantly better than
radiosensitive patients without radiotherapy, independent of stage
(Figs. 5 and 6). The overlap of results from cluster
analysis and predicted radiosensitive and non-radiosensitive
patients also validated the radiosensitive gene signature (Fig. 7). Taken together, these validation
results reveal that the identified radiosensitive gene signature is
a powerful biomarker for predicting which GC patients would benefit
from radiotherapy.
Our analysis not only developed a radiosensitive
gene signature, but also detected genes that may be potentially
associated with the molecular basis of GC. Based on the results, we
find that several genes, including C9orf16, DNAL4, SPSB2 and ACD,
are highly expressed in radiosensitive patients. Among these,
C9orf16 is considered to be related to ovarian cancer (26), but it has not been studied in
gastric cancer. DNAL4 encodes an axonemal dynein light chain that
functions as a component of the outer dynein arms complex, acting
as the molecular motor that provides the force to move cilia in an
ATP-dependent manner. Furthermore, the protein encoded by ACD plays
a key role in the assembly and stabilization of the
telosome/shelter-intelomeric complex, which functions to maintain
telomere length and to protect telomere ends. SPSB2 encodes a
member of a subfamily of proteins containing a central SPRY
(repeats in splA and RyR) domain and a C-terminal suppressor of
cytokine signaling (SOCS) box. This protein plays a role in cell
signaling. Despite the lack of studies on these genes in gastric
cancer, our results show that these genes may play a key role in
the progression of gastric cancer. The detailed mechanism of how
these genes function in gastric cancer will be the main aim of our
next study.
Acknowledgments
We acknowledge the contributions of the TCGA
Research Network. This study was supported by the National Natural
Science Foundation of China (no. 81573253 and 81773541) to Z.-X.T.,
a project funded by Jiangsu Provincial Medical Youth Talent to
J.Z., a project funded by Suzhou Science and Technology Bureau (no.
SYS201672) to H.-G.H., and a project funded by the Priority
Academic Program Development of Jiangsu Higher Education
Institutions at Soochow University.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ajani JA, D'Amico TA, Almhanna K, Bentrem
DJ, Chao J, Das P, Denlinger CS, Fanta P, Farjah F, Fuchs CS, et
al: Gastric cancer, version 3.2016, NCCN Clinical Practice
Guidelines in Oncology. J Natl Compr Canc Netw. 14:1286–1312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang ZX, Gu XZ, Yin WB, Huang GJ, Zhang
DW and Zhang RG: Randomized clinical trial on the combination of
preoperative irradiation and surgery in the treatment of
adenocarcinoma of gastric cardia (AGC) - report on 370 patients.
Int J Radiat Oncol Biol Phys. 42:929–934. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hazard L, O'Connor J and Scaife C: Role of
radiation therapy in gastric adenocarcinoma. World J Gastroenterol.
12:1511–1520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Macdonald JS, Smalley SR, Benedetti J,
Hundahl SA, Estes NC, Stemmermann GN, Haller DG, Ajani JA,
Gunderson LL, Jessup JM, et al: Chemoradiotherapy after surgery
compared with surgery alone for adenocarcinoma of the stomach or
gastroesophageal junction. N Engl J Med. 345:725–730. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee J, Lim DH, Kim S, Park SH, Park JO,
Park YS, Lim HY, Choi MG, Sohn TS, Noh JH, et al: Phase III trial
comparing capecitabine plus cisplatin versus capecitabine plus
cisplatin with concurrent capecitabine radiotherapy in completely
resected gastric cancer with D2 lymph node dissection: The ARTIST
trial. J Clin Oncol. 30:268–273. 2012. View Article : Google Scholar
|
|
8
|
Minn AY, Hsu A, La T, Kunz P, Fisher GA,
Ford JM, Norton JA, Visser B, Goodman KA, Koong AC, et al:
Comparison of intensity-modulated radiotherapy and 3-dimensional
conformal radiotherapy as adjuvant therapy for gastric cancer.
Cancer. 116:3943–3952. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Freidlin B, Jiang W and Simon R: The
cross-validated adaptive signature design. Clin Cancer Res.
16:691–698. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Freidlin B and Simon R: Adaptive signature
design: an adaptive clinical trial design for generating and
prospectively testing a gene expression signature for sensitive
patients. Clin Cancer Res. 11:7872–7878. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang Z, Zeng Q, Li Y, Zhang X, Ma J, Suto
MJ, Xu B and Yi N: Development of a radiosensitivity gene signature
for patients with soft tissue sarcoma. Oncotarget. 8:27428–27439.
2017.PubMed/NCBI
|
|
12
|
Molinaro AM, Simon R and Pfeiffer RM:
Prediction error estimation: A comparison of resampling methods.
Bioinformatics. 21:3301–3307. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moertel CG, Childs DS Jr, Reitemeier RJ,
Colby MY Jr and Holbrook MA: Combined 5-fluorouracil and
supervoltage radiation therapy of locally unresectable
gastrointestinal cancer. Lancet. 2:865–867. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lowy AM, Feig BW, Janjan N, Rich TA,
Pisters PW, Ajani JA and Mansfield PF: A pilot study of
preoperative chemoradiotherapy for resectable gastric cancer. Ann
Surg Oncol. 8:519–524. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rödel C, Liersch T, Becker H, Fietkau R,
Hohenberger W, Hothorn T, Graeven U, Arnold D, Lang-Welzenbach M,
Raab HR, et al German Rectal Cancer Study Group: Preoperative
chemo-radiotherapy and postoperative chemotherapy with fluorouracil
and oxaliplatin versus fluorouracil alone in locally advanced
rectal cancer: Initial results of the German CAO/ARO/AIO-04
randomised phase 3 trial. Lancet Oncol. 13:679–687. 2012.
View Article : Google Scholar
|
|
16
|
Hong YS, Nam BH, Kim KP, Kim JE, Park SJ,
Park YS, Park JO, Kim SY, Kim TY, Kim JH, et al: Oxaliplatin,
fluorouracil, and leucovorin versus fluorouracil and leucovorin as
adjuvant chemotherapy for locally advanced rectal cancer after
preoperative chemoradiotherapy (ADORE): An open-label, multicentre,
phase 2, randomised controlled trial. Lancet Oncol. 15:1245–1253.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dikken JL, Jansen EP, Cats A, Bakker B,
Hartgrink HH, Kranenbarg EM, Boot H, Putter H, Peeters KC, van de
Velde CJ, et al: Impact of the extent of surgery and postoperative
chemoradiotherapy on recurrence patterns in gastric cancer. J Clin
Oncol. 28:2430–2436. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smalley SR, Benedetti JK, Haller DG,
Hundahl SA, Estes NC, Ajani JA, Gunderson LL, Goldman B, Martenson
JA, Jessup JM, et al: Updated analysis of SWOG-directed intergroup
study 0116: A phase III trial of adjuvant radiochemotherapy versus
observation after curative gastric cancer resection. J Clin Oncol.
30:2327–2333. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu GF, Bair RJ, Bair E, Liauw SL and
Koshy M: Clinical outcomes for gastric cancer following adjuvant
chemoradiation utilizing intensity modulated versus
three-dimensional conformal radiotherapy. PLoS One. 9:e826422014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Li G, Zhang Y, Bai S, Xu F, Wei Y
and Gong Y: Single-arc volumetric-modulated arc therapy (sVMAT) as
adjuvant treatment for gastric cancer: Dosimetric comparisons with
three-dimensional conformal radiotherapy (3D-CRT) and
intensity-modulated radiotherapy (IMRT). Med Dosim. 38:395–400.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dahele M, Skinner M, Schultz B, Cardoso M,
Bell C and Ung YC: Adjuvant radiotherapy for gastric cancer: A
dosimetric comparison of 3-dimensional conformal radiotherapy,
tomotherapy and conventional intensity modulated radiotherapy
treatment plans. Med Dosim. 35:115–121. 2010. View Article : Google Scholar
|
|
22
|
Jameson JL and Longo DL: Precision
medicine - personalized, problematic, and promising. N Engl J Med.
372:2229–2234. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martens FK, Kers JG and Janssens AC:
External validation is only needed when prediction models are worth
it (Letter commenting on: J Clin Epidemiol. 2015;68:25–34). J Clin
Epidemiol. 69:249–250. 2016. View Article : Google Scholar
|
|
24
|
Bleeker SE, Moll HA, Steyerberg EW,
Donders AR, Derksen-Lubsen G, Grobbee DE and Moons KG: External
validation is necessary in prediction research: A clinical example.
J Clin Epidemiol. 56:826–832. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bijlsma MW, Brouwer MC, Bossuyt PM,
Heymans MW, van der Ende A, Tanck MW and van de Beek D: Risk scores
for outcome in bacterial meningitis: Systematic review and external
validation study. J Infect. 73:393–401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Chen C, Li HF, Jiang XL and Zhang
L: Investigating key genes associated with ovarian cancer by
integrating affinity propagation clustering and mutual information
network analysis. Eur Rev Med Pharmacol Sci. 20:2532–2540.
2016.PubMed/NCBI
|