Introduction
Esophageal cancer (EC) is the sixth leading cause of
cancer-related mortality worldwide (1). In China, EC is ranked as the fourth
leading cause of cancer-related mortality, and an estimated 218,957
individuals succumbed to the disease in China in 2011 (2). In China, the majority of EC cases are
esophageal squamous cell carcinoma (ESCC) (3). ESCC is usually diagnosed at an
advanced stage and its prognosis is poor (4). Approximately 80% of ESCC cases occur
in the Central and South-East Asian region. China alone contributes
to more than half of the global cases (53%, 210,000 cases)
(5). A distinctive characteristic
of EC in China is its uneven burden between rural and urban areas.
Indeed, the rates of EC are 2- to 10-fold higher in rural areas
compared with urban areas (2).
Kazakh patients show particularly high morbidity
(68.88/100,000) and mortality rates from ESCC, particularly in the
Tuoli (Xinjiang) Province, which exhibits the highest incidence
(155.8/100,000) of EC worldwide. The incidence of ESCC among Kazakh
patients is 4-fold higher than the general incidence in China
(14.95/100,000) (6).
The treatment of ESCC involves a multidisciplinary
approach combining surgery, chemotherapy and radiation therapy
(7). Nevertheless, even in
developed countries, the 5-year overall survival (OS) of patients
with ESCC is only 15% (4),
stressing the need for novel markers for screening and prognosis of
ESCC.
The ubiquitin-proteasome pathway (UPP) consists of
enzyme 1 (E1; or ubiquitin activating enzyme), enzyme 2 (E2; or
ubiquitin-conjugating enzyme), enzyme 3 (E3; or ubiquitin ligase)
and 26S proteasome (8). The UPP
influences and regulates apoptosis and plays a central role in the
regulation of ubiquitin binding and substrate specificity (9). To date, only E3 ligases of the
RING-type have been found to participate in Snail and Twist
ubiquitination, which play important roles in
epithelial-mesenchymal transition (EMT) (10,11).
The degradation of most EMT factors is strictly dependent upon
multi-subunit RING-type E3s (11).
Ring finger protein 113A (RNF113A), previously known
as ZNF183, is located on chromosome Xq24.9 (12). RNF113A has two conserved zinc
finger domains: AC3H1-type zinc finger domain and a C3HC4 zinc
finger domain. Zinc finger domains are shared by various tumor
suppressors, DNA repair proteins and cytokine receptor-associated
molecules (13). The C3H1-type
RNF113A zinc finger domain is often found in RNA-binding proteins
involved in splicing, while zf-C3HC4 is considered to be an
ubiquitin-related structural domain (14) and is often found in E3 ubiquitin
ligases (15).
A number of studies have indicated that there are
close associations between ubiquitin ligase and the occurrence,
development and metastasis of cancer (16,17).
Previous studies have found that the disruption of RNF113A in
zebrafish (Danio rerio) by transgenic insertional
mutagenesis results in a small and slightly necrotic head, small
eyes, pericardial edema and an underdeveloped liver and gut
(12,18). In Caenorhabditis elegans,
the knockdown of RNF113 has been shown to sensitize the cells to
ultraviolet A (UVA)-induced DNA damage and is required for RAD51
focus formation during DNA interstrand crosslink repair (19). Corbett et al (12) and Pellagatti et al (20) reported that RNF113A mutations are
associated with non-photosensitive trichothiodystrophy (TTD).
RNF113A may regulate neuronal differentiation in the human central
nervous system (15). Pellagatti
et al (20) studied gene
expression profiles in the neutrophils of 21 patients with
myelodysplastic syndrome (MDS) using cDNA microarrays comprising
6,000 human genes, and found that ZNF183 was one of the most
upregulated genes in MDS (20).
Therefore, it can be hypothesized RNF113A is associated with the
development and progression of ESCC.
To date, and at least to the best of our knowledge,
there are no available studies on the role of RNF113A in cancer.
Therefore, this study aimed to explore the role of RNF113A in the
development of ESCC by comparing the expression of RNF113A in
Kazakh patients with ESCC and paired adjacent non-tumor tissues.
The biological roles of RNF113A were also investigated by inducing
either the overexpression or knockdown of RNF113A in the ESCC cell
lines, Eca109 and EC9706. Finally, xenograft tumors in athymic nude
mice were created out to assess the tumorigenic effect of RNF113A
in vivo.
Materials and methods
Study design, patients and specimens
This study was approved by the Medical Ethics
Committee of the First Affiliated Hospital of Xinjiang Medical
University, Urumqi, China. Informed consent was obtained from all
subjects prior to obtaining the samples. Paraffin-embedded samples
from 117 Kazakh patients with ESCC treated at our hospital from
March 2010 to December 2014, were selected. Paired ESCC and
adjacent normal tissues were available for 41 patients. None of the
patients had received chemoradiotherapy or radiotherapy prior to
surgery. ESCC was staged and graded according to the International
Union Against Cancer (21). The
patients were followed-up by consulting their charts and through
telephonic monitoring. Follow-up data were available for periods
ranging from 3 months to 6.8 years (median, 20.8 months).
Immunohistochemistry (IHC)
The samples were incubated with anti-human RNF113A
antibody (1:50 dilution, HPA000160; Sigma, St. Louis, MO, USA)
overnight at 4°C, and then with biotinylated secondary antibody
(PV6001; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.,
Beijing, China). IHC analysis was carried out according to
previously described methods (22). The staining intensity was assessed
using 5 randomly selected high-power fields (×400 magnification).
The staining was evaluated by two independent pathologists who were
blinded to the clinical characteristics of the patients. Any
differences in opinion were resolved by consensual agreement, based
on the criteria by Sinicrope et al (23). Staining was assessed with regard to
intensity (0, no intensity; 1, weak intensity; 2, moderate
intensity; and 3, strong intensity) and the percentage of positive
cells (0, <5%; 1, between 5 and 25%; 2, between 26 and 50%; and
3, >50%). The patients were classified into 2 groups according
to the total score (i.e., staining intensity score + positive cell
score): the low expression group (total score of 0–2) and the high
expression group (total score of 3–9).
Cell culture
The human ESCC cell lines, EC9706 and Eca109, were
provided by Wuhan University (originally obtained from the China
Center for Type Culture Collection, Wuhan, China). These two cell
lines are frequently used to study the the malignant behavior of
ESCC cells (3,16,24–28).
The cells were cultured in Roswell Park Memorial Institute
(RPMI)-1640 medium supplemented with 10% fetal bovine serum (FBS)
(both from Thermo Fisher Scientific, Waltham, MA, USA) and 1%
penicillin/streptomycin at 37°C in a 5% CO2 humidified
incubator.
shRNA synthesis and lentivirus packaging
and transduction
The shRNA sequences against RNF113A (GenBank no.
NC_000023) (Table I) and the
scramble sequence (5′-TTC TCC GAA CGT GTC ACG T-3′) were
synthesized by GeneChem (Shanghai, China). Lentiviral vectors of
RNF113A were constructed by GeneChem using the GV248 vector
(GeneChem). This vector (batch #CON077) contains hU6, MCS,
ubiquitin, EGFP, IRES and puromycin sequences. The ESCC cells
(3–5×104/ml) were transduced with lentivirus-mediated
RNF113A shRNA or scramble-shRNA [multiplicity of infection (MOI) of
20], according to the manufacturer's instructions. The cells were
harvested 72 h after transduction for western blot analysis. For
the subsequent experiments, we selected RNF113A-shRNA2 as it was
more effective than RNF113A-shRNA1 in decreasing the expression of
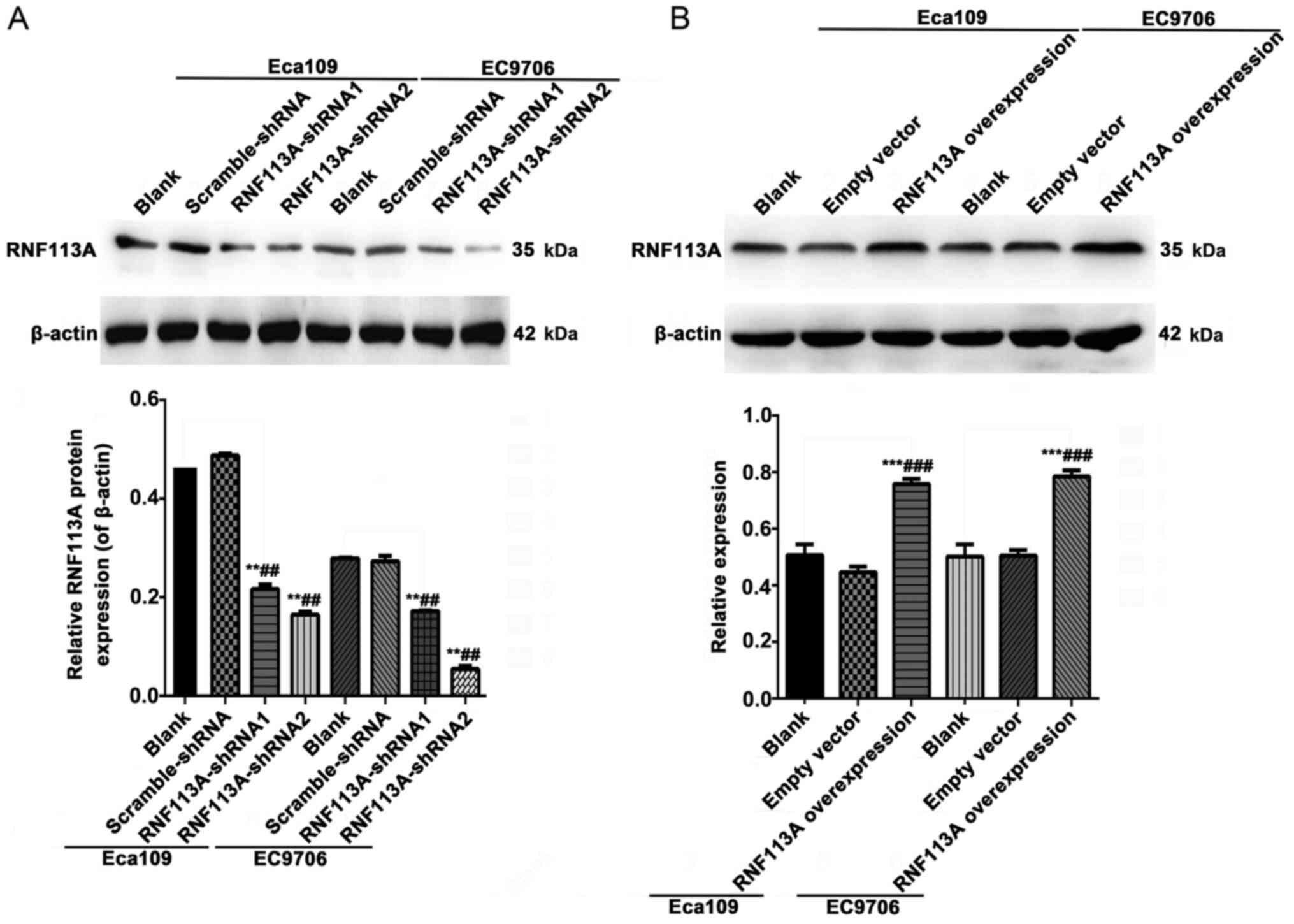
RNF113A (Fig. 2A).
 | Table IOligonucleotide shRNA sequences of
RNF113A. |
Table I
Oligonucleotide shRNA sequences of
RNF113A.
| RNF113A-shRNA | 5′ | Stem | Loop | Stem | 3′ |
|---|
| RNF113A-shRNA1 | Ccgg |
GCGTCTTCAATCCAGCGAAAGAATT | CTCGAG |
AATTCTTTCGCTGGATTGAAGACGC | TTTTTg |
| RNF113A-shRNA2 | aattcaaaaa |
GCGTCTTCAATCCAGCGAAAGAATT | CTCGAG |
AATTCTTTCGCTGGATTGAAGACGC | |
pcDNA3.1-RNF113A overexpression
The eukaryotic expression vector, pcDNA3.1,
overexpressing RNF113A (sense, 5′-TCC
GCTCGAGATGATGGACTTGGAGCTGCC-3′ and antisense,
5′-ATGGGGTACCGAGTTTTTCTTAACATCTGGC-3′) was constructed by GeneChem.
Polymerase chain reaction (PCR) was used to identify the
recombinant clones. Sequencing was used to confirm the presence of
the insert as well as the sequence and orientation of the insert.
The ESCC cells were transfected with pcDNA3.1 (empty vector) and
pcDNA3.1-RNF113A over-expression vector using Lipofectamine 2000
(Thermo Fisher Scientific).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from 41 pairs of frozen ESCC
and adjacent normal tissue samples, and from tumor xenografts
(please see description of mouse model below) using TRIzol reagent
(Thermo Fisher Scientific). The concentration and purity of the RNA
in each sample was determined by measuring he absorbance at 260 and
280 nm using a NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific). Total RNA was reverse transcribed into single-stranded
cDNA using the RT reagent kit (Takara Bio, Otsu, Japan). The PCR
primers were as follows: RNF113A forward, 5′-TCC GCT CGA GAT GAT
GGA CTT GGA GCT GCC-3′ and reverse, 5′-ATG GGG TAC CGA GTT TTT CTT
AAC ATC TGG C-3′. Quantitative PCR was performed using the IQ5
system (Bio-Rad, Hercules, CA, USA) and SYBR Premix Ex Taq (Takara
Bio), according to the manufacturer's instructions. The relative
expression levels of RNF113A were normalized to those of β-actin
(primers used were: forward, 5′-CTA AGT CAT AGT CCG CCT AGA AGC
A-3′ and reverse, 5′-TGG CAC CCA GCA CAA TGA A-3′). The reaction
conditions for RNF113A were as follows: 95°C for 3 min, and 40
cycles at 95°C for 10 sec followed by 59.2°C for 30 sec. The
2−ΔΔCt method was used to calculate the relative mRNA
expression of the target gene.
Western blot analysis
The cells were harvested 72 h after transduction in
radio-immunoprecipitation assay (RIPA) lysis buffer (Bioteke,
Beijing, China). Proteins were quantified by BCA assay (Beyotime
Institute of Biotechnology, Shanghai, China), and 50 µg of
proteins were subjected to 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins
were transferred onto polyvinylidene fluoride (PVDF) microporous
membranes (Millipore Corp., Billerica, MA, USA) and the blots were
probed with rabbit polyclonal antibody against RNF113A (ab85797;
Abcam, Cambridge, MA, USA) at a concentration of 1 µg/ml.
β-actin (BA2305; Wuhan Boster Bio-Engineering Co., Ltd., Wuhan,
China) was used as an internal control. Mouse anti-rabbit secondary
antibody (31464; Thermo Fisher Scientific). Chemiluminescence
substrate (Thermo Fisher Scientific) was added to visualize the
bands, as previously described (22). The integrated optical density of
the target proteins was quantified using ImageJ software (National
Institutes of Health, Bethesda, MD, USA).
Analysis of the cell cycle and
apoptosis
Cell cycle progression and apoptosis were analyzed
by flow cytometry (FCM). For cell cycle analysis, the cells were
washed with pre-iced phosphate-buffered saline (PBS) twice and
fixed with 70% cold ethanol at 4°C overnight. Following incubation
with RNase for 1 h at 4°C, the cells were stained with 2 µl
of propidium iodide (PI) (500 mg/ml) for 15 min, and analyzed using
a Cytomics FC500 flow cytometer (Beckman Coulter, Brea, CA, USA).
For apoptosis analysis, an Annexin V-FITC apoptosis detection kit
was used (Thermo Fisher Scientific). Briefly, the cells were washed
twice with iced PBS and resuspended in 400 µl of 1X binding
buffer at a concentration of 1×106 cells/ml. A volume of
5 µl of the Annexin V-FITC solution and 2 µl of PI
was then mixed with the cells. The cells were then incubated 15 min
at 4°C in the dark. Cell staining was detected using a Cytomics
FC500 flow cytometry system (Beckman Coulter).
Cell invasion and migration
The cells at 5×105 cells/well were seeded
on a fibronectin-coated polycarbonate membrane insert in a
Transwell chamber (Corning Inc., Corning, NY, USA). RPMI-1640
supplemented with 10% FBS was added to the lower chamber. The cells
that adhered to the upper surface of the chamber were wiped with a
cotton swab. The number of migrated cells was counted under a light
microscope (Olympus, Tokyo, Japan) in 5 random fields. For the
Matrigel invasion assay, inserts pre-coated with Matrigel (40
µl, 1 mg/ml; BD Biosciences, Franklin Lakes, NJ, USA) were
used for invasion assays and incubated at 37°C for 6 h.
Wound healing assay
The wound healing assay was carried out to analyze
cell migration. The cells were plated in a 6-well plate at
5×105 cells/well. Upon confluent growth, scrape wounds
were made using a 10-µl pipette tip. The cells were
photographed at 0, 24, 48 and 72 h using an inverted fluorescence
microscope (Olympus). The wound closure rate was calculated as
follows: [Cell-free distance (0 h)-cell-free distance (24/48/72
h)]/cell-free distance (0 h), as previously described (29,30).
Xenograft tumor assays using athymic nude
mice
All animal experiments were approved by the Animal
Ethics Committee of our hospital. Female 4–6-week-old BALB/c nude
mice (weighing, 14–16 g) (n=12) were purchased from Charles River
Laboratories (Beijing, China) and raised in a sterile environment.
The mice were divided into 3 groups (n=4/group) as follows: the
group injected with Eca109 cells, the group injected with Eca109
cells transduced with RNF113A-shRNA2, and the group injected with
Eca109 cells transduced with scramble-shRNA. The cells were
injected at 4×106 cells/mouse into the right flank.
Tumor dimensions were measured by calipers every 7 days, and tumor
volume (TV) was calculated each week according to the formula: TV
(mm3) = length × width2 × 0.5. Tumors were
removed and weighed 28 days after cell injection. Part of the tumor
tissue was cryopreserved in liquid nitrogen for the analysis of
RNF113A mRNA expression by RT-qPCR. The remaining tissues were
paraffin-embedded for immunohistochemical analysis of RNF113A.
Statistical analysis
Data were analyzed using SPSS 17.0 software (IBM,
Armonk, NY, USA). The results are presented as the means ± standard
deviation (SD). The Student's t test and one-way ANOVA were used to
evaluate the differences among groups. The LSD test was used
following ANOVA. The Chi-square test and Fisher's exact test were
used to analyze the association between RNF113A expression and
clinicopatho-logical characteristics. Survival curves were plotted
using the Kaplan-Meier method and compared with the log-rank test.
Univariate and multivariate analyses of survival were performed by
Cox regression. Two-sided P-values <0.05 were considered to
indicate statistically significant differences.
Results
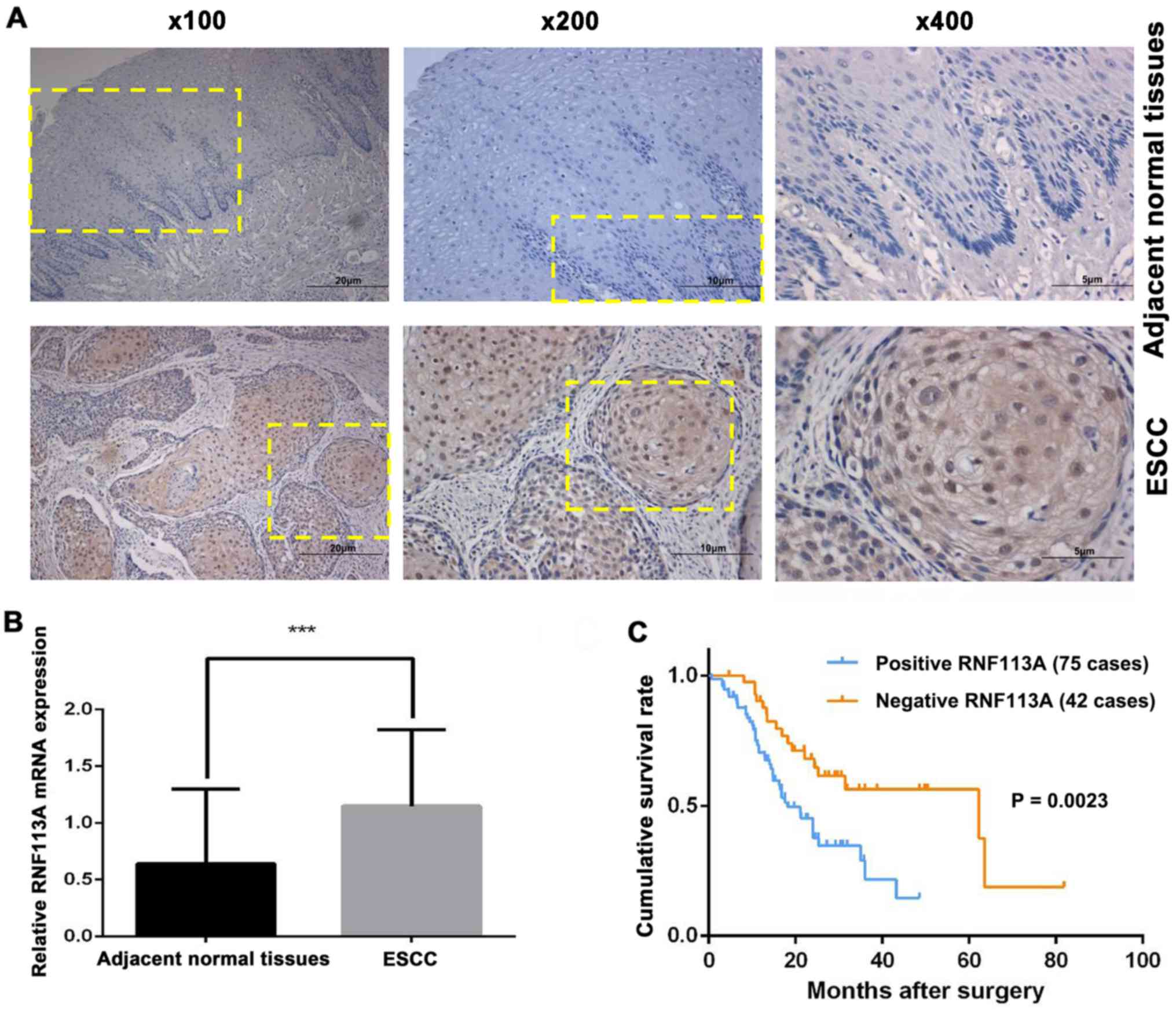
RNF113A is significantly upregulated
inESCC and is associated with a poor prognosis
RNF113A was mainly localized in the cytoplasm and
nucleus of the cancer cells (Fig.
1A). RNF113A expression was positive in 64.1% (75/117) of the
ESCC samples. Moreover, RNF113A expression was extremely low or
hardly detectable in the adjacent non-tumor tissues. In addition,
RNF113A mRNA expression in the 41 fresh-frozen ESCC cases was
higher (cancer/normal ratio >1.0) in 73% (30/41) of the samples
when compared with the adjacent non-tumor tissues, with the mean
expression level of 1.15±0.66 in ESCC and 0.67±0.66 in their
counterparts (P<0.001) (Fig.
1B).
The associations between RNF113A expression and the
clinicopathological characteristics were analyzed (Table II). Tumor differentiation
(P=0.008) and T classification (P<0.001) were significantly
associated with RNF113A expression. RNF113A was predominantly
upregulated in late-stage, but not early-stage tumor tissues. No
statistically significant association was observed between RNF113A
expression and sex, age, histological type, tumor location and
lymph node metastasis (N classification) (all P>0.05).
 | Table IIAssociation between RNF113A
expression and clinicopathological characteristics of patients with
ESCC. |
Table II
Association between RNF113A
expression and clinicopathological characteristics of patients with
ESCC.
| Variable | RNF113A expression
| P-value |
|---|
| No. | Positive
(n=75) | Negative
(n=42) |
|---|
| Age (years) | | | | 0.086 |
| <60 | 53 | 38 (71.7) | 15 (28.3) | |
| ≥60 | 64 | 37 (57.8) | 27 (42.2) | |
| Sex | | | | 0.457 |
| Male | 87 | 55 (63.2) | 32 (36.8) | |
| Female | 30 | 20 (66.7) | 10 (33.3) | |
|
Differentiation | | | | 0.008 |
| Well | 26 | 12 (46.2) | 14 (53.8) | |
| Moderate | 57 | 34 (59.6) | 23 (40.4) | |
| Poor | 24 | 21 (87.5) | 3 (12.5) | |
| T
classification | | | |
<0.001 |
| T1-T2 | 54 | 19 (35.2) | 35 (64.8) | |
| T3-T4 | 63 | 56 (88.9) | 7 (11.1) | |
| N
classification | | | | 0.11 |
| N0 | 76 | 43 (56.6) | 33 (43.4) | |
| N1 | 24 | 15 (62.5) | 9 (37.5) | |
| N2 | 11 | 10 (90.9) | 1 (9.1) | |
| N3 | 6 | 5 (83.3) | 1 (16.7) | |
| Histological
type | | | | 0.416 |
| Protruding
type | 17 | 8 (47.1) | 9 (52.9) | |
| Ulcerative
type | 69 | 45 (65.2) | 24 (34.8) | |
| Fungating
type | 13 | 9 (69.2) | 4 (30.8) | |
| Medullary
type | 18 | 13 (72.2) | 5 (27.8) | |
| Tumor
locationa | | | | 0.762 |
| Upper | 11 | 8 (72.7) | 3 (27.3) | |
| Middle | 59 | 39 (66.1) | 20 (33.9) | |
| Lower | 47 | 29 (61.7) | 18 (38.3) | |
Fig. 1C shows that
survival was improved among patients with a low RNF113A expression
in their tumors compared with patients with a high RNF113A
expression in their tumors (P=0.002). Multivariate analysis
revealed that RNF113A expression (HR=2.406; 95% CI, 1.301-4.449,
P=0.005) and lymph node metastasis (HR=3.219, 95% CI, 1.894–5.469,
P<0.001) were independently associated with overall survival in
Kazakh patients with ESCC (Table
III).
 | Table IIIUnivariate and multivariate Cox
regression analyses of the clinicopathological variables and
cumulative survival of the patients with ESCC. |
Table III
Univariate and multivariate Cox
regression analyses of the clinicopathological variables and
cumulative survival of the patients with ESCC.
| Covariant | Univariate analysis
| Multivariate
analysis
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| RNF113A
expression | 2.449 | 1.351–4.439 | 0.003 | 2.406 | 1.301–4.449 | 0.005 |
| Sex | 0.882 | 0.465–1.670 | 0.699 | 0.826 | 0.420–1.627 | 0.581 |
| Age | 0.685 | 0.410–1.144 | 0.148 | 1.387 | 0.720–2.675 | 0.328 |
| Tumor location | 1.624 | 0.716–3.685 | 0.457 | 1.068 | 0.578–1.970 | 0.593 |
| Histological
type | 1.187 | 0.476–2.956 | 0.711 | 0.079 | 0.205–1.191 | 0.254 |
| Tumor
differentiation | 1.491 | 0.840–3.012 | 0.132 | 1.254 | 0.563–2.793 | 0.748 |
| T
classification | 1.798 | 1.040–3.110 | 0.036 | 1.205 | 0.564–2.577 | 0.63 |
| Lymph node
metastasis | 3.154 | 1.882–5.285 |
<0.001 | 3.219 | 1.894–5.469 |
<0.001 |
Effects of knockdown or the
overexpression of RNF113A on cell cycle distribution and the
apoptosis of Eca109 and EC9706 cells
Fig. 2 shows the
effects of the knockdown or overexpression of RNF113A on RNF113A
protein expression in human Eca109 and EC9706ESCC cells. RNF113A
protein expression was lower in the cells transfected with
RNF113A-shRNA1 and RNF113A-shRNA2 than in the untransfected Eca109
and EC9706 cells and scramble-shRNA cells (all P<0.01) (Fig. 2A). RNF113A protein expression was
higher in the RNF113A-overexressing cells than in the untransfected
Eca109 and EC9706 cells and empty vector-transfected cells (all
P<0.001) (Fig. 2B).
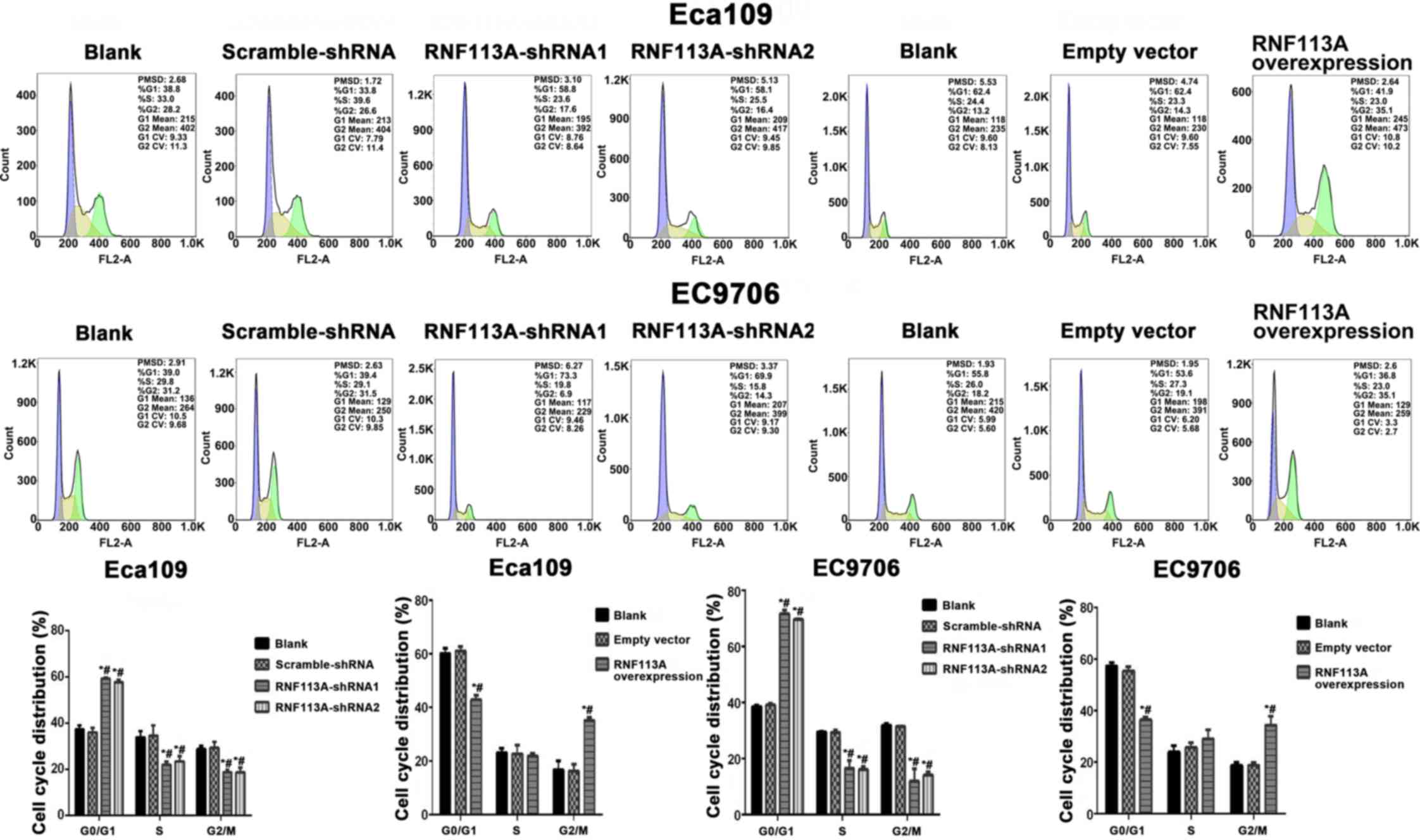
Fig. 3 shows that
cell numbers were significantly increased in the G0/G1 phase and
decreased in the S phase and G2/M phase in the cells transfected
with RNF113A-shRNA compared with the untransfected Eca109 and
EC9706 cells and scramble-shRNA cells (all P<0.05). It can be
seen that the cell numbers were significantly decreased in the
G0/G1 phase and increased in the G2/M phase in the cells
transfected with the pCDNA3.1-RNF113A overexpression vector
compared with the empty vector-transfected group and untransfected
Eca109 and EC9706 cells. Therefore, the overexpression of RNF113A
promoted ESCC cell proliferation.
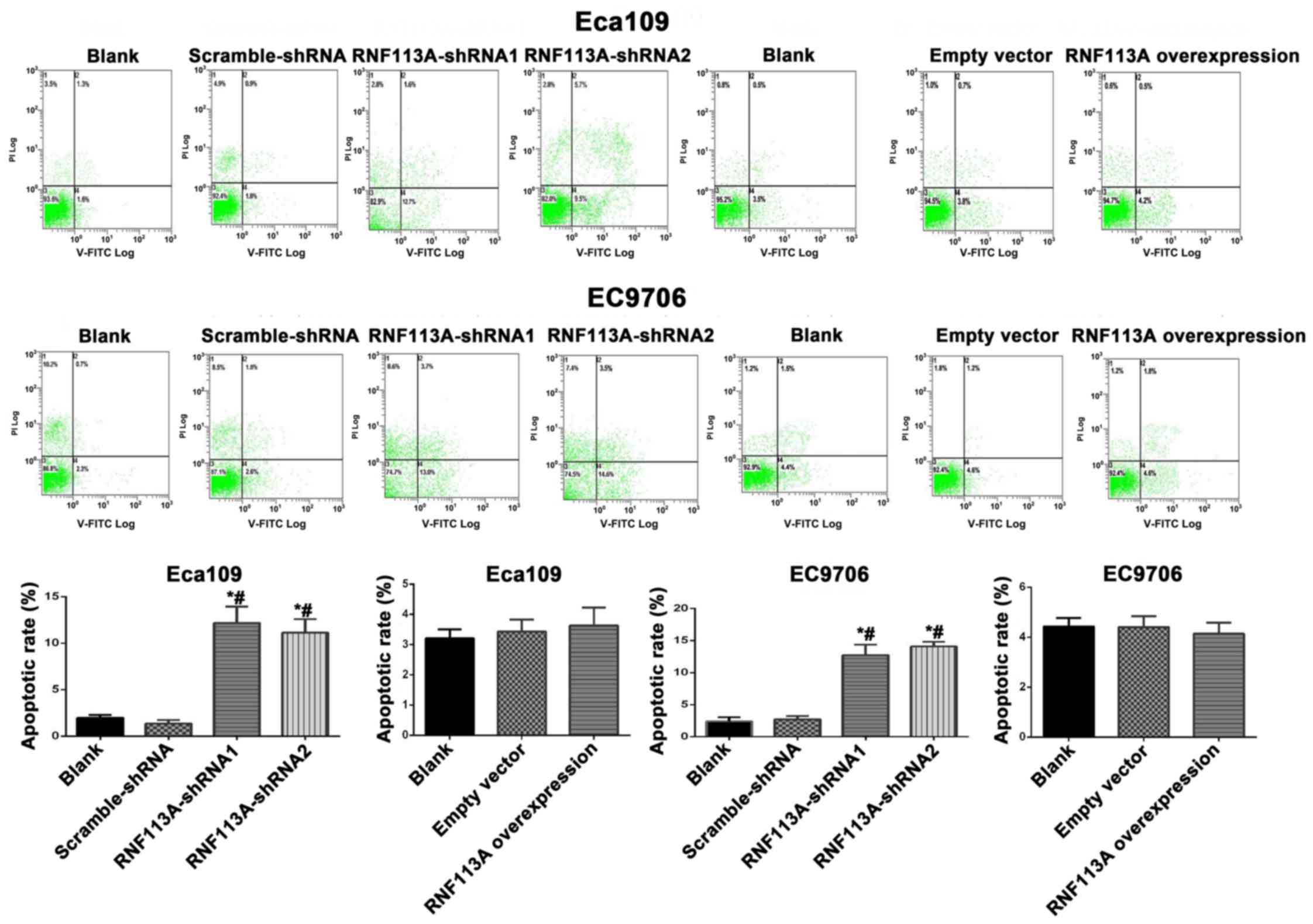
Apoptosis was also increased when RNF113A was
knocked down (all P<0.050) (Fig.
4). The overexpression of RNF113A did not lead to changes in
apoptosis compared with the controls (P>0.05) (Fig. 4).
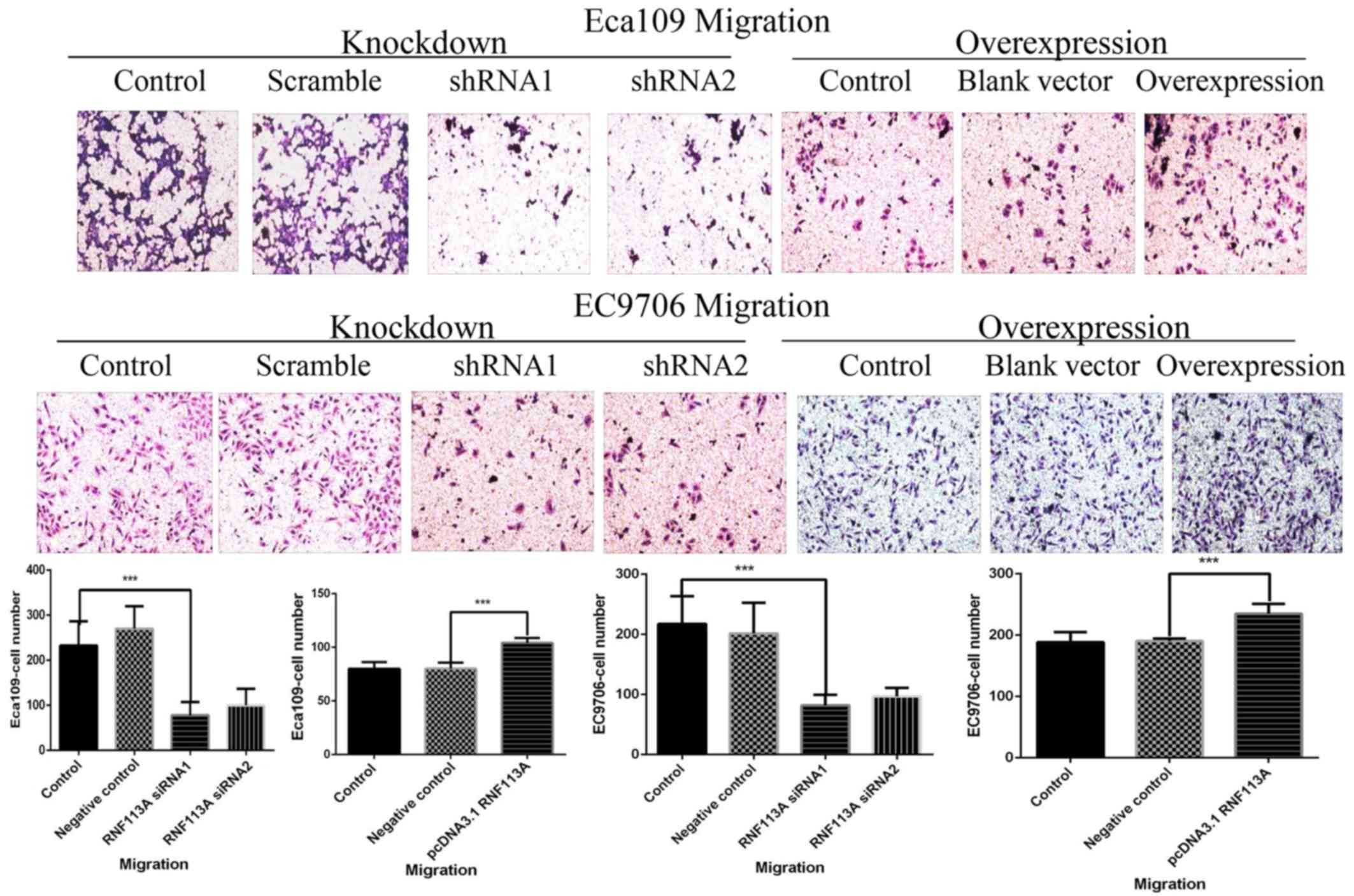
Effects of the knockdown and
overexpression of RNF113A on the migration and invasion of ESCC
cells in vitro
In the Transwell assay, the numbers of migrating
cells were decreased in the RNF113A-shRNA1 and RNF113A-shRNA2
groups compared with the untransfected Eca109 and EC9706 cells and
scramble-shRNA-transfected cells (all P<0.001) (Fig. 5). However, the numbers of migrating
cells were increased in the RNF113A-overexressing cells compared
with the untransfected Eca109 and EC9706 cells and empty
vector-transfected cells (all P<0.001) (Fig. 5).
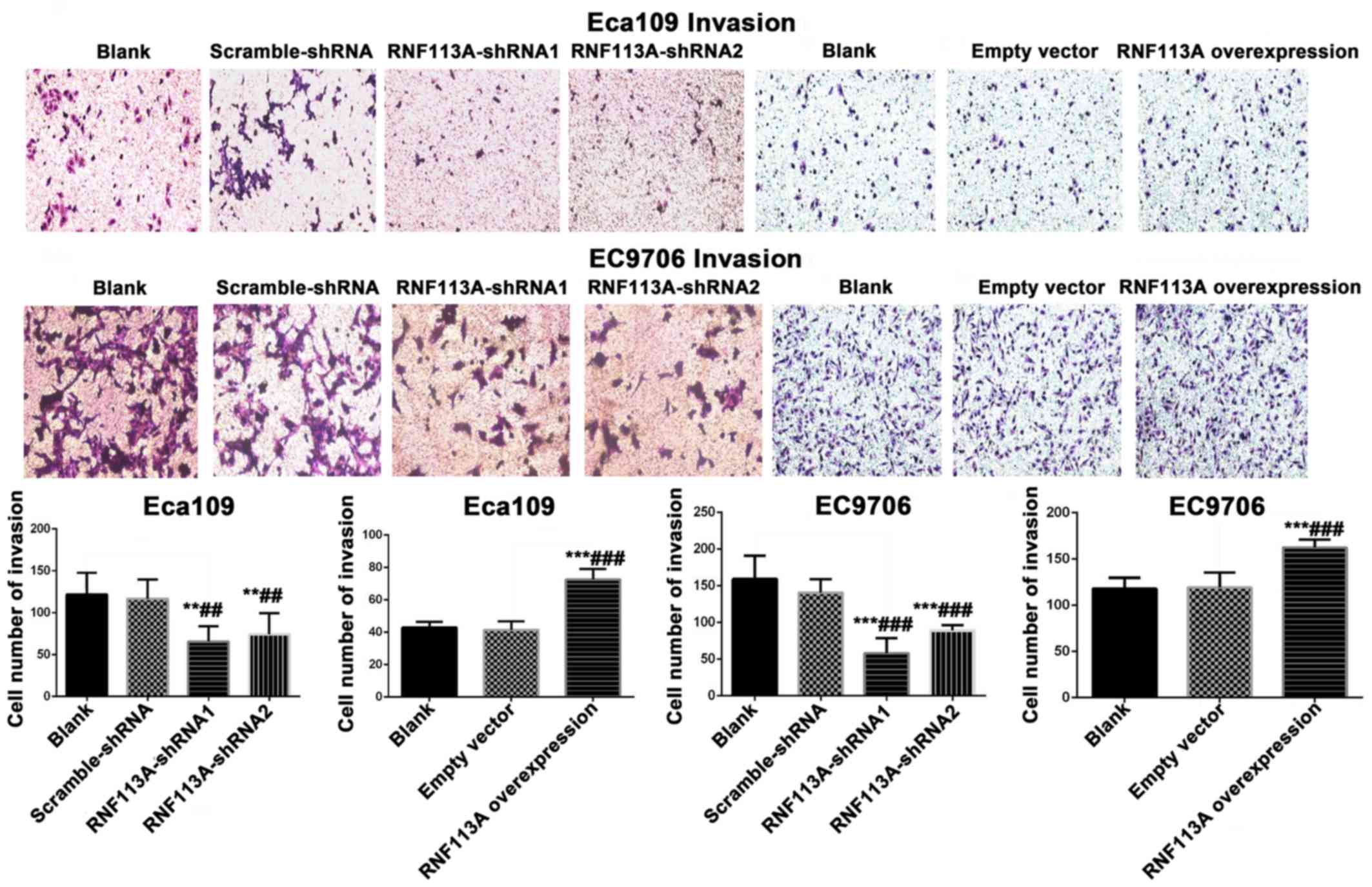
The knockdown of RNF113A led to a marked decrease in
the invasiveness of the Eca109 (P<0.01) and EC9706 (P<0.001)
cells (Fig. 6). The overexpression
of RNF113A significantly increased the invasiveness of the Eca109
and in EC9706 cells (all P<0.001) (Fig. 6).
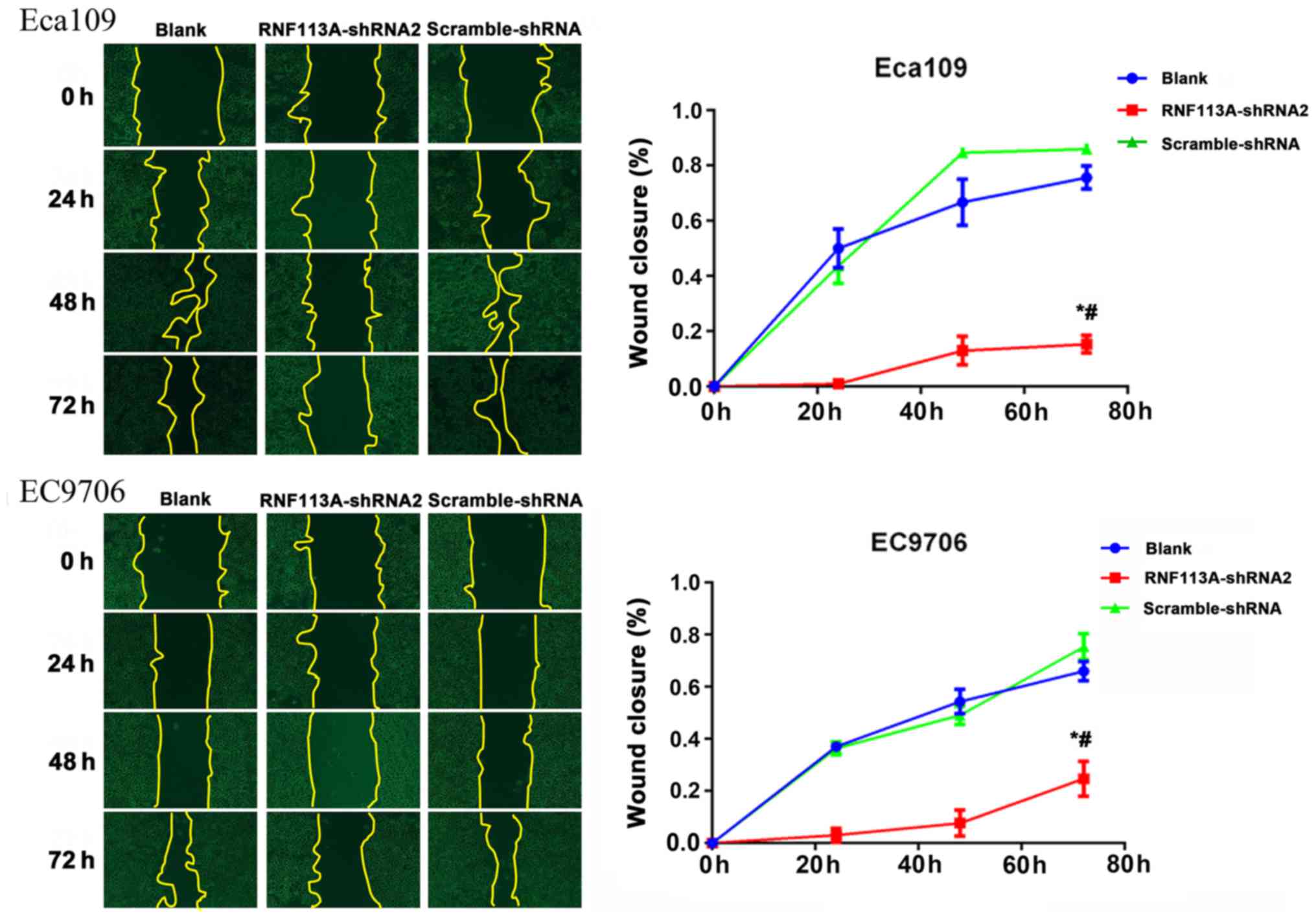
The wound healing assay also revealed that the
knockdown of RNF113A inhibited the migration of the Eca109
(P<0.05) and EC9706 (P<0.05) cells, compared with the
untransfected Eca109 and EC9706 cells and
scramble-shRNA-transfected cells (Fig.
7).
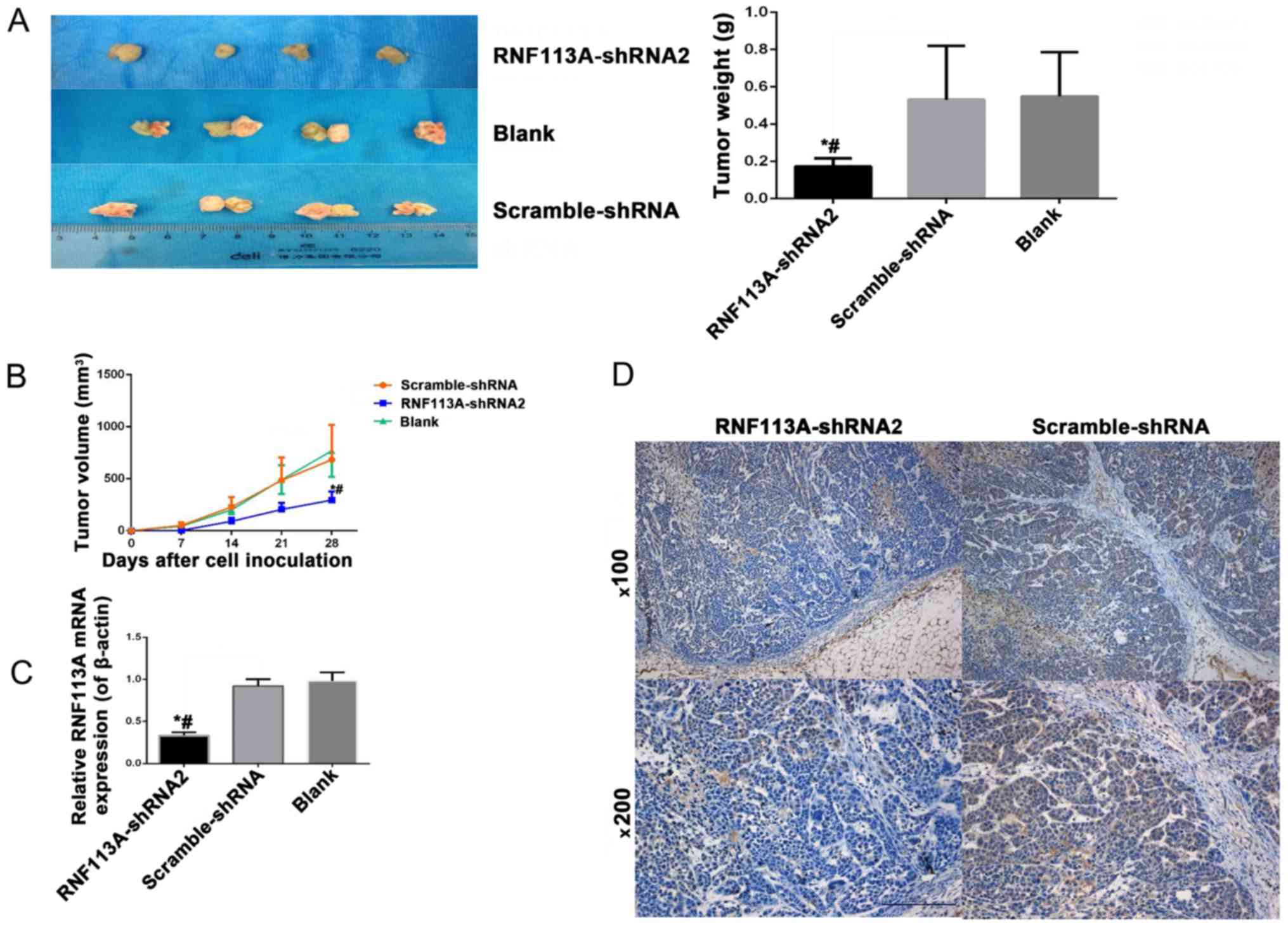
Knockdown of RNF113A suppresses tumor
growth in a nude mouse tumor xenograft model with Eca109 cells
All nude mice developed tumors; however, the tumors
formed from the scramble-shRNA-transfected cells grew more rapidly
than those from the RNF113A-shRNA-transfected cells. The tumors
formed from the RNF113A-shRNA-transfected cells were significantly
lighter and smaller than those from the scramble-shRNA-transfected
cells and Eca109-transfected cells (all P<0.05) (Fig. 8A and B). RT-qPCR and IHC confirmed
that the RNF113A mRNA and protein levels were downregulated in the
sections of the subcutaneous tumors of nude mice inoculated with
RNF113A-shRNA-transfected cells, compared with the untransfected
Eca109 cells and scramble-shRNA-transfected cells (Fig. 8C and D).
Discussion
The UPP influences and regulates apoptosis and plays
a central role in the binding of ubiquitin and substrate
specificity (9). E3 ligases are
involved in Snail and Twist ubiquitination, which play important
roles in EMT (10,11). RNF113A has a RING domain (14), which is frequently found in E3
ubiquitin ligases. Previous studies have indicated that there are
close associations between ubiquitin ligase and the occurrence,
development and metastasis of cancer (16,17).
In C. elegans, the knockdown of RNF113 sensitizes the cells
to UVA-induced DNA damage and is required for RAD51 during repair
(19). Pellagatti et al
(20) showed that ZNF183 was one
of the most upregulated genes in MDS (20). Therefore, it can be hypothesized
that RNF113A is associated with the progression/development of
ESCC. Nevertheless, little is known about the expression of RNF113A
in ESCC and its significance.
The present study demonstrated that RNF113A
expression was associated with the T stage, suggesting that it may
be associated with tumorigenesis, as well as tumor progression.
RT-qPCR revealed that the overexpression of RNF113A in 73% of ESCC
samples, supporting the results of IHC. Kaplan-Meier analysis
revealed a poorer survival rate in patients with ahigh RNF113A
expression. Furthermore, multivariate analysis revealed that the
RNF113A levels could be used as an independent prognostic factor
for patients with ESCC. Therefore, RNF113A may be a novel
diagnostic and prognostic biomarker for ESCC, although this
requires further validation. Nevertheless, these results are
supported by previous studies of various RING finger proteins in
various cancer types (31–34).
Cell cycle progression, the inhibition of apoptosis,
and cell migration and invasion are central features of ESCC
progression (26,35–37).
Supporting the role of RNF113A in ESCC aggressiveness, the present
study demonstrated that the overexpression of RNF113A was
associated with cell cycle progression and an increased migration
and invasiveness of Eca109 and EC9706 cells. These results are
similar to those observed with various RING finger proteins in
various cancer types. Yonemori et al (38) showed that ZFP36L2 promoted cancer
cell proliferation, migration and invasion in pancreatic ductal
adenocarcinoma. Zhang and Yang (39) showed that silencing tripartite
motif containing 59 (TRIM59) inhibited the proliferation, migration
and invasion of breast cancer cells. Similar results were also
observed in gastric cancer with RNF180 (40) and colorectal cancer with RNF216
(41). These results support the
roles of RING finger proteins in carcinogenesis. Tumor metastasis
is one of the most important biological characteristics of
malignant tumors, and metastasis is a strong independent prognostic
factor for ESCC (42). In this
study, RNF113A expression was not associated with lymph node
metastasis as revealed the examination of tumor samples; however,
in vitro experiments suggested that RNF113A may promote the
migration and invasiveness of ESCC cell lines. The exact role of
RNF113A in the advanced stage of ESCC remains to be through
analyzed in the future, based on clinical studies with an enlarged
sample size and in vitro studies using a greater number of
cell lines.
The exact mechanisms responsible for the effects of
RNF113A on cancer progression are poorly understood and further
investigations are warranted. Nevertheless, some previous studies
may provide some clues. The normal expression of RNF113A plays an
important role in the differentiation of cells of the central
nervous system during embryogenesis (15). RNF113A participates in the
regulation of the stability of proteins of the E2 and E3 families
(15). EMT is a hallmark of cancer
invasiveness and the regulation of the stability of the
transcription factors involved in EMT is essential to this process
(10,11). Among others, ubiquitin ligases
controlling Snail and Twist stability plays crucial role in EMT
(11). Indeed, Snail and Twist are
short-lived due to rapid polyubiquitination in normal cells and E3
is involved in this process (11).
The degradation of most EMT factors depends upon RING-type E3s. In
addition, RNF113A has a C3HC4 zinc finger domain, which is found in
E3 ubiquitin ligase and is involved in tumorigenesis (11,43,44).
Furthermore, other proteins of the family are known to be involved
in tumorigenesis. Indeed, RNF183 promotes the proliferation and
metastasis of colorectal cancer cells via the activation of the
NF-κB-IL-8 axis (45). RNF125 is a
potential biomarker for the highly aggressive and unfavorable
prognosis of gallbladder cancers by promoting the invasion and
metastasis of human gallbladder cancers via the activation of the
TGF-β1-SMAD3-ID1 signaling pathway (46). Cancer progression and cell
proliferation, migration and invasion are orchestrated by a very
complex and interwoven array of molecules. Two approaches are
available to study these interactions: Either the use of
bioinformatics to grasp a global view of the processes, or the
study of a single specific factor. The present study aimed to
assess the effect of RNF113A on the progression of ESCC. The
results indeed demonstrated that RNF113A plays an important role in
ESCC progression; however, the mechanisms through which this factor
interacts with other factors remain to be elucidated.
The present study is not without limitations. The
sample size was small and from a single center. In addition, all
patients were Kazakh, which is a Chinese minority that has the
highest incidence of ESCC worldwide (6); however, this limits the
generalizability of the results. It is probable that this high
incidence is due to a number of environmental and genetic factors
that may influence the results of the present study. Additional
studies on various populations are warranted to confirm these
results. The mechanisms through which RNF113A promotes malignant
behaviors in vitro required further investigation.
In conclusion, the present study strongly suggests
that RNF113A promotes the proliferation, migration and invasion of
ESCC cells, and that RNF113A is associated with a poor prognosis of
ESCC in Kazakh patients.
Abbreviations:
|
EC
|
esophageal cancer
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
OS
|
overall survival
|
|
UPP
|
ubiquitin-proteasome pathway
|
|
EMT
|
epithelial-mesenchymal transition
|
|
TTD
|
trichothiodystrophy
|
|
MDS
|
myelodysplastic syndrome
|
|
IHC
|
immunohistochemistry
|
|
FBS
|
fetal bovine serum
|
|
MOI
|
multiplicity of infection
|
|
RIPA
|
radio-immunoprecipitation assay
|
|
PVDF
|
polyvinylidene fluoride
|
|
FCM
|
flow cytometry
|
|
PBS
|
phosphate-buffered saline
|
|
PI
|
propidium iodide
|
|
TV
|
tumor volume
|
|
UVA
|
ultraviolet A
|
Acknowledgments
This study was funded by Major Projects in the
Xinjiang Uygur Autonomous Region (no. 2016B03054).
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Parkin DM, Pisani P and Ferlay J: Global
cancer statistics. CA Cancer J Clin. 49:33–64. 311999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang CS, Chen X and Tu S: Etiology and
prevention of esophageal cancer. Gastrointest Tumors. 3:3–16. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arnold M, Soerjomataram I, Ferlay J and
Forman D: Global incidence of oesophageal cancer by histological
subtype in 2012. Gut. 64:381–387. 2015. View Article : Google Scholar
|
|
6
|
Ainiwaer J, Li DS and Zhang L:
Investigation on prevalence of esophageal cancer during 2005–2008
in Yili of Xinjiang, China. Xinjiang Medical J. 41:112–114.
2011.
|
|
7
|
NCCN Clinical Practice Guidelines in
Oncology (NCCN Guidelines): Esophageal and Esophagogastric Junction
Cancers. (Version 2.2017). National Comprehensive Cancer Network;
Fort Washington: 2017
|
|
8
|
Mukhopadhyay D and Riezman H:
Proteasome-independent functions of ubiquitin in endocytosis and
signaling. Science. 315:201–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sullivan JA, Shirasu K and Deng XW: The
diverse roles of ubiquitin and the 26S proteasome in the life of
plants. Nat Rev Genet. 4:948–958. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue Z, Wu X, Chen X, Liu Y, Wang X, Wu K,
Nie Y and Fan D: Mesenchymal stem cells promote epithelial to
mesenchymal transition and metastasis in gastric cancer though
paracrine cues and close physical contact. J Cell Biochem.
116:618–627. 2015. View Article : Google Scholar
|
|
11
|
Díaz VM, Viñas-Castells R and García de
Herreros A: Regulation of the protein stability of EMT
transcription factors. Cell Adhes Migr. 8:418–428. 2014. View Article : Google Scholar
|
|
12
|
Corbett MA, Dudding-Byth T, Crock PA,
Botta E, Christie LM, Nardo T, Caligiuri G, Hobson L, Boyle J,
Mansour A, et al: A novel X-linked trichothiodystrophy associated
with a nonsense mutation in RNF113A. J Med Genet. 52:269–274. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Frattini A, Faranda S, Bagnasco L,
Patrosso C, Nulli P, Zucchi I and Vezzoni P: Identification of a
new member (ZNF183) of the Ring finger gene family in Xq24-25.
Gene. 192:291–298. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Korneta I, Magnus M and Bujnicki JM:
Structural bioinformatics of the human spliceosomal proteome.
Nucleic Acids Res. 40:7046–7065. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carney TD, Struck AJ and Doe CQ: midlife
crisis encodes a conserved zinc-finger protein required to maintain
neuronal differentiation in Drosophila. Development. 140:4155–4164.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bromberg KD, Kluger HM, Delaunay A, Abbas
S, DiVito KA, Krajewski S and Ronai Z: Increased expression of the
E3 ubiquitin ligase RNF5 is associated with decreased survival in
breast cancer. Cancer Res. 67:8172–8179. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen C, Sun X, Guo P, Dong XY, Sethi P,
Zhou W, Zhou Z, Petros J, Frierson HF Jr, Vessella RL, et al:
Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate
cancer. Oncogene. 26:2386–2394. 2007. View Article : Google Scholar
|
|
18
|
Amsterdam A, Nissen RM, Sun Z, Swindell
EC, Farrington S and Hopkins N: Identification of 315 genes
essential for early zebrafish development. Proc Natl Acad Sci USA.
101:12792–12797. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee H, Alpi AF, Park MS, Rose A and Koo
HS: C. elegans ring finger protein RNF-113 is involved in
interstrand DNA crosslink repair and interacts with a RAD51C
homolog. PLoS One. 8:e600712013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pellagatti A, Esoof N, Watkins F, Langford
CF, Vetrie D, Campbell LJ, Fidler C, Cavenagh JD, Eagleton H,
Gordon P, et al: Gene expression profiling in the myelodysplastic
syndromes using cDNA microarray technology. Br J Haematol.
125:576–583. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heath-Brown N: International Union Against
Cancer. The Stateman's Yearbook 2016. Palgrave MacMillan; London:
2015
|
|
22
|
Ma JQ, Tuersun H, Jiao SJ, Zheng JH, Xiao
JB and Hasim A: Functional role of NRF2 in cervical carcinogenesis.
PLoS One. 10:e01338762015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sinicrope FA, Ruan SB, Cleary KR, Stephens
LC, Lee JJ and Levin B: Bcl-2 and p53 oncoprotein expression during
colorectal tumorigenesis. Cancer Res. 55:237–241. 1995.PubMed/NCBI
|
|
24
|
Zhu J, Li H, Ma J, Huang H, Qin J and Li
Y: PTPN9 promotes cell proliferation and invasion in Eca109 cells
and is negatively regulated by microRNA-126. Oncol Lett.
14:1419–1426. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen HX, Wang S, Wang Z, Zhang ZP and Shi
SS: Overexpression of RUNX3 inhibits malignant behaviour of Eca109
cells in vitro and in vivo. Asian Pac J Cancer Prev. 15:1531–1537.
2014. View Article : Google Scholar
|
|
26
|
Li S, Wang F, Qu Y, Chen X, Gao M, Yang J,
Zhang D, Zhang N, Li W and Liu H: HDAC2 regulates cell
proliferation, cell cycle progression and cell apoptosis in
esophageal squamous cell carcinoma EC9706 cells. Oncol Lett.
13:403–409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun Z, Ji N, Bi M, Wang S, Liu X and Wang
Z: PTEN gene is infrequently hypermethylated in human esophageal
squamous cell carcinoma. Tumour Biol. 36:5849–5857. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang T, Xuan X, Pian L, Gao P, Hu H, Zheng
Y, Zang W and Zhao G: Notch-1-mediated esophageal carcinoma EC-9706
cell invasion and metastasis by inducing epithelial-mesenchymal
transition through Snail. Tumour Biol. 35:1193–1201. 2014.
View Article : Google Scholar
|
|
29
|
Cory G: Scratch-wound assay. Methods Mol
Biol. 769:25–30. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jonkman JE, Cathcart JA, Xu F, Bartolini
ME, Amon JE, Stevens KM and Colarusso P: An introduction to the
wound healing assay using live-cell microscopy. Cell Adhes Migr.
8:440–451. 2014. View Article : Google Scholar
|
|
31
|
Sugiura T, Yamaguchi A and Miyamoto K: A
cancer-associated RING finger protein, RNF43, is a ubiquitin ligase
that interacts with a nuclear protein, HAP95. Exp Cell Res.
314:1519–1528. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu J, Zhao C, Zhuang T, Jonsson P, Sinha
I, Williams C, Strömblad S and Dahlman-Wright K: RING finger
protein 31 promotes p53 degradation in breast cancer cells.
Oncogene. 35:1955–1964. 2016. View Article : Google Scholar :
|
|
33
|
Horie K, Urano T, Ikeda K and Inoue S:
Estrogen-responsive RING finger protein controls breast cancer
growth. J Steroid Biochem Mol Biol. 85:101–104. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oyanagi H, Takenaka K, Ishikawa S, Kawano
Y, Adachi Y, Ueda K, Wada H and Tanaka F: Expression of LUN gene
that encodes a novel RING finger protein is correlated with
development and progression of non-small cell lung cancer. Lung
Cancer. 46:21–28. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Zhang Y, Yun H, Lai R and Su M:
Correlation of STAT1 with apoptosis and cell-cycle markers in
esophageal squamous cell carcinoma. PLoS One. 9:e1139282014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bozzuto G, Ruggieri P and Molinari A:
Molecular aspects of tumor cell migration and invasion. Ann Ist
Super Sanita. 46:66–80. 2010.PubMed/NCBI
|
|
37
|
Friedl P and Wolf K: Tumour-cell invasion
and migration: Diversity and escape mechanisms. Nat Rev Cancer.
3:362–374. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yonemori K, Seki N, Kurahara H, Osako Y,
Idichi T, Arai T, Koshizuka K, Kita Y, Maemura K and Natsugoe S:
ZFP36L2 promotes cancer cell aggressiveness and is regulated by
antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer
Sci. 108:124–135. 2017. View Article : Google Scholar :
|
|
39
|
Zhang Y and Yang WB: Down-regulation of
tripartite motif protein 59 inhibits proliferation, migration and
invasion in breast cancer cells. Biomed Pharmacother. 89:462–467.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Deng J, Liang H, Zhang R, Hou Y, Liu Y,
Ying G, Pan Y and Hao X: Clinical and experimental role of ring
finger protein 180 on lymph node metastasis and survival in gastric
cancer. Br J Surg. 103:407–416. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang H, Wang Y, Qian L, Wang X, Gu H, Dong
X, Huang S, Jin M, Ge H, Xu C, et al: RNF216 contributes to
proliferation and migration of colorectal cancer via suppressing
BECN1-dependent autophagy. Oncotarget. 7:51174–51183.
2016.PubMed/NCBI
|
|
42
|
Ando N, Kato H, Igaki H, Shinoda M, Ozawa
S, Shimizu H, Nakamura T, Yabusaki H, Aoyama N, Kurita A, et al: A
randomized trial comparing postoperative adjuvant chemotherapy with
cisplatin and 5-fluorouracil versus preoperative chemotherapy for
localized advanced squamous cell carcinoma of the thoracic
esophagus (JCOG9907). Ann Surg Oncol. 19:68–74. 2012. View Article : Google Scholar
|
|
43
|
Lazzari E and Meroni G: TRIM32 ubiquitin
E3 ligase, one enzyme for several pathologies: From muscular
dystrophy to tumours. Int J Biochem Cell Biol. 79:469–477. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cai W and Yang H: The structure and
regulation of Cullin 2 based E3 ubiquitin ligases and their
biological functions. Cell Div. 11:72016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Geng R, Tan X, Wu J, Pan Z, Yi M, Shi W,
Liu R, Yao C, Wang G, Lin J, et al: RNF183 promotes proliferation
and metastasis of colorectal cancer cells via activation of
NF-κB-IL-8 axis. Cell Death Dis. 8:e29942017. View Article : Google Scholar
|
|
46
|
Liu ZY, Cao J, Zhang JT, Xu GL, Li XP,
Wang FT, Ansari KH, Mohamed H and Fan YZ: Ring finger protein 125,
as a potential highly aggressive and unfavorable prognostic
biomarker, promotes the invasion and metastasis of human
gallbladder cancers via activating the TGF-β1-SMAD3-ID1 signaling
pathway. Oncotarget. 8:49897–49914. 2017.PubMed/NCBI
|