Introduction
Propolis is a complex mixture of resinous material,
produced by bees, created by mixing their saliva with the botanical
sources they live on. The color (yellowish green to dark brown) and
odor (odorless to aromatic) of propolis vary and depend on its
botanical source, origin of place and bee characteristics, such as
strain and age (1,2). Apart from the structural and
functional attributes of propolis for beehives (1), propolis has been reported to possess
a variety of disease-preventive and therapeutic potentials for the
human population (3). There are
mainly two types of propolis known that differ in their
constituents: New Zealand propolis that possesses caffeic acid
phenethyl ester (CAPE) and Brazilian green propolis that possesses
artepillin C (ARC) as predominant bioactive ingredients. Besides
these, over 150 constituents, including flavonoids, phenolic acids,
esters, terpenoids, steroids, amino acids and cinnamic acid
derivatives have been identified (3), and are hence considered as popular
pharmacological research material (4). A broad spectrum of biological
activities identified in propolis include antitumor (5–12),
anti-inflammatory (13–15), anti-bacterial (16–18),
anti-viral (16,19,20)
and anti-fungal (16) activities.
Propolis is used in cosmetic products, such as body lotions,
ointments, face creams and in functional food in various forms,
such as tablets, capsules, toothpaste and mouthwash preparations
(21). Molecular studies on the
anticancer activity of propolis have revealed that its phenolic
components cause cell cycle arrest and apoptosis (11), mitochondrial stress (22) and the inhibition of tumor growth
(11,22,23).
CAPE-based propolis extract (Bio-30) has been reported to block p21
(RAC1) activated kinase 1 (PAK1) signalling and to suppress tumors
in neurofibromatosis (9,24,25).
ARC (3,5-diprenyl-4-hydroxycinnamic acid) is one of
the active phenolic acid components of Brazilian propolis generated
by bees from materials from a Brazilian plant, Baccharis
dracunculifolia. It has been shown to possess various
biological activities, such as anti-viral (14), anti-bacterial (14,26),
antioxidant (26) and
anti-carcinogenic (14,26–28)
activities. ARC has been reported to inhibit the growth of
transplanted solid human and mouse tumors, including malignant
melanoma in athymic and thymic mice, respectively (27). CAPE and ARC have been reported to
differ in their bioavailability profile that in turn is determined
by the stability of the compounds to the digestive enzymes and
absorption through the intestinal lining. CAPE alone becomes
degraded by secreted esterases (12); however, when combined with γ
cyclodextrin (γCD) it is protected and has improved activity in
in vitro and in vivo antitumor assays (12). On the other hand, ARC has been
reported to possess extremely low absorption efficiency and
bioavailability. Whereas CAPE is absorbed and distributed by the
monocarboxylic acid transporter (MCT)-mediated transport system
(29), ARC is mainly permeated
across by transcellular passive diffusion (30). We previously performed a cDNA array
of CAPE-treated normal human cells and found that the cytotoxicity
of CAPE was mediated by the activation of p53-growth arrest and DNA
damage-inducible 45 (GADD45). Bioinformatics and experimental
analyses revealed that CAPE targeted mortalin-p53 interactions,
resulting in the nuclear translocation and re-activation of p53,
leading to growth arrest in cancer cells (12). In the present study, we report that
ARC possesses similar capabilities; however its cytotoxic efficacy
is low. In order to increase the potential of ARC, we prepared the
following: i) supercritical extract [green propolis supercritical
extract (GPSE)]; and ii) its complex with γCD (GPSE-γCD). We then
tested their cytotoxicity in human cancer cells. We report that
GPSE contains 9.6% ARC and exerted cytotoxic effects at 0.5% (16.6
µM) and was equivalent to ~500 µM of pure ARC in
in vitro toxicity assays. The extract exhibited marked
anti-migratory activity. Furthermore, the in vivo efficacy
of GPSE was significantly enhanced by its complex with γCD.
Materials and methods
Docking of mortalin and p53 with ARC
The crystal structures of human mortalin (PDB ID:
4KBO) and p53 (PDB ID: 1OLG) were obtained from Protein Data Bank
(PDB) (31), while the ligand, ARC
(Compound ID: 5472440) was from PubChem Compound (32) database. ArgusLab 4.0.1 (M 2004), a
freely available molecular modelling, graphics and drug design
package, was used for docking analyses. The structures of protein
and the ligand molecules were prepared and the receptor grid around
the binding site residues selected from the receptor was defined.
The grid resolution was kept as 0.4 Å and an exhaustive search
docking was performed. The docking process was carried out using
the 'ArgusDock' docking engine, calculation type as 'Dock' and the
ligand was kept in rigid mode. The scoring function used was an
empirical scoring function, 'AScore', which takes into account van
der Waals energy, hydrophobic component, hydrogen bond and
deformation penalty. The parameter file, 'AScore.prm' was used to
compute the binding energies. A total of 150 docking poses were
generated, ranked according to the scoring function, and the
highest scoring pose was used in further analyses.
Molecular dynamics simulations of
ARC-docked mortalin and p53 structure
MD simulations were carried out using the GROMACS
package (33,34). The force field Gromos43a1 (35) was used for ARC-docked mortalin
structure (33,36,37).
The GROMACS topology file was generated using the antechamber
python parser interface (ACPYPE) script. The docked protein
structure was solvated in a cubic box. Water molecules and
appropriate counter-ions were added to neutralize the system. The
solvated system was minimized using steepest descent and conjugate
gradient methods until the force on each atom was less than 100
kJ/mol/nm. These geometry minimized systems were used for 10 nsec
for carrying out isobaric (constant pressure-temperature) MD
simulations. The temperature and pressure of the system was
maintained at 300 K and 1 atmosphere pressure, respectively, with a
time constant of 5 psec. A 2-fsec time step was used for
integrating the equations of motion. Particle Mesh Ewald summation
method along with periodic boundary conditions were also applied
throughout to calculate the electrostatic potential between partial
charges on atoms. Visual Molecular Dynamics (VMD) version 1.9.2 was
used to calculate the root mean square deviations (RMSD) and
hydrogen bond dynamics.
Cell culture
HT1080 (fibrosarcoma), A549 (lung carcinoma) and
U2OS (osteosarcoma) human cell lines were purchased from DS Pharma
Biomedical Co., Ltd., Osaka, Japan, and cultured in complete
Dulbecco's modified Eagle's medium (DMEM; Life Technologies) medium
using normal cell culture conditions with 10% fetal bovine serum as
a supplement at 37°C, 5% CO2 and 95% air in a humidified
incubator. The cultured cells were maintained for 6–10 passages.
ARC (Wako Pure Chemical Industries, Ltd., Osaka, Japan), GPSE and
GPSE:50%γCD complex were dissolved in dimethylsulfoxide (DMSO), and
directly added to the cell culture medium to obtain the working
concentrations (as indicated in the respective figures).
Preparation of the GPSE-50%γCD
complex
The complex of green propolis supercritical extract
(GPSE) with γCD was prepared by a conventional kneading method.
GPSE and γCD were mixed in a small amount of water. The slurry was
kneaded until it became a homogeneous paste. After freeze-drying of
the paste, the GPSE-γCD complex thus obtained was used in the
present study. The levels of artepillin in GPSE, and its complex
with γCD, were determined by high-performance liquid chromatography
(HPLC) using the Shimadzu HPLC system (LC-2010C; Shimadzu Corp.,
Kyoto, Japan) as previously described (38). A Phenomenex HPLC column [Luna 5u
C18(2) 100A: 4.60 mm I.D. ×150 mm]
was used and the fractionation was performed at 40°C using solution
A: H2O (0.5% acetic acid) and solution B: Acetonitrile
with gradient program as follows: Isocratic 70% A (0–5 min), linear
gradient 70% → 0% A (5–30 min); flow rate, 1 ml/min; injection
volume, 10 µl. Detection was performed at 320 nm (Shimadzu
HPLC system LC-2010C; Shimadzu Corp.).
Cell proliferation assay
Cytotoxicity assay was performed using
3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT)
assay (Life Technologies/Thermo Fisher Scientific, Waltham, MA,
USA) in which cell viability was observed by the conversion of
yellow MTT by the mitochondrial dehydrogenases of living cells into
purple formazon (39). The
statistical significance of the results was determined from 3–4
independent experiments including triplet or quadruplet sets in
each experiment. IC50 values were calculated by the
linear regression methods in MS excel.
Morphological observation
The cells were seeded in 12-well plates and treated
with various concentrations of ARC (100–400 µM, as indicated
in Fig. 2C), GPSE and GPSE-50%γCD
(0.5%). Morphological changes were observed under a phase contrast
microscope (Nikon Eclipse TE300; Nikon, Tokyo, Japan) after 48
h.
Colony formation assay
The cells (500 cells/well) were seeded in a 6-well
plate. Following 24 h of incubation, when the cells had attached to
the surface of the dish they were treated with ARC, GPSE or
GPSE-50%γCD (0.1%) in culture medium (DMEM). The cultures were
maintained for 10–15 days (when colonies formed in the control
cultures) with a regular change of medium every 3rd day. The
colonies were fixed in acetone: methanol (1:1), and stained with
crystal violet (Wako Pure Chemical Industries Ltd.) at the end of
experiments.
Immunofluorescence staining
The cells were seeded on glass coverslips placed in
12-well plates and treated with either ARC (300 µM), GPSE or
GPSE-50%γCD (0.5%) in DMEM. After 48 h of treatment, cells were
fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS),
permeabilized with 0.1% Triton X-100 for 10 min, blocked with 0.2%
bovine serum albumin (BSA)/PBS for 1 h and incubated with specific
primary antibodies including anti-mortalin (clone 088: raised in
our laboratory) (3–5 µg/ml) and p53 (DO-1: SC-126) (3–5
µg/ml) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at
4°C overnight. The cells were then incubated with either Alexa-488
or Alexa-594 conjugated secondary antibodies (1 µg/ml)
(A11034 or A11032, Molecular Probes, Eugene, OR, USA) for 1 h.
Counterstaining was performed with Hoechst 33342 (Sigma, St. Louis,
MO, USA) in the dark for 10 min. The cells were examined on a Zeiss
Axiovert 200 M microscope and analyzed using AxioVision 4.6
software (Carl Zeiss, Oberkochen, Germany).
Wound healing assay
The cells were plated in 6-well plates and allowed
to make monolayers following which a scratch was made using a
100-ml pipette tip. The cells were then washed with PBS and
cultured with the control or test compounds [GPSE or GPSE-50%γCD or
γCD, (0.1%) as indicated in Fig.
4C]. The movement of the cells to the scratch area was followed
for the following 24–48 h. Images were acquired under a phase
contrast microscope (Nikon) with a X10 phase objective lens at the
0, 24 and 48 h time-points. The movement was quantitated by
measuring the empty surface area in the control and test samples by
ImageJ software. The results are expressed as the means ± standard
deviation of the mean (SEM). Statistical significance of the data
was determined using a Student's t-test. Values of P<0.05,
P<0.01 and P<0.001 were considered to indicate significant,
very significant and highly significant differences,
respectively.
In vivo tumor progression
The effects of GPSE and GPSE-50%γCD on in
vivo tumor progression were investigated in a nude mouse
subcutaneous tumor xenograft model. BALB/c nude mice (4 weeks old,
female, 3 mice in each group/average weight 21.8 g) were obtained
from Nihon Clea (Tokyo, Japan). The animals were allowed to
acclimatize in our laboratory for 1 week. The cells were injected
subcutaneously (1×107 suspended in 0.2 ml of growth
medium) into the abdomen of the nude mice and the tail vein
(1×106 suspended in 0.2 ml of growth medium). GPSE and
GPSE 50%γCD were administered (by oral gavage) every alternate day
beginning at 1 day after the injection of the cells for 3 weeks.
Tumor formation and the body weight of the mice were monitored
every alternate day. The volume of the subcutaneous tumors was
calculated as V = L × W2/2, where 'L' was the length and 'W' was
the width of the tumor, respectively. Statistical significance of
the data was calculated from 3 independent experiments (n=3 per
experiment). All procedures were carried out in accordance with the
Animal Experiment and Ethics Committee, Safety and Environment
Management Division, National Institute of Advanced Industrial
Science and Technology (AIST), Tsukuba, Japan.
Statistical analysis
All the experiments were performed in triplicate and
data are expressed as the means ± SEM. An unpaired t-test (GraphPad
Prism GraphPad Software, San Diego, CA, USA) and one-way ANOVA
followed by Tukey's honestly significant difference (HSD) post hoc
test were used to determine the degree of significance between the
control and experimental samples. Statistical significance was
defined as P-values as follows: P<0.05 (significant), P<0.01
(very significant) and P<0.001 (highly significant) for the
t-test and P<0.05 for ANOVA followed by a post hoc test, as
indicated in the figure legends.
Results
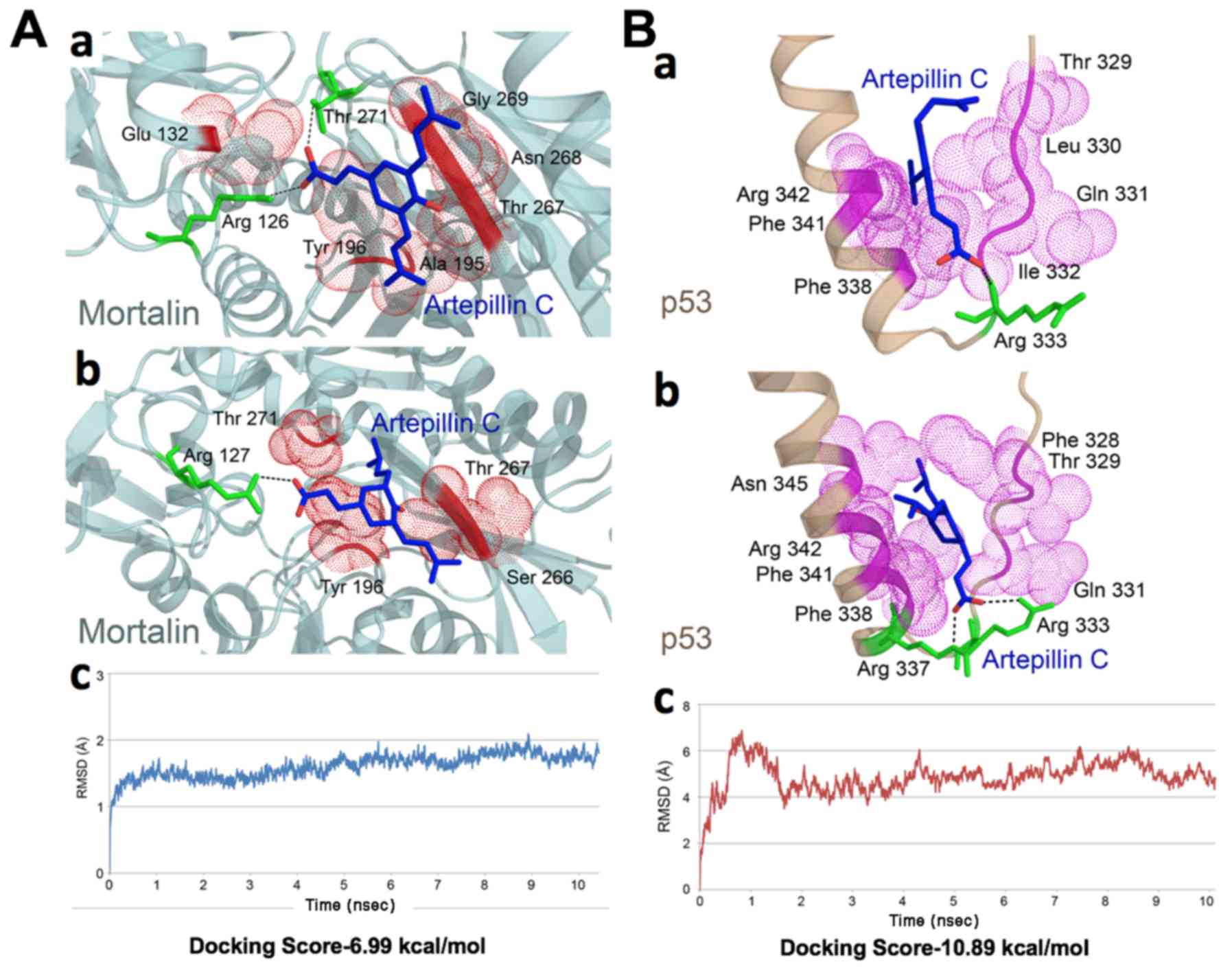
ARC docks into the mortalin-p53 complexes
and induces the activation of p53
The interaction of mortalin and p53 occurs at
N-terminal amino acids residues (253–282) of mortalin and
C-terminal residues (323–337) of p53 (12,40–42).
Based on this information, we docked ARC with mortalin or p53 or at
mortalin-p53 complex interface. ARC interacted with the crystal
structure of mortalin and p53 at the binding regions. It showed a
docking score of −6.99 kcal/mol with mortalin. The docked complex
exhibited multiple hydrogen bonds and hydrophobic interactions
(Fig. 1A–a). The ligand formed 3
hydrogen bonds, 2 with Arg 126 and 1 with Thr 271 of mortalin,
while residues Glu 132, Ala 195, Tyr 196, Thr 267, Asn 268 and Gly
269 played a role in hydrophobic interactions. The second oxygen
atom of ARC formed 2 hydrogen bonds with nitrogen atoms (NH1 and
NH2) of Arg 126. The third hydrogen bond was formed between the
third oxygen atom of ARC and the third oxygen atom of Thr 271.
Together the hydrogen bonds and hydrophobic interactions formed a
stable protein-ligand complex.
The docking of ARC with p53 exhibited a score of
−10.89 kcal/mol. The ligand was observed to form 1 hydrogen bond
and various hydrophobic interactions (Fig. 1B–a). The nitrogen atom of Arg 333
was observed to form a hydrogen bond with the third oxygen atom of
ARC. Residues Thr 329, Leu 330, Gln 331, Ile 332, Phe 338, Phe 341
and Arg 342 were involved in the hydrophobic interaction with p53.
ARC docked in the cavity of p53 forming a stable complex.
Molecular dynamics simulations and energy
stabilization of ARC in the mortalin-p53 complex
The stability of the ARC-docked mortalin structure
was further analyzed by simulating the complex for 10 nsec under
conditions mimicking the bodily environment. The complex was
observed to be stable for a period of 5 nsec from 5 to 10 nsec, as
seen in the RMSD trajectory. Changes observed in the interacting
residues of mortalin are shown in Fig.
1A–b. One hydrogen bond was observed between the oxygen atom of
Arg 127 and the third oxygen atom of ARC, while residues Tyr 196,
Ser 266, Thr 267 and Thr 271 were involved in hydrophobic
interaction following simulations. Despite the changes in the
interaction pattern of ligand-docked protein complex, the structure
was more stable following simulations. The RMSD trajectory is shown
Fig. 1A–c. A change in the
interaction pattern of ARC docked p53 complex was observed
(Fig. 1B–b). The number of
hydrogen bonds between ARC and p53 increased to 2 post-simulations.
One bond was formed between the second oxygen atom of ARC and
nitrogen atom (NH1) of Arg 333, while the second was observed to be
formed between nitrogen atom (NE) of Arg 337. The number of
residues involved in hydrophobic interactions were the same, with 2
residues (Leu 330 and Ile 332) replaced with Phe 328 and Asn 345.
The complex was more stable post-simulations. The ARC-docked p53
structure was observed to be stable from 5 to 10 nsec that can be
seen in the RMSD trajectory (Fig.
1B–c). Bioinformatics analyses revealed that the ARC docked
into the mortalin-p53 complexes (Fig.
1), similar to CAPE, and hence may cause abrogation of these
complexes, and the re-activation of p53 function in cancer
cells.
Anticancer activity of ARC, GPSE and the
GPSE-γCD complex
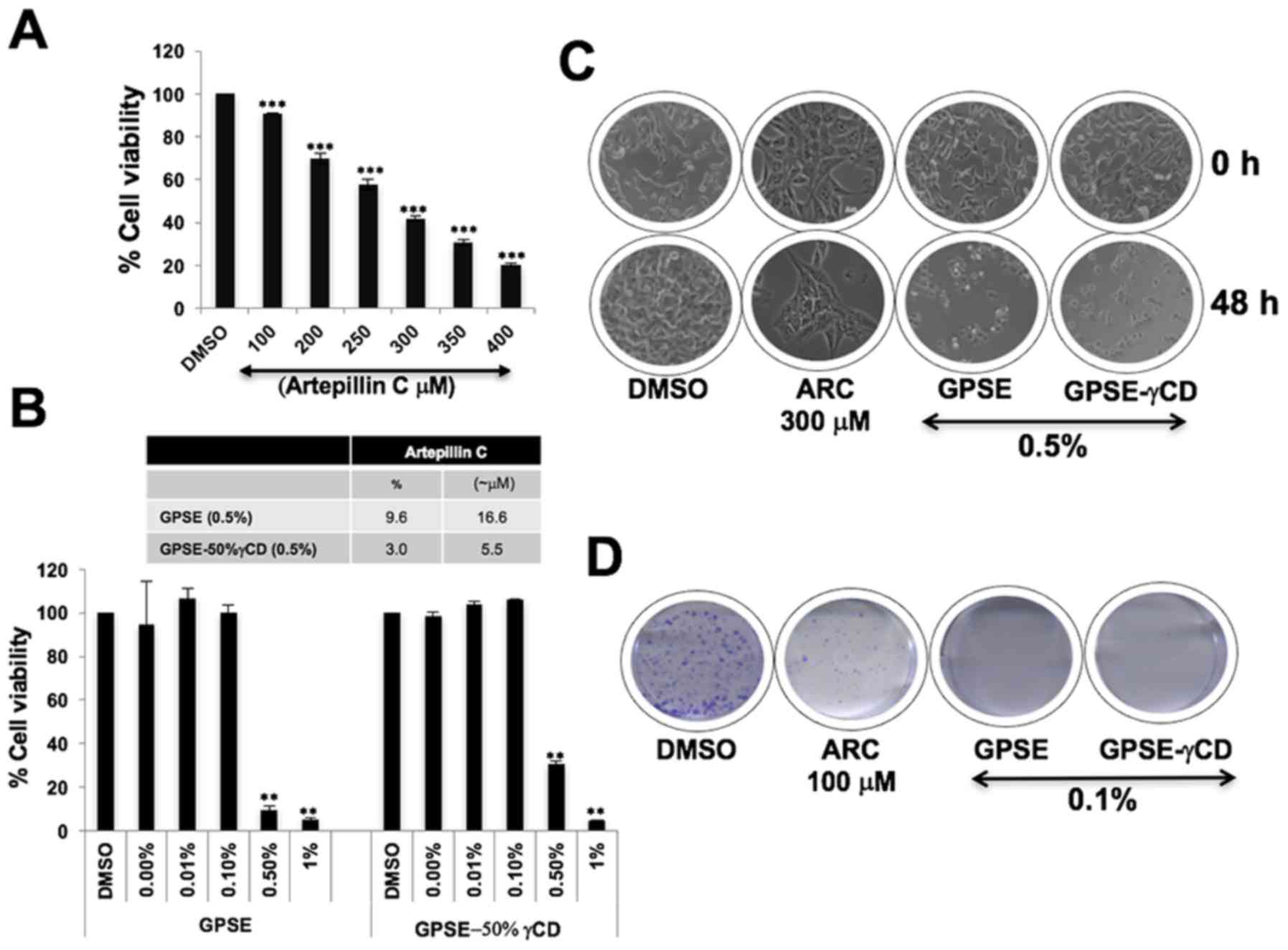
We then examined the cytotoxicity of ARC in HT1080
cells and found that it induced a considerable (>20%) decrease
in cell viability at concentrations of 250 µM
(IC50, 275 µM) (Fig.
2A). Based on such high IC50 values, we considered
whether the extract of green propolis could be more effective than
purified ARC for the reasons including that: i) it may provide a
complex mixture of bioactives with multi-module action; and ii) it
may protect the bioactive components from degradation. We prepared
the GPSE and its conjugate with γCD (GPSE-γCD) and HPLC analyses
revealed that they contained 9.6 and 3.0% ARC, respectively. The
IC50 values were 0.2–0.5% for GPSE and GPSE-γCD, whereas
they were 250–300 µM for ARC (Fig. 2B). Similar results were obtained
with the A549 and U2OS cells (data not shown). Cell morphological
analysis revealed that the cytotoxicity induced by 0.5% GPSE and
GPSE-γCD was comparable to that induced by 300 µM ARC
(Fig. 2C). Long-term colony
forming assays revealed that ARC (100 µM) induced a marked
decrease in the number and size of the colonies; a more potent
decrease was observed by treatment with 0.1% GPSE and GPSE-γCD
(Fig. 2D).
We then examined the above-mentioned concentrations
of ARC, GPSE and GPSE-γCD, and determined their effects on
mortalin-p53 interactions as predicted by bioinformatics analysis
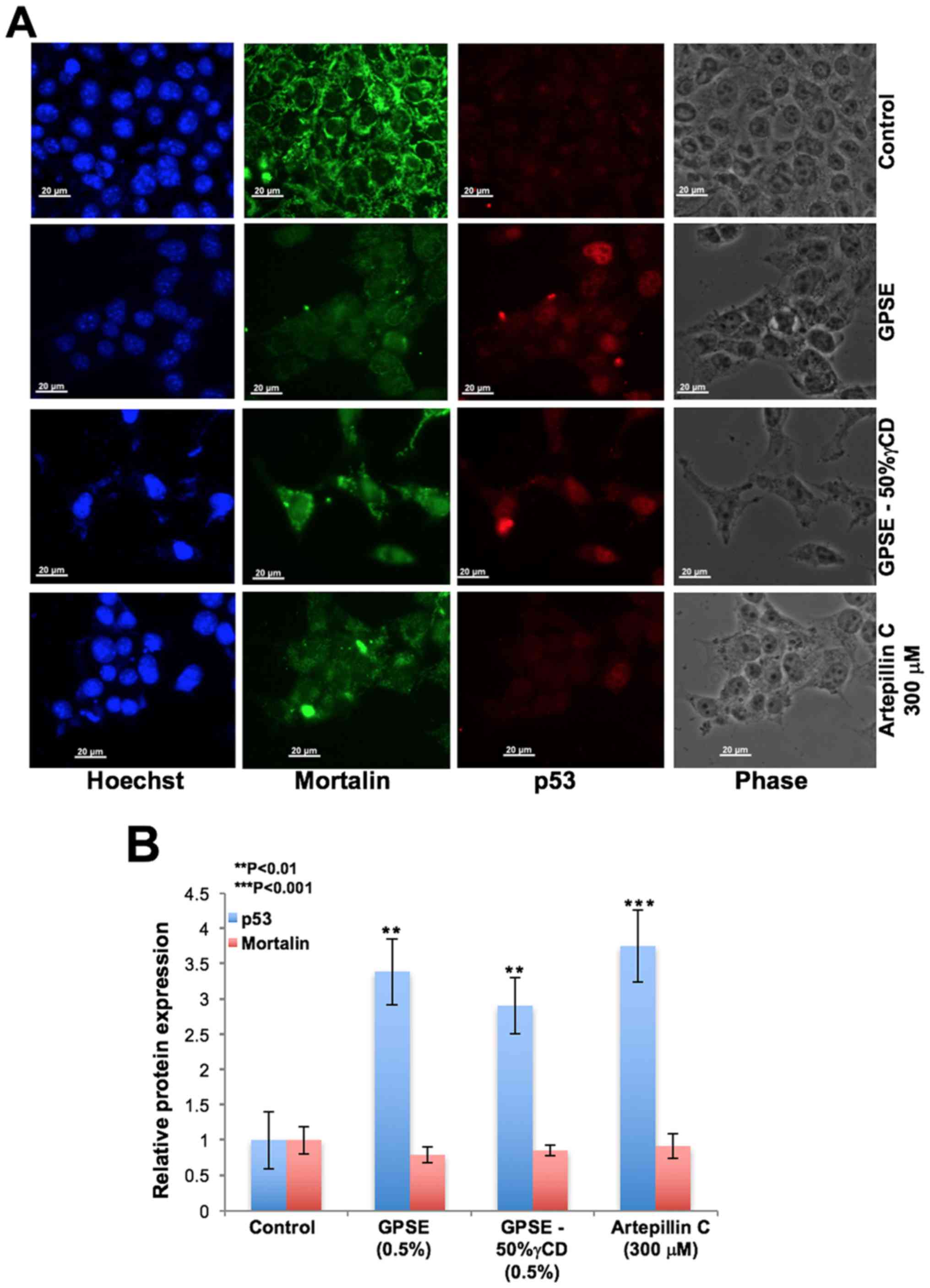
(Fig. 1). In several independent
experiments, we found that treatment with GPSE and ARC led to the
nuclear trans-location of p53 (Fig.
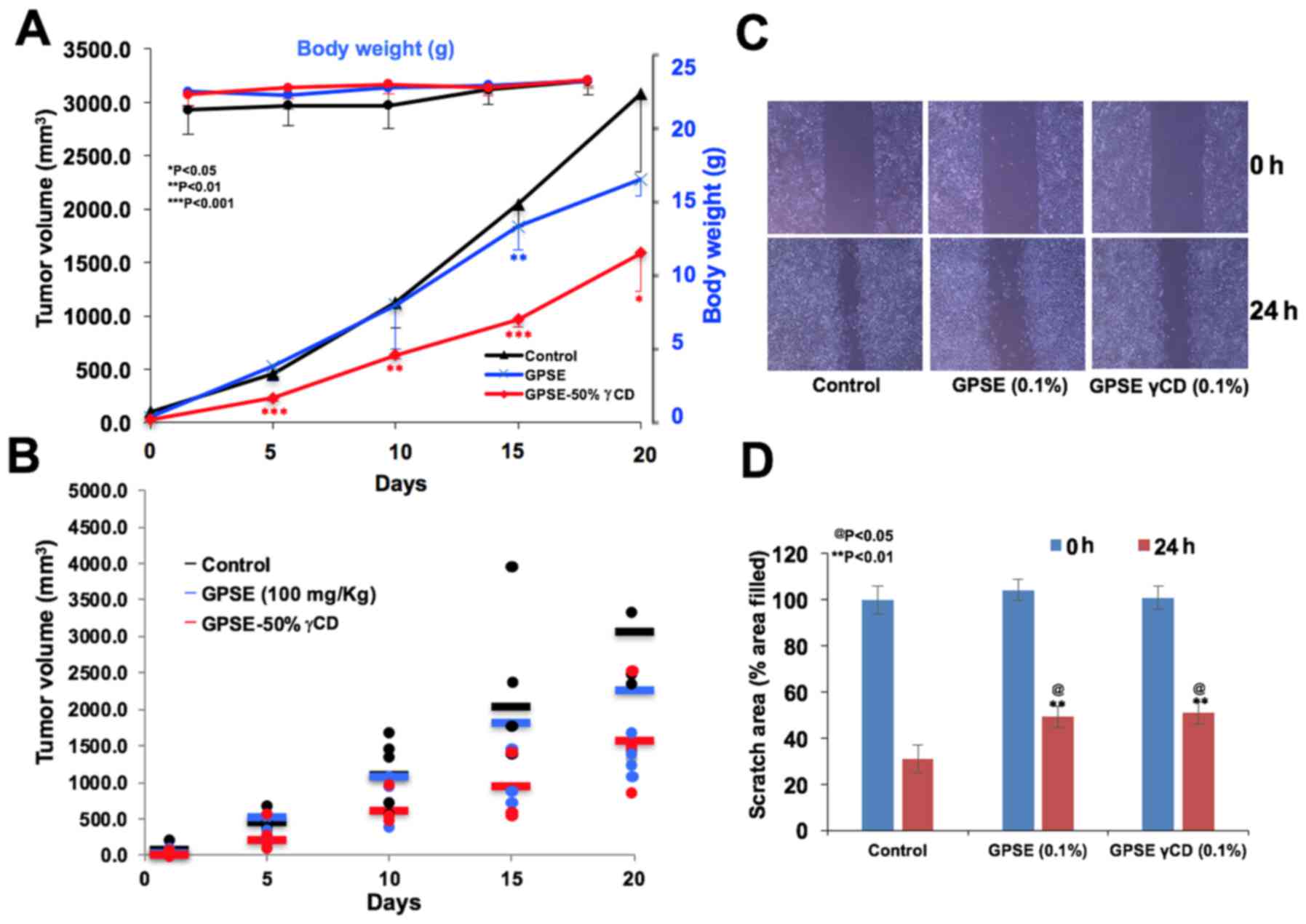
3A), accompanied by an increase in p53 expression (Fig. 3B). We performed antitumor assays
using a subcutaneous tumor xenograft model with nude mice. As shown
in Fig. 4, mice fed the GPSE or
GPSE-γCD exhibited no change in body weight (Fig. 4A) or physical activity (data not
shown) with respect to the control group. A reduction in tumour
growth was observed in the mice fed GPSE or GPSE-γCD as compared to
the control group over the 3 weeks of treatment (Fig. 4B). The tumor growth data were
strongly suggestive that the GPSE-γCD induced a more marked
suppressive effect on tumor growth, endorsing its potential as an
antitumor natural compound. We also performed in vitro wound
healing assays to examine whether the GPSE and GPSE-γCD extracts
possess anti-migratory activity and to determine whether they can
be used to inhibit cancer metastasis. As shown in Fig. 4C and D, a considerable inhibition
of cell migration was detected in the cells cultured in the
presence of non-toxic doses of GPSE and GPSE-γCD.
Discussion
A number of different methods have been used to
extract active components from propolis, including solvent
extraction, e.g., ethanol (3),
water or diluted aqueous ethanol (43) and supercritical carbon dioxide
extracts (8,44,45).
Apart from differences in the yield due to the function of
temperature, pressure and solvent concentration, the different
extraction methods generate mixtures with different components and
also show different activities. Supercritical extracts have
non-polar characteristics that are tolerant to oxidation and
provide high yields (8,44,46).
However, due to their poor solubility in aqueous solvents, their
use is limited in in vitro and in vivo studies.
Cyclodextrins (CDs), with inner lipophilic cavities and hydrophilic
outer surfaces, are capable of encapsulating molecules by
non-covalent inclusion (47,48).
We previously used γCD to conjugate with CAPE to generate a
CAPE-γCD complex that showed significant anticancer activity
(12). In the present study, we
demonstrate that ARC, an active component of Brazilian green
propolis, activates p53 tumor suppressor protein by abrogating its
complex with mortalin (p53 inactivating protein), and thus
possesses anticancer activity. However, it turned out to be a low
efficacy compound (IC50, 275 µM). We hypothesized
that crude extracts of green propolis may possess better efficacy.
Hence, we generated its supercritical extract (GPSE) and its
complex with γCD (GPSE-γCD). We herein report that the GPSE
containing 9.6% ARC possesses high cytotoxicity to cancer cells,
and was effective at concentrations of 0.25 to 0.5% (8.3–16.6
µM ARC). Furthermore, the GPSE-γCD complex (containing 3%
ARC) exhibited greater cytotoxicity in vitro (0.25 to 0.5% =
2.7–5.5 µM ARC) and greater antitumor activity in
vivo. Anti-migration assays revealed that GPSE, as well as the
GPSE-50%γCD complex, at non-toxic concentrations, caused the
delayed migration of cells to the scratch in wound healing assays,
suggesting its potential for the treatment of metastatic cancers.
Based on these data, GPSE-γCD is proposed as a Natural Efficient
and Welfare (NEW) antitumor composite. Further studies are
warranted to elucidate the molecular mechanism(s) of the potent
anticancer activity of GPSE and its complex with γCD.
Abbreviations:
|
ARC
|
artepillin C
|
|
GPSE
|
green propolis supercritical
extract
|
|
γCD
|
γ cyclodextrin
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
CAPE
|
caffeic acid phenethyl ester
|
|
GADD45
|
growth arrest and DNA damage-inducible
45
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,
5-diphenyltetrazolium bromide
|
|
BSA
|
bovine serum albumin
|
|
PBS
|
phosphate-buffered saline
|
Acknowledgments
Priyanshu Bhargava is a recipient of the Ministry of
Education, Culture, Sports, Science and Technology (MEXT)
Scholarship, Japan. This study was supported by AIST (Japan) and
DBT (Government of India) funds.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Bankova V: Chemical diversity of propolis
and the problem of standardization. J Ethnopharmacol. 100:114–117.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuropatnicki AK, Szliszka E and Krol W:
Historical aspects of propolis research in modern times. Evid Based
Complement Alternat Med. 2013:9641492013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Burdock GA: Review of the biological
properties and toxicity of bee propolis (propolis). Food Chem
Toxicol. 36:347–363. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bankova V, Popova M and Trusheva B:
Propolis volatile compounds: chemical diversity and biological
activity: A review. Chem Cent J. 8:282014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fauzi AN, Norazmi MN and Yaacob NS:
Tualang honey induces apoptosis and disrupts the mitochondrial
membrane potential of human breast and cervical cancer cell lines.
Food Chem Toxicol. 49:871–878. 2011. View Article : Google Scholar
|
|
6
|
Ghashm AA, Othman NH, Khattak MN, Ismail
NM and Saini R: Antiproliferative effect of Tualang honey on oral
squamous cell carcinoma and osteosarcoma cell lines. BMC Complement
Altern Med. 10:492010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khacha-ananda S, Tragoolpua K,
Chantawannakul P and Tragoolpua Y: Antioxidant and anti-cancer cell
proliferation activity of propolis extracts from two extraction
methods. Asian Pac J Cancer Prev. 14:6991–6995. 2013. View Article : Google Scholar
|
|
8
|
Machado BA, Silva RP, Barreto GA, Costa
SS, Silva DF, Brandão HN, Rocha JL, Dellagostin OA, Henriques JA,
Umsza-Guez MA, et al: Chemical composition and biological activity
of extracts obtained by supercritical extraction and ethanolic
extraction of brown, green and red propolis derived from different
geographic regions in Brazil. PLoS One. 11:e01459542016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Messerli SM, Ahn MR, Kunimasa K,
Yanagihara M, Tatefuji T, Hashimoto K, Mautner V, Uto Y, Hori H,
Kumazawa S, et al: Artepillin C (ARC) in Brazilian green propolis
selectively blocks oncogenic PAK1 signaling and suppresses the
growth of NF tumors in mice. Phytother Res. 23:423–427. 2009.
View Article : Google Scholar
|
|
10
|
Rao CV, Desai D, Simi B, Kulkarni N, Amin
S and Reddy BS: Inhibitory effect of caffeic acid esters on
azoxymethane-induced biochemical changes and aberrant crypt foci
formation in rat colon. Cancer Res. 53:4182–4188. 1993.PubMed/NCBI
|
|
11
|
Sawicka D, Car H, Borawska MH and
Nikliński J: The anticancer activity of propolis. Folia Histochem
Cytobiol. 50:25–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wadhwa R, Nigam N, Bhargava P, Dhanjal JK,
Goyal S, Grover A, Sundar D, Ishida Y, Terao K and Kaul SC:
Molecular characterization and enhancement of anticancer activity
of caffeic acid phenethyl ester by γ cyclodextrin. J Cancer.
7:1755–1771. 2016. View Article : Google Scholar :
|
|
13
|
Gao W, Wu J, Wei J, Pu L, Guo C, Yang J,
Yang M and Luo H: Brazilian green propolis improves immune function
in aged mice. J Clin Biochem Nutr. 55:7–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khayyal MT, el-Ghazaly MA and el-Khatib
AS: Mechanisms involved in the antiinflammatory effect of propolis
extract. Drugs Exp Clin Res. 19:197–203. 1993.PubMed/NCBI
|
|
15
|
Wang L, Yang L, Debidda M, Witte D and
Zheng Y: Cdc42 GTPase-activating protein deficiency promotes
genomic instability and premature aging-like phenotypes. Proc Natl
Acad Sci USA. 104:1248–1253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kujumgiev A, Tsvetkova I, Serkedjieva Y,
Bankova V, Christov R and Popov S: Antibacterial, antifungal and
antiviral activity of propolis of different geographic origin. J
Ethnopharmacol. 64:235–240. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pepeljnjak S, Jalsenjak I and Maysinger D:
Flavonoid content in propolis extracts and growth inhibition of
Bacillus subtilis. Pharmazie. 40:122–123. 1985.PubMed/NCBI
|
|
18
|
Velikova M, Bankova V, Tsvetkova I,
Kujumgiev A and Marcucci MC: Antibacterial ent-kaurene from
Brazilian propolis of native stingless bees. Fitoterapia.
71:693–696. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jafarzadeh Kashi TS, Kasra Kermanshahi R,
Erfan M, Vahid Dastjerdi E, Rezaei Y and Tabatabaei FS: Evaluating
the in vitro antibacterial effect of Iranian propolis on oral
microorganisms. Iran J Pharm Res. 10:363–368. 2011.PubMed/NCBI
|
|
20
|
Sartori G, Pesarico AP, Pinton S,
Dobrachinski F, Roman SS, Pauletto F, Rodrigues LC Jr and Prigol M:
Protective effect of brown Brazilian propolis against acute vaginal
lesions caused by herpes simplex virus type 2 in mice: Involvement
of antioxidant and anti-inflammatory mechanisms. Cell Biochem
Funct. 30:1–10. 2012. View
Article : Google Scholar
|
|
21
|
Choudhari MK, Haghniaz R, Rajwade JM and
Paknikar KM: Anticancer activity of Indian stingless bee propolis:
An in vitro study. Evid Based Complement Alternat Med.
2013.928280:2013.
|
|
22
|
Benguedouar L, Boussenane HN, Wided K,
Alyane M, Rouibah H and Lahouel M: Efficiency of propolis extract
against mitochondrial stress induced by antineoplasic agents
(doxorubicin and vinblastin) in rats. Indian J Exp Biol.
46:112–119. 2008.PubMed/NCBI
|
|
23
|
Chen CN, Weng MS, Wu CL and Lin JK:
Comparison of radical scavenging activity, cytotoxic effects and
apoptosis induction in human melanoma cells by Taiwanese propolis
from different sources. Evid Based Complement. Alternat Med.
1:175–185. 2004.
|
|
24
|
Hirokawa Y, Levitzki A, Lessene G, Baell
J, Xiao Y, Zhu H and Maruta H: Signal therapy of human pancreatic
cancer and NF1-deficient breast cancer xenograft in mice by a
combination of PP1 and GL-2003, anti-PAK1 drugs (Tyr-kinase
inhibitors). Cancer Lett. 245:242–251. 2007. View Article : Google Scholar
|
|
25
|
Maruta H: Effective neurofibromatosis
therapeutics blocking the oncogenic kinase PAK1. Drug Discov Ther.
5:266–278. 2011. View Article : Google Scholar
|
|
26
|
Veiga RS, De Mendonça S, Mendes PB,
Paulino N, Mimica MJ, Lagareiro Netto AA, Lira IS, López BG, Negrão
V and Marcucci MC: Artepillin C and phenolic compounds responsible
for antimicrobial and antioxidant activity of green propolis and
Baccharis dracunculifolia DC. J Appl Microbiol. 122:911–920. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kimoto T, Arai S, Kohguchi M, Aga M,
Nomura Y, Micallef MJ, Kurimoto M and Mito K: Apoptosis and
suppression of tumor growth by artepillin C extracted from
Brazilian propolis. Cancer Detect Prev. 22:506–515. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matsuno T, Jung SK, Matsumoto Y, Saito M
and Morikawa J: Preferential cytotoxicity to tumor cells of
3,5-diprenyl-4-hydroxycinnamic acid (artepillin C) isolated from
propolis. Anticancer Res. 17A:3565–3568. 1997.
|
|
29
|
Konishi Y, Hitomi Y, Yoshida M and
Yoshioka E: Absorption and bioavailability of artepillin C in rats
after oral administration. J Agric Food Chem. 53:9928–9933. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Konishi Y: Transepithelial transport of
artepillin C in intestinal Caco-2 cell monolayers. Biochim Biophys
Acta. 1713:138–144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berman HM, Battistuz T, Bhat TN, Bluhm WF,
Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et
al: The Protein Data Bank. Acta Crystallogr D Biol Crystallogr.
58:899–907. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bolton EE, Wang Y, Thiessen PA and Bryant
SH: Chapter 12-PubChem: Integrated platform of small molecules and
biological activities. Annu Rep Comput Chem. 4:217–241. 2008.
View Article : Google Scholar
|
|
33
|
Van Der Spoel D, Lindahl E, Hess B,
Groenhof G, Mark AE and Berendsen HJ: GROMACS: Fast, flexible, and
free. J Comput Chem. 26:1701–1718. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
van der Spoel D, van Maaren PJ and Caleman
C: GROMACS molecule & liquid database. Bioinformatics.
28:752–753. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lindorff-Larsen K, Piana S, Palmo K,
Maragakis P, Klepeis JL, Dror RO and Shaw DE: Improved side-chain
torsion potentials for the Amber ff99SB protein force field.
Proteins. 78:1950–1958. 2010.PubMed/NCBI
|
|
36
|
van der Spoel D, Lindahl E, Hess B, Van
Buuren AR, Apol E, Meulenhoff PJ, Tieleman DP, Sijbers ALTM,
Feenstra KA, van Drunen R, et al: GROMACS user manual version 3.3.
2008, http://ftp.gromacs.org/pub/manual/manual-3.3.pdfurisimpleftp://ftp.gromacs.org/pub/manual/manual-3.3.pdf.
|
|
37
|
van Gunsteren W, Billeter S, Eising A,
Hünenberger P, Krüger P, Mark A, Scott W and Tironi I: Gromos43a1.
Hochschulverlag AG an der ETH Zürich; Zürich: 1996
|
|
38
|
Nobushi Y, Oikawa N, Okazaki Y, Tsutsumi
S, Park YK, Kurokawa M and Yasukawa K: Determination of
Artepillin-C in Brazilian propolis by HPLC with photodiode array
detector. J Pharm Nutr Sci. 2:127–131. 2012.
|
|
39
|
Ryu J, Kaul Z, Yoon AR, Liu Y, Yaguchi T,
Na Y, Ahn HM, Gao R, Choi IK, Yun CO, et al: Identification and
functional characterization of nuclear mortalin in human
carcinogenesis. J Biol Chem. 289:24832–24844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grover A, Priyandoko D, Gao R, Shandilya
A, Widodo N, Bisaria VS, Kaul SC, Wadhwa R and Sundar D: Withanone
binds to mortalin and abrogates mortalin-p53 complex: Computational
and experimental evidence. Int J Biochem Cell Biol. 44:496–504.
2012. View Article : Google Scholar
|
|
41
|
Lu WJ, Lee NP, Kaul SC, Lan F, Poon RT,
Wadhwa R and Luk JM: Mortalin-p53 interaction in cancer cells is
stress dependent and constitutes a selective target for cancer
therapy. Cell Death Differ. 18:1046–1056. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nagpal N, Goyal S, Dhanjal JK, Ye L, Kaul
SC, Wadhwa R, Chaturvedi R and Grover A: Molecular dynamics-based
identification of novel natural mortalin-p53 abrogators as
anticancer agents. J Recept Signal Transduct Res. 37:8–16. 2017.
View Article : Google Scholar
|
|
43
|
Markiewicz-Żukowska R, Car H, Naliwajko
SK, Sawicka D, Szynaka B, Chyczewski L, Isidorov V and Borawska MH:
Ethanolic extract of propolis, chrysin, CAPE inhibit human
astroglia cells. Adv Med Sci. 57:208–216. 2012. View Article : Google Scholar
|
|
44
|
Machado BA, Barreto GA, Costa AS, Costa
SS, Silva RP, da Silva DF, Brandão HN, da Rocha JL, Nunes SB,
Umsza-Guez MA, et al: Determination of parameters for the
supercritical extraction of antioxidant compounds from green
propolis using carbon dioxide and ethanol as co-solvent. PLoS One.
10:e01344892015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Takara K, Fujita M, Matsubara M, Minegaki
T, Kitada N, Ohnishi N and Yokoyama T: Effects of propolis extract
on sensitivity to chemotherapeutic agents in HeLa and resistant
sublines. Phytother Res. 21:841–846. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Akanda MJ, Sarker MZ, Ferdosh S, Manap MY,
Ab Rahman NN and Ab Kadir MO: Applications of supercritical fluid
extraction (SFE) of palm oil and oil from natural sources.
Molecules. 17:1764–1794. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Szente L and Szejtli J: Highly soluble
cyclodextrin derivatives: Chemistry, properties, and trends in
development. Adv Drug Deliv Rev. 36:17–28. 1999. View Article : Google Scholar
|
|
48
|
Charumanee S, Okonogi S, Sirithunyalug J,
Wolschann P and Viernstein H: Effect of cyclodextrin types and
co-solvent on solubility of a poorly water soluble drug. Sci Pharm.
84:694–704. 2016. View Article : Google Scholar : PubMed/NCBI
|