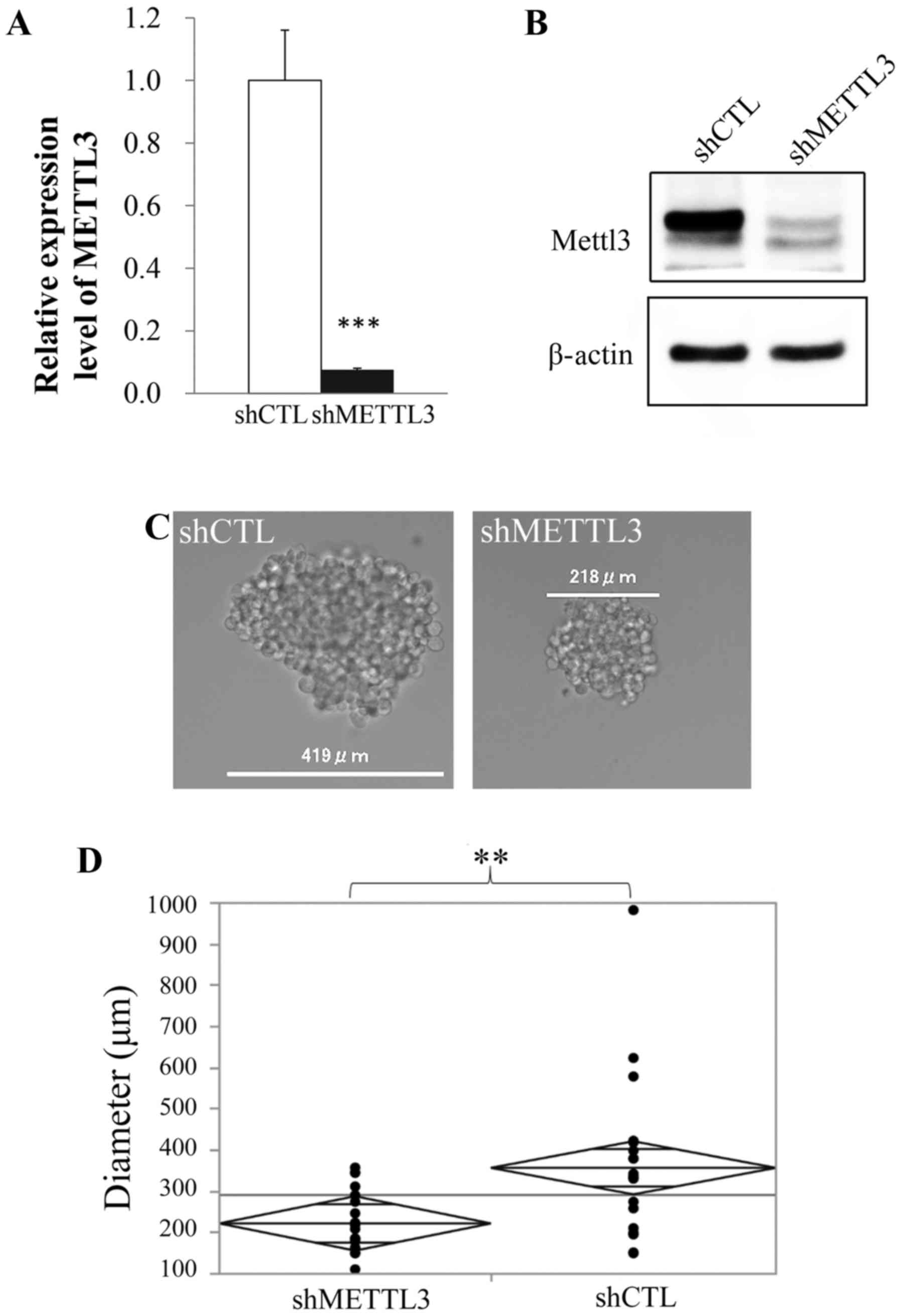
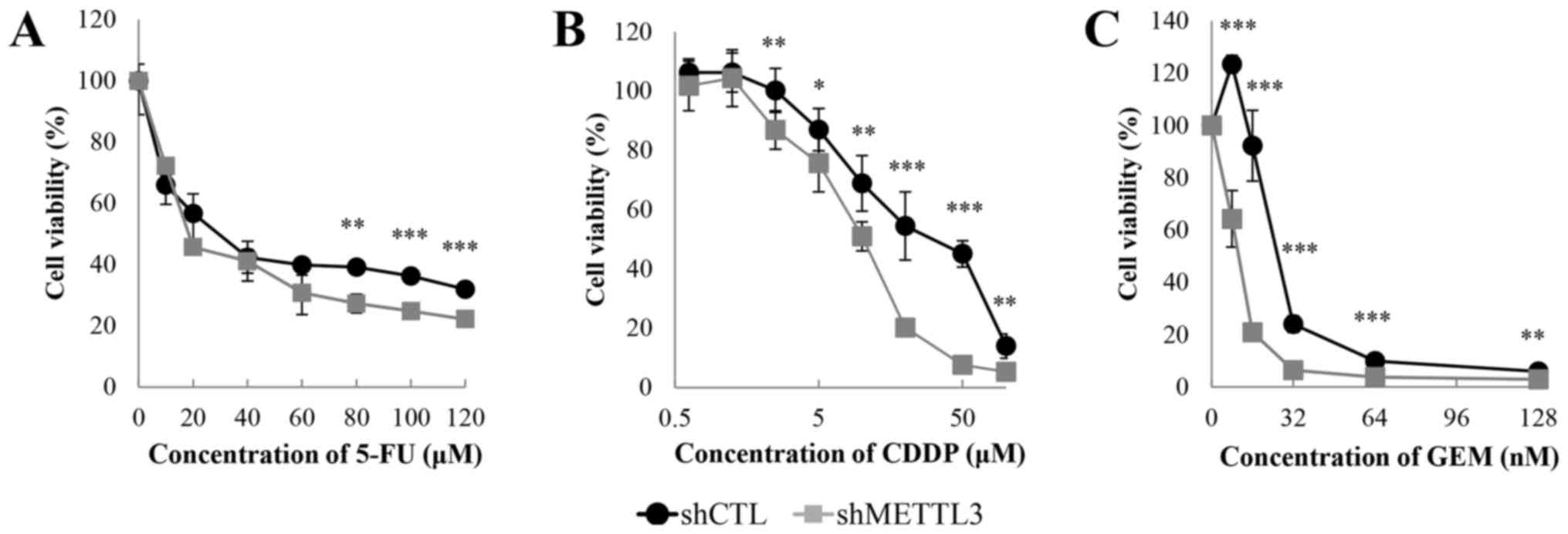
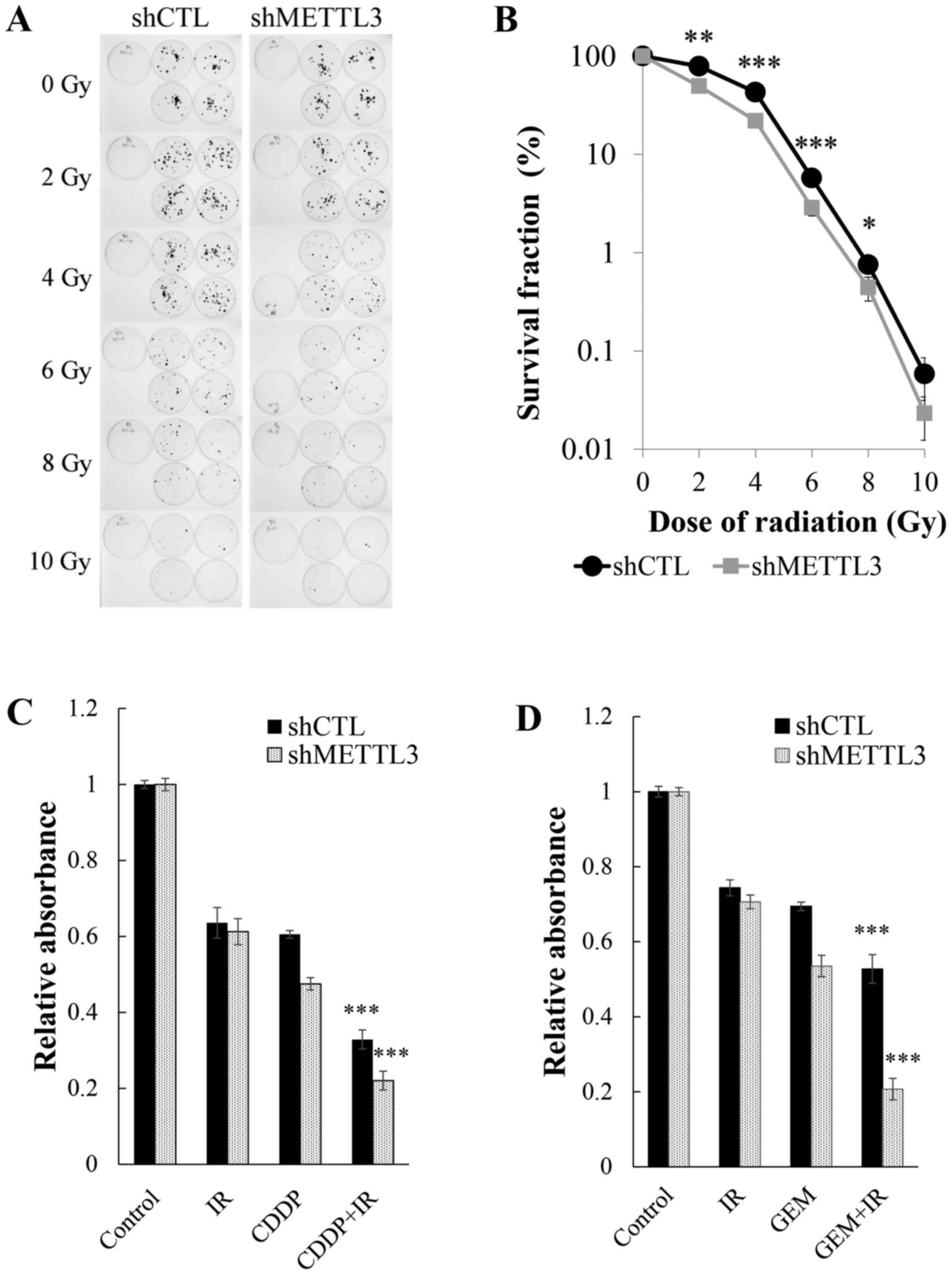
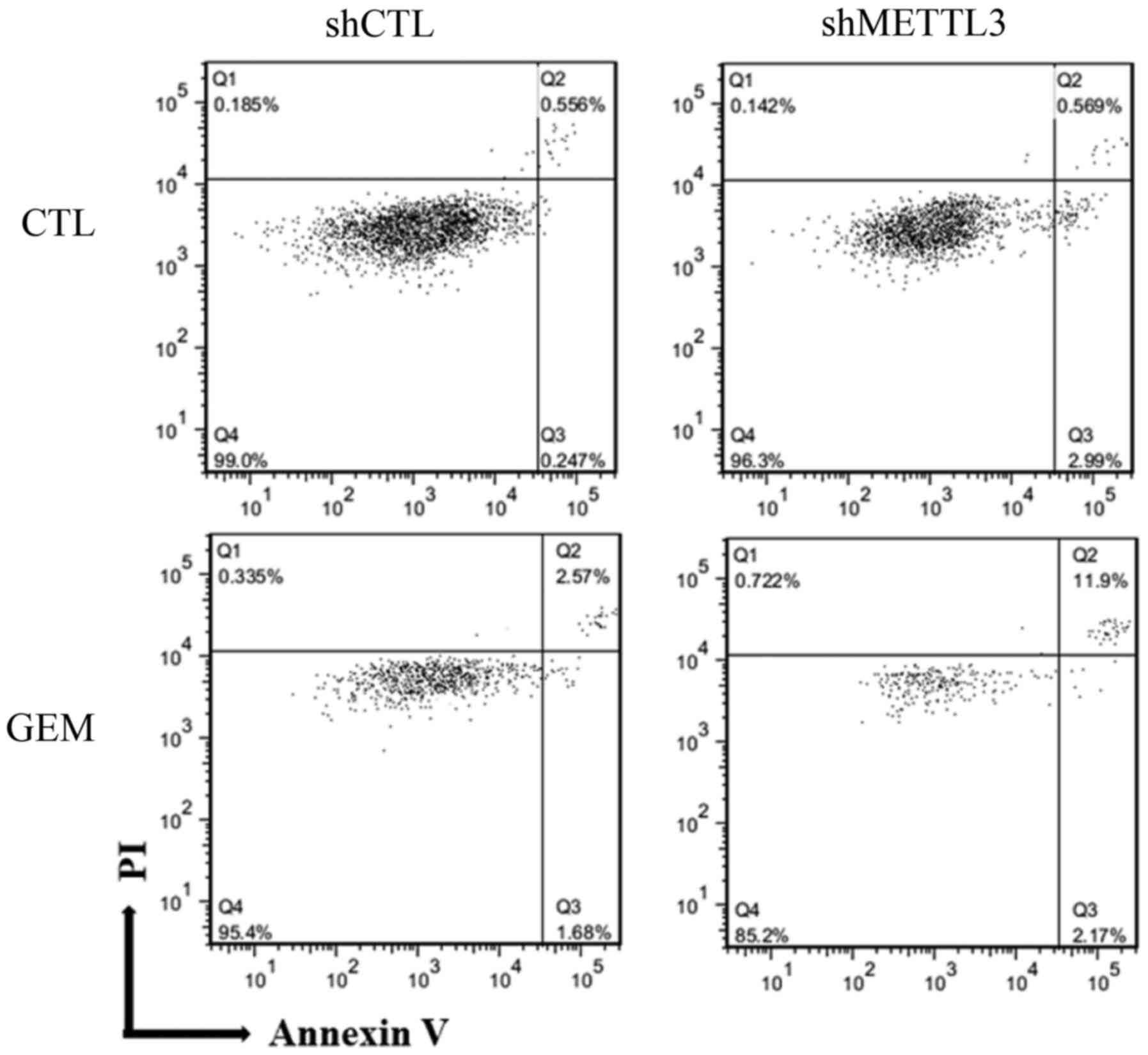
|
1
|
Waddington CH: The epigenotype. Endeavour.
1:18–20. 1942.
|
|
2
|
Cantara WA, Crain PF, Rozenski J,
McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D and Agris
PF: The RNA Modification Database, RNAMDB: 2011 update. Nucleic
Acids Res. 39(Database): D195–D201. 2011. View Article : Google Scholar :
|
|
3
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltrans-ferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
5
|
Perry RP and Kelley DE: Existence of
methylated messenger RNA in mouse L cells. Cell. 1:37–42. 1974.
View Article : Google Scholar
|
|
6
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by
m6A-seq. Nature. 485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA
stability. Nature. 505:117–120. 2014. View Article : Google Scholar
|
|
10
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m6A Methyltransferase METTL3 promotes
translation in human cancer cells. Mol Cell. 62:335–345. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Batista PJ, Molinie B, Wang J, Qu K, Zhang
J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al:
m6A RNA modification controls cell fate transition in
mammalian embryonic stem cells. Cell Stem Cell. 15:707–719. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG, et al:
N6-methyladenosine in nuclear RNA is a major substrate
of the obesity-associated FTO. Nat Chem Biol. 7:885–887. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar :
|
|
14
|
Liu N, Dai Q, Zheng G, He C, Parisien M
and Pan T: N6-methyladenosine-dependent RNA structural
switches regulate RNA-protein interactions. Nature. 518:560–564.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fustin JM, Doi M, Yamaguchi Y, Hida H,
Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I,
et al: RNA-methylation-dependent RNA processing controls the speed
of the circadian clock. Cell. 155:793–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M,
Ma J and Wu L: YTHDF2 destabilizes m6A-containing RNA
through direct recruitment of the CCR4-NOT deadenylase complex. Nat
Commun. 7:126262016. View Article : Google Scholar
|
|
17
|
Wang Y, Li Y, Toth JI, Petroski MD, Zhang
Z and Zhao JC: N6-methyladenosine modification
destabilizes developmental regulators in embryonic stem cells. Nat
Cell Biol. 16:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m6A promotes cap-independent translation. Cell.
163:999–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR
and Qian SB: Dynamic m6A mRNA methylation directs
translational control of heat shock response. Nature. 526:591–594.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear
m6A reader YTHDC1 regulates mRNA splicing. Mol Cell.
61:507–519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary
microRNAs for processing. Nature. 519:482–485. 2015. View Article : Google Scholar
|
|
22
|
Alarcón CR, Goodarzi H, Lee H, Liu X,
Tavazoie S and Tavazoie SF: HNRNPA2B1 is a mediator of
m6A-dependent nuclear RNA processing events. Cell.
162:1299–1308. 2015. View Article : Google Scholar
|
|
23
|
Chen T, Hao YJ, Zhang Y, Li MM, Wang M,
Han W, Wu Y, Lv Y, Hao J, Wang L, et al: m6A RNA
methylation is regulated by microRNAs and promotes reprogramming to
pluripotency. Cell Stem Cell. 16:289–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Geula S, Moshitch-Moshkovitz S,
Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V,
Peer E, Mor N, Manor YS, et al: Stem cells m6A mRNA
methylation facilitates resolution of naïve pluripotency toward
differentiation. Science. 347:1002–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m6A RNA methylation
promotes XIST-mediated transcriptional repression. Nature.
537:369–373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiang Y, Laurent B, Hsu CH, Nachtergaele
S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al: RNA
m6A methylation regulates the ultraviolet-induced DNA
damage response. Nature. 543:573–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang SY, Zhang SW, Liu L, Meng J and
Huang Y: m6A-Driver: Identifying context-specific mRNA
m6A methylation-driven gene interaction networks. PLOS
Comput Biol. 12:e10052872016. View Article : Google Scholar
|
|
28
|
Franken NA, Rodermond HM, Stap J, Haveman
J and van Bree C: Clonogenic assay of cells in vitro. Nat Protoc.
1:2315–2319. 2006. View Article : Google Scholar
|
|
29
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
30
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar
|
|
31
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tempero MA, Malafa MP, Al-Hawary M, Asbun
H, Bain A, Behrman SW, Benson AB III, Binder E, Cardin DB, Cha C,
et al: Pancreatic adenocarcinoma, version 2.2017, NCCN Clinical
Practice Guidelines in Oncology. J Natl Compr Cancer Netw.
15:1028–1061. 2017. View Article : Google Scholar
|
|
34
|
Dhillon AS, Hagan S, Rath O and Kolch W:
MAP kinase signalling pathways in cancer. Oncogene. 26:3279–3290.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Corre I, Paris F and Huot J: The p38
pathway, a major pleiotropic cascade that transduces stress and
metastatic signals in endothelial cells. Oncotarget. 8:55684–55714.
2017.PubMed/NCBI
|
|
36
|
Gong F and Miller KM: Mammalian DNA
repair: HATs and HDACs make their mark through histone acetylation.
Mutat Res. 750:23–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shan W, Jiang Y, Yu H, Huang Q, Liu L, Guo
X, Li L, Mi Q, Zhang K and Yang Z: HDAC2 overexpression correlates
with aggressive clinicopathological features and DNA-damage
response pathway of breast cancer. Am J Cancer Res. 7:1213–1226.
2017.PubMed/NCBI
|
|
38
|
Livingstone C: IGF2 and cancer. Endocr
Relat Cancer. 20:R321–R339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Moens U, Kostenko S and Sveinbjørnsson B:
The role of mitogen-activated protein kinase-activated protein
kinases (MAPKAPKs) in inflammation. Genes (Basel). 4:101–133. 2013.
View Article : Google Scholar
|
|
41
|
Koltai T: Clusterin: A key player in
cancer chemoresistance and its inhibition. Onco Targets Ther.
7:447–456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Albany C and Hahn NM: Heat shock and other
apoptosis-related proteins as therapeutic targets in prostate
cancer. Asian J Androl. 16:359–363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Somasagara RR, Spencer SM, Tripathi K,
Clark DW, Mani C, Madeira da Silva L, Scalici J, Kothayer H,
Westwell AD, Rocconi RP, et al: RAD6 promotes DNA repair and stem
cell signaling in ovarian cancer and is a promising therapeutic
target to prevent and treat acquired chemoresistance. Oncogene. Aug
14–2017.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rui L, Yuan M, Frantz D, Shoelson S and
White MF: SOCS-1 and SOCS-3 block insulin signaling by
ubiquitin-mediated degradation of IRS1 and IRS2. J Biol Chem.
277:42394–42398. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Beaurivage C, Champagne A, Tobelaim WS,
Pomerleau V, Menendez A and Saucier C: SOCS1 in cancer: An oncogene
and a tumor suppressor. Cytokine. 82:87–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cano F, Rapiteanu R, Sebastiaan Winkler G
and Lehner PJ: A non-proteolytic role for ubiquitin in
deadenylation of MHC-I mRNA by the RNA-binding E3-ligase MEX-3C.
Nat Commun. 6:86702015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Burrell RA, McClelland SE, Endesfelder D,
Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM,
Gronroos E, et al: Replication stress links structural and
numerical cancer chromosomal instability. Nature. 494:492–496.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Eblen ST: Regulation of chemoresistance
via alternative messenger RNA splicing. Biochem Pharmacol.
83:1063–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang J and Manley JL: Misregulation of
pre-mRNA alternative splicing in cancer. Cancer Discov.
3:1228–1237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Savage KI, Gorski JJ, Barros EM, Irwin GW,
Manti L, Powell AJ, Pellagatti A, Lukashchuk N, McCance DJ,
McCluggage WG, et al: Identification of a BRCA1-mRNA splicing
complex required for efficient DNA repair and maintenance of
genomic stability. Mol Cell. 54:445–459. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Perry WL III, Shepard RL, Sampath J, Yaden
B, Chin WW, Iversen PW, Jin S, Lesoon A, O'Brien KA, Peek VL, et
al: Human splicing factor SPF45 (RBM17) confers broad multidrug
resistance to anticancer drugs when overexpressed - a phenotype
partially reversed by selective estrogen receptor modulators.
Cancer Res. 65:6593–6600. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Correa BR, de Araujo PR, Qiao M, Burns SC,
Chen C, Schlegel R, Agarwal S, Galante PA and Penalva LO:
Functional genomics analyses of RNA-binding proteins reveal the
splicing regulator SNRPB as an oncogenic candidate in glioblastoma.
Genome Biol. 17:1252016. View Article : Google Scholar : PubMed/NCBI
|