Introduction
Pancreatic cancer is one of the most devastating
malignancies of the gastrointestinal tract as well as one of the
most common causes of cancer-associated mortality worldwide
(1). Surgery is the optimal method
of treating patients with pancreatic cancer; however, the
application of surgical therapy in patients with pancreatic cancer
is limited, as the majority of patients are at an advanced stage of
the disease at diagnosis. Indeed, ≤20% of patients with pancreatic
cancer are suitable to undergo such surgery (2). For the majority of patients with
pancreatic cancer, the primary method of treatment is chemotherapy
and gemcitabine is the most common chemotherapeutic drug used to
treat patients with inoperable pancreatic cancer. Although progress
has been made in the treatment of pancreatic cancer over the past
few decades, the median survival time of such patients remains
<1 year (3,4). The failure of chemotherapy is
attributed to the complete resistance against chemotherapeutic
drugs and chemotherapy resistance presents a major clinical
challenge in the treatment of cancer. Therefore, it is important to
understand the molecular mechanisms underlying the development of
chemoresistance in pancreatic cancer in order to develop effective
therapeutic targets.
The resistance of cancer cells to a wide spectrum of
chemotherapeutic drugs may be associated with the intrinsic
resistance of cancer cells and/or acquired resistance following
several cycles of chemotherapy (5). The most common mechanism by which
chemoresistance arises is the development of efflux pumps on the
surface of tumor cells that eject anticancer drugs from inside
tumor cells to the outside environment (6). Other anti-drug mechanisms include the
upregulation of DNA repair enzymes, insensitivity to drug-induced
apoptosis, stromal proliferation, induction of drug-detoxifying
mechanism and reduced angiogenesis, all of which serve a crucial
role in the development of drug resistance of cancer cells
(7–11). Therefore, it is crucial to develop
comprehensive understanding of the specific mechanism by which
chemoresistance arises in pancreatic cancer cells in order to
identify a novel therapeutic strategy to overcome chemoresistance
and improve the clinical therapeutic response of patients with
pancreatic cancer.
Micro (mi)RNAs post-transcriptionally regulate a
variety of target genes by binding to the 3′-untranslated region
(3′-UTR) of their target genes (12). The aberrant expression of miRNAs
serves either oncogenic or tumor-suppressive roles in the
tumorigenesis, progression and metastasis of various types of
cancer (13–19). Previous studies have demonstrated
that miRNAs are important mediators of chemoresistance in
pancreatic cancer. Chaudhary et al (20) reported that miR-205-5p was
differentially downregulated in pancreatic cancer cells and
tissues. It was also demonstrated that the ectopic expression of
miR-205-3p in combination with gemcitabine significantly reduced
the proliferation and tumor growth of pancreatic cancer cells in
mouse models (20). Furthermore
the oncogenic miRNA miR-181c is dramatically elevated in pancreatic
cancer tissues and this high expression of miR-181c induces
chemoresistance in pancreatic cancer by inactivating the Hippo
signaling pathway (21). These
results indicate that the dysregulation of miRNAs serves an
important role in the development of chemoresistance in pancreatic
cancer.
The present study measured miR-374b-5p expression by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and evaluated whether there was an association between
miR-374b-5p levels, clinicopathological characteristics and overall
and progression-free survival of patients with pancreatic cancer.
The effects of miR-374b-5p expression on chemoresistance were
examined by performing gain or loss of function assays in
vitro and in vivo and the potential targets of
miR-374b-5p were identified using bioinformatics analysis, RT-qPCR,
western blotting, luciferase reporter and RNA immunoprecipitation
assays. The results revealed that the decreased expression of
miR-374b-5p promoted the resistance of pancreatic cancer cells to
the chemotherapeutic reagent gemcitabine by enhancing the
expression of several anti-apoptotic proteins, including B-cell
lymphoma 2 (BCL2), Baculoviral IAP Repeat Containing 3 (BIRC3) and
X-linked inhibitor of apoptosis (XIAP). These results suggest that
the delivery of miR-374b-5p may be developed as a novel therapeutic
strategy to treat patients with chemotherapy resistant pancreatic
cancer.
Materials and methods
Cell lines and cell culture
The human pancreatic cancer cell lines BxPC-3,
PANC-1, AsPC-1, SW1990, Capan-1, Capan-2, CFPAC-1 and MIA PaCa-2
were all obtained from the American Type Culture Collection (ATCC;
Manassas, VA, USA). All human pancreatic cancer cell lines were
maintained in RPMI-1640 (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(HyClone; GE Healthcare, Logan, UT, USA) and 100 U/ml penicillin
and 100 µg/ml streptomycin. Cells were maintained under a
humidified atmosphere of 5% CO2 at 37°C.
Patients and tumor tissues
A total of 87 pancreatic cancer tissues and 8
adjacent normal tissues were obtained from 87 patients (51 males
and 36 females) with a median age of 65 years old, undergoing
surgery at the First Hospital of Jilin University (Changchun,
China) between January 2007 and December 2015. Adjacent normal
tissues were taken from 8 of these patients (5 males and 3
females), with median age of 60. Information regarding the response
of patients to chemotherapy was obtained for only 63 patients with
pancreatic cancer. Patients were diagnosed with pancreatic cancer
based on clinicopathological evidence. Specimens were immediately
snap-frozen and stored in liquid nitrogen tanks. Written informed
consent from each patient enrolled in the study was obtained and
ethical approval for the use of human tissues was obtained from the
Institutional Research Ethics Committee at the First Hospital of
Jilin University (ethics no. 2005-001). Clinicopathological
information regarding the patients is presented in Tables I and II. Tumors were graded and classified
according to the AJCC Cancer Staging Manual (22). The present study examined the
expression of miR-374b-5p in the 87 pancreatic tissues and 8
adjacent normal tissues. Levels of miR-374b-5p expression in all
pancreatic cancer tissues were normalized to that in the pancreatic
cancer tissue with the lowest level of miR-374b-5p expression.
Subsequently, the median miR-374b-5p expression in the pancreatic
cancer tissues was used as the cutoff to distinguish between
tissues exhibiting high and low expression of miR-374b-5p.
 | Table IClinicopathological information of
the 87 patients with pancreatic cancer for microRNA-374b-5p
expression analysis. |
Table I
Clinicopathological information of
the 87 patients with pancreatic cancer for microRNA-374b-5p
expression analysis.
| Cases (n) | Percentage (%) |
|---|
| Sex | | |
| Male | 51 | 58.6 |
| Female | 36 | 41.4 |
| Age (years) | | |
| ≤60 | 28 | 32.2 |
| >60 | 59 | 67.8 |
| Histopathology | | |
| Ductal
adenocarcinoma | 70 | 80.5 |
| Other | 17 | 19.5 |
| Location | | |
| Head of
pancreas | 71 | 81.6 |
| Other | 16 | 18.4 |
| Grade | | |
| G1 | 16 | 18.4 |
| G2 | 47 | 54.0 |
| G3 | 24 | 27.6 |
| T
classification | | |
| T1 | 8 | 9.2 |
| T2 | 12 | 13.8 |
| T3 | 60 | 69.0 |
| T4 | 7 | 8.0 |
| N
classification | | |
| N0 | 25 | 28.7 |
| N1 | 62 | 71.3 |
| M
classification | | |
| M0 | 83 | 95.4 |
| M1 | 4 | 4.6 |
| Stage | | |
| Stage I | 13 | 14.9 |
| Stage II | 64 | 73.6 |
| Stage III | 6 | 6.9 |
| Stage IV | 4 | 4.6 |
| Chemotherapeutic
response | | |
| Sensitivity | 29 | 33.3 |
| Resistance | 34 | 39.1 |
| Not available | 24 | 27.6 |
 | Table IIClinicopathological information of
the 8 patients with pancreatic cancer that underwent
microRNA-374b-5p expression analysis. |
Table II
Clinicopathological information of
the 8 patients with pancreatic cancer that underwent
microRNA-374b-5p expression analysis.
| Cases (n) | Percentage (%) |
|---|
| Sex | | |
| Male | 5 | 62.5 |
| Female | 3 | 37.5 |
| Age (years) | | |
| ≤60 | 3 | 37.5 |
| >60 | 5 | 62.5 |
| Histopathology | | |
| Ductal
adenocarcinoma | 6 | 75.0 |
| Other | 2 | 25.0 |
| Location | | |
| Head of
pancreas | 7 | 87.5 |
| Other | 1 | 12.5 |
| Grade | | |
| G1 | 1 | 12.5 |
| G2 | 3 | 37.5 |
| G3 | 4 | 50.0 |
| Stage | | |
| Stage I | 2 | 25 |
| Stage II | 5 | 62.5 |
| Stage III | 1 | 12.5 |
| Stage IV | 0 | 0 |
RT-qPCR
Total RNA from tissues or cells was extracted using
an RNA Isolation kit (Qiagen, Inc., Valencia, CA, USA) following
the manufacturer's protocol. mRNA and miRNA were reverse
transcribed using the RevertAid First Strand cDNA Synthesis kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Reverse transcription was performed at 42°C for 60 min
and 70°C for 10 min. qPCR was performed using
LightCycler® 480 SYBR-Green I Master (Roche Diagnostics,
Basel, Switzerland). And was performed at 50°C for 3 min, 95°C for
10 min, followed by 45 cycles of 95°C for 10 sec, 60°C for 20 sec
and 70°C for 10 sec and then increased from 66°C to 95°C to obtain
the melting curve. Each sample was analyzed in triplicate. qPCR was
performed according to a standard method, as previously described
(23). The following primers were
used in the qPCR reaction: BCL2, forward,
5′-TCCGCATCAGGAAGGCTAGA-3′ and reverse, 5′-AGGACCAGGCCTCCAAGCT-3′;
BIRC3, forward, 5′-TTTCCGTGGCTCTTATTCAAACT-3′ and reverse,
5′-GCACAGTGGTAGGAACTTCTCAT-3′; XIAP, forward,
5′-AATAGTGCCACGCAGTCTACA-3′ and reverse,
5′-CAGATGGCCTGTCTAAGGCAA-3′; GAPDH, forward,
5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse,
5′-GCCATCACGCCACAGTTTC-3′. Primers for U6 (patent no. MQP-0202;
http://www.ribobio.com/sitecn/product_info.aspx?id=51)
and miR-374b-5p (patent no. miRQ0004955-1-2, http://www.ribobio.com/sitecn/product_info.aspx?id=203689)
were synthesized and purified by Guangzhou RiboBio (Guangzhou,
China). U6 or GAPDH were used as endogenous controls for miRNA and
mRNA, respectively. Relative fold expressions were calculated using
the comparative threshold cycle (2−ΔΔCq) method
(24).
Plasmid, small interfering (si) RNA and
transfection
The human miR-374b-5p expression plasmid was
generated by cloning the genomic pre-miR-374b gene into the
retroviral transfer plasmid pMSCV-puro (Takara Bio, Inc., Kusatsu,
Japan) to generate the plasmid pMSCV-miR-374b-5p. pMSCV-miR-374b-5p
was then cotransfected with the pIK packaging plasmid (cat no.
75483) into 293FT cells (cat. no. CRL-1573) (both from ATCC) using
the standard calcium phosphate transfection method, as previously
described (25). A total of 36 h
following co-transfection, supernatants were collected and
incubated with cells to be infected for 24 h in the presence of
polybrene (2.5 µg/ml). Following infection, puromycin (1.5
µg/ml) was used to select stably transduced cells over a
10-day period. The 3′-UTR regions of BCL2, BIRC3 and XIAP were
amplified from genomic DNA using qPCR with 2× Phanta HS Master mix
(cat. no. P512-01; Vazyme Biotech Co., Ltd, Nanjing, China) and
using the thermocycling conditions described in the preceding
paragraph. The primers used in the PCR clone reactions are listed
in Table III. They were then
cloned into a pmirGLO luciferase reporter vector (Promega
Corporation, Madison, WI, USA). The miArrest plasmids for
anti-miR-374b-5p and negative controls were constructed and cloned
into phU6 plasmids (Shanghai GeneChem Co., Ltd., Shanghai, China).
The sequence of the anti-miR-374b-5p is CACTTAGCAGGTTGTAT TATAT.
siRNAs for BCL2 (patent no. stB0003729A-1-5, http://www.ribobio.com/sitecn/product_info.aspx?id=278148),
BIRC3 (patent no. stB0001202A-1-5, http://www.ribobio.com/sitecn/product_info.aspx?id=275621)
and XIAP (patent no. stB0002398A-1-5, http://www.ribobio.com/sitecn/product_info.aspx?id=276817)
knockdown and siRNA negative control (part of the BCL2, BIRC2 and
XIAP siRNA kits) were obtained from Guangzhou Ribobio. Transfection
of siRNAs and plasmids was performed using Lipofectamine 3000
(Thermo Fisher Scientific, Inc.) in BxPC-3 and SW1990 cells
following the manufacturer's protocol. The concentration of each
siRNA used in transfection was 50 nmol/l, temperature was 37°C and
the duration of transfection was 48 h. The period of time between
transfection and subsequent experimentation was <7 days.
 | Table IIIPrimer sequences used in the
reactions for clone polymerase chain reaction. |
Table III
Primer sequences used in the
reactions for clone polymerase chain reaction.
| Gene | Sequence
(5′–3′) |
|---|
|
miR-374b-5p-clone-F |
CGGATTAGGCACTGTGAATACAAAG |
|
miR-374b-5p-clone-R |
TCTGCCAGGTAGAGTGGGAAAC |
| BCL2-3′-UTR-F |
GGGGGTTATCTGTACATCCTGGG |
| BCL2-3′-UTR-R |
AACGATCACCTTTGCTCACAAATAG |
| BIRC3-3′-UTR-F |
GTGGAAGAACAATTGCGGAGAC |
| BIRC3-3′-UTR-R |
CCTGGCTTCATGTTCCCGGTATTAG |
| XIAP-3′-UTR-F |
AGAACGTCCAGGGTTTACATTACAAG |
| XIAP-3′-UTR-R |
GTGGCACCTGCCTGTAATTCC |
Western blotting
Western blotting was performed following a
previously described method (26).
Briefly, cells were seeded in 100-mm tissue culture dishes.
Following 24 h, when cell confluence reached 60–70%, cells were
washed with pre-chilled PBS and proteins were extracted using
sample buffer [62.5 mmol/l Tris-HCl (pH 6.8), 2% SDS, 10% glycerol
and 5% 2-β-mercaptoethanol]. The concentration of protein was
determined using the BCA method. Equal amounts (30 µg) of
protein from the supernatant were loaded per lane and resolved
using 9% SDS-PAGE. Proteins were transferred onto a PVDF membrane
(EMD Millipore, Billerica, MA, USA), blocked with 5% nonfat milk
for 1 h at room temperature and probed with primary antibodies
against BCL2 (cat. no. ab194583; Abcam, Cambridge, UK), BIRC3 (cat.
no. 3130), XIAP (cat. no. 14334), cytochrome c (cat. no.
11940, all dilution 1:1,000) (all from Cell Signaling Technology,
Inc., Danvers, MA, USA) and mouse anti-α-tubulin antibody (cat. no.
ab7291, dilution, 1:5,000; Abcam) overnight at 4°C. Membranes were
washed three times (10 min/wash) in TBS-T buffer and incubated for
40 min at room temperature with horseradish peroxidase-conjugated
anti-mouse (cat. no. 7076) or anti-rabbit (cat. no. 7074; Cell
Signaling Technology, Inc.) secondary antibodies (both, dilution
1:5,000). Blots were then washed three times (10 min/wash) in TBS-T
and developed using an enhanced chemiluminescence system (BeyoECL
Plus, cat. no. P0018; Beyotime Institute of Biotechnology, Haimen,
China). Protein loading was normalized by reprobing the blots with
the mouse anti-α-tubulin antibody.
Analysis of cell apoptosis
Flow cytometric analysis of apoptosis was performed
using the fluorescein isothiocyanate (FITC) Annexin V Apoptosis
Detection kit I (BD Biosciences, San Jose, CA, USA) and performed
as previously described (27).
Briefly, cells were dissociated with trypsin and resuspended at
1×106 cells/ml in binding buffer with 50 µl/ml
FITC Annexin V and 50 µl/ml propidium iodide (PI). Cells
were subsequently incubated for 15 min at room temperature and
analyzed using a Gallios flow cytometer (Beckman Coulter, Inc.,
Brea, CA, USA). The cell's inner mitochondrial membrane potential
(Δψm) was detected by flow cytometry using a MitoScreen JC-1
staining kit (BD Biosciences) following a previously described
protocol (27). Briefly, cells
were dissociated with trypsin, resuspended at 1×106
cells/ml in assay buffer and then incubated at 37°C for 15 min with
10 µl/ml JC-1. Cells were washed twice with the assay buffer
prior to analysis with the flow cytometer. Flow cytometric data
were analyzed using FlowJo 7.6 software (FlowJo LLC, Ashland, OR,
USA).
Quantification of pancreatic cell
viability following treatment with gemcitabine
All pancreatic cancer cells, including BxPC-3,
PANC-1, AsPC-1, SW1990, Capan-1, Capan-2, CFPAC-1 and MIA PaCa-2,
were cultured in 96-well plates and treated with dimethyl sulfoxide
(vehicle) or 10 µM gemcitabine (cat. no. 1288463;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 36 h under a
humidified atmosphere of 5% CO2 at 37°C. The number of
live cells was quantified using the CellTiter-Glo®
Luminescent Cell Viability Assay (cat. no. G7570; Promega
Corporation). Surviving cells were calculated as the fraction of
vehicle controls and the results were averaged between six wells
per dose in three independent experiments.
Caspase-9 and -3 activity assays
Caspase-9 and -3 activity was analyzed by
spectrophotometry using the caspase-9 (cat. no. KGA403) and
caspase-3 (cat. no. KGA202) colorimetric assay kits, respectively
(Nanjing Keygen Biotech, Co., Ltd., Nanjing, China), following the
manufacturer's protocol. Briefly, 5×106 cells or 100 mg
fresh tumor tissues were washed with cold PBS, resuspended in lysis
buffer and incubated on ice for 30 min. A total of 50 µl
cell suspension, 50 µl reaction buffer and 5 µl
caspase-3/-9 substrate was mixed and then incubated at 37°C for 4
h. Absorbance was then measured at 405 nm and the results of BCA
protein quantitative analysis were used as the reference to
normalize expression in each experimental group.
High throughput data processing and
visualization
Datasets regarding miRNA expression and the clinical
profile of pancreatic cancer were downloaded from The Cancer Genome
Atlas (TCGA: https://cancergenome.nih.gov/) and ArrayExpress
(http://www.ebi.ac.uk/arrayexpress/).
The log2 values of miRNAs in each sample were analyzed
using Excel 2010 (Microsoft Corporation, Redmond, WA, USA) and
GraphPad 5 (GraphPad, Inc., La Jolla, CA, USA). Differences in the
miRNA expression of all pancreatic cancer tissues between the two
groups were assessed using paired or unpaired t-tests. Datasets
were downloaded from GSE24279 and GSE29352 (https://www.ncbi.nlm.nih.gov/geo/) to assess the
expression of miRNAs in pancreatic cancer. The expression of miRNAs
in each sample were analyzed as previously described (28). The expression of miR-374b-5p in
pancreatic cancer datasets taken from TCGA and GSE was analyzed.
Targetscan (http://www.targetscan.org/vert_71/) and miRanda
(http://34.236.212.39/microrna/home.do) were used to
predict the potential targets of miR-374b-5p and assessed using a
previously described protocol (29,30).
Tumor xenografts
A total of 20, 6-week-old female BALB/c-nu mice
weighing 10–12 g were randomly divided into 4 groups (each, n=5).
Mice were kept at a temperature of 18–29°C, humidity of 40–70% and
under a 12-h light-dark cycle. Mice also had ad libitum
access to food and water mice. Cells (2×106) were
inoculated subcutaneously into the inguinal folds of the nude mice.
In 2 groups of mice, 2×106 control AsPC-1 cells were
inoculated subcutaneously in the left dorsal flank; in the other 2
groups, the mice were subcutaneously injected with the same number
of miR-374b-5p overexpressing-AsPC-1 cells. At 7 days after the
cell inoculation, one group of mice injected with the control cells
and one group of mice injected with the anti-miR-149-5p cells
received 50 µg/g gemcitabine (cat. no. 1288463,
Sigma-Aldrich; Merck KGaA) via intraperitoneal injection twice a
week for 4 weeks. Tumor volume was determined using an external
caliper and calculated using the following equation: Length ×
(Weight)2/2. Tumor volume was assessed using the IVIS
Spectrum In Vivo Imaging System (Perkin Elmer, Inc., Waltham, MA,
USA) at 5-day intervals. On day 40, animals were euthanized via
cervical dislocation. Tumors were excised, weighed and stored in
liquid nitrogen tanks. Ethical approval for the animal study was
obtained from the Institutional Research Ethics Committee from the
First Hospital of Jilin University (ethics no. 2005-001).
Dual-luciferase reporter assay
Cells (4×104) were seeded in triplicate
in 24-well plates and cultured for 24 h. Subsequently, a
dual-luciferase reporter assay was performed following a previously
described protocol (31). Briefly,
cells were plated in 60-mm cell culture dishes and cultured for 24
h until they reached 60–80%. Subsequently, 100 ng BCL2-3′-UTR,
BIRC3-3′-UTR or XIAP-3′-UTR luciferase plasmids, plus 5 ng pRL-TK
Renilla plasmid (Promega Corporation) were transfected into
cells using Lipofectamine 3000. Following 12-h incubation, the
transfection medium was replaced; cells were harvested and washed
with PBS and lysed using passive lysis buffer (Promega
Corporation). Cell lysates were analyzed immediately using a
Synergy™ 2 microplate system (BioTek Instruments, Inc., Winooski,
VT, USA). Luciferase and Renilla luciferase were measured
using a dual-Luciferase Reporter assay system (Promega Corporation)
following the manufacturer's protocol. The luciferase activity of
each lysate was normalized to Renilla luciferase activity.
The relative transcriptional activity was converted into fold
induction above the vehicle control value.
miRNA immunoprecipitation (IP)
Cells were co-transfected with pIRESneo-FLAG/human
influenza hemagglutinin (HA)-Ago2 plasmid (cat. no. 10822; Addgene,
Inc., Cambridge, MA, USA), followed by HA-Ago2 IP using HA-antibody
(dilution, 1:1,000; cat. no. H3663, Sigma-Aldrich; Merck KGaA),
following a previously described protocol (32). Briefly, cells (4×104)
were plated in 60-mm cell culture dishes and cultured for 24 h
until they reached 60–80% confluence. pIRESneo-FLAG/HA-Ago2
plasmids were transfected into cells using Lipofectamine 3000.
Following 48-h transfection, cells were washed and lysed in
radioimmunoprecipitation buffer (Sigma-Aldrich; Merck KGaA)
containing 10% proteinase inhibitor cocktail and 1 mM
phenylmethylsulfonyl fluoride. A fraction of the whole cell lysate
was used for RNA isolation and the remaining lysate was subjected
to IP using antibodies against HA (dilution, 1:1,000; cat. no.
H3663; Sigma-Aldrich; Merck KGaA) or immunoglobulin G (dilution,
1:2,000; cat. no. ab109489; Abcam). RNA from whole cell lysates and
RNA IP (RIP) fractions were extracted using TRIzol (Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. Relative
levels of mRNA were determined using RT-qPCR, following the
aforementioned protocol. The relative mRNA enrichment in the RIP
fractions was computed based on the ratio of relative mRNA levels
in the RIP fractions and the relative mRNA levels in the whole cell
lysates.
Statistical analysis
All values are presented as the mean ± standard
deviation. Significant differences were determined using GraphPad
5.0 software (GraphPad, Inc.). One-way analysis of variance was
used to determine statistical differences between ≥2 groups,
followed by Tukey's post hoc test. Unpaired or paired t-tests were
used to determine statistical differences between 2 groups. The
χ2 test was used to analyze the association between
miR-374b-5p expression and patient clinicopathological
characteristics. Survival curves were plotted using the Kaplan
Meier method and compared using the log-rank test using X-tile
software version 3.6.1 (Yale University, New Haven, CT, USA).
Pearson's correlation coefficient was used to analyze the
correlation between miR-374b-5p expression levels and the apoptotic
ratio following treatment with gemcitabine. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were repeated three times.
Results
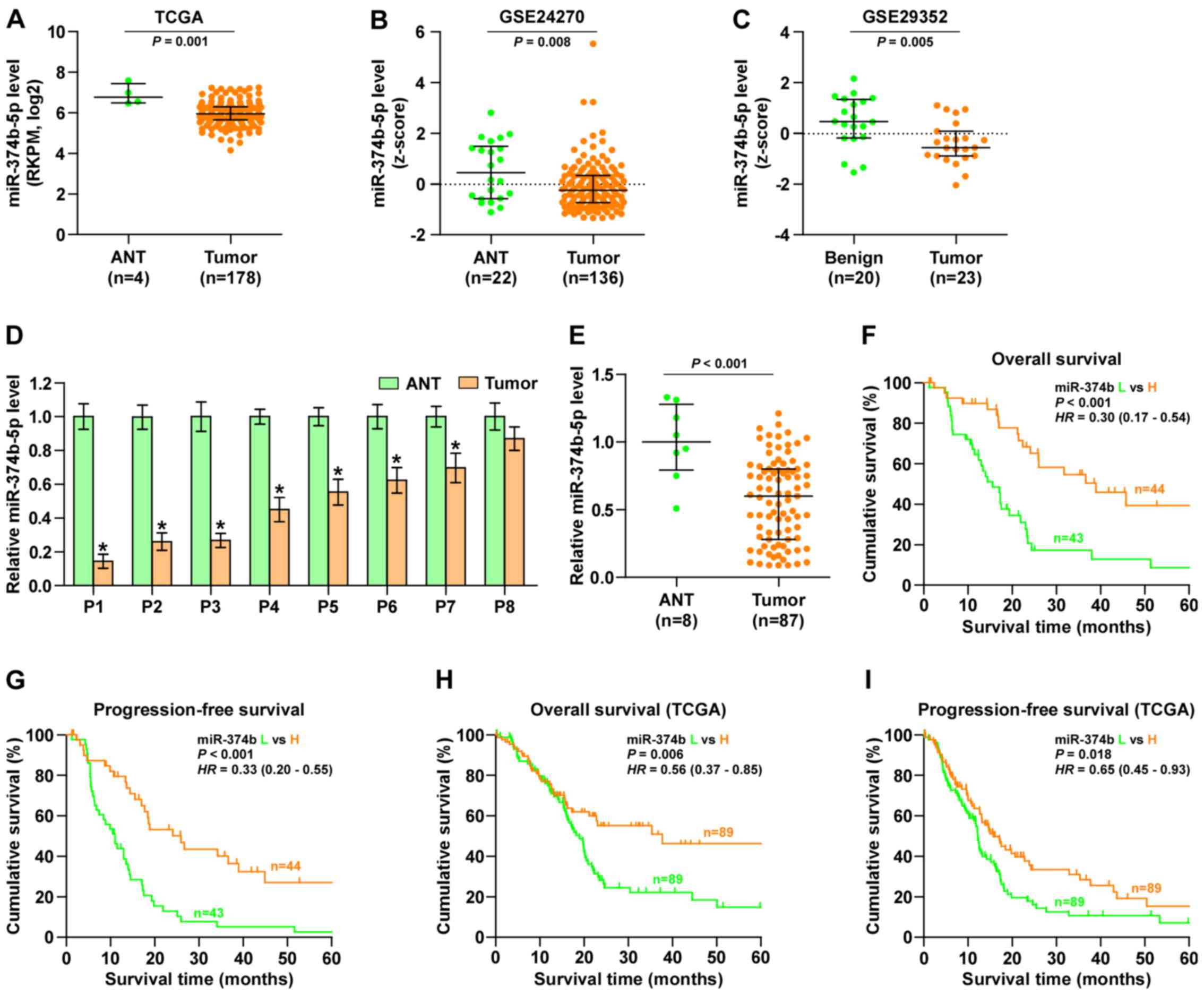
miR-374b-5p expression is reduced in
pancreatic cancer tissues
By analyzing several miRNA sequencing datasets of
pancreatic cancer from TCGA and GSE using ArrayExpress, it was
demonstrated that miR-374b-5p expression was significantly
decreased in pancreatic cancer tissues compared with adjacent
normal tissues (Fig. 1A–C).
Subsequently, miR-374b-5p expression levels in the pancreatic
cancer tissues taken from 8 patients recruited in the current study
were measured and compared with miR-374-5p expression in the
adjacent normal tissues. It was demonstrated that that miR-374b-5p
expression was significantly decreased in 7 of the pancreatic
cancer tissues compared with the respective adjacent normal tissue
(Fig. 1D). Furthermore, miR-374-5p
expression in the 87 pancreatic cancer tissues was significantly
decreased compared with the normal tissues (Fig. 1E). The association between
miR-374b-5p expression and the clinicopathological characteristics
of patients with pancreatic cancer was then assessed and it was
demonstrated that miR-374b-5p expression was inversely associated
with node classification in patients with pancreatic cancer
(Table IV). Kaplan-Meier survival
analysis indicated that patients with pancreatic cancer that
exhibited low expression of miR-374b-5p had significantly poorer
overall and progression-free survival than those exhibiting high
expression of miR-374b-5p (Fig. 1F and
G). This was also the case in patients from the TCGA dataset
(Fig. 1H and I). These results
indicate that miR-374b-5p expression is sigificantly reduced in
pancreatic cancer tissues and that this is assiciated with
increased mortality in patients with pancreatic cancer.
 | Table IVThe association between miR-374b-5p
expression and the clinicopathological characteristics of the 87
patients with pancreatic cancer. |
Table IV
The association between miR-374b-5p
expression and the clinicopathological characteristics of the 87
patients with pancreatic cancer.
| Parameters | No. of cases | miR-374b-5p
expression
| P-values |
|---|
| Low | High |
|---|
| Sex | | | | |
| Male | 51 | 29 | 22 | 0.099 |
| Female | 36 | 14 | 22 | |
| Age (years) | | | | |
| ≤60 | 28 | 16 | 12 | 0.321 |
| >60 | 59 | 27 | 32 | |
| Histopathology | | | | |
| Ductal
adenocarcinoma | 70 | 35 | 35 | 0.828 |
| Other | 17 | 8 | 9 | |
| Location | | | | |
| Head of
pancreas | 71 | 38 | 33 | 0.108 |
| Other | 16 | 5 | 11 | |
| Grade | | | | |
| G1–G2 | 63 | 31 | 32 | 0.947 |
| G3 | 24 | 12 | 12 | |
| T
classification | | | | |
| T1-2 | 20 | 7 | 13 | 0.142 |
| T3-4 | 67 | 36 | 31 | |
| N
classification | | | | |
| N0 | 25 | 6 | 19 | 0.003a |
| N1-2 | 62 | 37 | 25 | |
| M
classification | | | | |
| M0 | 83 | 39 | 44 | 0.055 |
| M1 | 4 | 4 | 0 | |
| Stage | | | | |
| I–II | 77 | 36 | 41 | 0.167 |
| III–IV | 10 | 7 | 3 | |
| Chemotherapeutic
response | | | | |
| Sensitivity | 29 | 9 | 20 | <0.001a |
| Resistance | 34 | 25 | 9 | |
The low expression of miR-374b-5p is
positively correlated with chemoresistance in pancreatic
cancer
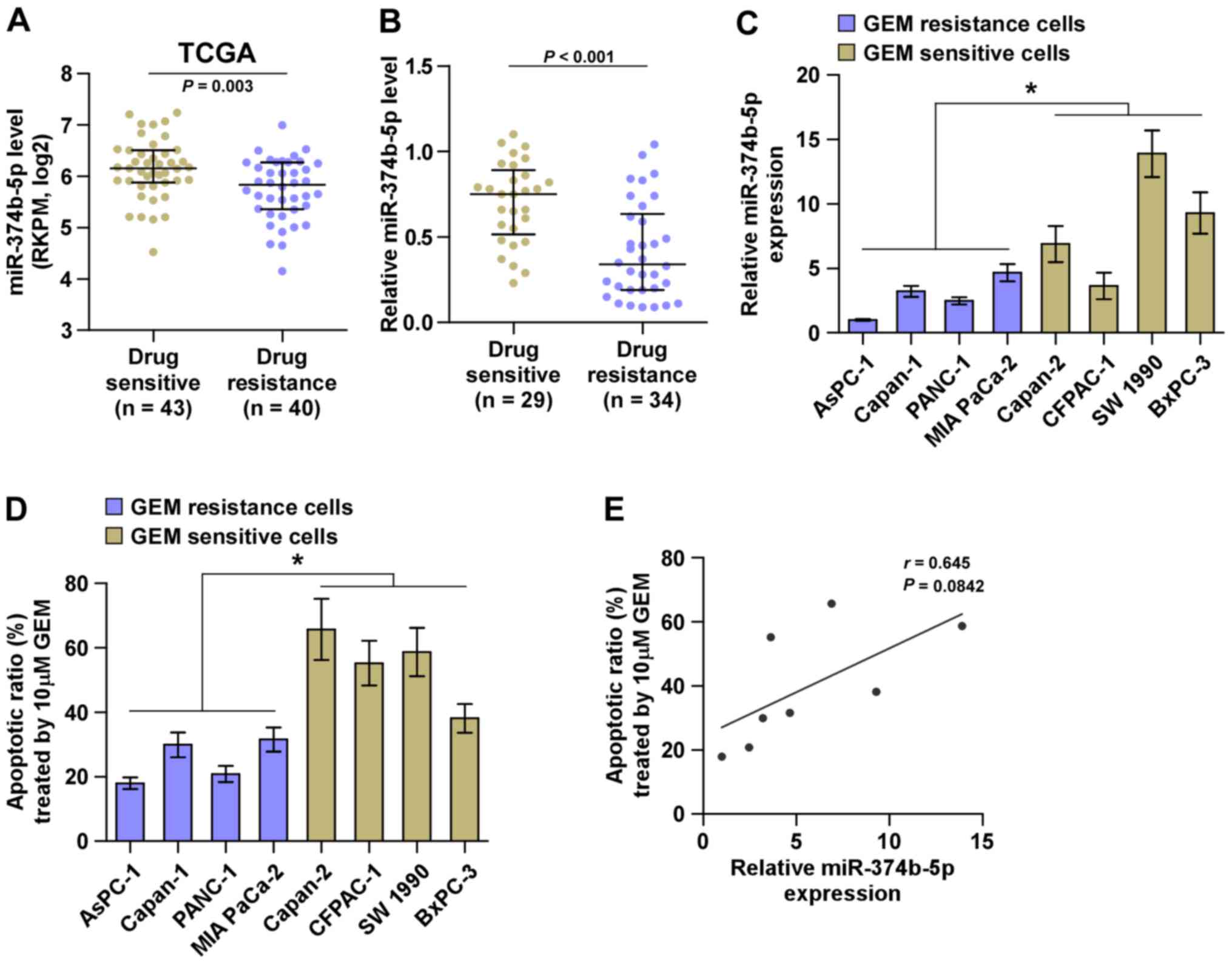
miRNA sequencing of the TCGA dataset containing
patients with pancreatic cancer indicated that miR-374b-5p
expression was significantly decreased in tissues from patients
with drug-resistant pancreatic cancer compared with those from
patients with drug-sensitive pancreatic cancer (Fig. 2A). This was also the case in
patients recruited in the current study (Fig. 2B). The association between
miR-374b-5p expression and the chemotherapeutic response of
pancreatic cancer cells was evaluated by calculating the apoptotic
ratio of pancreatic cancer cells treated with gemcitabine. As
presented in Fig. 2C and D,
pancreatic cancer cells with low expression of miR-374b-5p
exhibited significantly increased resistance to gemcitabine
compared with those that had high expression of miR-374b-5p.
Furthermore, there was a positive correlation between miR-374b-5p
expression and the apoptotic ratio of pancreatic cancer cells
(Fig. 2E). Collectively, these
results indicate that the low expression of miR-374b-5p is strongly
associated with chemoresistance in pancreatic cancer.
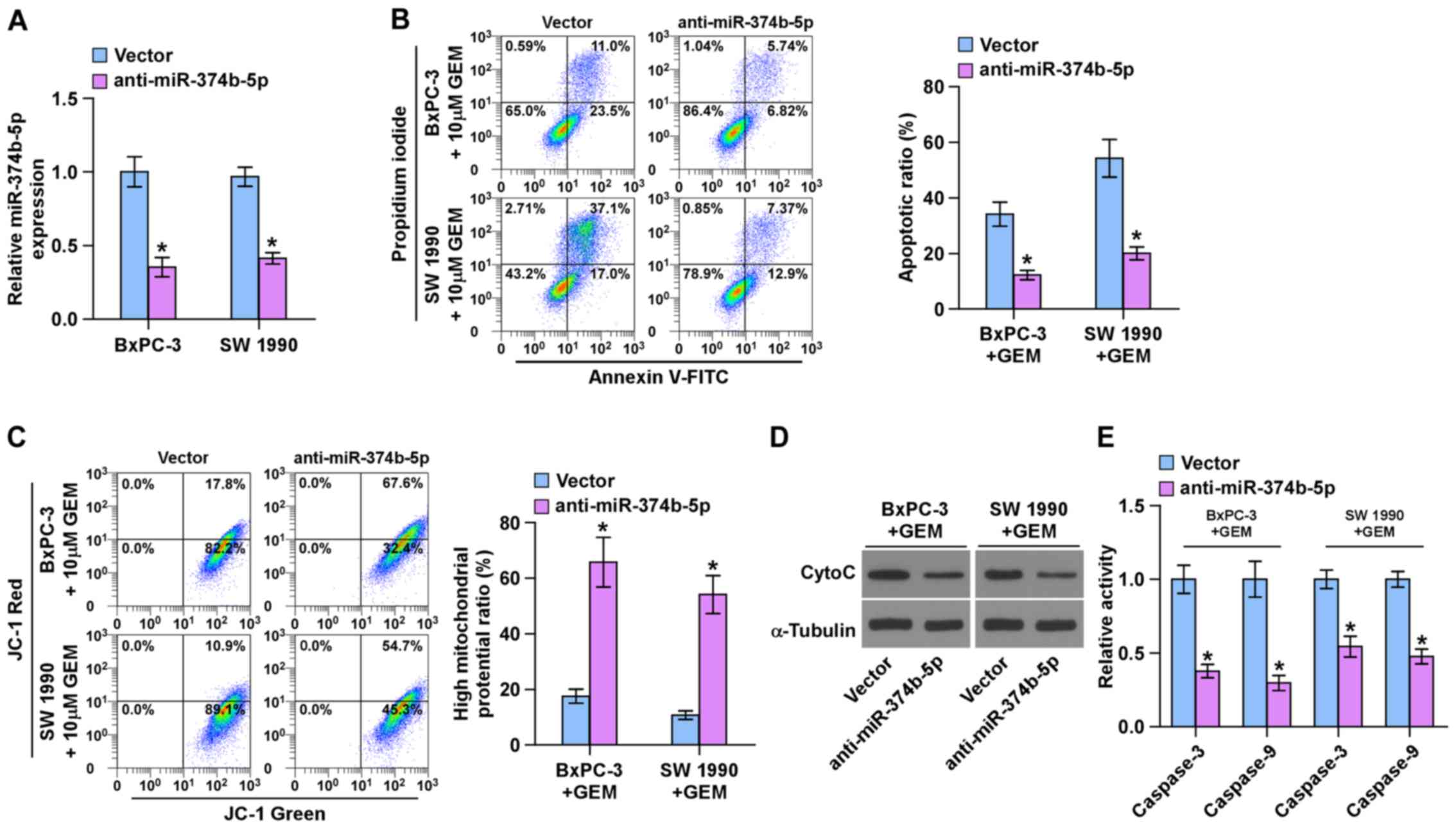
Silencing miR-374b-5p promotes
chemoresistance in pancreatic cancer cells
The role of miR-374b-5p in the chemoresistance of
pancreatic cancer was further investigated. The BxPC-3 and SW 1990
chemosensitive pancreatic cancer cells exhibited the highest
expression of miR-374-5p (Fig.
2C); therefore, miR-374b-5p expression was knocked down by
transfecting anti-miR-374b-5p into BxPC-3 and SW 1990 cells
(Fig. 3A). An Annexin V apoptosis
assay was performed and the results demonstrated that silencing
miR-374b-5p expression significantly decreased the apoptotic ratio
in BxPC-3 and SW 1990 cells following treatment with gemcitabine
(Fig. 3B). A mitochondrial
membrane potential assay was then performed and the results
indicated that silencing miR-374b-5p significantly enhanced the
mitochondrial potential of BxPC-3 and SW 1990 cells following
treatment with gemcitabine (Fig.
3C). Furthermore, silencing miR-374b-5p decreased the
expression of cytochrome c (Fig. 3D) and significantly decreased
caspase-3 and -9 activity in BxPC-3 and SW 1990 cells following
treatment with GEM (Fig. 3E).
These results indicate that silencing miR-374b-5p induces the
chemoresistance of pancreatic cancer cells to gemcitabine.
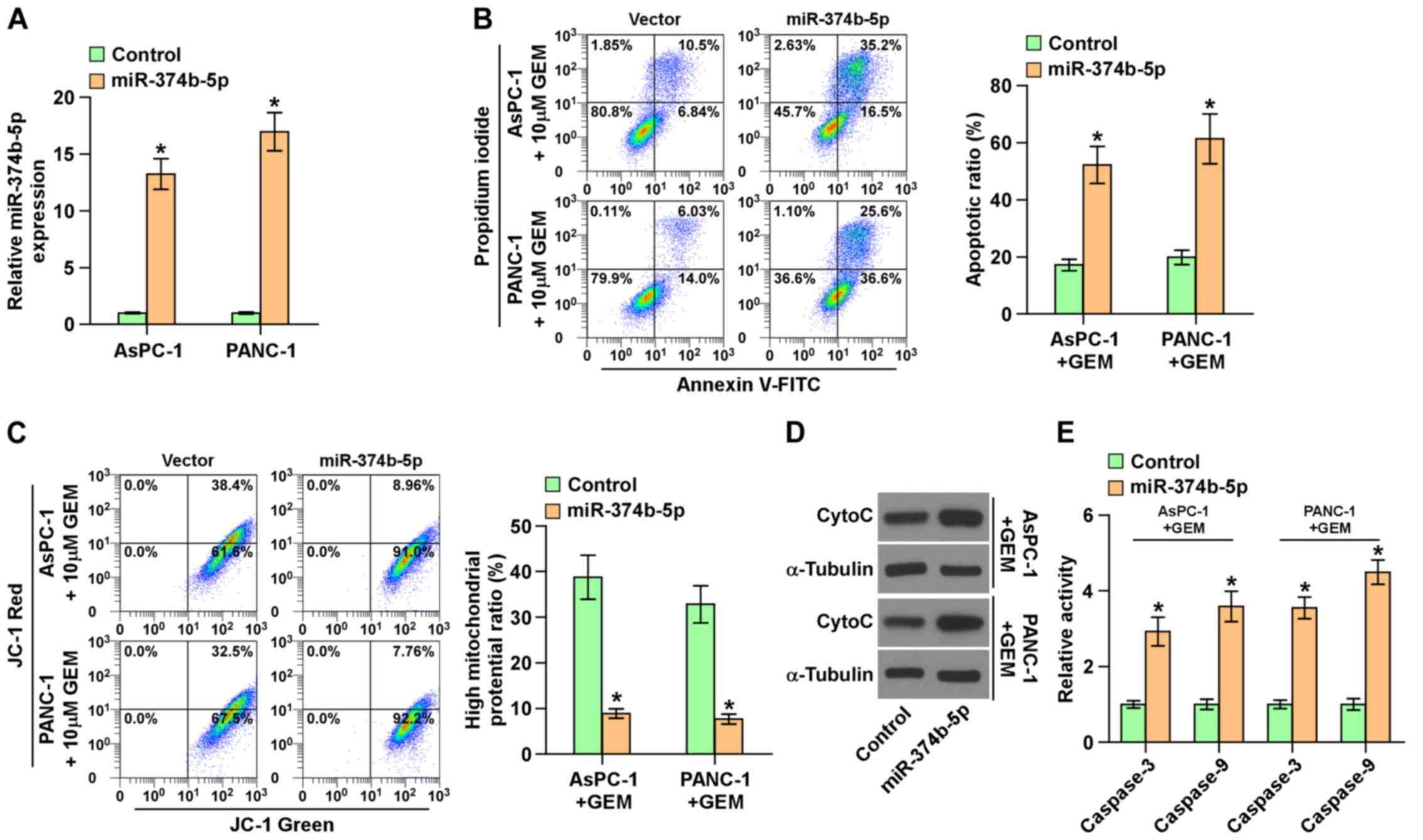
Ectopic expression of miR-374b-5p
attenuates chemoresistance in pancreatic cancer cells
Subsequently, miR-374b-5p was overexpressed in
AsPC-1 and PANC-1 cells via viral transduction (Fig. 4A), as these two cell lines
expressed the lowest levels of miR-374b-5p (Fig. 2C). The upregulation of miR-374b-5p
significantly increased the apoptotic ratio, the expression of
cytochrome c and caspase-3 and -9 activity, but
significantly reduced the mitochondrial potential in AsPC-1 and
PANC-3 cells following treatment with gemcitabine (Fig. 4B–E). These results demonstrate that
the upregulation of miR-374b-5p abrogates the resistance of
pancreatic cancer cells to gemcitabine.
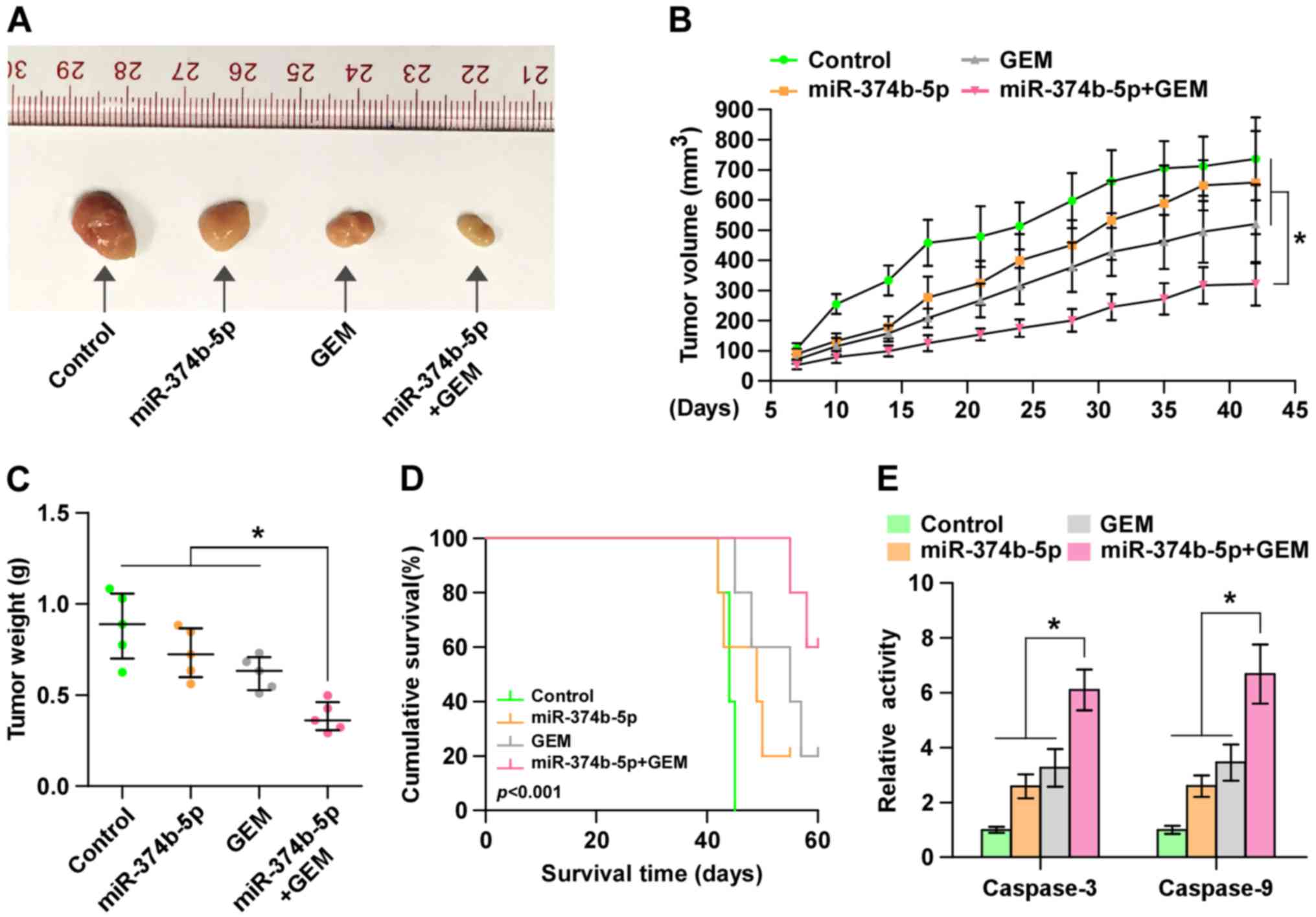
Upregulation of miR-374b-5p ameliorates
the resistance of pancreatic cancer cells to gemcitabine in
vivo
To investigate the effect of miR-374b-5p on the
chemoresistance of pancreatic cancer cells in vivo, mice
were randomly divided into 4 groups (n=5/group). In the first two
groups of mice, 2×106 control AsPC-1 cells were
inoculated subcutaneously into the left dorsal flank; in the other
two groups, mice were subcutaneously injected with the same number
of miR-374b-5p-overex-pressing-AsPC-1 cells. Following 1 week, one
control and one miR-374-5p-overexpressing group received an
intraperitoneal injection of 50 µg/g gemcitabine twice per
week over a 4-week period. Mice that received
miR-374b-5p-overexpressing cells and gemcitabine treatment
exhibited significantly smaller tumor volumes and weights (Fig. 5A–C) and significantly longer
survival times compared with other three groups (Fig. 5D). Furthermore, caspase-3 and -9
activity were significantly increased in the mice injected with
miR-374b-5p-overex-pressing cells and receiving gemcitabine
treatment (Fig. 5E). These results
indicate that the upregulation of miR-374b-5p restores the
sensitivity of pancreatic cancer cells to gemcitabine in
vivo.
miR-374b-5p targets multiple
anti-apoptotic proteins in pancreatic cancer cells
By analyzing several public databases, including
TargetScan and miRanda, it was determined that several
anti-apoptotic proteins, including BCL2, BIRC3 and XIAP were
potential targets of miR-374b-5p (Fig.
6A). The results of western blotting demonstrated that
silencing miR-374b-5p increased the expression of BCL2, BIRC3 and
XIAP in GEM-sensitive pancreatic cancer cell lines (Fig. 6B). Furthermore, the upregulation of
miR-374b-5p significantly repressed the luciferase reporter
activity of the 3′-UTRs of BCL2, BIRC3 and XIAP in pancreatic
cancer cells (Fig. 6C), whereas
silencing miR-374b-5p significantly elevated it (Fig. 6D). The results of the RNA IP assay
identified a direct association between miR-374b-5p and BCL2, BIRC3
and XIAP transcripts (Fig. 6E and
F). Collectively, these results demonstrate that BCL2, BIRC3
and XIAP are direct targets of miR-374b-5p in pancreatic cancer
cells.
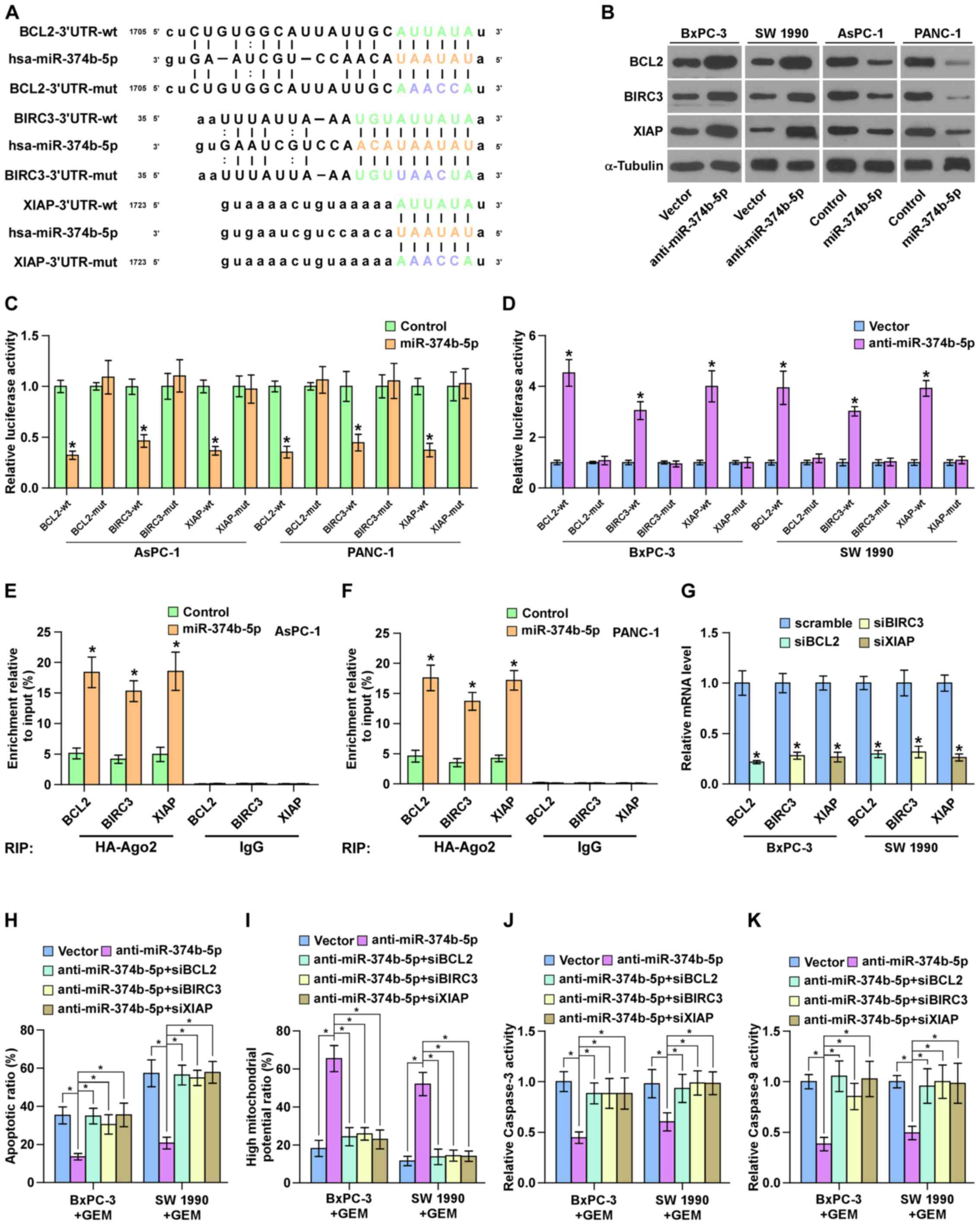 | Figure 6miR-374b-5p targets BCL2, BIRC3 and
XIAP in pancreatic cancer cells. (A) Predicted miR-374b-5p target
sequence in the 3′-UTRs of BCL2, BIRC3 and XIAP. (B) Western
blotting measuring BCL2, BIRC3 and XIAP expression. α-tubulin
served as the loading control. (C and D) Luciferase assay of the
cells transfected with the indicated reporter following
transfection with (C) miR-374b-5p mimic or (D) anti-miR-374b-5p.
*P<0.05 vs. control or vector, respectively. (E and
F) RNA immunoprecipitation assay indicating the association between
miR-374b-5p and BCL2, BIRC3 and XIAP transcripts in the indicated
cells. *P<0.05 vs. vector (G) Reverse
transcription-quantitative polymerase chain reaction analysis of
BCL2, BIRC3 and XIAP expression in the indicated pancreatic cancer
cells. Transcript levels were normalized to U6 expression.
*P<0.05 vs. scramble. (H) The protective effect of
anti-miR-374b-5p on the apoptotic rate of pancreatic cancer cells
was attenuated by the individual silencing of BCL2, BIRC3 and XIAP.
(I) The stimulatory effect of anti-miR-374b-5p on the mitochondrial
potential of pancreatic cancer cells was attenuated by the
individual silencing of BCL2, BIRC3 and XIAP. (J and K) The
stimulatory effects of anti-miR-374b-5p on caspase-3 and -9
activity of pancreatic cancer cells was attenuated by the silencing
of BCL2, BIRC3 and XIAP. Data are presented as the mean ± standard
deviation of three independent experiments. *P<0.05.
miR-374b-5p, microRNA-374b-5p; BCL2, B-cell lymphoma 2; BRIC3,
Baculoviral IAP Repeat Containing 3; XIAP, X-linked inhibitor of
apoptosis; UTR, untranslated region; mut, mutant; wt, wild-type;
si, small interfering; GEM, gemcitabine. |
BCL2, BIRC3 and XIAP are crucial for the
pro-chemoresistance effects of anti-miR-374b-5p on pancreatic
cancer cells
It was then investigated whether BCL2, BIRC3 and
XIAP mediate the functional effects of anti-miR-374b-5p on the
chemoresistance of ovarian cancer cells. BCL2, BIRC3 and XIAP
expression were silenced following transfection of siRNAs against
BCL2, BIRC3 and XIAP into pancreatic cancer cells. The results of
RT-qPCR demonstrated that silencing BCL2, BIRC3 and XIAP
significantly reduced BCL2, BIRC3 and XIAP expression compared with
the respective scramble controls (Fig.
6G). The silencing of BCL2, BIRC3 and XIAP expression
significantly attenuated the stimulatory effects of
anti-miR-374b-5p on the mitochondrial potential and caspase-3 and
-9 activity, and reversed the inhibition of apoptosis induced by
anti-miR-374b-5p in pancreatic cancer cells (Fig. 6H–K). Taken together, these results
indicate that BCL2, BIRC3 and XIAP are crucial for the effects of
anti-miR-374b-5p on the chemoresistance of pancreatic cancer
cells.
Discussion
The present study indicates that miR-374b-5p serves
a tumor suppressive role in chemotherapeutic resistant pancreatic
cancer. miR-374b-5p expression was decreased in pancreatic cancer
tissues, particularly in chemoresistant pancreatic cancer tissues,
and the low expression of miR-374b-5p was strongly associated with
chemotherapeutic resistance, as well as poor overall and
progression-free survival in patients with pancreatic cancer. The
upregulation of miR-374b-5p decreased, whereas miR-374b-5p
silencing increased, the chemoresistance of pancreatic cancer cells
to gemcitabine in vitro and in vivo. Furthermore, the
results of the current study revealed that the upregulation of
miR-374b-5p restored the chemotherapeutic sensitivity of pancreatic
cancer cells to gemcitabine via the direct targeting of several
anti-apoptotic proteins, including BCL2, BIRC3 and XIAP. Therefore,
the results of the current study indicate that miR-374b-5p serves a
tumor suppressive role in the development of chemotherapeutic
resistance in pancreatic cancer.
BCL2 belongs to the Bcl-2 family and is an inhibitor
of apoptosis. Previous studies have demonstrated that the
upregulation of BCL2 contributes to intrinsic and acquired drug
resistance in different types of human cancer (33–35).
In pancreatic cancer, the upregulation of BCL2 by the transcription
factor cut like homeobox 1 significantly increased drug resistance
(36). IAP proteins are another
well-known family that act as endogenous inhibitors of apoptosis by
directly binding to and inhibiting caspase proteins (37). The most well characterized IAP
proteins include XIAP and BIRC3, which have been implicated in the
chemoresistance of different types of cancer (38–40).
Targeting Bcl-2 or IAP proteins markedly enhance the therapeutic
effects of chemotherapeutic agents in various types of cancer
(41,42); however, it remains unknown how
these anti-apoptotic proteins are simultaneously repressed in
cancer. The results of the current study demonstrated that
miR-374b-5p simultaneously targets BCL2, BIRC3 and XIAP, thus
contributing to the chemoreistance of pancreatic cancer cells to
gemcitabine. Therefore, the results of the current study identified
a novel mechanism by which miR-374b-5p improves the
chemotherapeutic response of pancreatic cancer to gemcitabine.
The dysregulation of miR-374b-5p has been implicated
several disorders, including obesity, calcific aortic stenosis,
ischemic stroke, IgA nephropathy and infertility (43–47).
In cancer, the overexpression of miR-374b-5p has been identified in
breast, head and neck, gastric and prostate cancer, as well as in
melanoma (48–53). However, the downregulation of
miR-374b-5p has also been identified in T-cell lymphoblastic
lymphoma and colorectal cancer (54,55).
These results indicate that miR-374b-5p serves different roles in
different types of tumor. Notably, a study by Schreiber et
al (56) indicated that,
following step-wise treatment with increasing concentrations of
cisplatin over >20 passages, the downregulation of miR-374b-5p
was directly involved in the development of the cisplatin-resistant
phenotype. This suggests that the decreased expression of
miR-374b-5p is associated with chemoresistance in pancreatic
cancer. However, the clinical significance and underlying mechanism
mediating the functional role of miR-374b-5p in pancreatic cancer
are yet to be elucidated. The current study demonstrated that
miR-374b-5p was down-regulated in pancreatic cancer tissues and was
associated with poor responses to chemotherapy, as well as poor
overall and progression-free survival in patients with pancreatic
cancer. Furthermore, the results of the current study indicate that
the upregulation of miR-374b-5p reduces the chemotherapeutic
resistance of pancreatic cancer cells to gemcitabine by inhibiting
the expression of several anti-apoptotic proteins, including BCL2,
BIRC3 and XIAP. This clarifies that miR-374b-5p serves a tumor
suppressive role during the development of chemotherapeutic
resistance in pancreatic cancer.
It has been suggested that miR-374b-5p may be used
as a diagnostic marker in different types of cancer. Hanniford
et al (50) reported that
miR-374-b-5p has a diagnostic signature in patients with melanoma
and brain metastasis compared with those without brain metastasis.
Analysis of the TCGA dataset indicated that miR-374b-5p may be a
prognostic marker of breast cancer (49). The results of the current study
indicated that miR-374b-5p expression was dramatically reduced in
pancreatic cancer tissues and the low expression of miR-374b-5p
predicted the poor overall and progression-free survival of
patients with pancreatic cancer. This indicates that miR-374b-5p
may be used as a diagnostic marker in patients with pancreatic
cancer. Importantly, Summerer et al (51) reported that high levels of
miR-374b-5p in the plasma predicted the poor prognosis of patients
with head and neck cancer, suggesting that miR-374b-5p may be used
as a minimally invasive diagnostic marker in patients with cancer.
However, further studies are required to determine whether
miR-374b-5p levels in the serum or plasma may be used as a
minimally invasive diagnostic or prognostic marker for patients
with pancreatic cancer.
In conclusion, the results of the current study
clarify that the upregulation of miR-374b-5p restores the
chemotherapeutic resistance of pancreatic cancer cells to
gemcitabine by directly repressing BCL2, BIRC3 and XIAP expression.
Comprehensive understanding of the underlying mechanism of
miR-374b-5p in the chemoresistance of pancreatic cancer may provide
insights into the development of chemoresistance in pancreatic
cancer, which may facilitate the development of novel therapeutic
methods to treat patients with pancreatic cancer.
Acknowledgments
Not applicable.
Notes
[1]
Funding
The present study was supported by grants from the
Finance Department Foundation of Jilin Province
(120170627191002).
[2] Availability
of data and materials
The datasets generated and analyzed in the current
study are available at TCGA (https://cancergenome.nih.gov/) and ArrayExpress
(http://www.ebi.ac.uk/arrayexpress/)
[3] Authors'
Contributions
PZ conceived the experiments and drafted the
manuscript. DS and XW conducted the experiments and helped to
analyze the data. GS performed data analysis. SC and MY performed
data analysis and revised the manuscript. All authors contributed
to revise the manuscript and approved the final version for
publication.
[4] Ethics
approval and consent to participate
Written informed consent from each patient enrolled
in the study was obtained and ethical approval for the use of human
tissues and for the animal study was obtained from the
Institutional Research Ethics Committee at The First Hospital of
Jilin University (ethics no. 005-001).
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no conflicts of
interest.
References
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J,
Smigal C and Thun MJ: Cancer statistics, 2006. CA Cancer J Clin.
56:106–130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bond-Smith G, Banga N, Hammond TM and
Imber CJ: Pancreatic adenocarcinoma. BMJ. 344:e24762012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Merl MY, Abdelghany O, Li J and Saif MW:
First-line treatment of metastatic pancreatic adenocarcinoma: Can
we do better? Highlights from the '2010 ASCO Annual Meeting'.
Chicago, IL, USA. June 4–8, 2010. JOP. 11:317–320. 2010.PubMed/NCBI
|
|
4
|
Kindler HL: Front-line therapy of advanced
pancreatic cancer. Semin Oncol. 32(Suppl 9): S33–S36. 2005.
View Article : Google Scholar
|
|
5
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gottesman MM and Pastan IH: The role of
multidrug resistance efflux pumps in cancer: Revisiting a JNCI
publication exploring expression of the MDR1 (P-glycoprotein) gene.
J Natl Cancer Inst. 107:1072015. View Article : Google Scholar
|
|
7
|
Radin D, Lippa A, Patel P and Leonardi D:
Lifeguard inhibition of Fas-mediated apoptosis: A possible
mechanism for explaining the cisplatin resistance of
triple-negative breast cancer cells. Biomed Pharmacother.
77:161–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ojini I and Gammie A: Rapid identification
of chemoresistance mechanisms using yeast DNA mismatch repair
mutants. G3 (Bethesda). 5:1925–1935. 2015. View Article : Google Scholar
|
|
9
|
Aldinucci D, Celegato M and Casagrande N:
Microenvironmental interactions in classical Hodgkin lymphoma and
their role in promoting tumor growth, immune escape and drug
resistance. Cancer Lett. 380:243–252. 2016. View Article : Google Scholar
|
|
10
|
Mastri M, Rosario S, Tracz A, Frink RE,
Brekken RA and Ebos JM: The challenges of modeling drug resistance
to antiangiogenic therapy. Curr Drug Targets. 17:1747–1754. 2016.
View Article : Google Scholar
|
|
11
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ren D, Wang M, Guo W, Huang S, Wang Z,
Zhao X, Du H, Song L and Peng X: Double-negative feedback loop
between ZEB2 and miR-145 regulates epithelial-mesenchymal
transition and stem cell properties in prostate cancer cells. Cell
Tissue Res. 358:763–778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ren D, Wang M, Guo W, Zhao X, Tu X, Huang
S, Zou X and Peng X: Wild-type p53 suppresses the
epithelial-mesenchymal transition and stemness in PC-3 prostate
cancer cells by modulating miR 145. Int J Oncol. 42:1473–1481.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Jin Y, Mu Z, Chen W and Jiang S:
MicroRNA 146a 5p enhances cisplatin induced apoptosis in ovarian
cancer cells by targeting multiple anti apoptotic genes. Int J
Oncol. 51:327–335. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feng C, Sun P, Hu J, Feng H, Li M, Liu G,
Pan Y, Feng Y, Xu Y, Feng K, et al: miRNA-556-3p promotes human
bladder cancer proliferation, migration and invasion by negatively
regulating DAB2IP expression. Int J Oncol. 50:2101–2112. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Liu J, Zang D, Wu S, Liu A, Zhu
J, Wu G, Li J and Jiang L: Upregulation of miR-572
transcriptionally suppresses SOCS1 and p21 and contributes to human
ovarian cancer progression. Oncotarget. 6:15180–15193.
2015.PubMed/NCBI
|
|
18
|
Ren D, Yang Q, Dai Y, Guo W, Du H, Song L
and Peng X: Oncogenic miR-210-3p promotes prostate cancer cell EMT
and bone metastasis via NF-κB signaling pathway. Mol Cancer.
16:1172017. View Article : Google Scholar
|
|
19
|
Guo W, Ren D, Chen X, Tu X, Huang S, Wang
M, Song L, Zou X and Peng X: HEF1 promotes epithelial mesenchymal
transition and bone invasion in prostate cancer under the
regulation of microRNA-145. J Cell Biochem. 114:1606–1615. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chaudhary AK, Mondal G, Kumar V, Kattel K
and Mahato RI: Chemosensitization and inhibition of pancreatic
cancer stem cell proliferation by overexpression of microRNA-205.
Cancer Lett. 402:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen M, Wang M, Xu S, Guo X and Jiang J:
Upregulation of miR-181c contributes to chemoresistance in
pancreatic cancer by inactivating the Hippo signaling pathway.
Oncotarget. 6:44466–44479. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The Eighth Edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
'personalized' approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang M, Ren D, Guo W, Huang S, Wang Z, Li
Q, Du H, Song L and Peng X: N-cadherin promotes
epithelial-mesenchymal transition and cancer stem cell-like traits
via ErbB signaling in prostate cancer cells. Int J Oncol.
48:595–606. 2016. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Hahn WC, Dessain SK, Brooks MW, King JE,
Elenbaas B, Sabatini DM, DeCaprio JA and Weinberg RA: Enumeration
of the simian virus 40 early region elements necessary for human
cell transformation. Mol Cell Biol. 22:2111–2123. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Ren D, Guo L, Wang L, Wu S, Lin
C, Ye L, Zhu J, Li J, Song L, et al: Thymosin beta 10 is a key
regulator of tumorigenesis and metastasis and a novel serum marker
in breast cancer. Breast Cancer Res. 19:152017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y,
Liang Y, Cao L, Li X, Li R, et al: Maintenance of cancer stemness
by miR-196b-5p contributes to chemoresistance of colorectal cancer
cells via activating STAT3 signaling pathway. Oncotarget.
8:49807–49823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheadle C, Vawter MP, Freed WJ and Becker
KG: Analysis of microarray data using Z score transformation. J Mol
Diagn. 5:73–81. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar :
|
|
30
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. Aug 12–2015.Epub ahead of print. View Article : Google Scholar
|
|
31
|
Zhang X, Zhang L, Lin B, Chai X, Li R,
Liao Y, Deng X, Liu Q, Yang W, Cai Y, et al: Phospholipid
phosphatase 4 promotes proliferation and tumorigenesis, and
activates Ca2+-permeable cationic channel in lung
carcinoma cells. Mol Cancer. 16:1472017. View Article : Google Scholar
|
|
32
|
Li X, Liu F, Lin B, Luo H, Liu M, Wu J, Li
C, Li R, Zhang X, Zhou K, et al: miR 150 inhibits proliferation and
tumorigenicity via retarding G1/S phase transition in
nasopharyngeal carcinoma. Int J Oncol. Mar 10–2017.Epub ahead of
print. View Article : Google Scholar
|
|
33
|
Johnstone RW, Ruefli AA and Lowe SW:
Apoptosis: A link between cancer genetics and chemotherapy. Cell.
108:153–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Reed JC: Bcl-2 and the regulation of
programmed cell death. J Cell Biol. 124:1–6. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Reed JC: Dysregulation of apoptosis in
cancer. J Clin Oncol. 17:2941–2953. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ripka S, Neesse A, Riedel J, Bug E, Aigner
A, Poulsom R, Fulda S, Neoptolemos J, Greenhalf W, Barth P, et al:
CUX1: Target of Akt signalling and mediator of resistance to
apoptosis in pancreatic cancer. Gut. 59:1101–1110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Deveraux QL, Takahashi R, Salvesen GS and
Reed JC: X-linked IAP is a direct inhibitor of cell-death
proteases. Nature. 388:300–304. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mendoza-Rodríguez M, Arévalo Romero H,
Fuentes-Pananá EM, Ayala-Sumuano JT and Meza I: IL-1β induces
up-regulation of BIRC3, a gene involved in chemoresistance to
doxorubicin in breast cancer cells. Cancer Lett. 390:39–44. 2017.
View Article : Google Scholar
|
|
39
|
Cillessen SA, Reed JC, Welsh K, Pinilla C,
Houghten R, Hooijberg E, Deurhof J, Castricum KC, Kortman P, Hess
CJ, et al: Small-molecule XIAP antagonist restores caspase-9
mediated apoptosis in XIAP-positive diffuse large B-cell lymphoma
cells. Blood. 111:369–375. 2008. View Article : Google Scholar
|
|
40
|
Fakler M, Loeder S, Vogler M, Schneider K,
Jeremias I, Debatin KM and Fulda S: Small molecule XIAP inhibitors
cooperate with TRAIL to induce apoptosis in childhood acute
leukemia cells and overcome Bcl-2-mediated resistance. Blood.
113:1710–1722. 2009. View Article : Google Scholar
|
|
41
|
Kashkar H, Deggerich A, Seeger JM,
Yazdanpanah B, Wiegmann K, Haubert D, Pongratz C and Krönke M:
NF-kappaB-independent down-regulation of XIAP by bortezomib
sensitizes HL B cells against cytotoxic drugs. Blood.
109:3982–3988. 2007. View Article : Google Scholar
|
|
42
|
Kater AP, Dicker F, Mangiola M, Welsh K,
Houghten R, Ostresh J, Nefzi A, Reed JC, Pinilla C and Kipps TJ:
Inhibitors of XIAP sensitize CD40-activated chronic lymphocytic
leukemia cells to CD95-mediated apoptosis. Blood. 106:1742–1748.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jones A, Danielson KM, Benton MC, Ziegler
O, Shah R, Stubbs RS, Das S and Macartney-Coxson D: miRNA
signatures of insulin resistance in obesity. Obesity (Silver
Spring). 25:1734–1744. 2017. View Article : Google Scholar
|
|
44
|
Xu HX, Wang Y, Zheng DD, Wang T, Pan M,
Shi JH, Zhu JH and Li XF: Differential expression of microRNAs in
calcific aortic stenosis. Clin Lab. 63:1163–1170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tan JR, Tan KS, Yong FL, Armugam A, Wang
CW, Jeyaseelan K and Wong PT: MicroRNAs regulating cluster of
differentiation 46 (CD46) in cardioembolic and non-cardioembolic
stroke. PLoS One. 12:e01721312017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hu S, Bao H, Xu X, Zhou X, Qin W, Zeng C
and Liu Z: Increased miR-374b promotes cell proliferation and the
production of aberrant glycosylated IgA1 in B cells of IgA
nephropathy. FEBS Lett. 589:4019–4025. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang C, Yang C, Chen X, Yao B, Yang C, Zhu
C, Li L, Wang J, Li X, Shao Y, et al: Altered profile of seminal
plasma microRNAs in the molecular diagnosis of male infertility.
Clin Chem. 57:1722–1731. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang K, Wang YW, Wang YY, Song Y, Zhu J,
Si PC and Ma R: Identification of microRNA biomarkers in the blood
of breast cancer patients based on microRNA profiling. Gene.
619:10–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chang JT, Wang F, Chapin W and Huang RS:
Identification of microRNAs as breast cancer prognosis markers
through the cancer genome atlas. PLoS One. 11:e01682842016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hanniford D, Zhong J, Koetz L,
Gaziel-Sovran A, Lackaye DJ, Shang S, Pavlick A, Shapiro R, Berman
R, Darvishian F, et al: A miRNA-based signature detected in primary
melanoma tissue predicts development of brain metastasis. Clin
Cancer Res. 21:4903–4912. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Summerer I, Unger K, Braselmann H,
Schuettrumpf L, Maihoefer C, Baumeister P, Kirchner T, Niyazi M,
Sage E, Specht HM, et al: Circulating microRNAs as prognostic
therapy biomarkers in head and neck cancer patients. Br J Cancer.
113:76–82. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xie J, Tan ZH, Tang X, Mo MS, Liu YP, Gan
RL, Li Y, Zhang L and Li GQ: MiR-374b-5p suppresses RECK expression
and promotes gastric cancer cell invasion and metastasis. World J
Gastroenterol. 20:17439–17447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin
ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al: Global analysis of the
differentially expressed miRNAs of prostate cancer in Chinese
patients. BMC Genomics. 14:7572013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Qian D, Chen K, Deng H, Rao H, Huang H,
Liao Y, Sun X, Lu S, Yuan Z, Xie D, et al: MicroRNA-374b suppresses
proliferation and promotes apoptosis in T-cell lymphoblastic
lymphoma by repressing AKT1 and Wnt-16. Clin Cancer Res.
21:4881–4891. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wu X, Li S, Xu X, Wu S, Chen R, Jiang Q,
Li Y and Xu Y: The potential value of miR-1 and miR-374b as
biomarkers for colorectal cancer. Int J Clin Exp Pathol.
8:2840–2851. 2015.PubMed/NCBI
|
|
56
|
Schreiber R, Mezencev R, Matyunina LV and
McDonald JF: Evidence for the role of microRNA 374b in acquired
cisplatin resistance in pancreatic cancer cells. Cancer Gene Ther.
23:241–245. 2016. View Article : Google Scholar : PubMed/NCBI
|