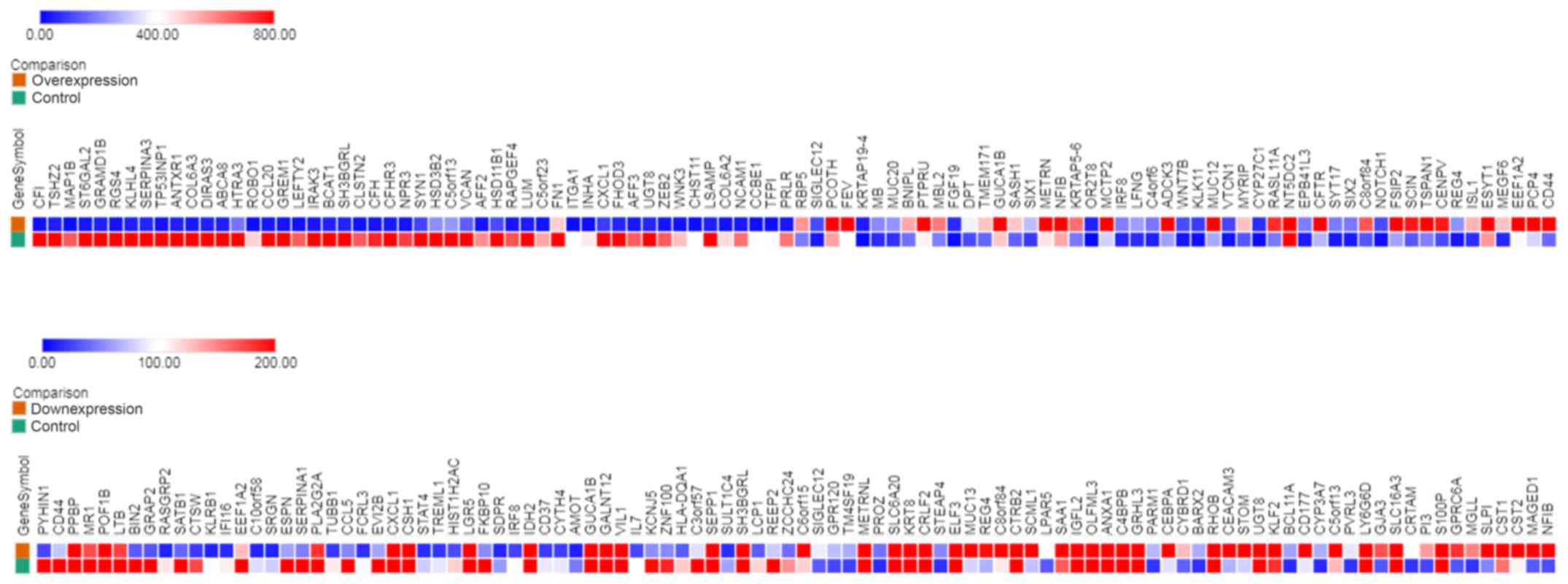
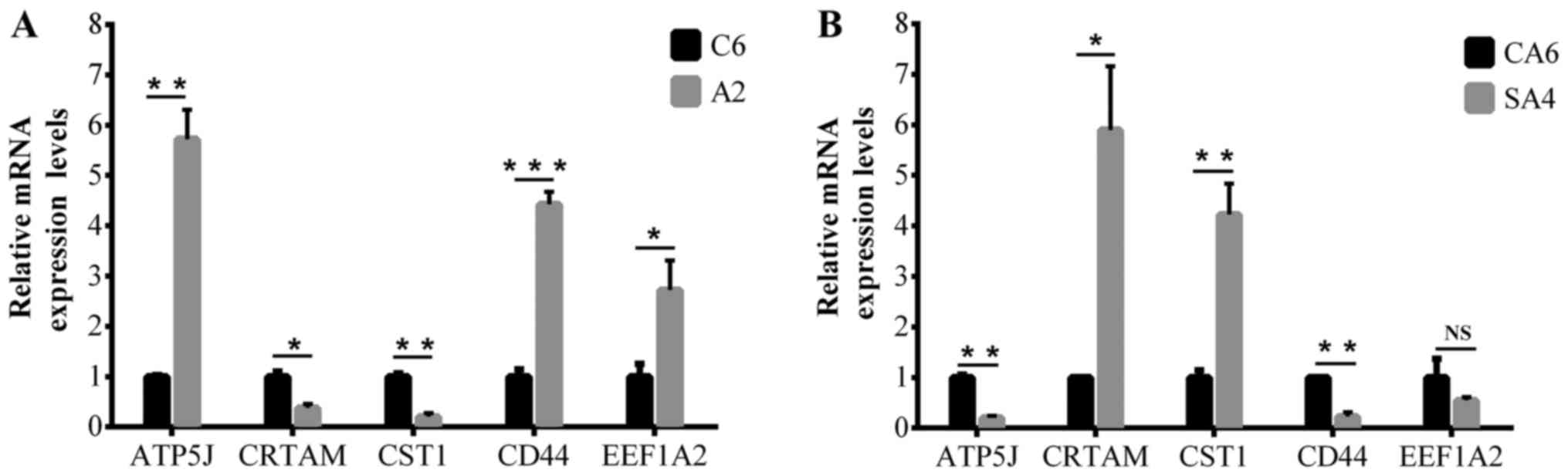
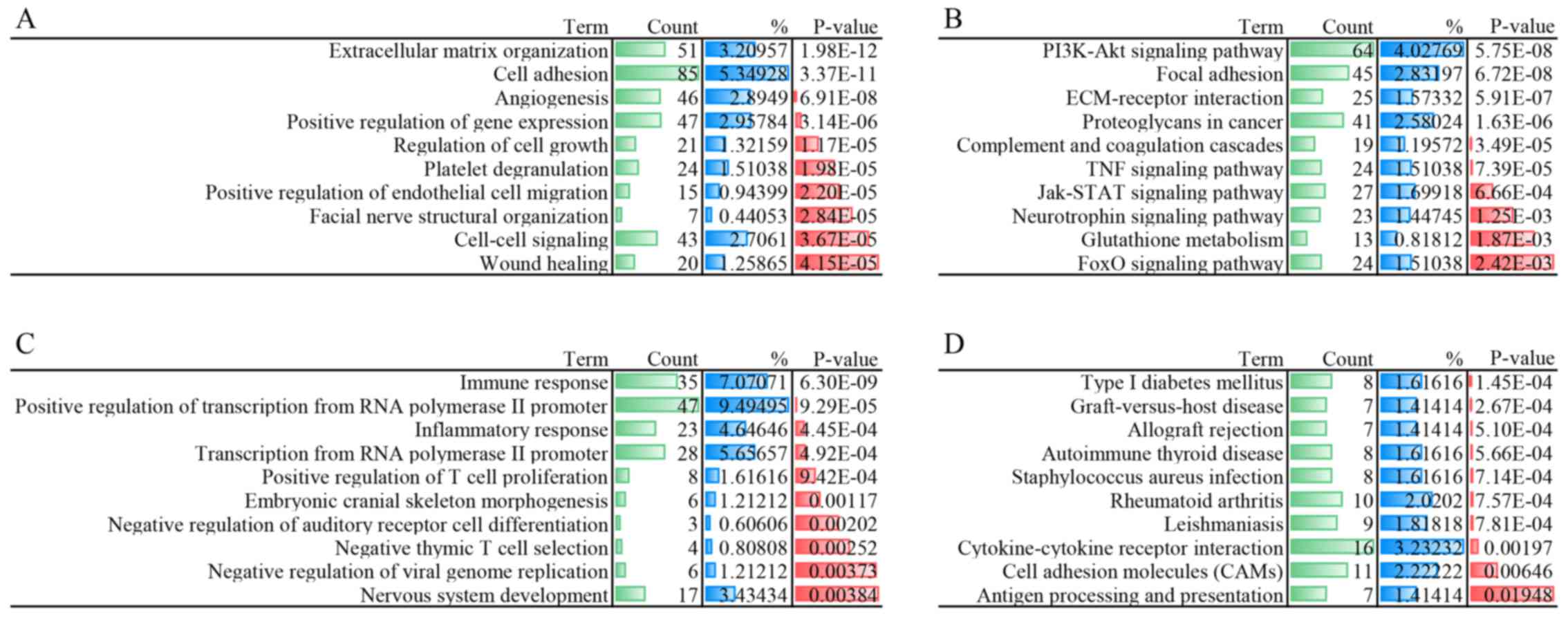
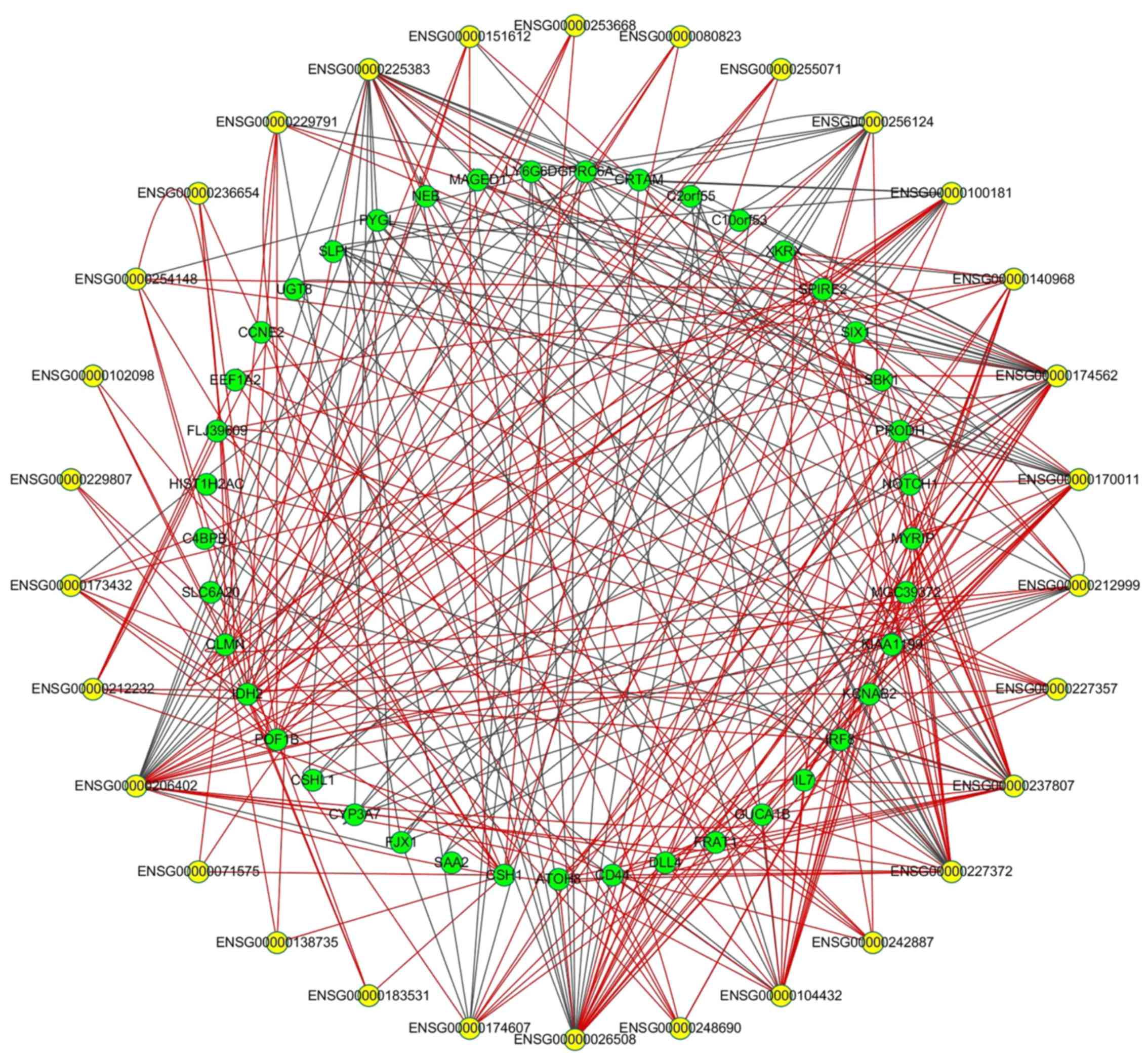
|
1
|
Chau I and Cunningham D: Chemotherapy in
colorectal cancer: New options and new challenges. Br Med Bull.
64:159–180. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shao YC, Chang YY, Lin JK, Lin CC, Wang
HS, Yang SH, Jiang JK, Lan YT, Lin TC, Li AF, et al: Neoadjuvant
chemotherapy can improve outcome of colorectal cancer patients with
unresectable metastasis. Int J Colorectal Dis. 28:1359–1365. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu HY and Zhang CJ: Identification of
differentially expressed genes and their upstream regulators in
colorectal cancer. Cancer Gene Ther. 24:244–250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pedersen PL: Bioenergetics of cancer cells
- A brief orientation to this minireview series. J Bioenerg
Biomembr. 29:301–302. 1997. View Article : Google Scholar
|
|
5
|
Cuezva JM, Krajewska M, de Heredia ML,
Krajewski S, Santamaría G, Kim H, Zapata JM, Marusawa H, Chamorro M
and Reed JC: The bioenergetic signature of cancer: A marker of
tumor progression. Cancer Res. 62:6674–6681. 2002.PubMed/NCBI
|
|
6
|
Shin YK, Yoo BC, Chang HJ, Jeon E, Hong
SH, Jung MS, Lim SJ and Park JG: Down-regulation of mitochondrial
F1F0-ATP synthase in human colon cancer cells with induced
5-fluorouracil resistance. Cancer Res. 65:3162–3170. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Collinson IR, van Raaij MJ, Runswick MJ,
Fearnley IM, Skehel JM, Orriss GL, Miroux B and Walker JE: ATP
synthase from bovine heart mitochondria. In vitro assembly of a
stalk complex in the presence of F1-ATPase and in its absence. J
Mol Biol. 242:408–421. 1994.PubMed/NCBI
|
|
8
|
Joshi S and Pringle MJ: ATP synthase
complex from bovine heart mitochondria. Passive H+
conduction through mitochondrial coupling factor 6-depleted F0
complexes. J Biol Chem. 264:15548–15551. 1989.PubMed/NCBI
|
|
9
|
Zhu H, Chen L, Zhou W, Huang Z, Hu J, Dai
S, Wang X, Huang X and He C: Over-expression of the ATP5J gene
correlates with cell migration and 5-fluorouracil sensitivity in
colorectal cancer. PLoS One. 8:e768462013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang W, Lu Y, Xu Y, Xu L, Zheng W, Wu Y,
Li L and Shen P: Estrogen represses hepatocellular carcinoma (HCC)
growth via inhibiting alternative activation of tumor-associated
macrophages (TAMs). J Biol Chem. 287:40140–40149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bjerregaard H, Pedersen S, Kristensen SR
and Marcussen N: Reference genes for gene expression analysis by
real-time reverse transcription polymerase chain reaction of renal
cell carcinoma. Diagn Mol Pathol. 20:212–217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
13
|
Carson MB and Lu H: Network-based
prediction and knowledge mining of disease genes. BMC Med Genomics.
8(Suppl 2): S92015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zamani-Ahmadmahmudi M, Najafi A and
Nassiri SM: Reconstruction of canine diffuse large B-cell lymphoma
gene regulatory network: Detection of functional modules and hub
genes. J Comp Pathol. 152:119–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu P, Weaver VM and Werb Z: The
extracellular matrix: A dynamic niche in cancer progression. J Cell
Biol. 196:395–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hynes RO: The extracellular matrix: Not
just pretty fibrils. Science. 326:1216–1219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gilkes DM, Semenza GL and Wirtz D: Hypoxia
and the extracellular matrix: Drivers of tumour metastasis. Nat Rev
Cancer. 14:430–439. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Groner B and von Manstein V: Jak Stat
signaling and cancer: Opportunities, benefits and side effects of
targeted inhibition. Mol Cell Endocrinol. 451:1–14. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheung M and Testa JR: Diverse mechanisms
of AKT pathway activation in human malignancy. Curr Cancer Drug
Targets. 13:234–244. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Agarwal E, Brattain MG and Chowdhury S:
Cell survival and metastasis regulation by Akt signaling in
colorectal cancer. Cell Signal. 25:1711–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chau NM and Ashcroft M: Akt2: A role in
breast cancer metastasis. Breast Cancer Res. 6:55–57. 2004.
View Article : Google Scholar :
|
|
24
|
Rychahou PG, Kang J, Gulhati P, Doan HQ,
Chen LA, Xiao SY, Chung DH and Evers BM: Akt2 overexpression plays
a critical role in the establishment of colorectal cancer
metastasis. Proc Natl Acad Sci USA. 105:20315–20320. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Agarwal E, Robb CM, Smith LM, Brattain MG,
Wang J, Black JD and Chowdhury S: Role of Akt2 in regulation of
metastasis suppressor 1 expression and colorectal cancer
metastasis. Oncogene. 36:3104–3118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ding Z, Xu F, Li G, Tang J, Tang Z, Jiang
P and Wu H: Knockdown of Akt2 expression by shRNA inhibits
proliferation, enhances apoptosis, and increases chemosensitivity
to paclitaxel in human colorectal cancer cells. Cell Biochem
Biophys. 71:383–388. 2015. View Article : Google Scholar
|
|
27
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Huang Y, Huang Y, Fu Y, Tang D, Kang
R, Zhou R and Fan XG: The long non-coding RNA TP73-AS1 modulates
HCC cell proliferation through miR-200a-dependent HMGB1/RAGE
regulation. J Exp Clin Cancer Res. 36:512017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu G, Wang S, Chen J, Wang Z, Liang X,
Wang X, Jiang J, Lang J and Li L: Long noncoding RNA HAS2-AS1
mediates hypoxia-induced invasiveness of oral squamous cell
carcinoma. Mol Carcinog. 56:2210–2222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang H, Xiong Y, Xia R, Wei C, Shi X and
Nie F: The pseudogene-derived long noncoding RNA SFTA1P is
downregulated and suppresses cell migration and invasion in lung
adenocarcinoma. Tumour Biol. 39:10104283176914182017.
|
|
31
|
Song H, He P, Shao T, Li Y, Li J and Zhang
Y: Long non-coding RNA XIST functions as an oncogene in human
colorectal cancer by targeting miR-132-3p. J BUON. 22:696–703.
2017.PubMed/NCBI
|
|
32
|
Fang J, Sun CC and Gong C: Long noncoding
RNA XIST acts as an oncogene in non-small cell lung cancer by
epigenetically repressing KLF2 expression. Biochem Biophys Res
Commun. 478:811–817. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J,
Chen L, Xi Z, Teng H, Wang Z, et al: Knockdown of long non-coding
RNA XIST exerts tumor-suppressive functions in human glioblastoma
stem cells by up-regulating miR-152. Cancer Lett. 359:75–86. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu X, Dinglin X, Wang X, Luo W, Shen Q, Li
Y, Gu L, Zhou Q, Zhu H, Li Y, et al: Long noncoding RNA XIST
promotes malignancies of esophageal squamous cell carcinoma via
regulation of miR-101/EZH2. Oncotarget. 8:76015–76028.
2017.PubMed/NCBI
|
|
35
|
Wang H, Shen Q, Zhang X, Yang C, Cui S,
Sun Y, Wang L, Fan X and Xu S: The long non-coding RNA XIST
controls non-small cell lung cancer proliferation and invasion by
modulating miR-186-5p. Cell Physiol Biochem. 41:2221–2229. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kara M, Yumrutas O, Ozcan O, Celik OI,
Bozgeyik E, Bozgeyik I and Tasdemir S: Differential expressions of
cancer-associated genes and their regulatory miRNAs in colorectal
carcinoma. Gene. 567:81–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bione S, Rizzolio F, Sala C, Ricotti R,
Goegan M, Manzini MC, Battaglia R, Marozzi A, Vegetti W, Dalprà L,
et al: Mutation analysis of two candidate genes for premature
ovarian failure, DACH2 and POF1B. Hum Reprod. 19:2759–2766. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Crespi A, Bertoni A, Ferrari I, Padovano
V, Della Mina P, Berti E, Villa A and Pietrini G: POF1B localizes
to desmosomes and regulates cell adhesion in human intestinal and
keratinocyte cell lines. J Invest Dermatol. 135:192–201. 2015.
View Article : Google Scholar
|
|
39
|
Marzinke MA and Clagett-Dame M: The
all-trans retinoic acid (atRA)-regulated gene Calmin (Clmn)
regulates cell cycle exit and neurite outgrowth in murine
neuroblastoma (Neuro2a) cells. Exp Cell Res. 318:85–93. 2012.
View Article : Google Scholar
|