Introduction
Esophageal squamous cell carcinoma (ESCC) is one of
the most malignant tumors worldwide (1–3) and
the sixth most fatal cause of cancer-related death. Squamous cell
carcinoma (SCC) is the main histological type of this cancer.
Northern and central China is part of the 'Asian belt' which has a
very high incidence of ESCC; there are >100 cases per 100,000
annually. The urgency of preventing and curing ESCC is obvious
(4,5).
Similar to other types of cancer, the development of
ESCC is believed to be a multi-step process caused by the
accumulation of activated oncogenes and inactivated tumor
suppressor genes (TSGs) (6). TSGs
can be inactivated by both genetic and epigenetic mechanisms.
Genetic deletions and point mutations disrupt TSG functions, and
epigenetic mechanisms, including CpG island promoter methylation
and histone modifications, frequently lead to the loss of TSG
functions and are involved in tumor development and progression
(7). The aberrant methylation of
CpG islands leads to gene silencing, resulting in TSG inactivation,
which can increase the rate of tumor formation by disabling
multiple normal cellular processes, such as apoptosis and cell
cycle progression (7). In breast
cancer and renal clear cell carcinoma (8), methylation of erythrocyte membrane
protein band 4.1 like 3 (EPB41L3) disables its tumor suppressive
functions (9,10), suggesting that this gene is a
potential example of TSG inactivation in human cancers. However,
the role of EPB41L3 in ESCC remains unclear. EPB41L3, which belongs
to the protein 4.1 family, is a membrane skeletal protein 4 that is
commonly expressed in various human tissues. It is localized to
sites of cell-cell contact and functions as an adapter protein,
linking the plasma membrane to the cytoskeleton or associated
cytoplasmic signaling effectors. Subsequently, EPB41L3 facilitates
their activities in different pathways (11). Using a methylation microarray
analysis, our group has demonstrated that EPB41L3 is methylated in
ESCC tissue (12). Our subsequent
study revealed that EPB41L3 suppressed tumor cell invasion and
inhibited matrix metallopeptidase (MMP)2 and MMP9 expression in
ESCC cells (13). However, the
underlying mechanism by which EPB41L3 displays a tumor suppressive
role has not been fully elucidated. Thus, aiming to improve
clinical management and prolong the life expectancy of ESCC
patients, the present study investigated the tumor suppressive
functions of EPB41L3 in ESCC and its potential as a prognostic
indicator. To explore the molecular function of EPB41L3 in ESCC,
the expression of EPB41L3 in seven cell lines was measured. Then a
tissue microarray with 97 pairs of tumor and paired normal tissues
was used in order to assess its methylation status. The results
demonstrated that expression of EPB41L3 was decreased in both ESCC
cells and tissues and its methylation rate was increased compared
with normal cells and tissues. Multiple tumor suppressors have been
reported to function by inducing apoptosis, and loss of tumor
suppression results in reduced apoptotic activity and increased
tumor growth (14–17). Therefore the role of EPB41L3 in
ESCC apoptosis and growth was examined. The results suggested that
EPB41L3 may be a potential tumor suppressor and prognostic
indicator in ESCC.
Materials and methods
Tissue microarray and
immunohistochemistry
A ESCC tissue microarray was used to examine the
expression of EPB41L3. The tissue microarray was purchased from
Shanghai Outdo Biotech Co., Ltd. (Shanghai, China) and contained 97
pairs of ESCC samples and their para-carcinoma tissues. The
patients were selected based on a clear pathological diagnosis of
relatively early stage (Stages IA-IIIA) ESCC and follow-up data
dated back to July 2007. Their clinical features were listed in
Table I. None of the patients had
received chemotherapy or radiotherapy prior to surgery.
Immunohistochemistry on these tissues was performed following a
previously published protocol (9).
Scoring of EPB41L3 expression was based on the ratio of positive
cells (score 0, 0–5% positive cells; 1, 6–35%; 2, 36–70%; 3,
>70%) and staining intensity (score 0, no staining; 1, weak
staining; 2, moderate staining; 3, strong staining). The final
score was calculated as follows: positive cell score x staining
intensity score. The samples were marked as follows: '−' for a
score of 0–1, '+' for a score of 2–3, '++' for a score of 4–6, and
'+++' for a score of >6. Low expression was defined as a total
score of <4 and high expression as a total score ≥4. For further
analysis, '−' and '+' were considered as low expression, while '++'
and '+++' as high expression. These scores were determined
independently by two senior pathologists (Yu Wang and Jingyu Li;
Department of Pathology, Zhujiang Hospital) in a blinded manner
(18).
 | Table ICorrelation between
clinicopathological characteristics and EPB41L3 protein expression
in esophageal squamous cell carcinoma. |
Table I
Correlation between
clinicopathological characteristics and EPB41L3 protein expression
in esophageal squamous cell carcinoma.
| Characteristic | Low | High | Total | P-value |
|---|
| Sex | | | | |
| Male | 49 | 21 | 70 | 0.125 |
| Female | 23 | 4 | 27 | |
| Age (years) | | | | |
| <65 | 34 | 12 | 46 | 0.947 |
| >65 | 38 | 13 | 51 | |
| Pathology | | | | |
| G1-G2 | 55 | 22 | 77 | 0.216 |
| G3 | 17 | 3 | 20 | |
| Lymph node
metastasis | | | | |
| Negative | 39 | 11 | 50 | 0.381 |
| Positive | 33 | 14 | 47 | |
| Tumor infiltration
depth | | | | |
| T1-T2 | 12 | 10 | 22 | 0.016 |
| T3 | 60 | 15 | 75 | |
| TNM stage | | | | |
| C1-C2 | 38 | 17 | 55 | 0.186 |
| C3 | 34 | 8 | 42 | |
Cell lines
The non-neoplastic esophageal epithelial cell line
Het-1a and six ESCC cell lines (Eca-109, TE-1, CaEs17, KYSE-150,
KYSE-450 and KYSE-510) were obtained from the Research Center of
Clinical Medicine at Nanfang Hospital in Guangzhou, China. Het-1a
was cultured in RPMI-1640 medium (Life Technologies; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (Merck Millipore, Billerica, MA, USA) and 2 mM L-glutamine.
CaEs17 and Eca-109 were cultured in DMEM (Life Technologies; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(Merck Millipore), and the other four cell lines were cultured in
RPMI-1640 supplemented with 10% fetal bovine serum.
DNA and RNA extraction
Total DNA and RNA were extracted using the Omega
Tissue DNA kit and the Omega Total RNA kit I (both from Omega
Bio-Tek, Inc., Norcross, GA, USA) respectively, according to the
manufacturer's instructions.
Reverse transcription-polymerase chain
reaction (RT-PCR) and reverse transcription -quantitative PCR
(RT-qPCR)
Using the total RNA extracted from cells described
above, reverse transcription was performed using the PrimeScript RT
Reagent kit with gDNA Eraser (Takara Bio Inc., Otsu, Japan). qPCR
was performed using SYBR Premix Ex Taq reagent (Takara Bio Inc.).
The sequences of the reverse transcription-PCR primers were as
follows: EPB41L3 forward, 5′-GTAGTGGTCCATAA AGAGACAGAGA-3 and
reverse, 5′-GATACAAGTCAGTTGG GTTAGAAGA-3; GAPDH forward,
5′-TGCTGAGTATGTCGT GGAGTCT-3′ and reverse, 5′-CCCTGTTGCTGTAGCCATA
TTC-3′. The gel concentration was 1% and the results were
visualized under a UV lamp. qPCR was performed using a LightCycler
480 system (Roche, Basel, Switzerland). The qPCR reaction protocol
was 30 sec at 95°C followed by 40 cycles of 5 sec at 95°C and 20
sec at 60°C. Quantification was performed using the
2−ΔΔCq method (19).
Methylation-specific PCR (MSP)
Total DNA was extracted as aforementioned, and
bisulfite-modified using the BisulFlash DNA Modification kit
(EpiGentek, Farmingdale, NY, USA) according to the manufacturer's
protocol. The modified DNA was maintained at −20°C until PCR
amplification. MSP was performed with 0.25 µl Taq HS,
primers (500 nM), 50 ng total genomic DNA, and dNTP Mixture, and
10X PCR Buffer (both from Takara Biotechnology Co., Ltd., Dalian,
China) in a total reaction volume of 20 µl. The primer pairs
for EPB41L3 were as follows: to detect unmethylated sites, US,
5′-TTTGTGTATTGT TGTTGAGGAGTG-3′ and UAS, 5′-CACAATCCCCCACTCCA
AAAAACA-3′; and to detect methylated sites, MS, 5′-GCAGTG
CAAAGTGATACTTC-3′ and MAS, 5′-TCTGGTGGATAAA ATTTCACAT-3′. The
thermocycling conditions for the MSP were: 95°C for 10 min, then 38
cycles of 94°C for 30 sec, 58°C or 60°C or 55°C for 30 sec and 72°C
for 30 sec, followed by 72°C for 5 min, as previously described
(20). The results were visualized
using a BiQ Analyzer (Max Planck Institute for Informatics,
Saarbrücken, Germany).
Bisulfite treatment and pyrosequencing
analysis
Genomic DNA was extracted from seven cell lines
using the Tissue DNA kit (Omega Bio-Tek, Inc.). Extracted DNA was
bisulfite-modified using the BisulFlash DNA Modification kit
(EpiGentek). The average methylation rates of 33 CpG sites that are
located at −141551, −141494, −141439 and −141379 bp in the EPB41L3
promoter were detected. For EPB41L3, methylation rates of >20%,
10–20% and <10% were defined as hypermethylation, partial
methylation, and non-methylation respectively (8).
For the methylation analysis, the extracted seven
DNA samples were subjected to bisulfite conversion using the
Bisulfite Conversion kit (Qiagen GmbH, Hilden, Germany), according
to the manufacturer's instructions. Then, the purified
bisulfate-converted DNA was used to perform MSP with biotinylated
primers and Platinum Taq DNA polymerase (Kapa Biosystems, Inc.,
Wilmington, MA, USA). All the primers used for PCR were designed
using PyroMark Assay Design 2.0 (Qiagen GmbH), synthesized by BGI
(Shenzhen, China), and presented in Table II. The PCR products were purified
using a Qiaquick PCR purification kit (Qiagen GmbH), and
single-stranded DNA was prepared using Dynabeads M280 streptavidin
(Thermo Fisher Scientific, Inc.). The samples were analyzed using
PyroMark Q96 ID (Qiagen GmbH). Analysis of the results was
performed with the PyroMark Q96-CpG software (Qiagen GmbH).
 | Table IISequences of primers used for
pyrosequencing. |
Table II
Sequences of primers used for
pyrosequencing.
| Primer | Sequence | 5′ labeling |
|---|
| 122-01-1F |
AGTTTAGGGAGGAGGTTTGTAAG | |
| 122-01-1R |
ACACCTCCAAATTACCACCTACAA | Biotin |
| 122-01-1S |
GTTTGTAAGGAGATTTATATTTTG | |
| 122-01-2F |
GGAGGAGGTTTGTAAGGAGAT | |
| 122-01-2R |
CACCTCCAAATTACCACCTACA | Biotin |
| 122-01-2S |
TTTAGGGAGGGGTAGA | |
| 122-01-3F |
GAGGAGGTTTGTAAGGAGATTTAT | |
| 122-01-3R |
ACCTCCAAATTACCACCTAC | Biotin |
| 122-01-3S |
GGGTTAGAGAGGGTTGA | |
| 122-01-4F |
GGGAGGAGGTTTGTAAGGAGATTT | |
| 122-01-4R |
ATCCCTCCCCAAAAACTCTTTCCTT | Biotin |
| 122-01-4S |
GAAGAGAGTTGTAGGTGGTAAT | |
5-Aza-2-deoxygcytidine treatment
To determine whether demethylation could restore the
expression of EPB41L3 in ESCC cells, 2×105 cells were
seeded in each well of a 6-well plate and treated with 50
µmol/l 5-Aza-2′-deoxycytidine (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 3 days. The medium was changed with fresh
medium containing 5-Aza-2′-deoxycytidine every 24 h. Total RNA and
protein was then extracted, and EPB41L3 expression was measured by
RT-PCR and western blotting (6).
Cell counting kit-8 (CCK-8) cell
viability assay
Cell viability was assessed by a CCK-8 assay
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Vec-150,
epb-150, vec-510, epb-510 cells, si-510 and si-NC (5×103
cells/well) were plated separately into 96-well plates. After being
plated for 24, 48 or 72 h, the CCK-8 reagent was added to each
well, and cells were incubated for 2 h at 37°C. The absorbance was
measured at 450 nm by spectrophotometry.
Western blot analysis
Western blotting was performed according to the
standard protocol (21) with the
following antibodies: EPB41L3 (1:2,000; Abcam, Cambridge, MA, USA),
caspase-8 (1:1,000), caspase-9 (1:1,000), caspase-3 (1:1,000),
CyclinA (1:2,000), CyclinD (1:1,000), CyclinE (1:1,000) (all from
Cell Signaling Technology, Inc., Danvers, MA, USA), CyclinB1
(1:1,000), Cyclin-dependent kinase 1 (CDK1, also known as Cdc2;
1:1,000) (both from ProteinTech Wuhan Sanying Biotechnology, Wuhan,
China). GAPDH (1:8,000; ProteinTech Wuhan Sanying Biotechnology)
was used as loading control.
Flow cytometry
To measure dividing cells by flow cytometry,
KYSE-150 and KYSE-510 cells transfected with Vec-NC or Vec-epb were
fixed in 70% ethanol overnight, stained with propidium iodide the
next day, and DNA content was analyzed using an LSR FORTESSA (BD
Biosciences, Franklin Lakes, NJ, USA). To measure apoptosis, these
cells were further were stained and analyzed using the Annexin
V-allophycocyanin (APC)/7-am inoactinomycin D (7AAD) apoptosis kit
(KeyGen Biotech Co., Ltd., Nanjing, China), according to the
manufacturer's instructions. Briefly, 2×106 cells were
washed with PBS, stained with 7-AAD and Annexin V-APC for 15 min at
room temperature and analyzed using an LSR FORTESSA. The results
were analyzed using ModFit LT2.0 software (Verity Software House,
Topsham, ME, USA).
EPB41L3 plasmid construction and cell
transfection
The EPB41L3 expression vector pReceiver-M98-EPB41L3
was constructed by inserting the 417nt ORF sequence of EPB41L3
(MFIQIFPVIFLETSIAYSNVVWVYISYLHLLMKMFMR
DHFGCLMNYLPCFSTETFSLTPTVLAVGWLLERREVS
FSCSEWDKVASVVEQCLKYFFLSLFCSPFLSHLEHGIW TCQSSGNTLPLLVGTWIMWVSPEICV)
into the pReceiver-M98 vector (Genecopoeia Inc., Rockville, MD,
USA). One pair of specific oligonucleotides (EPB41L3 gene isoform
1) was annealed and then subcloned into the vector. Cell
transfection was performed with Lipofectamine 2000 and KYSE-150 and
KYSE-510 were selected with 500 µg/ml G418 sulfate (both
from Thermo Fisher Scientific, Inc.) for 2–3 weeks, according to
the manufacturer's protocol for stable transfection. The KYSE-150
and KYSE-510 cells transfected with the EPB41L3 overexpression
plasmid were termed vec-150-epb and vec-510-epb respectively. The
KYSE-150 and KYSE-510 cells transfected with the empty control
plasmid were termed vec-150-nc and vec-510-nc respectively.
Small interefering (si) RNA vector
construction and cell transfection
Three pairs of siRNA and a negative control siRNA
were purchased from GenePharma Co., Ltd. (Shanghai, China) and
subcloned into pcDNA6.2 GW/EmGFP vectors (Thermo Fisher Scientific,
Inc.). KYSE-510 cells were transfected with the vectors carrying
siRNA-EPB41L3 or the negative control siRNA. There were 3
siRNA-epb41l3 vectors were transfected separately. Silencing
efficiency was measured at 48 h post-transfection by western
blotting. The siRNA with the highest knockdown efficiency was
selected for subsequent experiments (sense, GCUGCGAAUAAACAGAUUUTT
and antisense, AAAUCUGUUUAUUCGCAGCTT). The negative control siRNA
(sense, CUGCGGAAAUAAUUUCAGATT and antisense,
AAUCAUUUUGACGUUAGCCTT). Cells with stable knockdown of EPB41L3
expression were established under selection with Blasticidin S HCl
(Thermo Fisher Scientific, Inc.). KYSE-519 cells transfected with
siRNA-EPB41L3 or the negative control siRNA were termed si-510-epb
and si-510-nc respectively.
Clonogenic assays
The effects of EPB41L3 expression were analyzed by
clonogenic assays using standard protocols. Eighteen h prior to
transfection, cells were seeded in 6-well plates at a density of
2×105 per well in complete RPMI-1640 or DMEM (Thermo
Fisher Scientific, Inc.). Cells were transfected with plasmid DNA
using the Lipofectamine 2000 transfection reagent. Twenty-four h
after transfection, cells were trypsinized and plated in 100 mm
dishes at identical densities for each transfection. Transfected
cells were selected in 500 µg/ml G418 for 10 to 14 days, and
surviving colonies were stained with crystal violet and counted
(22). Transgene expression was
confirmed for each experiment. Each experiment was conducted at
least three times.
In vivo experiments
To assess the role of EPB41L3 in inhibiting tumor
growth in vivo, suspensions of 1×106
EPB41L3-overexpressing KYSE-150 cells in 0.1 ml of PBS were
injected subcutaneously into the backs of 4–5-week-old male
immunodeficient SCID-Beige mice weighing 18–20 g (obtained from the
Laboratory Animal Center, Southern Medical University, Guangzhou,
China). As a control, suspensions of 1×106 KYSE-150
cells transfected with control vector were injected into the same
mice on a different site. To assess the role of EPB41L3 in
promoting tumor growth in vivo, suspensions of
1×106 EPB41L3-knockdown or control KYSE-510 cells in 0.1
ml PBS were injected subcutaneously into two sites of the backs of
4–5-week-old male immunodeficient SCID-Beige mice. The slightly
upper position of the waist was selected as the optimal injection
site. Four mice were used in each experimental group. Tumor growth
was assessed by measuring the xenografts in two dimensions once a
week. Tumor volumes were calculated according to the following
formula: (volume) = 1/2 × (long axis) × (short axis)2.
At 28 days post-injection, mice were sacrificed and the tumors were
carefully dissected. All animal experiments were performed in
accordance with the principles and procedures outlined in the
Southern Medical University Guide for the Care and Use of Animals
under the assurance number SCXK (Guangdong) 2008–0002. Approval was
obtained from the Nanfang Hospital Animal Ethics Committee.
Statistical analysis
Statistical analysis was performed using the SPSS
version 13.0 software (SPSS, Inc., Chicago, IL, USA). Each
experiment was performed in triplicate. The results were expressed
as the mean ± standard deviation. Significant differences were
determined using Student's t-test to compare two groups. The
χ2 test was used to analyze the correlations between
EPB41L3 expression and clinicopathological features in patients
with ESCC. Survival curves were evaluated using the Kaplan-Meier
method, and differences between survival curves were assessed by
the log-rank test. The Cox proportional hazards regression model
was used to examine univariate and multivariate hazard ratios for
the study variables that were dichotomized. Only significantly
different variables in univariate analysis were entered into the
subsequent multivariate analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
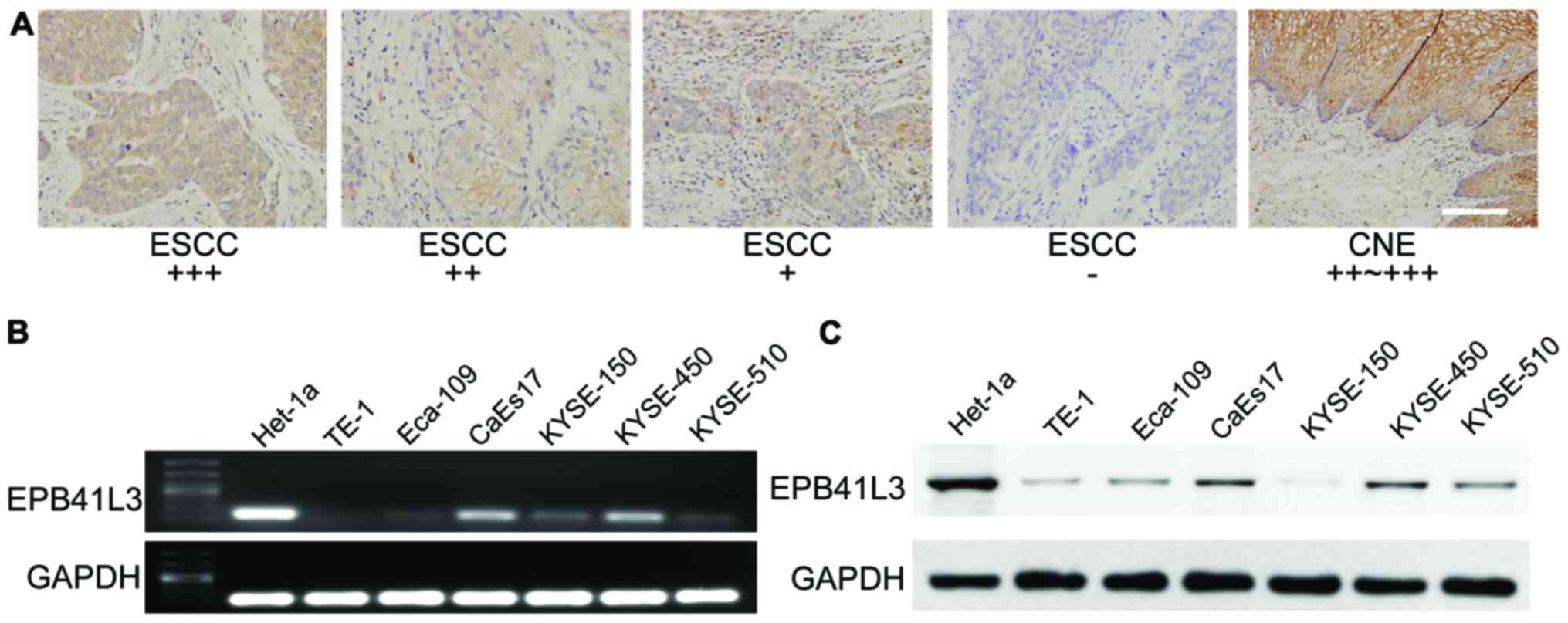
EPB41L3 expression is downregulated in
ESCC tissues
To explore the expression status of EPB41L3 in ESCC
tissues, immunohistochemistry analysis was performed using a tissue
microarray which contained 97 pairs of ESCC samples and their
adjacent non-tumor tissue samples. In general, the results
demonstrated that EPB41L3 expression was significantly lower in
tumor tissues compared with their matched adjacent non-tumor
tissues (Table III; Fig. 1A). High immune reactivity to
EPB41L3 was detected along cell membranes and cytoplasm in 91 out
of 97 non-tumor tissue samples, while only 25 out of 97 ESCC tissue
samples revealed high expression. Only 6 out of 97 adjacent
non-tumor tissue samples exhibited low EPB41L3 expression or no
expression, while 72 out of 97 ESCC tissue samples exhibited low
expression or no expression (Table
III; Fig. 1A).
 | Table IIIEPB41L3 protein expression in NE and
ESCC samples. |
Table III
EPB41L3 protein expression in NE and
ESCC samples.
| Group | Cases | EPB41L3 protein
expression
| P-value |
|---|
| Low | High |
|---|
| NE | 97 | 6 | 91 | <0.001 |
| ESCC | 97 | 72 | 25 | |
EPB41L3 expression is lower in ESCC cells
compared with Het-1a cells due to methylation
RT-PCR analysis was performed to dete2ct EPB41L3
mRNA expression in six ESCC cell lines and in one non-neoplastic
esophagus cell line, Het-1a. The results revealed that the
non-neoplastic Het-1a cell line had markedly higher expression of
EPB41L3 compared with all six esophagus cancer cell lines (Fig. 1B). Western blotting was then used
to detect EPB41L3 expression at the protein level. Similar to the
RT-PCR results, higher EPB41L3 protein expression was observed in
the normal esophagus cell line compared with the ESCC cell lines
(Fig. 1C).
In order to explore the reason why EPB41L3 was
down-regulated in ESCC cells, methylation patterns were examined
using MSP in the six esophagus cancer cell lines and the
non-neoplastic esophagus Het-1a cell line. All six esophagus cancer
cell lines developed both methylated and unmethylated bands,
suggesting that EPB41L3 had partial methylation in the cancer cell
lines. By contrast, Het-1a developed only an unmethylated band,
demonstrating that EPB41L3 was unmethylated in the normal
esophageal cells (Fig. 2A).
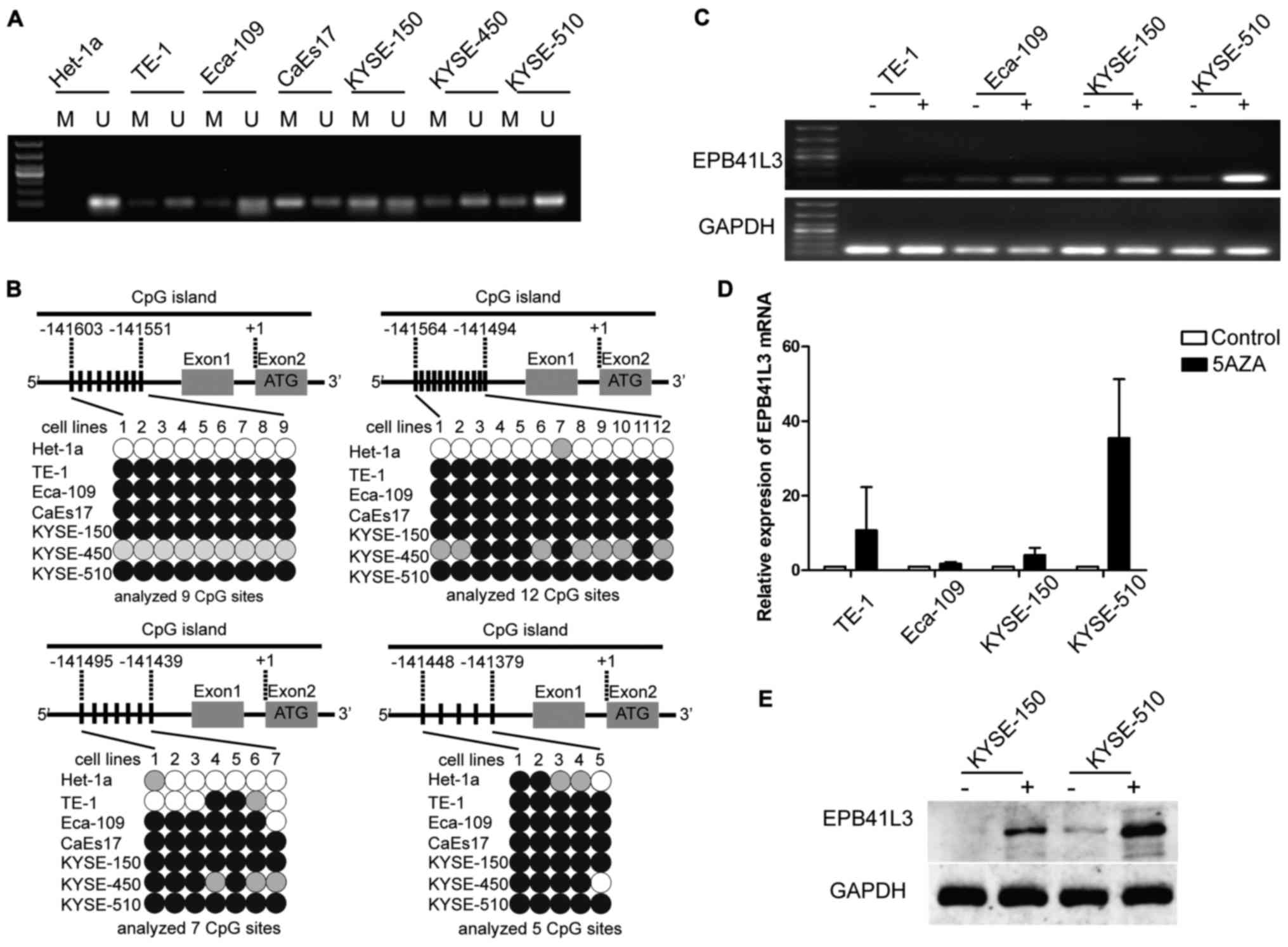 | Figure 2Reduced expression of EPB41L3 in ESCC
is due to methylation. (A) Methylation-specific polymerase chain
reaction analysis of the seven cell lines revealed that EPB41L3 was
partially methylated in all six ESCC cell lines while it was
unmethylated in the normal Het-1a cells. (B) Representation
schematic of the pyrosequencing results for the methylation status
of the CpG promoter in the seven cell lines tested. Gray boxes
indicate exons. Vertical bars indicate CpG sites examined for
methylation. Black, gray, and white circles represent
hypermethylation, partial methylation and nonmethylation,
respectively. The schematic illustrates the representative results
of pyrosequencing of a cytosine residue(s) at −141551, −141494,
−141439 and −141379 bp of the EPB41L3 promoter. (C) Treatment with
5-Aza-2-deoxygcitidine restored the expression of EPB41L3 in ESCC
lines, suggesting that downregulation of EPB41L3 in ESCC is due to
methylation. (D) Reverse transcription-quantitative polymerase
chain reaction analyses revealed that EPB41L3 expression was
increased in ESCC cell lines following 5-Aza-2-deoxygcitidine
treatment. The increase was most evident in the KYSE-510 cells. In
the other cell lines, although an increase was observed, this was
not as evident as in the KYSE-510 cells. (E) Western blot analyses
revealed that EPB41L3 expression was increased in KYSE-150 and
KYSE-510 cells following 5-Aza-2-deoxygcitidine treatment. EPB41L3,
erythrocyte membrane protein band 4.1 like 3; ESCC, esophageal
squamous cell carcinoma; M, methylated primer; U, unmethylated
primer; 5aza, 5-Aza-2-deoxygcitidine. |
Next, the different promoter methylation levels were
explored by pyrosequencing. A quantitative analysis of DNA
methylation at 33 CpG sites of the EPB41L3 gene promoter was
performed. The pyrosequencing results revealed high levels of
EPB41L3 methylation in five cell lines, KYSE-150, KYSE-510,
Eca-109, CaEs17 and TE-1, which were represented by dark circles in
Fig. 2B. For Het-1a, the
methylation rate was <20%, which was considered to be
unmethylated (represented by white circles in Fig. 2B). These results were in accordance
with the MSP results.
To identify whether the downregulated EPB41L3 mRNA
and protein expression levels in ESCC were due to methylation,
5-aza-2-deoxycytidine, a potent inhibitor of DNA methyltransferase,
was used to treat the TE-1, Eca-109, KYSE-150 and KYSE-510 cell
lines. If the downregulation of EPB41L3 was due to methylation,
treatment with 5-aza-2-deoxycytidine would be expected to increase
EPB41L3 expression. Indeed, treating the cells with the inhibitor
restored EPB41L3 mRNA expression, as identified by RT-PCR (Fig. 2C) and qPCR (Fig. 2D). Furthermore, western blotting
confirmed restoration of EPB41L3 protein expression in KYSE-150 and
KYSE-510 (Fig. 2E). These results
indicated that the decreased EPB41L3 expression in esophageal
cancer cell lines occurred via promoter methylation.
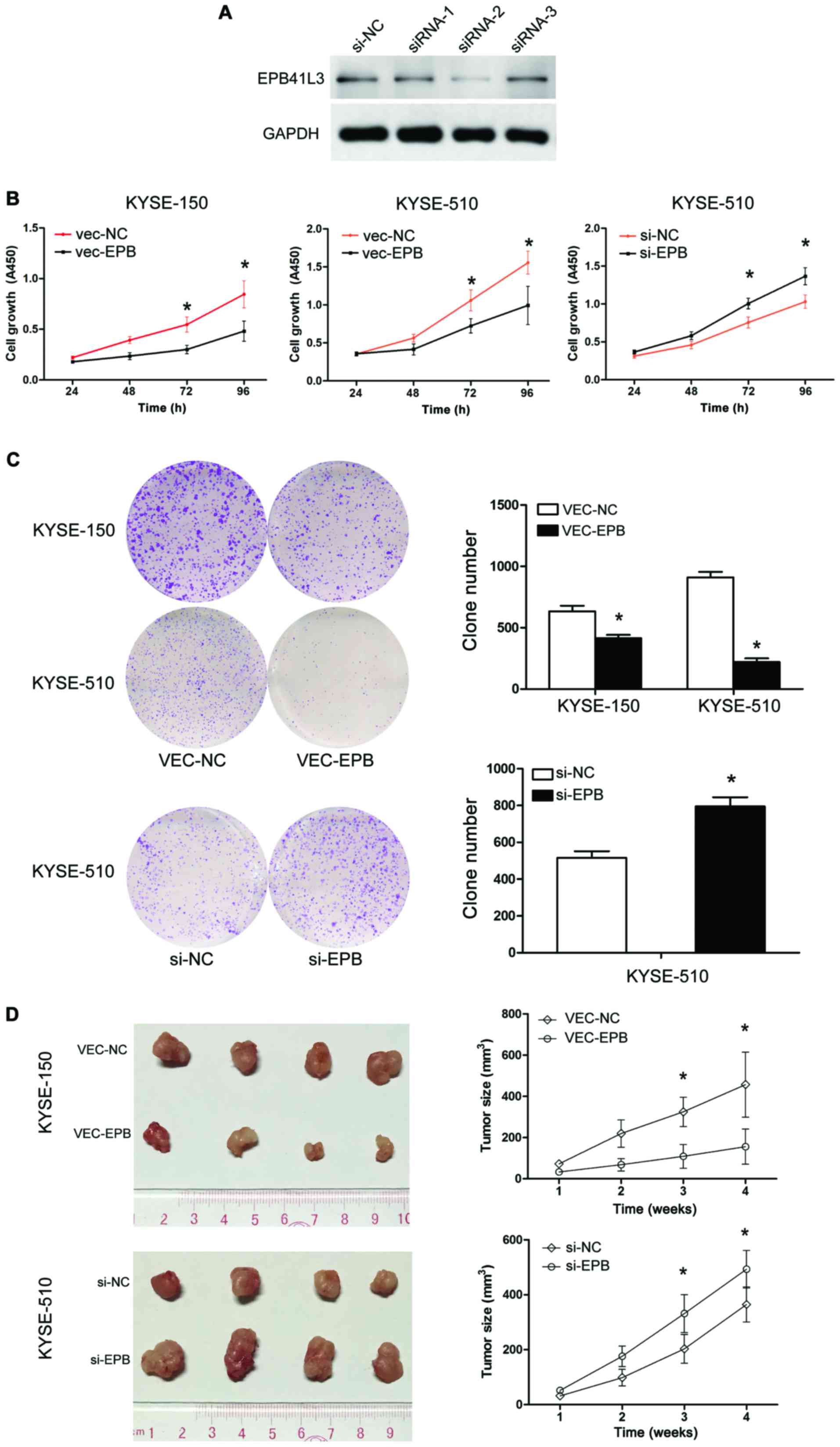
EPB41L3 inhibits ESCC cell proliferation
and reduces colony formation in vitro
In order to examine the functional role of EPB41L4
in ECC, its expression was silenced in ESCC cells by siRNA. First,
the knockdown efficiency of the siRNAs was confirmed by western
blotting. The results demonstrated that siRNA2 resulted in the
highest knockdown efficiency (Fig.
3A), and therefore siRNA2 as selected for subsequent
experiments.
To investigate the effects of overexpressing EPB41L3
on human ESCC cell growth in vitro, the KYSE-150 and
KYSE-510 cell lines were used to perform CCK-8 assays. EPB41L3
overexpression in KYSE-150 and KYSE-510 cells resulted in
significant inhibition of cell proliferation compared with control
cells (P<0.05; Fig. 3B). By
contrast, the transfection of EPB41L3 siRNA in KYSE-510 cells
resulted in increased cell proliferation compared with control
cells (P<0.05; Fig. 3B). The
ability of these two cell lines to form colonies in soft agar was
then examined for two weeks. Overexpression of EPB41L3
significantly reduced the cell colony numbers in KYSE-150 and
KYSE-510 cells, while EPB41L3 knockdown by siRNA in KYSE-510 cells
had the opposite effect (Fig. 3C).
Collectively, these results indicated that EPB41L3 inhibited the
growth of ESCC cells in vitro.
Epb41l3 inhibits tumor formation in
vivo
KYSE-510 cells transfected with EPB41L3
overexpression plasmid or control plasmid were termed vec-510-epb
and vec-510-nc, respectively. KYSE-510 cells transfected with
siRNA-EPB41L3 or control siRNA were termed si-510-epb or si-510-nc,
respectively. To further examine the role of EPB41L3 in tumor
growth in vivo, tumor xenografts were generated in mice by
injecting vec-510-epb, vec-510-nc, si-510-epb and si-510-nc cells
(four nude mice per group). Visible tumors began to form at 10 days
post-injection. The volume of the tumors was measured twice a week
and the mice were sacrificed at day 28 post-injection. At the end
of the experiment, the mice weighted 168–180 g. The maximum
diameter of a single tumor was 1.5 cm. None of the mice developed
multiple tumors. The group injected with vec-510-epb cells formed
significantly smaller tumors compared with the group injected with
the control vec-510-nc cells (Fig.
3D). The group injected with si-510-EPB cells formed
significantly larger tumors compared with the group injected with
the control si-510-NC cells (Fig.
3D). Overall, these results data demonstrated that increased
expression of Epb41L3 inhibited tumor growth, while decreased
expression of Epb41L3 promoted tumor growth in vivo.
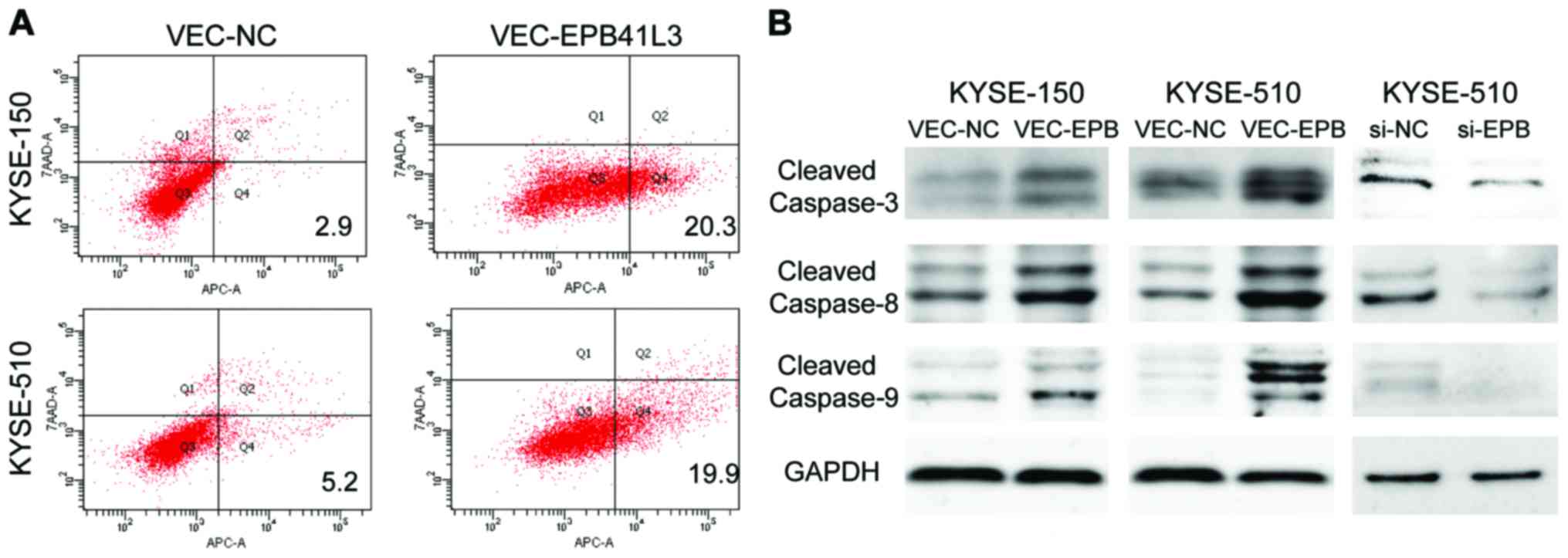
Ectopic expression of EPB41L3 promotes
apoptosis by activating the caspase-3/8/9 pathway
Apoptosis was detected using the 7-AAD/Annexin V-APC
double staining method followed by flow cytometry analysis.
Compared with the control group, KYSE-150 and KYSE-510 cells
overexpressing EPB41L3 had significantly higher apoptotic rates
compared with cells transfected with control plasmid (20.3 vs.
2.9%, and 19.9 vs. 5.2%, respectively) (Fig. 4A). To further investigate the
mechanisms underlying the higher apoptotic rates following EPB41L3
overexpression, western blot analysis was performed to determine
the expression levels of apoptosis-related proteins. The results
demonstrated that the expression levels of caspase-8, caspase-9 and
caspase-3 were elevated in KYSE-150 and KYSE-510 cells following
EPB41L3 over-expression compared with control cells (Fig. 4B). Then, the expression levels of
caspase-8, caspase-9 and caspase-3 were detected in KYSE-510 cells
following EPB41L3 knockdown by siRNA. The results demonstrated that
expression of caspase-8, caspase-9 and caspase-3 was markedly
decreased following EPB41L3 knockdown compared with control cells
(Fig. 4B).
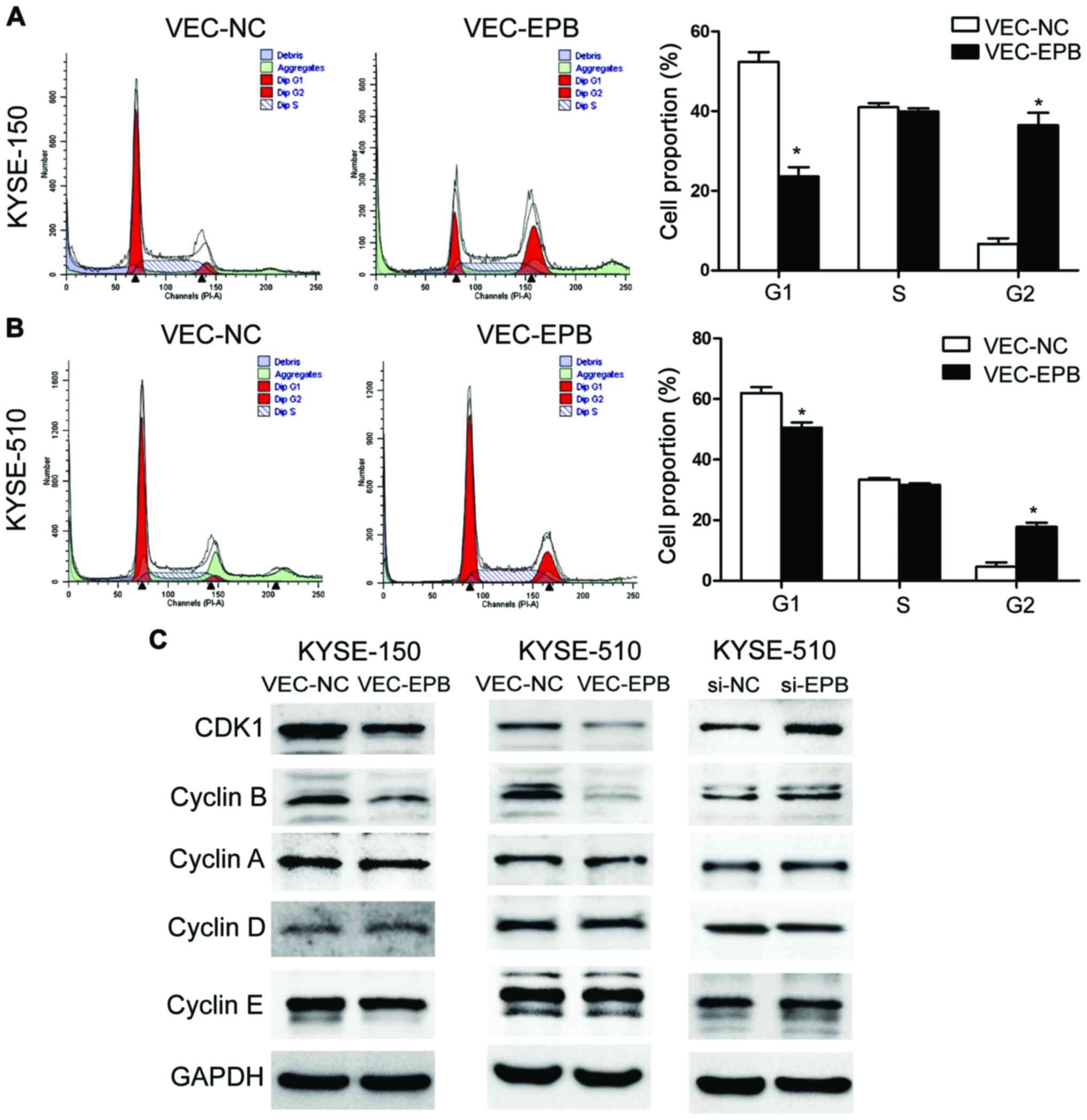
EPB41L3 causes G2/M cell cycle arrest by
activating the CDK1 pathway
Flow cytometry experiments revealed that the % of
cells in the G2 phase of the cell cycle was significantly increased
in the EPB41L3-overexpressing group compared with the control
group, in both the KYSE-150 (Fig.
5A) and KYSE-510 (Fig. 5B)
cell lines. The % of cells in G2 was 36.43 and 6.77% for the
EPB41L3-overexpressing and control KYSE150 cells respectively
(Fig. 5A), and 39.35 and 8.93% for
the EPB41L3-overexpressing and control KYSE-510 cells respectively
(Fig. 5B). Western blotting was
also performed in order to explore the mechanism of G2/M cell cycle
arrest. The results demonstrated that overexpression of EPB41L3
decreased the protein expression levels of CDK1 and CyclinB1 in
vec-150-epb and vec-510-epb cells, compared with their respective
controls (Fig. 5C). In addition,
the expression levels of these proteins were examined following
EPB41L3 knockdown in KYSE-510 cells. The expression levels of CDK1
and CyclinB1 were significantly increased in the knockdown cells
compared with control cells (Fig.
5C). Together with the flow cytometry results, these data
suggested that inhibition of CDK1/CyclinB1 might be the main
mechanism of the G2/M arrest induced by EPB41L3.
EPB41L3 is negatively correlated with
depth of tumor infiltration and may be a prognostic indicator
The relationship between clinicopathological
characteristics and EPB41L3 expression levels in patients with ESCC
is summarized in Table I. The
clinicopathological characteristics contained age, sex, pathology
classification, numbers of lymph nodes, tumor infiltration and
tumor-node-metastasis (TNM) stage (based on the 7th edition of
American Joint Committee on Cancer clinical staging (23). No significant association was
observed between EPB41L3 expression levels and the patients' age,
sex, pathologic type or lymph status in the 97 ESCC cases. However,
the expression levels of EPB41L3 were significantly negatively
correlated with depth of tumor infiltration (P=0.016; Table I). Next, univariate analysis was
used to explore the association between clinicopathological
characteristics and the overall survival. The results demonstrated
that tumor infiltration depth (P=0.01), TNM stage (P<0.001) and
EPB41L3 expression (P=0.004) were all positively correlated with
overall survival (Table IV).
These three factors were further investigated using multivariate
analysis, and the results revealed that only TNM stage (P=0.004)
and EPB41L3 expression (P=0.033) were statistically significant
(Table IV). To investigate the
prognostic value of EPB41L3 expression in ESCC, the association
between the expression levels of EPB41L3 and the patients' survival
was assessed using Kaplan-Meier analysis with the log-rank test.
The results demonstrated that patients with lower expression of
EPB41l3 had shorter survival time compared with the patients that
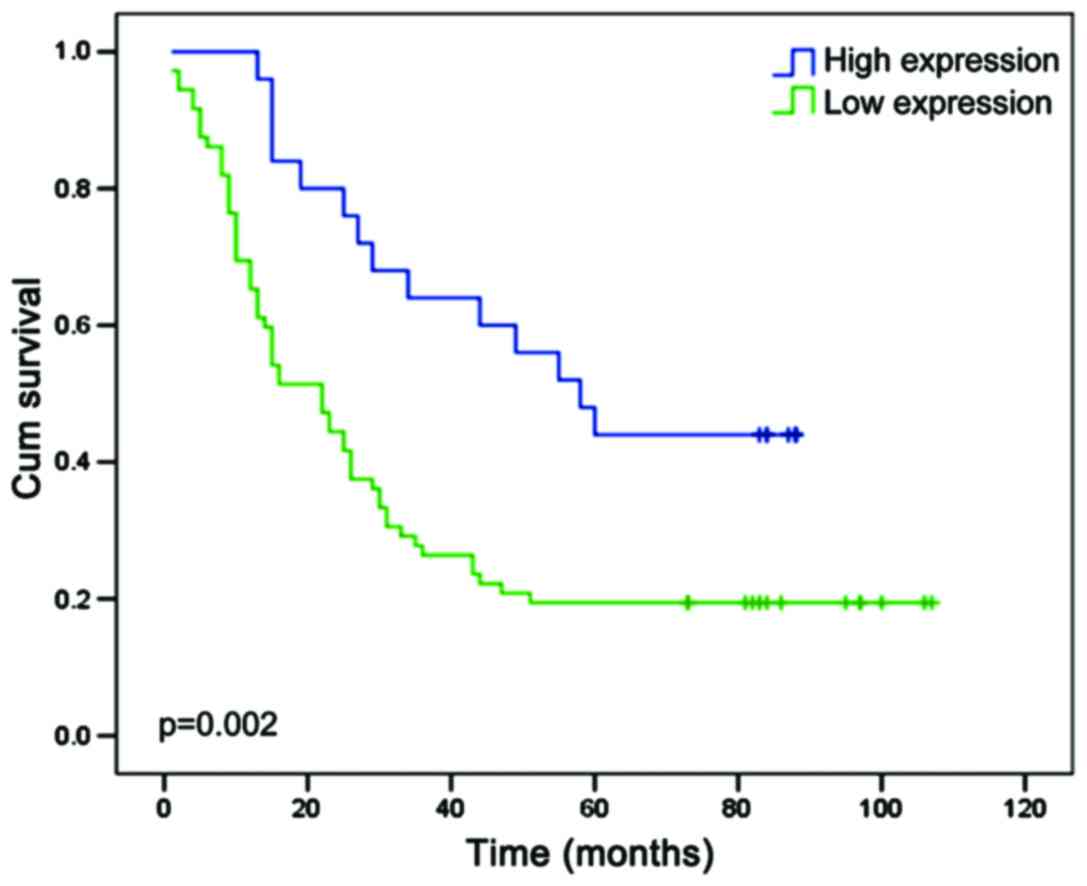
had higher EPB41l3 expression (P=0.002; Fig. 6). These findings revealed that
EPB41L3 may have the potential to serve as a prognosis predictor in
ESCC.
 | Table IVSummary of univariate and
multivariate Cox regression analyses of overall survival. |
Table IV
Summary of univariate and
multivariate Cox regression analyses of overall survival.
| Parameter | Univariate analysis
| Multivariate
analysis
|
|---|
| P-value | HR | 95% CI | P-value | HR | 95% CI |
|---|
| Sex | 0.26 | 1.368 | 0.793–2.36 | | | |
| Age (years) | 0.649 | 1.114 | 0.701–1.769 | | | |
| Pathology | 0.91 | 0.968 | 0.548–1.71 | | | |
| Lymph nodes
involved | 0.071 | 0.651 | 0.409–1.037 | | | |
| Tumor infiltration
depth | 0.01 | 0.425 | 0.222–0.812 | 0.301 | 0.689 | 0.34–1.395 |
| TNM stage | <0.001 | 0.398 | 0.247–0.643 | 0.004 | 0.478 | 0.289–0.79 |
| EPB41L3 expression
in cancer tissue | 0.004 | 2.392 | 1.329–4.304 | 0.033 | 1.942 | 1.054–3.577 |
| EPB41L3 expression
in para-cancerous tissue | 0.327 | 1.521 | 0.657–3.524 | | | |
Discussion
Esophageal carcinoma is a common malignancy with
poor prognosis. The main reason for low survival is that the
majority of ESCC are asymptomatic and undetected until they have
spread beyond the esophageal wall and thus become unresectable
(24). EPB41L3, also known as 4.1B
or DAL1, is located on chromosome 18p11.32, and it is expressed in
multiple adult tissues, including the brain, lung, kidney,
intestine and prostate. It has been demonstrated to serve as an
antitumor factor in some cancers, primarily in non-small-cell lung
carcinoma (NSCLC). Kikuchi et al (30) noted that EPB41L3 methylation was
involved in the development and progression of NSCLC and served as
a poor prognosis predictor. Nevertheless, previous studies have not
focused on the mechanisms underlying the tumor suppressive function
and the clinical significance of EPB41L3 in ESCC. Our group has
been studying the role of EPB41L3 in ESCC with the aim to explore
its methylation status and to identify potential plasma biomarkers
for early diagnosis. By using an Infinium Methylation 450k array,
the methylation frequency of EPB41L3 was demonstrated to be higher
in tumor tissues compared with normal surrounding tissues, and this
was correlated with large tumor size and advanced pT tumor stage
(12). EPB41L3 has been
demonstrated to be partly silenced due to hypermethylation
(13). Other genetic and
epigenetic mechanisms may result in the regulation of EPB41L3
expression, including histone modifications and microRNAs, and
these will need further investigation in the future.
There has been evidence suggesting that
hypermethylation has a critical role in the inactivation of
EPB41L3. Demethylation treatments were able to restore EPB41L3
expression in several types of cancer (9,25–27).
Aberrant DNA methylation is one of the best-characterized
epigenetic modifications, contributing to tumor initiation and
progression (28,29). This is because DNA methylation,
which usually occurs on the cytosine residues of CpG dinucleotides,
is key to tissue differentiation during early embryonic growth
(30). The present data provided
evidence that EPB41L3 expression was downregulated in ESCC cells
due to promoter methylation. First, EPB41L3 expression was
demonstrated to be lower in ESCC tissues and cell lines compared
with adjacent normal tissues and a non-neoplastic cell line.
Second, MSP and pyrosequencing confirmed higher promoter
methylation rate in ESCC cell lines compared with the normal
esophageal cell line. Notably, although methylation was evident in
all six cell lines, they all exhibited partial methylation rather
than full methylation of the promoter. The methylation rates were
different in different ESCC cell lines, demonstrating heterogeneity
in these cancer cells. Pyrosequencing has been recently regarded as
a better method for testing methylation in large-scale validation
studies, biomarker development and clinical diagnostics (31), while MSP has some disadvantages,
including its inability of detecting mosaic DNA methylation
(32,33).
DNA methylation provides an alternative way for
tumor suppressor gene function loss (12). Having detected EPB41L3 methylation
in ESCC cells, the present study aimed to explore the mechanisms
underlying the role of EPB41L3 as a tumor suppressor. Thus,
experiments related to proliferation, apoptosis and cell cycle
phase distribution were performed. CCK-8 cell viability and
clonogenic assay results indicated that EPB41L3 overexpression
inhibited tumor cell proliferation in vitro. Tumor xenograft
experiments revealed that EPB41L3 over-expression suppressed tumor
growth in vivo. Flow cytometry assay demonstrated higher
rates of apoptosis in EPB41L3-overexpressing cells compared with
control cells. These results offered a foundation for mechanistic
exploration. Previous study has revealed that apoptosis in MCF-7
cells was primarily associated with the activation of Caspase-8,
and inhibition of this activation blocked the ability of EPB41L3 to
induce cell death in MCF-7 (10).
Cleaved-caspase-8 and cleaved-caspase-9 activate pro-caspase 3 into
cleaved-caspase-3, resulting in apoptosis. Thus, the expression of
the caspase family was detected by western blotting in order to
explore the mechanism of apoptosis induction in ESCC. The results
demonstrated that expression of caspase-8, caspase-9 and caspase-3
were elevated in the EPB41L3-overexpressing cells. By contrast,
EPB41L3 knockdown by siRNA resulted in the opposite effects.
Kuns et al (31) reported that, during pregnancy, the
overexpression of EPB41L3 results in reduced CyclinA expression and
Rb phosphorylation, which is accompanied by decreased tyrosine
kinase receptor ErbB2 phosphorylation, causing the mammary
epithelial cells to be arrested in the G1 phase (34). The present data revealed no
significant change in CyclinA expression, while expression of
CyclinB1 and CDK1 decreased following EPB41L3 overexpression. High
expression of CyclinB1-CDK1 only occurs in the M phase, not the G2
phase (35). CDK1 controls entry
into and exit from the M phase of the cell cycle and is commonly
accompanied by CyclinA and CyclinB1 (36). This is in accordance with the
present cell cycle data. Cell death can occur during mitochondrial
membrane permeabilization with the release of cell death effectors
such as apoptosis-inducing factor (37). The G2/M phase arrest may limit the
proliferation of chromosomally unstable, pre-cancerous cells.
Downregulation of the G2/M checkpoint may lead to tumorigenesis
(38). The present findings
indicate that ectopic expression of EPB41L3 may inhibit cell
proliferation by upregulating apoptosis and causing G2/M phase
arrest.
To date, several genes have been reported to be
hypermethylated in ESCC, including cyclin-dependent kinase
inhibitor 2A (CDKN2A)/p16INK4a (39–41).
These methylated genes have the potential to be used as diagnostic
or prognostic molecular markers for ESCC. Recent study has focused
on the correlation of epigenetic changes and ESCC patient survival.
Zare et al (42) and Lee
et al (43) separately
reported that methylation of adenomatous polyposis coli (APC) or
fragile histidine triad protein (FHIT) was closely correlated to
poor outcome in patients with ESCC. The current study examined
EPB41L3 expression in ESCC and its correlation with
clinicopathological features and prognosis. A statistically
significant difference was observed between EPB41L3 expression in
ESCC tumor tissues and their matched adjacent non-tumor tissues.
This was consistent in both RT-PCR and western blotting results.
Survival analysis revealed that patients with higher expression of
EPB41L3 had improved prognosis. The relationship between EPB41L3
expression levels and certain clinical features were then analyzed.
Univariate analysis demonstrated that EPB41L3 expression was
significantly correlated with tumor infiltration depth and TNM
stage. These results indicate that EPB41L3 has a significant
correlation with tumor progression. Through a tissue microarray,
the current study revealed the clinical importance of EPB41L3 in
ESCC patients, which may have the potential to be prognostic
indicator. To date, only 20 types of tumor markers have been put
into clinical use. No specific tumor markers exist to date for ESCC
(44). The main obstacle to
developing clinically useful indicators is the lack of large
clinical trials in ESCC, which are hampered by a lack of sizeable
esophageal tissue repositories that include complete clinical
annotation (45). Furthermore,
because chemotherapy and radiation remain important in ESCC
treatment, predictive indicators may be critical for identifying
patients who may benefit from molecular targeted agents or are more
sensitive to radiotherapy (46).
Further studies are warranted to probe the clinical significance of
EPB41L3 in ESCC.
In summary, low levels of EPB41L3 may be an
unfavorable indicator of ESCC prognosis. Overexpressing EPB41L3
inhibited proliferation, and induced apoptosis and G2/M arrest in
ESCC cells through the Caspase-3/8/9 and CDK1/CyclinB1 pathways.
Detection of the tumor EPB41L3 methylation status may aid in
predicting prognosis in patients with ESCC.
Acknowledgments
Not applicable.
Notes
[1]
Funding
This study was supported by the Natural Science
Foundation of Guangdong Province, China (grant no.
2015A030313273).
[2] Availability
of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
[3] Author's
contributions
RZ performed the cell culture, MSP, western
blotting, transfection, CCK-8, clonogenic assays and was involved
in the writing of the article. YL performed qPCR analysis. ZJJ
performed the flow cytometry experiment. JPH performed the
statistical analysis; YW performed the IHC analysis; XFL, WBX and
XCW were involved in the animal experiments; JRZ and QEW performed
pyrosequencing; YFZ contributed to the whole design of this
research project. JRZ, QEW and YFZ all contributed to the writing
of the manuscript. All authors have read and approved the final
manuscript.
[4] Ethics
approval and consent to participate
All animal experiments were performed in accordance
with the principles and procedures outlined by the Southern Medical
University Guide for the Care and Use of Animals under the
assurance number SCXK (Guangdong) 2008–0002. Approval was obtained
from the Nanfang Hospital Animal Ethics Committee.
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhu YH, Fu L, Chen L, Qin YR, Liu H, Xie
F, Zeng T, Dong SS, Li J, Li Y, et al: Downregulation of the novel
tumor suppressor DIRAS1 predicts poor prognosis in esophageal
squamous cell carcinoma. Cancer Res. 73:2298–2309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Murray T, Ward E, Samuels A,
Tiwari RC, Ghafoor A, Feuer EJ and Thun MJ: Cancer statistics,
2005. CA Cancer J Clin. 55:10–30. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Offner FA: Etiology, molecular biology and
pathology of squamous cell carcinoma of the esophagus. Pathologe.
21:349–357. 2000.In German. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kong KL, Kwong DL, Fu L, Chan TH, Chen L,
Liu H, Li Y, Zhu YH, Bi J, Qin YR, et al: Characterization of a
candidate tumor suppressor gene uroplakin 1A in esophageal squamous
cell carcinoma. Cancer Res. 70:8832–8841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Li J, Cui Y, Li T, Ng KM, Geng H,
Li H, Shu XS, Li H, Liu W, et al: CMTM3, located at the critical
tumor suppressor locus 16q22.1, is silenced by CpG methylation in
carcinomas and inhibits tumor cell growth through inducing
apoptosis. Cancer Res. 69:5194–5201. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takahashi Y, Iwai M, Kawai T, Arakawa A,
Ito T, Sakurai-Yageta M, Ito A, Goto A, Saito M, Kasumi F, et al:
Aberrant expression of tumor suppressors CADM1 and 4.1B in invasive
lesions of primary breast cancer. Breast Cancer. 19:242–252. 2012.
View Article : Google Scholar
|
|
9
|
Yamada D, Kikuchi S, Williams YN,
Sakurai-Yageta M, Masuda M, Maruyama T, Tomita K, Gutmann DH,
Kakizoe T, Kitamura T, et al: Promoter hypermethylation of the
potential tumor suppressor DAL-1/4.1B gene in renal clear cell
carcinoma. Int J Cancer. 118:916–923. 2006. View Article : Google Scholar
|
|
10
|
Jiang W and Newsham IF: The tumor
suppressor DAL-1/4.1B and protein methylation cooperate in inducing
apoptosis in MCF-7 breast cancer cells. Mol Cancer. 5:42006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z, Zhang J, Ye M, Zhu M, Zhang B, Roy
M, Liu J and An X: Tumor suppressor role of protein 4.1B/DAL-1.
Cell Mol Life Sci. 71:4815–4830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Zhou F, Jiang C, Wang Y, Lu Y, Yang
F, Wang N, Yang H, Zheng Y and Zhang J: Identification of a DNA
methylome profile of esophageal squamous cell carcinoma and
potential plasma epigenetic biomarkers for early diagnosis. PLoS
One. 9:e1031622014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zeng R, Huang JP, Li XF, Xiong WB, Wu G,
Jiang ZJ, Song SJ, Li JQ, Zheng YF and Zhang JR: Epb41l3 suppresses
esophageal squamous cell carcinoma invasion and inhibits MMP2 and
MMP9 expression. Cell Biochem Funct. 34:133–141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saeki T, Mhashilkar A, Swanson X, Zou-Yang
XH, Sieger K, Kawabe S, Branch CD, Zumstein L, Meyn RE, Roth JA, et
al: Inhibition of human lung cancer growth following
adenovirus-mediated mda-7 gene expression in vivo. Oncogene.
21:4558–4566. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ji L, Fang B, Yen N, Fong K, Minna JD and
Roth JA: Induction of apoptosis and inhibition of tumorigenicity
and tumor growth by adenovirus vector-mediated fragile histidine
triad (FHIT) gene overexpression. Cancer Res. 59:3333–3339.
1999.PubMed/NCBI
|
|
16
|
Roz L, Gramegna M, Ishii H, Croce CM and
Sozzi G: Restoration of fragile histidine triad (FHIT) expression
induces apoptosis and suppresses tumorigenicity in lung and
cervical cancer cell lines. Proc Natl Acad Sci USA. 99:361–3620.
2002. View Article : Google Scholar
|
|
17
|
Mao X, Seidlitz E, Truant R, Hitt M and
Ghosh HP: Re-expression of TSLC1 in a non-small-cell lung cancer
cell line induces apoptosis and inhibits tumor growth. Oncogene.
23:5632–5642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang JY, Jiang SH, Liu DJ, Yang XM, Huo
YM, Li J, Hua R, Zhang ZG and Sun YW: Decreased LKB1 predicts poor
prognosis in pancreatic ductal adenocarcinoma. Sci Rep.
5:105752015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Nagata M, Sakurai-Yageta M, Yamada D, Goto
A, Ito A, Fukuhara H, Kume H, Morikawa T, Fukayama M, Homma Y, et
al: Aberrations of a cell adhesion molecule CADM4 in renal clear
cell carcinoma. Int J Cancer. 130:1329–1337. 2012. View Article : Google Scholar
|
|
21
|
Liu Y, Zhang R, Yan K, Chen F, Huang W, Lv
B, Sun C, Xu L, Li F and Jiang X: Mesenchymal stem cells inhibit
lipopolysaccharide-induced inflammatory responses of BV2 microglial
cells through TSG-6. J Neuroinflammation. 11:1352014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gerber MA, Bahr SM and Gutmann DH: Protein
4.1B/differentially expressed in adenocarcinoma of the lung-1
functions as a growth suppressor in meningioma cells by activating
Rac1-dependent c-Jun-NH(2)-kinase signaling. Cancer Res.
66:5295–5303. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rice TW, Rusch VW, Ishwaran H and
Blackstone EH: Cancer of the esophagus and esophagogastric
junction: Data-driven staging for the seventh edition of the
American Joint Committee on Cancer/International Union Against
Cancer Cancer Staging Manuals. Cancer. 116:3763–3773. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Al-Kaabi A, van Bockel LW, Pothen AJ and
Willems SM: p16INK4A and p14A RF gene promoter hypermethylation as
prognostic biomarker in oral and oropharyngeal squamous cell
carcinoma: A review. Dis Markers. 2014:2605492014. View Article : Google Scholar
|
|
25
|
Baylin SB and Chen WY: Aberrant gene
silencing in tumor progression: Implications for control of cancer.
Cold Spring Harb Symp Quant Biol. 70:427–433. 2005. View Article : Google Scholar
|
|
26
|
Wong SY, Haack H, Kissil JL, Barry M,
Bronson RT, Shen SS, Whittaker CA, Crowley D and Hynes RO: Protein
4.1B suppresses prostate cancer progression and metastasis. Proc
Natl Acad Sci USA. 104:12784–12789. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Xu R, Li G, Xie X, Long J and
Wang H: Loss of expression of the differentially expressed in
adenocarcinoma of the lung (DAL-1) protein is associated with
metastasis of non-small cell lung carcinoma cells. Tumour Biol.
33:1915–1925. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma X, Wang YW, Zhang MQ and Gazdar AF: DNA
methylation data analysis and its application to cancer research.
Epigenomics. 5:301–316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kikuchi S, Yamada D, Fukami T, Masuda M,
Sakurai-Yageta M, Williams YN, Maruyama T, Asamura H, Matsuno Y,
Onizuka M, et al: Promoter methylation of DAL-1/4.1B predicts poor
prognosis in non-small cell lung cancer. Clin Cancer Res.
11:2954–2961. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bock C, Halbritter F, Carmona FJ, Tierling
S, Datlinger P, Assenov Y, Berdasco M, Bergmann AK, Booher K,
Busato F, et al BLUEPRINT consortium: Quantitative comparison of
DNA methylation assays for biomarker development and clinical
applications. Nat Biotechnol. 34:726–737. 2016. View Article : Google Scholar
|
|
32
|
Reed K, Poulin ML, Yan L and Parissenti
AM: Comparison of bisulfite sequencing PCR with pyrosequencing for
measuring differences in DNA methylation. Anal Biochem. 397:96–106.
2010. View Article : Google Scholar
|
|
33
|
Hömig-Hölzel C and Savola S: Multiplex
ligation-dependent probe amplification (MLPA) in tumor diagnostics
and prognostics. Diagn Mol Pathol. 21:189–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuns R, Kissil JL, Newsham IF, Jacks T,
Gutmann DH and Sherman LS: Protein 4.1B expression is induced in
mammary epithelial cells during pregnancy and regulates their
proliferation. Oncogene. 24:6502–6515. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Deshpande A, Sicinski P and Hinds PW:
Cyclins and cdks in development and cancer: A perspective.
Oncogene. 24:2909–2915. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiang Q, Zhen Z, Deng DY, Wang J, Chen Y,
Li J, Zhang Y, Wang F, Chen N, Chen H, et al: Tivantinib induces
G2/M arrest and apoptosis by disrupting tubulin polymerization in
hepatocellular carcinoma. J Exp Clin Cancer Res. 34:1182015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Castedo M, Perfettini JL, Roumier T,
Andreau K, Medema R and Kroemer G: Cell death by mitotic
catastrophe: A molecular definition. Oncogene. 23:2825–2837. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Löbrich M and Jeggo PA: The impact of a
negligent G2/M checkpoint on genomic instability and cancer
induction. Nat Rev Cancer. 7:861–869. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo M, Ren J, House MG, Qi Y, Brock MV and
Herman JG: Accumulation of promoter methylation suggests epigenetic
progression in squamous cell carcinoma of the esophagus. Clin
Cancer Res. 12:4515–4522. 2006. View Article : Google Scholar
|
|
40
|
Salam I, Hussain S, Mir MM, Dar NA,
Abdullah S, Siddiqi MA, Lone RA, Zargar SA, Sharma S, Hedau S, et
al: Aberrant promoter methylation and reduced expression of p16
gene in esophageal squamous cell carcinoma from Kashmir valley: A
high-risk area. Mol Cell Biochem. 332:51–58. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Taghavi N, Biramijamal F, Sotoudeh M,
Khademi H, Malekzadeh R, Moaven O, Memar B, A'rabi A and
Abbaszadegan MR: p16INK4a hypermethylation and p53, p16 and MDM2
protein expression in esophageal squamous cell carcinoma. BMC
Cancer. 10:1382010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zare M, Jazii FR, Alivand MR, Nasseri NK,
Malekzadeh R and Yazdanbod M: Qualitative analysis of Adenomatous
Polyposis Coli promoter: Hypermethylation, engagement and effects
on survival of patients with esophageal cancer in a high risk
region of the world, a potential molecular marker. BMC Cancer.
9:242009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lee EJ, Lee BB, Kim JW, Shim YM, Hoseok I,
Han J, Cho EY, Park J and Kim DH: Aberrant methylation of Fragile
Histidine Triad gene is associated with poor prognosis in early
stage esophageal squamous cell carcinoma. Eur J Cancer. 42:972–980.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guo H, Zhou X, Lu Y, Xie L, Chen Q, Keller
ET, Liu Q, Zhou Q and Zhang J: Translational progress on tumor
biomarkers. Thorac Cancer. 6:665–671. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kaz AM and Grady WM: Epigenetic biomarkers
in esophageal cancer. Cancer Lett. 342:193–199. 2014. View Article : Google Scholar
|
|
46
|
Fareed KR, Kaye P, Soomro IN, Ilyas M,
Martin S, Parsons SL and Madhusudan S: Biomarkers of response to
therapy in oesophago-gastric cancer. Gut. 58:127–143. 2009.
View Article : Google Scholar
|