Introduction
At least 90% of the human genome is actively
transcribed into non-coding RNAs (ncRNAs), whereas <2% of genome
sequences encode proteins (1).
Over the last 10 years, one type of ncRNA, namely microRNA (miRNA),
has been increasingly investigated. miRNAs are ~21–23 nucleotides
in length and are derived from long hairpin precursors. miRNAs are
associated with Argonaute proteins, which post-transcriptionally
regulate target genes, usually by binding to partially
complementary sequences in the 3′-untranslated region (3′-UTR) of
their mRNAs (2). The dysregulation
of this process has been implicated in various biological disorders
and human diseases.
Another category of ncRNAs, long non-coding RNAs
(lncRNAs), has attracted considerable attention. The analysis of
high-throughput RNA sequencing data has provided a robust platform
for investigating transcriptomes and has led to the identification
of a large number of lncRNAs (3).
lncRNAs are defined as endogenous cellular RNA molecules >200
nucleotides in length, which resemble mRNAs but lack coding
potential and show poor sequence conservation between species
(4–6). Previous studies have indicated that
lncRNAs function as signal, decoy, guide, or scaffold RNAs
(7,8). However, the characteristics of
lncRNAs require further investigation. For example, lncRNAs can
guide cis- or trans-acting epigenetic-modifying
complexes to distinct genomic loci and can control the formation
and spread of heterochromatin domains, thereby activating or
repressing transcriptional activity (8). Additionally, lncRNAs possibly
function as adaptors that direct chromatin-remodeling complexes and
transcription factors to specific chromatin loci. lncRNAs can also
act as scaffolds that recruit multiple proteins simultaneously to
coordinate their activities (9).
Studies have shown that the aberrant expression of lncRNAs
including BC200 (10,11),
H19 (12–16), MALAT1 (17–20),
UCA1/CUDR (21–23), HOTAIR (24–26)
and GAS6 AS1 (27) can
cause various types of human cancer. These lncRNAs modulate gene
transcription and translation, and RNA processing and chromatin
remodeling, and have considerable potential as biomarkers, targets
and therapeutic agents (5).
One particular human pseudogene, PTENP1, has
been reported to regulate its corresponding protein-coding mRNA,
transcribed from the phosphatase and tensin homolog gene, by acting
as a decoy for miRNAs that bind to similar sequences in their
respective 3′-UTRs (28).
Furthermore, our previous study showed that the human pseudogene
ψPPM1K may generate endogenous small interfering (si)RNA to
suppress oncogenic cell growth in hepatocellular carcinoma
(29). These studies prompted the
hypothesis that there are further lncRNAs that require
identification. Consequently, the present study performed a
genome-wide bioinformatics screen and subsequent biological
validation experiments to test this hypothesis.
Materials and methods
Bioinformatics
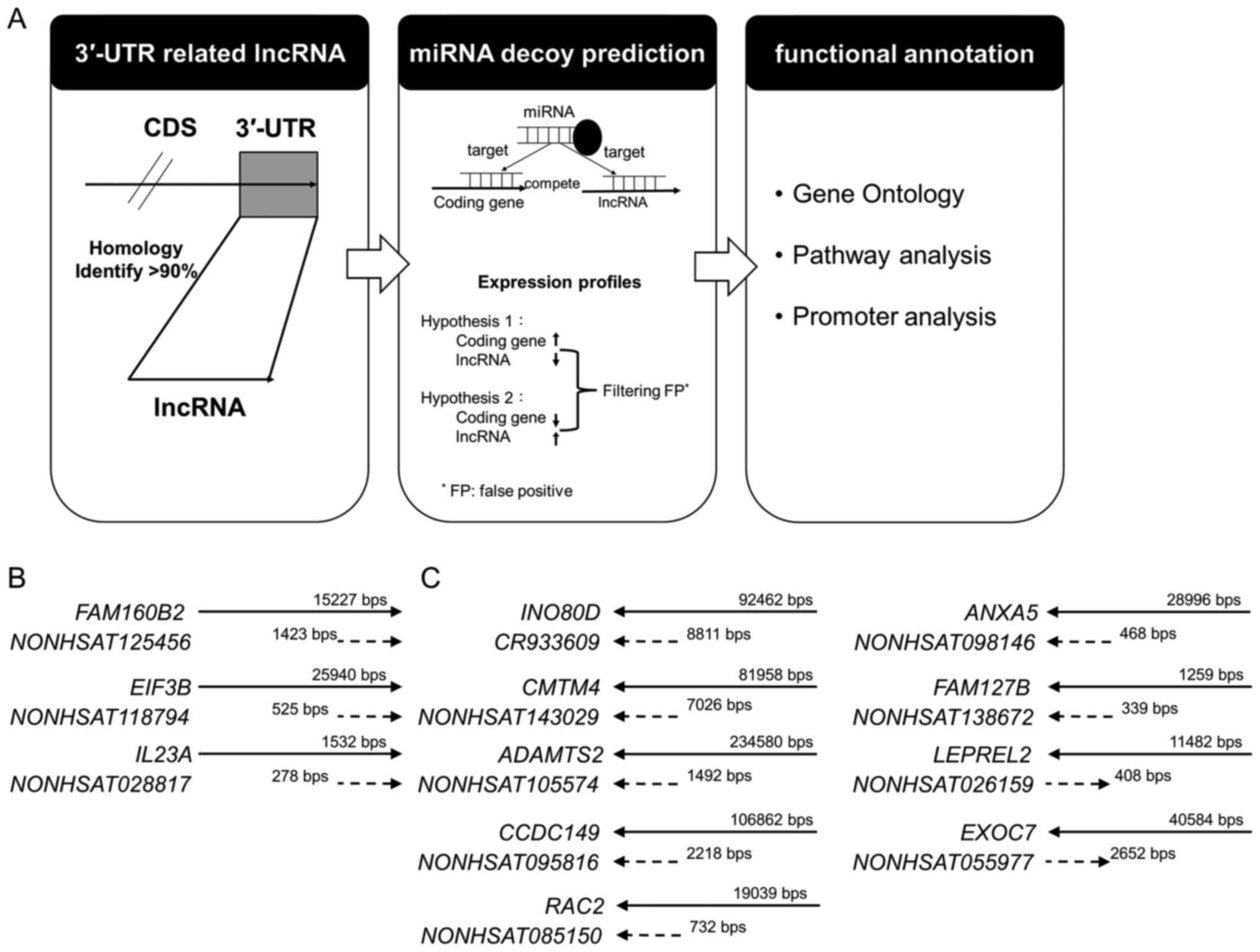
A flowchart for identifying lncRNAs that potentially
act as miRNA decoys is shown in Fig.
1. Over 20,000 human lncRNA transcripts were obtained from
NONCODE v.4 (30). The sequences
of miRNAs and 3′-UTRs of protein-coding genes were collected from
miRBase 20 (31) and UCSC hg19
(32), respectively. Previous
approaches were modified to assess whether lncRNAs behave as miRNA
decoys (29). Briefly, the
sequences of lncRNAs and 3′-UTRs of protein-coding genes were
aligned, and those lncRNAs (the same orientation as coding genes)
showing >90% identity, termed 3′-UTR-like lncRNAs, were
collected. These sequences were then analyzed using miRNA target
prediction tools, miRTar-Base 6.0 (33,34),
TargetScan 7.0 (35–37), miRanda 3.3a (38), and RNAhybrid 2.1.2 (20). The miRNA was selected according to
the score of four databases and the binding energy. The 3′-UTR-like
lncRNAs were all transcribed in the same direction as their
corresponding coding genes and thus had the potential to act as
miRNA decoys through reversed expression between lncRNA and its
corresponding coding gene. The expression profiles of these
lncRNA/coding-gene pairs in 460 squamous cell carcinomas were
obtained from The Cancer Genome Atlas (http://cancergenome.nih.gov/). To identify the
functions of these corresponding coding genes, the Database for
Annotation, Visualization and Integrated Discovery (DAVID)
(39) annotation tool was used to
assign Gene Ontology (GO) terms and to perform pathway analysis and
promoter analysis.
Clinical specimens
Resected primary tumor and adjacent non-tumor tissue
samples were obtained from 83 patients with NSCLC, 30 patients with
kidney cancer, 30 patients with colon cancer, 30 patients with oral
cancer, and 30 patients with liver cancer at either China Medical
University Hospital (Taichung, Taiwan; CMUH) or Changhua Christian
Hospital (Changhua, Taiwan; CCH). The tumor tissues were composed
of 90–100% cancer cells. They were frozen immediately following
surgical resection and then stored in liquid nitrogen until
extraction of either RNA or DNA. All participants provided their
written informed consent to participate in the study. The
institutional review boards of CMUH and CCH approved the study
(CMUH102-RECJ-015 and CCH-IRB-080322) and the consent procedure.
The baseline characteristics of the NSCLC population in the present
study are summarized in Table
I.
 | Table IClinical variables and relative gene
expression of the long non-coding RNA CR933609 in non-small
cell lung cancer. |
Table I
Clinical variables and relative gene
expression of the long non-coding RNA CR933609 in non-small
cell lung cancer.
| Characteristic | CR933609
| P-value | INO80D
| P-value |
|---|
| n | ΔCqa | n | ΔCqa |
|---|
| Age (years) | | | | | | |
| <65 | 33 | 5.35±1.56 | 0.469 | 33 | 5.91±1.31 | 0.129 |
| ≥65 | 50 | 5.07±1.73 | | 50 | 5.38±1.65 | |
| Sex | | | | | | |
| Men | 48 | 5.43±1.79 | 0.105 | 48 | 5.68±1.52 | 0.554 |
| Women | 35 | 4.84±1.42 | | 35 | 5.47±1.56 | |
| Smoking | | | | | | |
| Never | 46 | 4.96±1.45 | 0.197 | 46 | 5.51±1.59 | 0.607 |
|
Current/ex-smoker | 37 | 5.45±1.87 | | 37 | 5.69±1.48 | |
| ECOG PS | | | | | | |
| 0 | 38 | 4.99±1.50 | 0.336 | 38 | 5.36±1.59 | 0.213 |
| 1 | 45 | 5.34±1.78 | | 45 | 5.78±1.48 | |
| Histology | | | | | | |
|
Adenocarcinoma | 51 | 4.66±1.37 | 0.001 | 51 | 5.40±1.42 | 0.165 |
| Squamous cell
carcinoma | 32 | 6.01±1.77 | | 32 | 5.89±1.68 | |
| Grade | | | | | | |
| Well/moderate | 72 | 5.19±1.66 | 0.925 | 72 | 5.53±1.58 | 0.351 |
| Poor
differentiation | 11 | 5.14±1.73 | | 11 | 5.99±1.18 | |
| Stage | | | | | | |
| I+II | 65 | 5.19±1.67 | 0.973 | 65 | 5.51±1.59 | 0.350 |
| III+IV | 18 | 5.17±1.67 | | 18 | 5.89±1.31 | |
Reverse transcription quantitative
polymerase chain reaction (RT qPCR) analysis
Total RNA was extracted using the REzol reagent
(Protech Technology Enterprise Co., Ltd., Taipei, Taiwan) according
to the manufacturer's protocol and treated with RQ1 RNase-Free
DNase (Promega Corporation, Madison, WI, USA). cDNA was generated
from total RNA using the High-Capacity cDNA Reverse Transcription
kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). The RT-qPCR analysis was performed in a total reaction
volume of 10 μl, including 1 μl of cDNA, 5 μl
of LightCycler FastStart DNA Master HybProbe (Roche Diagnostics,
Indianapolis, IN, USA), 0.6 μl of mixed primers (Table II), 1.2 μl of universal
probes (Table II) (Roche
Diagnostics), and 2.2 μl of double-distilled water. A
LightCycler 480 (Roche Diagnostics) was used with the following
program: Preincubation for 10 min at 95°C; followed by 50 cycles of
10 sec at 95°C, 30 sec at 60°C, and 1 sec at 72°C; and finally
cooling to 4°C. Glyceraldehyde-3- phosphate dehydrogenase
(GAPDH) served as an internal reference. The relative RNA
levels were calculated using the comparative quantification cycle
method (39). The RT-qPCR product
was verified through Sanger sequencing to ensure the primers
specificity. Different target sites were verified to ensure primer
specificity. INO80 Complex Subunit D (INO80D): Primer target on
exon 5-exon 6 (ΔCq, 9.92±0.33), primer target on exon 11 used in
the present study (ΔCq, 9.45±0.36), primer target on exon 11 (ΔCq,
9.41±0.27). CR933609: primer used in the present study (ΔCq,
7.38±0.16), primer target on other site (ΔCq, 7.86±0.61).
 | Table IIPrimer sequences. |
Table II
Primer sequences.
| Name | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Probe no. |
|---|
|
NONHSAT143029 |
ggccaggtctgggagataag |
atagcagacaggtggggtgt | 38 |
| CMTM4 |
tgcggcatatgcagtgaa |
gctcggatgtagtcattggtg | 72 |
|
NONHSAT095816 |
cctgcaaatgtccagcacta |
aaacttggctgtgttttccttc | 21 |
| CCDC149 |
ctcacctggactccttcgag |
atccctttgccgtcttcc | 66 |
|
NONHSAT085150 |
ccaatggaaaatctgggttc |
caggtaagtgcagctcagga | 84 |
| RAC2 |
cccagcctcttatgagaacg |
gtgcccaccaggatgatg | 43 |
|
NONHSAT026159 |
accccaccatcctcttcatt |
aacactgtgcctccctgaac | 75 |
| LEPREL2 |
cagggtgctaagctgcttct |
caggtgggtgaaggacagat | 7 |
|
NONHSAT118794 |
gcggttcctctgttgcag |
cagagccagagagcacagg | 4 |
| EIF3B |
ggtggacactgacgagctg |
gacgaagaactcaatggtctcc | 48 |
|
NONHSAT105574 |
tggaagcctttgggaagag |
cttcagcggaagacaggtg | 33 |
| ADAMTS2 |
cctatgactgcctgctggat |
attgctcgttcatggagtagtg | 43 |
|
NONHSAT028817 |
gctgacctatgataaggttgagtattt |
caataaataatcctccccaaactg | 79 |
| IL23A |
cttctccgcttcaaaatcctt |
tgctccatgggcaaagac | 5 |
|
NONHSAT138672 |
ctggcaaccaaatcgaatct |
aaaggcacccatcagagaga | 2 |
| FAM127B |
gaaggtgacgttcctcatcac |
cccggtaatcattgagcag | 12 |
|
NONHSAT055977 |
caccatgtcctcctccctta |
ggtcacctgggacttcagg | 43 |
| EXOC7 |
tgcacaagcagacggaga |
gcaggacagcgtcttctca | 2 |
|
NONHSAT098146 |
aagagctccctgctgtgtg |
tggcatacaaatgcagctaaag | 8 |
| ANXA5 |
tctgtttggcagggatcttc |
tcataaagccgagagggtttc | 21 |
|
NONHSAT125456 |
gagcagtgaatgggatcgtc |
ctgggagttttgcaacagttt | 4 |
|
FAM160B2 |
aggactgctcccacgatg |
agctcctcggtctcagca | 5 |
|
CR933609 |
gacaaaacaaactagtgaagcacct |
tatacaccttgacacggcaga | 47 |
| INO80D |
cctgatgacttacaagattttgattt |
ctcctcagcctcttcggtag | 52 |
| GAPDH |
agccacatcgctcagacac |
gcccaatacgaccaaatcc | 60 |
Northern blots
Total RNA was extracted from cells by using REzol
C&T RNA extraction reagent (Protech Technology Enterprise Co.,
Ltd.). Following extraction, 15 μg of total RNA was
dissolved in a loading buffer containing 10 mM EDTA (pH 8.0), 96%
(v/v) formamide, 0.01% xylene cyanol, and 0.01% bromophenol blue,
heated at 95°C for 5 min, loaded onto a 1% MOPS 3-(N-morpholino)
propanesulfonic acid gel with 2% formamide, separated for 2 h at
150 V, and then transferred onto nitrocellulose membranes (Pall
Corporation, East Hills, NY, USA). The subsequently underwent
cross-linking with UV irradiation for 5 min. The membranes with RNA
blots were prehybridized at 42°C for 3 h using digoxigenin (DIG)
Easy Hyb Granules (Roche Diagnostics) and subjected to
hybridization with a DIG probe for the lncRNA NONHSAT098146
(0.1 μg/ml) overnight at 42°C. Following hybridization, the
membranes were rinsed and then washed sequentially with 2X SSC/0.1%
sodium dodecyl sulfate (SDS) and 0.1X SSC/0.1% SDS at 42°C.
Detection was performed using the DIG Northern Starter kit (Roche
Diagnostics) according to the manufacturer's protocol. The blots
were detected with a chemiluminescent substrate (CDP-Star) and
visualized using the ChemiDoc-It imaging system (UVP, Inc., Upland,
CA, USA).
Cell culture, and siRNA and short hairpin
(sh)RNA transfec tion
Cells from the A549, H441, and H520 NSCLC lines
[Bioresource Collection and Research Center (BCRC), Hsinchu,
Taiwan] were cultured in Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific,) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C
in a 5% CO2 atmosphere. The oligonucleotides used for
three siRNA and INO80D/annexin A5 (ANXA5) shRNA
constructs are shown in Table
III. The INO80D and ANXA5 shRNAs were obtained
from the National Research Program for Biopharmaceuticals (Taipei,
Taiwan). The A549 cells were transfected with the shRNAs (6
μg of INO80D shRNA in 2×10 A549 cells/ml) using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). The efficiency
of transfection was determined using RT-qPCR analysis.
 | Table IIIOligonucleotides for siRNA and shRNA
constructs. |
Table III
Oligonucleotides for siRNA and shRNA
constructs.
| Symbol | Ref ID | Target sequence
(5′-3′) | Note |
|---|
| siRNA1 | FR273090 |
GGGCACGTGTGTGAGAGATATGAAT | Common region with
CR933609 and INO80D |
| siRNA2 | FR273090 |
CAGACATGAGCTGTGTTTATCTATT | Common region with
CR933609 and INO80D |
| siRNA3 | FR273090 |
GGGATCATTTCACCGTGCATATTTC | Common region with
CR933609 and INO80D |
| shRNA1 | NM_017759 |
GCACTTACTTTCAGCAGAAAT |
INO80D-specific |
| shRNA2 | NM_017759 |
CCATTTGCTTTCAATGAGGAA |
INO80D-specific |
| shRNA3 | NM_017759 |
CAACATCGTACTCTGGTGATA |
INO80D-specific |
| shRNA4 | NM_017759 |
CTATTGAATGGGCGTATAGTA |
INO80D-specific |
| shRNA5 | NM_017759 |
GAGCTATTGAATGGGCGTATA |
INO80D-specific |
| NONHSAT098146 | NONHSAT098146 |
GUGUACUUAAUGUUACUAAdTdT | Common region with
NONHSAT098146 and ANXA5 |
| ANXA5-homo-531 | NM_001154 |
GAGCCAUCAAACAAGUUUATT |
ANXA5-specific |
| ANXA5-homo-993 | NM_001154 |
GUGAGAUUGAUCUGUUUAATT |
ANXA5-specific |
| ANXA5-homo-844 | NM_001154 |
CGAGACUUCUGGCAAUUUATT |
ANXA5-specific |
Establishment of CR933609 overexpressing
and NONHSAT098146 overexpressing stable cell lines
The cDNA from the lncRNAs CR933609 and
NONHSAT098146 was cloned in pCDNA3.0 (Invitrogen; Thermo
Fisher Scientific, Inc.). The A549 cells (2×106) were
transfected with CR933609-overexpressing,
NONHSAT098146-overexpressing, and control vectors using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). Following 48 h
of recovery, the transfected cells were cultured in a medium
containing 500 μg/ml G418 for 2–3 weeks. Clones representing
stable cell lines were then selected and expanded to large-scale
cultures. The RNA extracts prepared from these clones were analyzed
for target RNA levels using RT-qPCR analysis.
Cell proliferation assay
To examine whether lncRNA knockdown altered the
viability of NSCLC cells, cell proliferation assays were performed.
Briefly, 10,000 cells from the NSCLC cell lines, stable
transfectants subjected to lncRNA knockdown or parental NSCLC cell
lines, were cultured in DMEM with 10% FBS (according to the BCRC).
After 72 h, cell proliferation and viability were assessed using
the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide
assay. All experiments were performed in triplicate.
Wound healing assay
A wound-healing assay was conducted to assess cell
migration. The basic steps involved creating a 'wound' in a cell
monolayer, capturing images at the beginning and at regular
intervals as cells migrated into the wound, and comparing the
images to quantify the cell migration rate. For each assay,
2×105 cells were plated in 48-well plates (Corning
Costar, Schiphol-Rijk, The Netherlands) with serum-free medium. A
200-μl plastic pipette tip was drawn across the center of
the well to produce a clean wound of ~1 mm in width in the cell
monolayer.
miRNA mediated knockdown of CR933609 and
INO80D
A stable negative control (siCon:
5′-FAM-UUCUCCGAACGUGUCACGUTT-3′) and has-miR-5096 were purchased
from GeneDireX, Inc. (Las Vegas, NV, USA) and transfected into
cells using Lipofectamine RNAiMax (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
efficacy of mRNA knockdown following miRNA transfection for 48 h
was determined using RT-qPCR analysis.
Luciferase reporter assay
Fragments of DNA from the lncRNA CR933609
promoter region were obtained through PCR amplification and cloned
into the pGL3-basic reporter vector (Promega Corporation). The
constructs were transfected into A549 cells (2×106)
using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). The
cells were lysed and assayed for luciferase activity using the
Steady-Glo Luciferase assay system (Promega Corporation) according
to the manufacturer's protocol at 24 h post-transfection.
Statistical analysis
All data are presented as the mean ± standard
deviation or as percentages. Statistical analyses, including the
χ2 test, independent t-test, one-way analysis of
variance followed by Fisher's least significant difference post hoc
test, and paired t-test, were performed using SPSS version 17.0
software (SPSS, Inc., Chicago, IL, USA. P<0.05 was considered to
indicate a statistically significant difference.
Results
Overview of lncRNAs
The bioinformatics analysis (Fig. 1A) showed that 12 human lncRNA
transcripts contained sequences with significant matches to the
3′-UTRs of protein-coding genes (Table IV). These lncRNAs were all
transcribed in the same direction as their corresponding coding
genes and thus have the potential to act as miRNA decoys (Fig. 1B and C). To identify the functions
of these protein-coding genes, the annotation tool DAVID (40) was used to assign GO terms. Certain
GO biological process terms that were significantly associated with
the coding genes, including regulation of translational initiation
[eukaryotic translation initiation factor (EIF)4G2
and EIF3B; P<0.03] and response to chemical stimulus
[ras-related C3 botulinum toxin substrate 2, transporter 2,
ATP-binding cassette, subfamily B (TAP2), RNA binding motif
protein 4, ANXA5 and CKLF-like MARVEL trans-membrane domain
containing 4 (CMTM4); P<0.03], and molecular-function
terms, including receptor binding [interleukin 23 α subunit p19
(IL23A), TAP2, relaxin 2, ANXA5 and
CMTM4; P<0.01] (data not shown).
 | Table IVBasic data of 12 human lncRNA
transcripts with high homology with 3′-UTRs of protein-coding
genes. |
Table IV
Basic data of 12 human lncRNA
transcripts with high homology with 3′-UTRs of protein-coding
genes.
| No. | Transcript ID | Type | Chromosome | Start site | End site | Strand | Exon | Length | 3′-UTR
similarity | ncRNA
similarity |
|---|
| 1 |
CR933609 | lncRNA | 2 | 206,858,445 | 206,867,214 | − | 1 | 8,811 | 0.99 | 1 |
| INO80D | Gene | 2 | 206,858,445 | 206,950,906 | − | 11 | 92,462 | | |
| 2 |
NONHSAT143029 | lncRNA | 16 | 66,648,652 | 66,655,678 | − | 1 | 7,026 | 0.96 | 1 |
| CMTM4 | Gene | 16 | 66,648,653 | 66,730,610 | − | 5 | 81,958 | | |
| 3 |
NONHSAT105574 | ncRNA | 5 | 178,537,853 | 178,540,736 | − | 1 | 1,492 | 0.96 | 1 |
| ADAMTS2 | Gene | 5 | 178,537,852 | 178,772,431 | − | 22 | 234,580 | | |
| 4 |
NONHSAT095816 | lncRNA | 4 | 24,807,739 | 24,809,957 | − | 1 | 2,218 | 0.98 | 1 |
| CCDC149 | Gene | 4 | 24,807,739 | 24,981,826 | − | 13 | 106,862 | | |
| 5 |
NONHSAT125456 | lncRNA | 8 | 21,960,467 | 21,961,890 | + | 1 | 1,423 | 0.98 | 1 |
|
FAM160B2 | Gene | 8 | 21,958,951 | 21,961,891 | + | 4 | 15,227 | | |
| 6 |
NONHSAT085150 | lncRNA | 22 | 37,621,303 | 37,622,035 | − | 1 | 732 | 0.94 | 1 |
| RAC2 | Gene | 22 | 37,621,310 | 37,640,305 | − | 7 | 19,039 | | |
| 7 |
NONHSAT118794 | lncRNA | 7 | 2,419,855 | 2,420,380 | + | 1 | 525 | 0.94 | 0.99 |
| EIF3B | Gene | 7 | 2,394,474 | 2,420,377 | + | 19 | 25,940 | | |
| 8 |
NONHSAT098146 | lncRNA | 4 | 122,589,152 | 122,589,620 | − | 1 | 468 | 0.99 | 1 |
| ANXA5 | Gene | 4 | 122,589,152 | 122,618,147 | − | 13 | 28,996 | | |
| 9 |
NONHSAT138672 | lncRNA | X | 134,184,962 | 134,185,301 | − | 1 | 339 | 0.9 | 1 |
| FAM127B | Gene | X | 134,184,963 | 134,186,221 | − | 2 | 1,259 | | |
| 10 |
NONHSAT028817 | lncRNA | 12 | 56,733,915 | 56,734,193 | + | 1 | 278 | 0.91 | 1 |
| IL23A | Gene | 12 | 56,732,663 | 56,734,194 | + | 4 | 1,532 | | |
| 11 |
NONHSAT026159 | lncRNA | 12 | 6,948,604 | 6,949,010 | + | 1 | 408 | 0.99 | 1 |
| LEPREL2 | Gene | 12 | 6,937,538 | 6,949,018 | − | 5 | 11,482 | | |
| 12 |
NONHSAT055977 | lncRNA | 17 | 74,097,193 | 74,099,845 | + | 1 | 2,652 | 0.99 | 1 |
| EXOC7 | Gene | 17 | 74,077,086 | 74,099,868 | − | 20 | 40,584 | | |
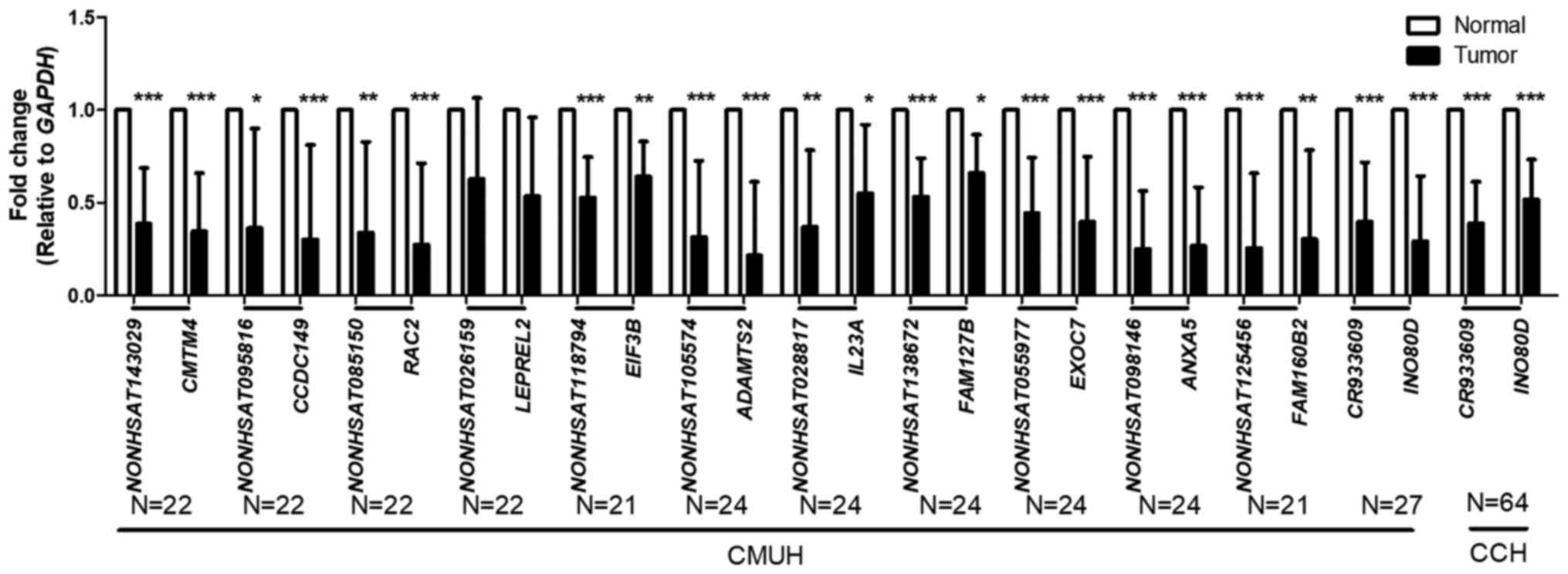
Expression profiles of lncRNAs and
parental coding genes in NSCLC
The average (Fig.
2) expression levels of lncRNAs and parental genes were
quantitatively analyzed through RT-qPCR analysis. The results
showed that the expression levels of lncRNAs and parental genes
were lower in cancer tissues than in non-tumor tissues. These
results were summarized from observations at either CMUH or CCH.
The expression levels of lncRNAs in a small number of samples were
undetectable, which is the reason for the difference in the number
of samples. The expression levels of INO80D and the
3′-UTR-like lncRNA CR933609 were significantly lower in the
cancerous tissues (P<0.01). The expression levels of
ANXA5 and the 3′-UTR-like lncRNA NONHSAT098146 showed
the most marked change. INO80D is involved in the
transcription-coupled nucleotide excision repair and DNA
double-strand break (DSB) repair pathways. In addition, DNA DSBs
contribute to the genomic instability driving cancer development.
Therefore, INO80D and CR933609 were selected for the
subsequent investigations. The INO80D and the 3′-UTR-like
lncRNA CR933609 pair, and the ANXA5 and the
3′-UTR-like lncRNA NONHSAT098146 pair were selected for
further examination.
Association between lncRNAs and parental
coding genes
INO80 Complex Subunit D (INO80D), a
protein-coding gene located on chromosome 2q33.3, is involved in
transcriptional regulation and possibly DNA repair (41). The computational results showed
that the sequence of the lncRNA CR933609, which maps to the
same region of chromosome 2, is 99% identical to the region of
8,762 nucleotides in the 3′-UTR of INO80D (Fig. 3A, top). This result suggested that
the CR933609 transcriptional unit is embedded in the
INO80D gene.
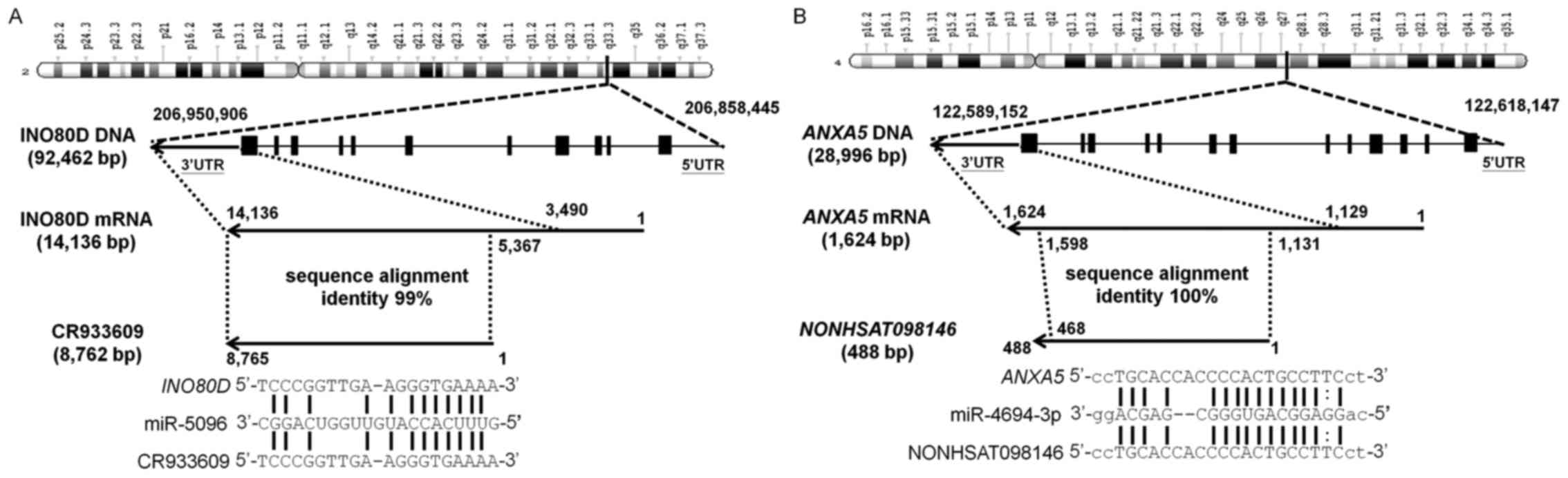 | Figure 3Associations between INO80D
and CR933609, and between ANXA5 and
NONHSAT098146. (A) INO80D is a protein-coding gene
located on chromosome 2q33.3. Computational results showed that the
lncRNA CR933609 matches the 3′-UTR of INO80D with 99%
identity. To examine whether CR933609 acts as an miRNA
decoy, four miRNA target prediction tools were used to investigate
potential miRNA interactions with INO80D and
CR933609. miRNA-5096 was found to be a likely candidate for
interaction with the common regions of CR933609 and
INO80D. (B) ANXA5, a protein-coding gene located on
chromosome 4q27, is implicated in membrane-related events along
exocytotic and endocytotic pathways. The computational results
showed that the sequence of the lncRNA NONHSAT098146, which
maps to the same region of chromosome 4, was 100% identical to a
468-nucleotide region in the 3′-UTR of ANXA5. To ascertain
whether NONHSAT098146 acts as an miRNA decoy, four miRNA
target prediction tools were used to investigate potential miRNA
interactions with ANXA5 and NONHSAT098146.
miRNA-4694-3p was found to be a likely candidate for interaction
with the common regions of ANXA5 and NONHSAT098146.
miRNA/miR, microRNA; lncRNA, long non-coding RNA; INO80D,
INO80 complex subunit D; ANXA5, annexin A5. |
ANXA5, a protein-coding gene located on
chromosome 4q27, is implicated in membrane-related events along
exocytotic and endocytotic pathways (42). The computational results showed
that the sequence of the lncRNA NONHSAT098146, which maps to
the same region of chromosome 4, is 100% identical to the region of
468 nucleotides in the 3′-UTR of ANXA5 (Fig. 3B, top). This result suggested that
the NONHSAT098146 transcriptional unit is embedded in the
ANXA5 gene.
To examine whether CR933609 and
NONHSAT098146 act as miRNA decoys, and thereby modulate
miRNA regulation in INO80D and ANXA5, four miRNA
target prediction tools were used to investigate possible target
sequences within the INO80D and CR933609 transcripts
and the ANXA5 and NONHSAT098146 transcripts. The
results indicated that miRNA-5096 interacts with a sequence within
the common regions of CR933609 and INO80D (Fig. 3A, bottom), and miRNA-4694-3p
potentially interacts with a sequence within the common regions of
NONHSAT098146 and ANXA5 (Fig. 3B, bottom). The microRNA,
hsa-mir-5096, was selected as a candidate according to the score of
four databases and the lowest binding energy (−16.34 kCal/mol), and
so was miRNA-4694-3p (-34.49 kCal/mol).
Expression levels of selected lncRNAs and
parental coding genes in five types of solid tumor
To determine the expression levels of the selected
lncRNAs and parental genes in cancer, RT-qPCR analysis was used to
measure the mRNA expression levels of CR933609 and
INO80D in five types of solid tumor. The CR933609 and
INO80D pair had significantly lower mRNA expression levels
in the tumors of lung, colon and kidney cancer, compared with their
adjacent non-tumor tissues (Fig.
4A). The NONHSAT098146 and ANXA5 pair exhibited
significantly lower mRNA expression levels in the tumors of lung,
colon, and kidney cancer, compared with their adjacent non-tumor
tissues (Fig. 4B). Notably, the
NONHSAT098146 and ANXA5 pair had significantly higher
mRNA expression levels in mucosal tumors. It was also found that,
in individual tumors and in A549 NSCLC cells, the expression levels
of CR933609 were higher than the expression levels of
INO80D (Fig. 4C). However,
the expression levels of NONHSAT098146 were lower than the
expression levels of ANXA5 (Fig. 4D). As lung cancer is the leading
cause of cancer-associated mortality worldwide, with NSCLC being
the most prominent subgroup accounting for >80% of all lung
cancer cases (43), the following
experiments focused on NSCLC. Among types of NSCLC, the expression
levels of CR933609 were significantly lower in squamous cell
carcinoma than in adenocarcinoma (P=0.001; Table I).
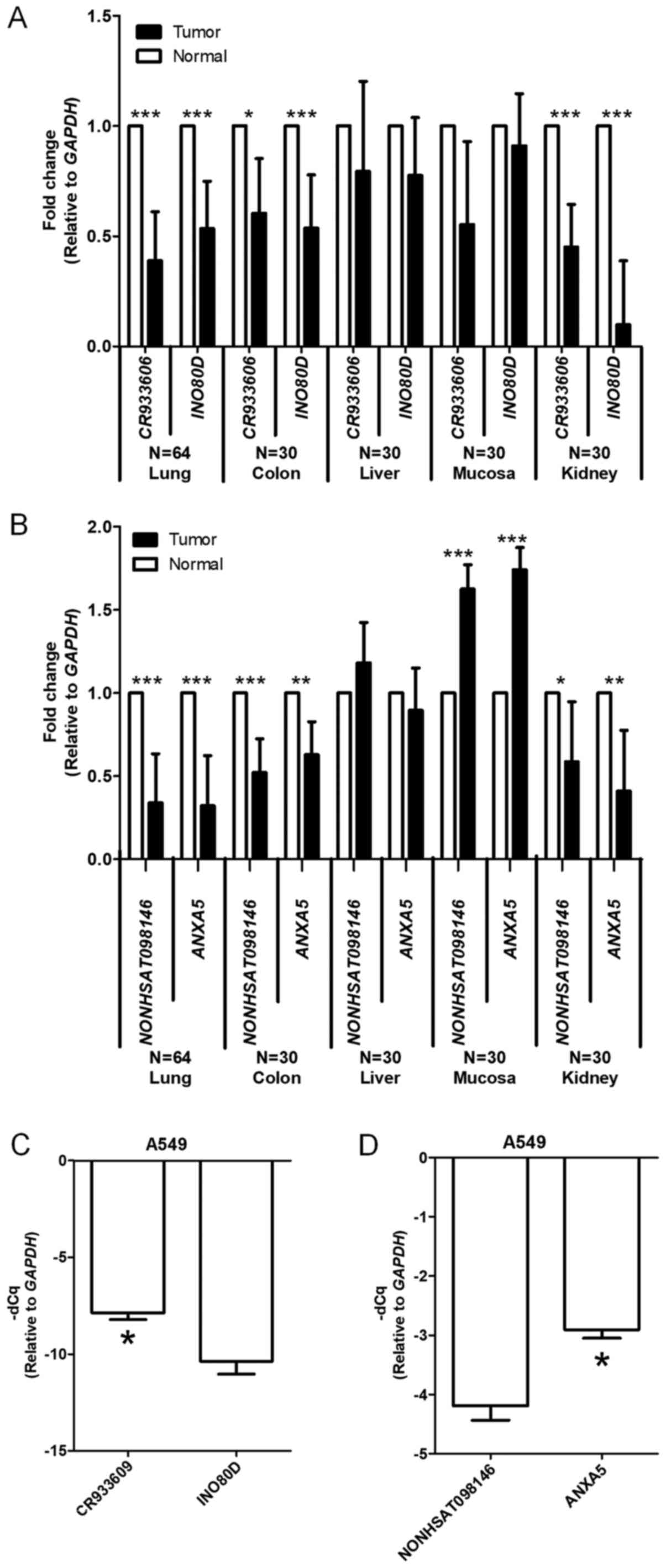 | Figure 4Gene expression levels of
CR933609 and INO80D, and NONHSAT098146 and
ANXA5 are similar between various solid tumors and A549
cells. (A) Expression levels of CR933609 and INO80D
genes were quantitatively analyzed by using RT-qPCR analysis. The
internal control was GAPDH. The white bar denotes the normal
region, the black bar denotes the tumor region. Error bars
represent the standard deviation. Student's t-test was used to
evaluate differences between the groups. *P<0.05 and
***P<0.001, compared with normal region. The results
are summarized from observations at CMUH. (B) Gene expression
levels of NONHSAT098146 and ANXA5 were quantitatively
analyzed using RT-qPCR analysis. The internal control was
GAPDH. The white bar denotes the normal region and the black
bar denotes the tumor region. Error bars represent the standard
deviation. Student's t-test was used to evaluate differences
between the groups. *P<0.05, **P<0.01
and ***P<0.001, compared with normal region. Results
are summarized from observations at CMUH. (C) Expression levels of
CR933609 and INO80D genes were quantitatively
analyzed using RT-qPCR analysis. The internal control was
GAPDH. Error bars represent the standard deviation.
Student's t-test was used to evaluate differences between the
groups. *P<0.05, compared with the INO80D
group. The results are summarized from observations of three
independent experiments. (D) Gene expression levels of
NONHSAT098146 and ANXA5 were quantitatively analyzed
using RT-qPCR analysis. The internal control was GAPDH.
Error bars represent the standard deviation. Student's t-test was
used to evaluate differences between the groups.
*P<0.05, compared with the ANXA5 group. The
results are summarized from observations of three independent
experiments. RT-qPCR, reverse transcription-quantitative polymerase
chain reaction analysis; INO80D, INO80 complex subunit D;
ANXA5, annexin A5; GAPDH, glyceraldehyde-3-phosphate
dehydrogenase; CMUH, China Medical University Hospital; CCH,
Changhua Christian Hospital. |
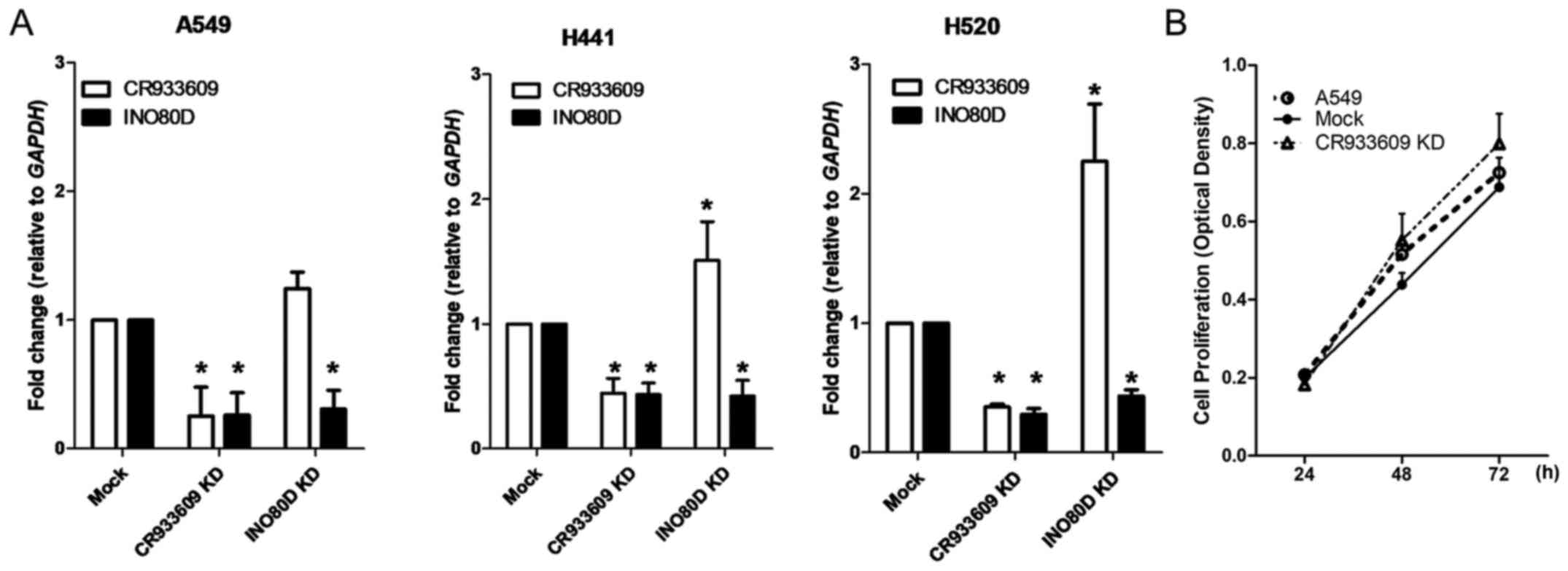
Associations between the expression of
CR933609 and INO80D, and expression of NONHSAT098146 and ANXA5 in
NSCLC cells
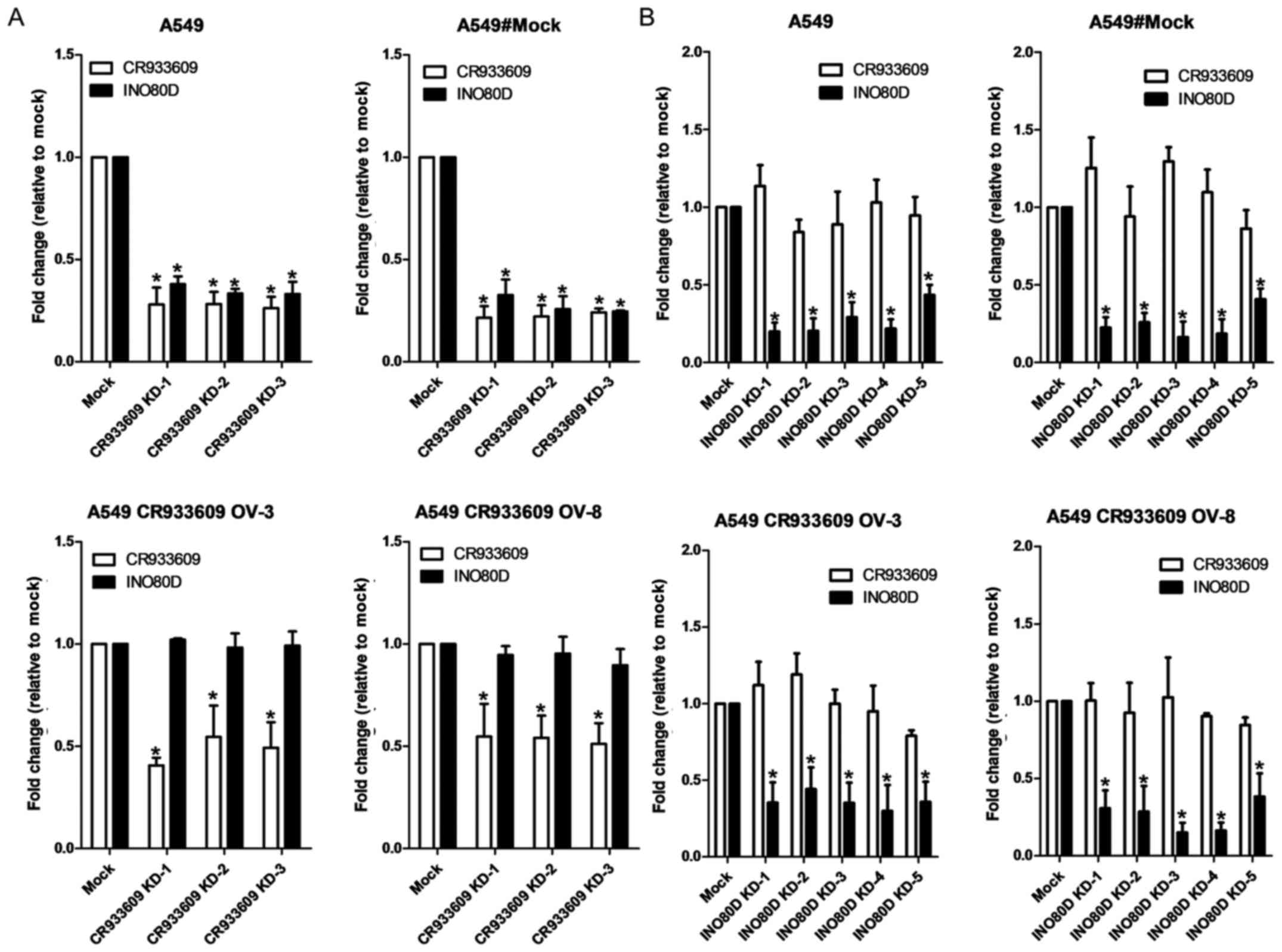
To examine the association between CR933609
and INO80D, cells from three NSCLC lines, namely A549, H441
and H520 cells, were transfected with siRNAs (representing their
common regions) and shRNAs (specifically targeted to
INO80D). The siRNAs downregulated CR933609 and
INO80D in NSCLC cells (Fig.
5A), whereas the cell proliferation assay revealed that the
siRNA-transfected cells showed a marginally higher proliferation
rate, compared with that in the control (Fig. 5B). Notably, the downregulation of
INO80D resulted in decreased expression levels only in
INO80D (Fig. 5A). These
results suggested that CR933609 and INO80D have
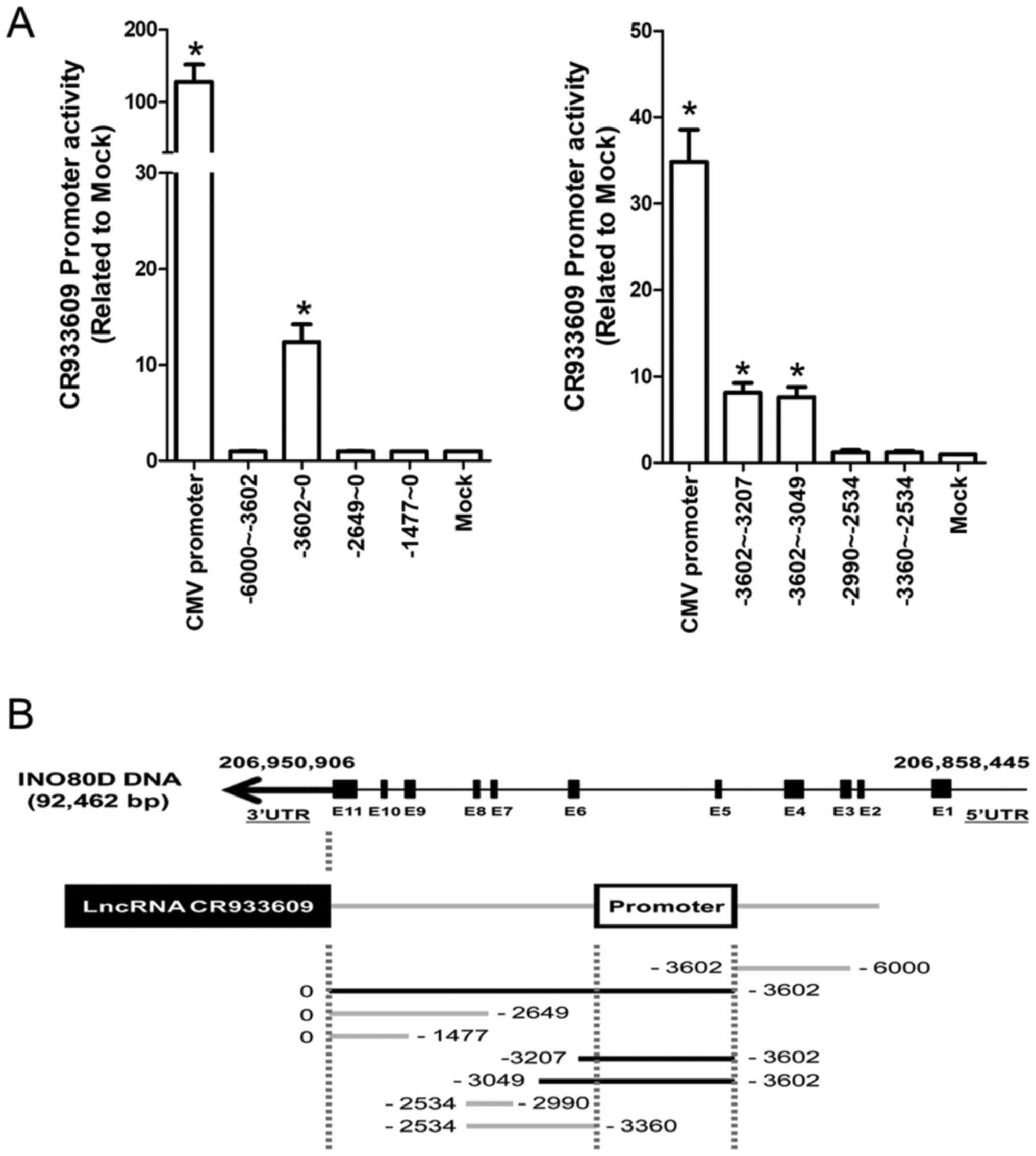
distinct transcripts. To determine whether CR933609 and
INO80D have different promoters, a luciferase reporter assay
was used to identify the promoter of CR933609. As shown in
Fig. 6A, a significant increase in
luciferase activity was detected in constructs containing DNA
fragments extending between −3602 and 0, −3602 and −3207, and −3602
and −3049 nucleotides upstream of the CR933609 transcription
initiation site, indicating a possible location of the
CR933609 promoter in the region between −3602 and −3360
nucleotides (Fig. 6B). Although
the CR933609 sequence is embedded within the INO80D
gene, it is transcribed from a different promoter.
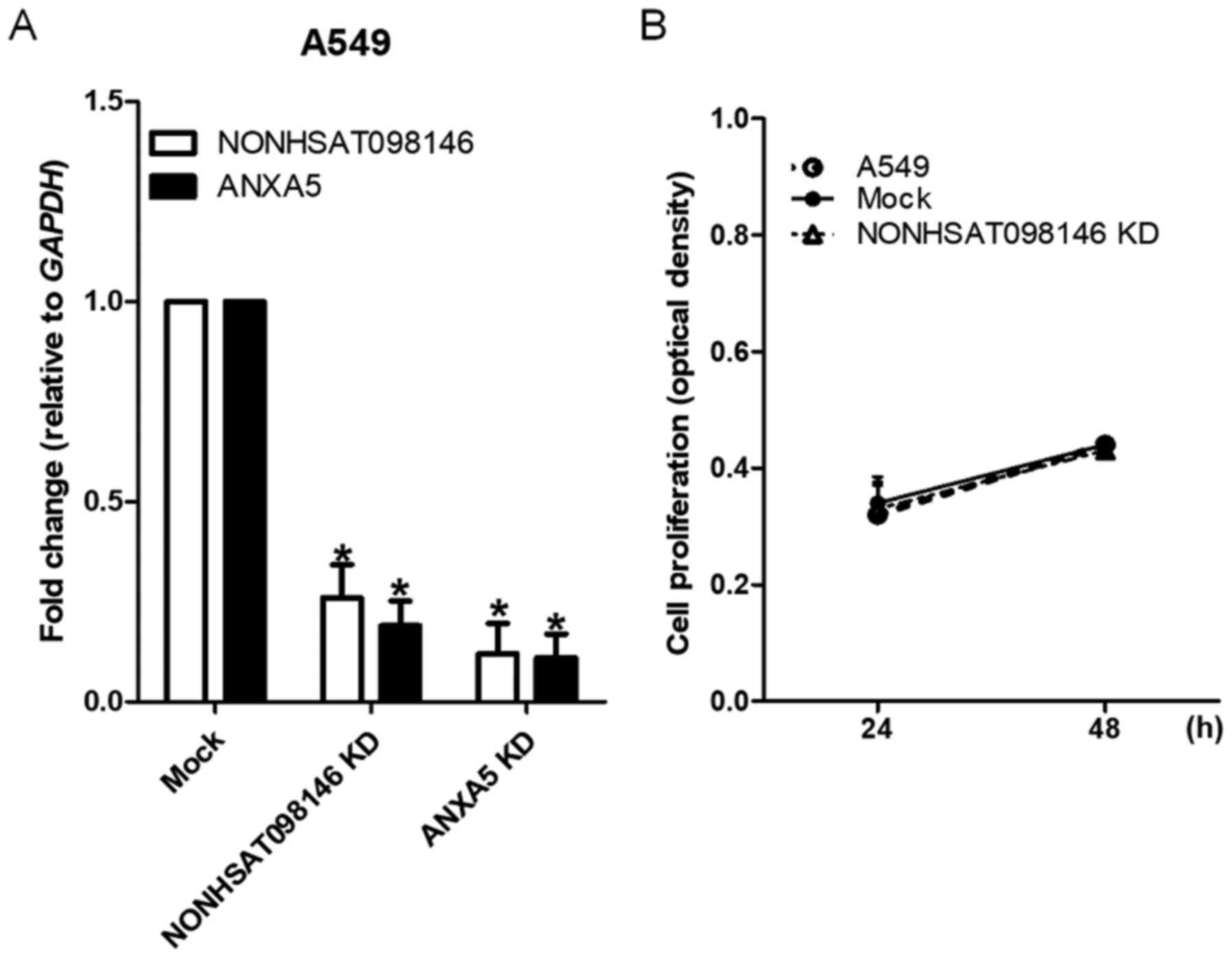
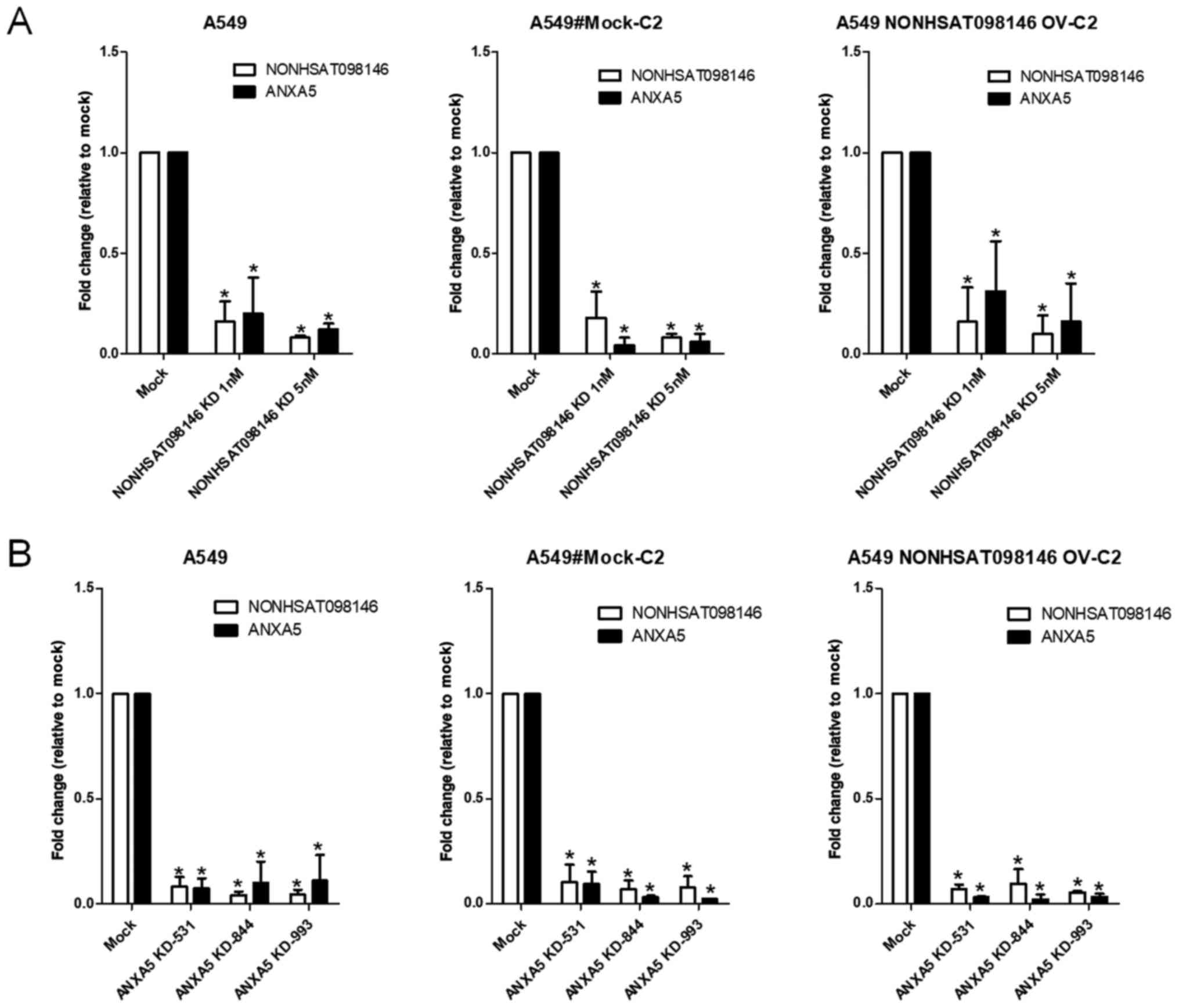
To examine the association between
NONHSAT098146 and ANXA5, the A549 cells were
transfected with artificial siRNAs (representing their common
regions) and shRNAs (specifically targeted to ANXA5).
NONHSAT098146 and ANXA5 were underexpressed in
NONHSAT098146 knockdown and ANXA5 knockdown cells,
respectively (Fig. 7A). Cell
proliferation was stable in the NONHSAT098146-knockdown
cells (Fig. 7B). These results
indicated that NONHSAT098146 and ANXA5 have the same
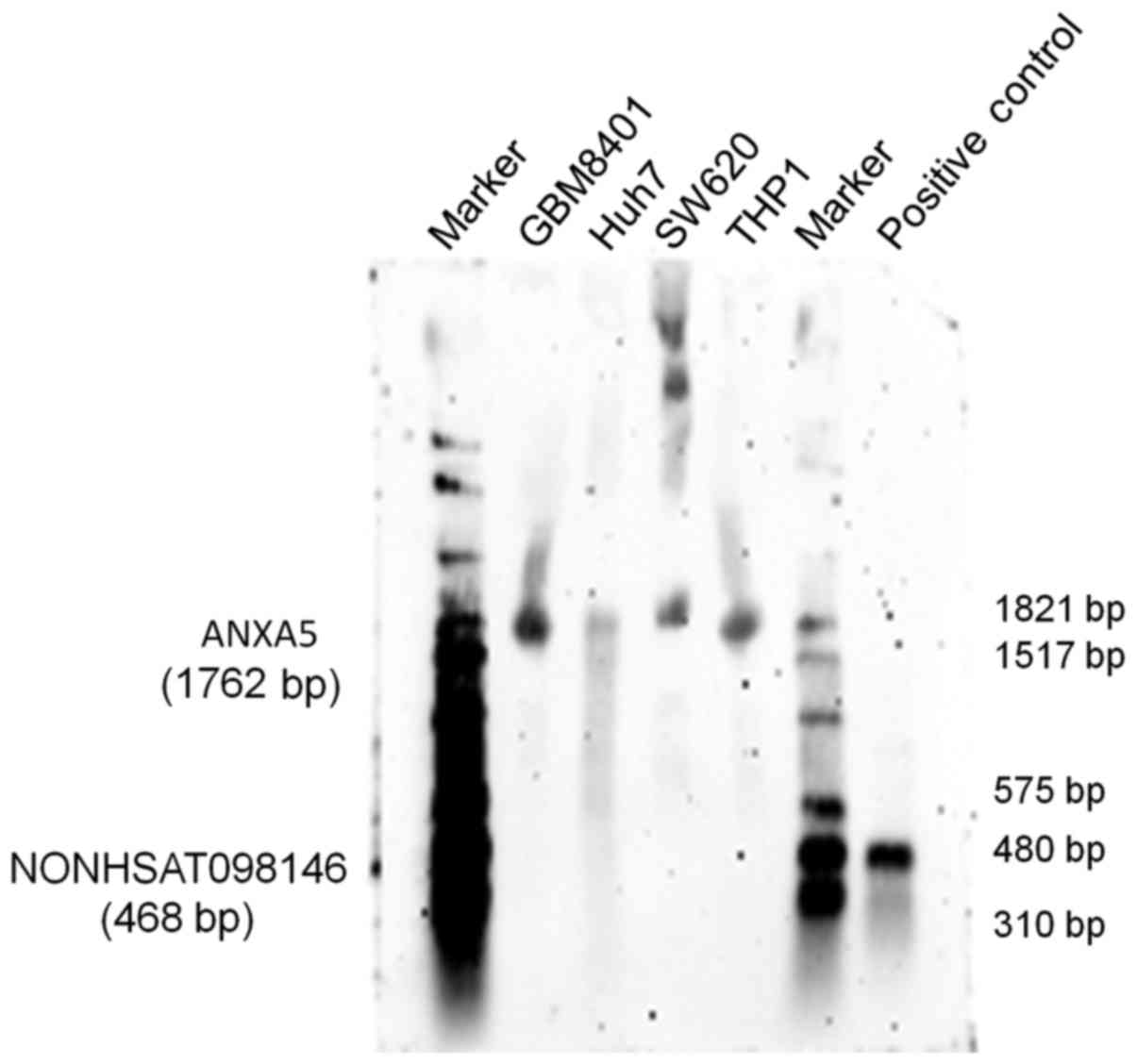
transcripts. The RNA expression of the lncRNA NONHSAT098146
was identified through northern blotting. The results showed that
NONHSAT098146 hybridization signals [468 base pairs (bp)]
were invisible, whereas ANXA5 hybridization signals (1,762
bp) were present in the cells (Fig.
8). Therefore, the lncRNA NONHSAT098146 may be an
artificial gene.
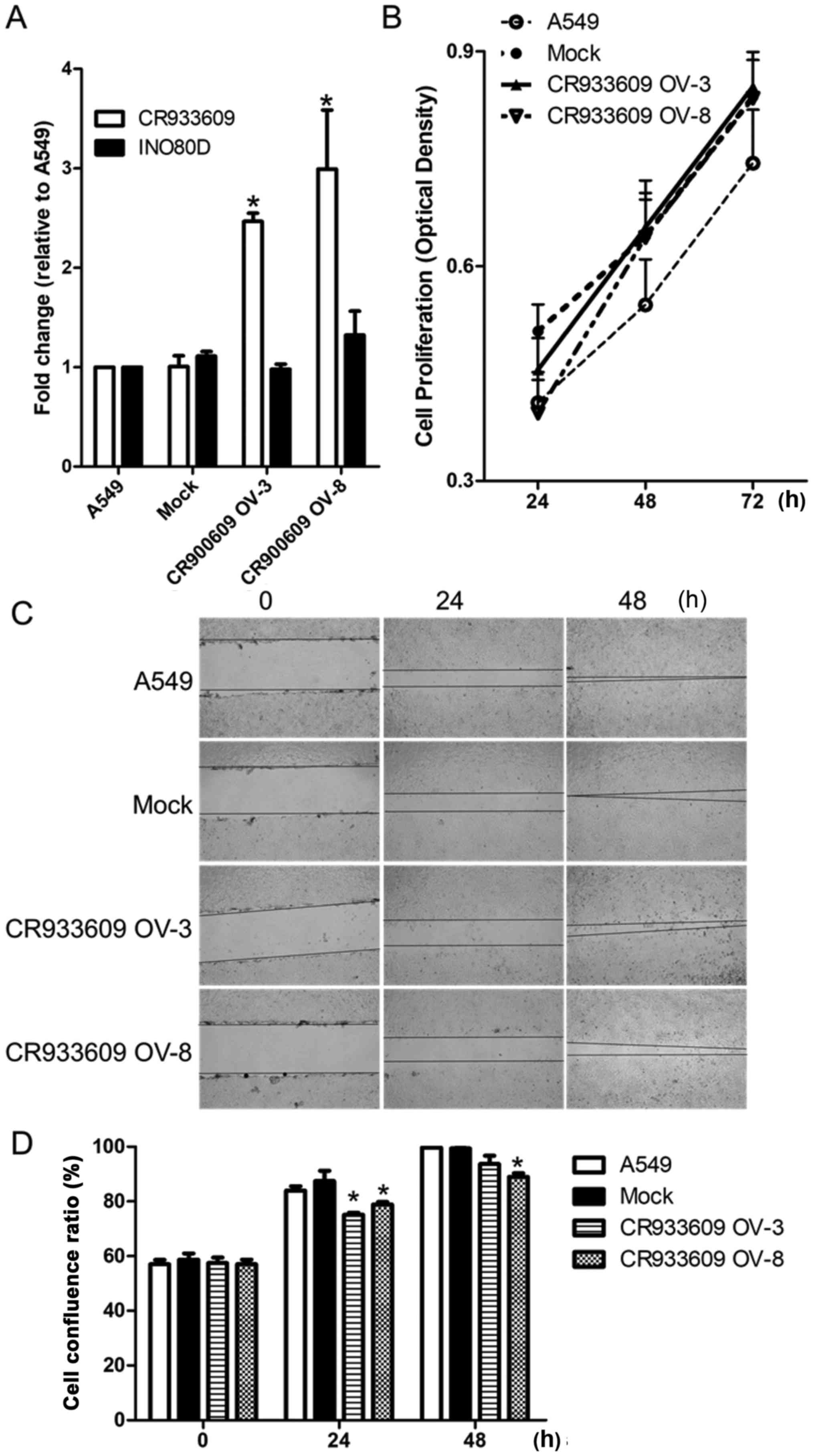
Established lncRNA overexpressing stable
cell lines
To examine the effect of the overexpression of
lncRNA CR933609, the A549 cells were transfected with a
CR933609 recombinant plasmid vector, and stable and control
clones were isolated. The gene expression of CR933609
increased by >2.5-fold in the CR933609-overexpressing
cells, compared the vector control cells, but no change in
INO80D was observed (Fig.
9A). No significant change in cell proliferation was observed
in the CR933609-overexpressing cells (Fig. 9B). It was found that cell migration
was significantly decreased in the CR933609-overexpressing
cells (Fig. 9C and D). Therefore,
CR933609 may act as a tumor suppressor gene.
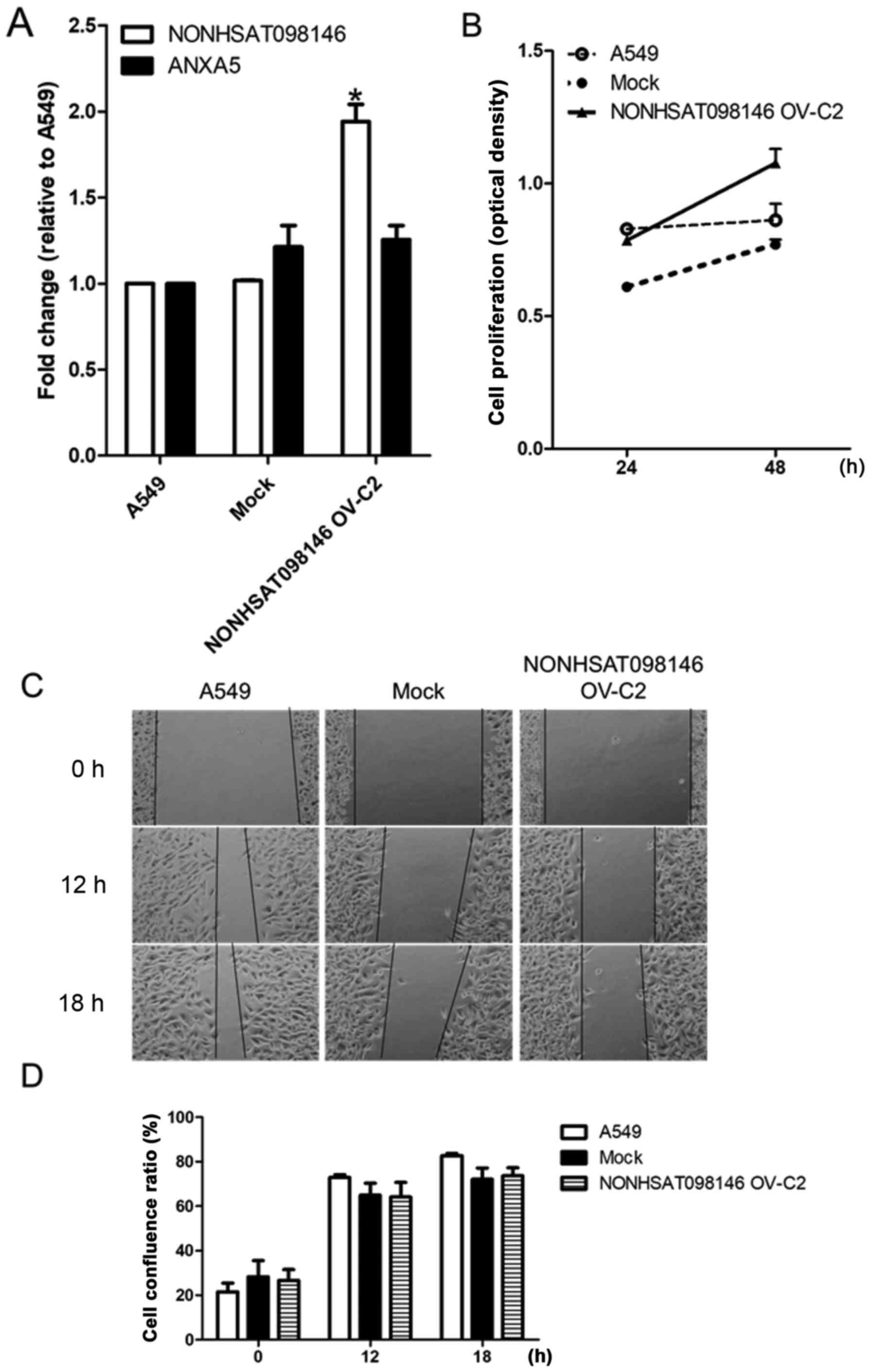
NONHSAT098146-overexpressing stable cell
lines were established to ascertain the effect of overexpression.
The expression of NONHSAT098146 increased by 2-fold in
NONHSAT098146-overexpressing cells, compared with the vector
control cells, but no change in ANXA5 was observed (Fig. 10A). No significant differences in
proliferation or migration were observed in the
NONHSAT098146-overexpressing cells (Fig. 10Bx–D). These established cell
lines were used in the subsequent experiment.
lncRNA CR933609 acts as a repressor decoy
to prevent downregulation of expression levels of INO80D
As shown in Fig.
11A (top), the expression levels of CR933609 and
INO80D were decreased in the A549 and mock (A549 transfected
entry vector) cells transfected with siRNAs targeting
CR936609, in accordance with the results in Fig. 5A. Only the expression levels of
CR933609 were decreased in the
CR933609-overexpressing cells transfected with siRNAs
(Fig. 11A, bottom). The
overexpression of CR933609 may counteract the inhibitory
effects of siRNAs on the expression levels of INO80D. There
were five target sites for INO80D knockdown, which decreased
the expression of INO80D only; the expression of
CR933609 remained stable (Fig.
11B). The expression levels of NONHSAT098146 and
ANXA5 were decreased in the NONHSAT098146-knockdown
(Fig. 12A) and in the
ANXA5-knockdown (Fig. 12B)
cells overexpressing NONHSAT098146. These results indicated
that the lncRNA CR933609 acts as a repressor decoy to
prevent downregulation of the expression of INO80D, whereas
NONHSAT098146 was not a repressor decoy.
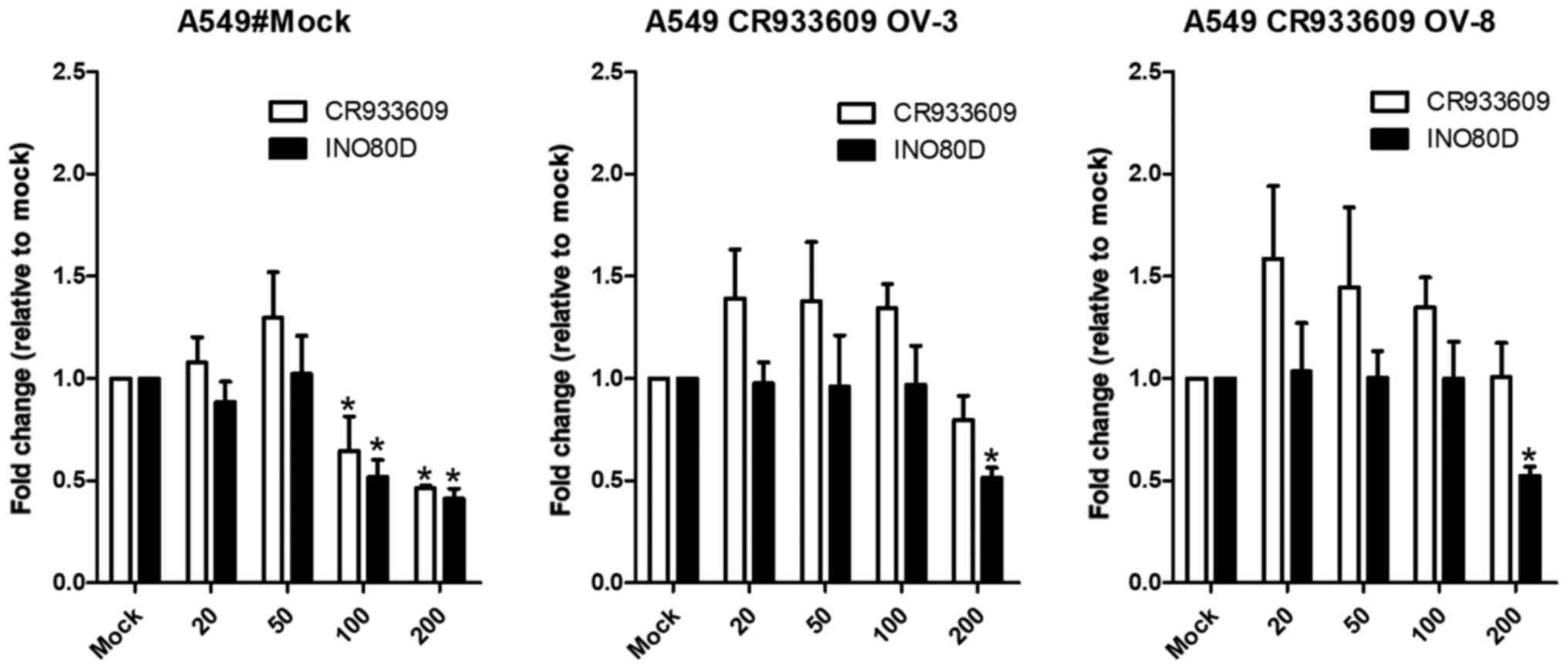
Overexpression of lncR NA CR933609
prevents downregulation of the expression levels of INO80D in endog
enous miRNA 5096 treatment
As the computational results suggested that
miRNA-5096 may target the INO80D and CR933609
transcripts at their common sequences (Fig. 3A, bottom), the A549 and
CR933609-overexpressing A549 cells were transfected with
miRNA-5096. The results indicated that miRNA-5096 at a
concentration of 100 nM downregulated the expression levels of
INO80D and CR933609 in the A549 cells (Fig. 13, left), but not in the
CR933609-overexpressing A549 cells (Fig. 13, middle and right). However,
miRNA-5096 at a concentration of 200 nM downregulated the
expression levels of INO80D and CR933609 in the A549
cells (Fig. 13, left), but
decreased only the expression levels of INO80D in the
CR933609-overexpressing A549 cells (Fig. 13, middle and right). Accordingly,
these results suggested that CR933609 acts as a repressor
decoy to protect INO80D from downregulation by miRNAs.
Discussion
In the present study, a bioinformatics pipeline was
constructed to identify lncRNA sequences that were similar to the
3′-UTRs of protein-coding genes, and to confirm that these lncRNA
and coding gene pairs contain the same miRNA target sites. The
expression levels of CR933609 and INO80D, and those
of NONHSAT098146 and ANXA5 were significantly
decreased in NSCLC. In addition, the expression levels of
CR933609 and INO80D were significantly decreased in
colon and kidney cancer, and the expression levels of
NONHSAT098146 and ANXA5 were significantly decreased
in colon, mucosal, and kidney cancer. The expression levels of
CR933609 and INO80D were decreased in
CR933609-knockdown NSCLC cells, whereas only the expression
levels of INO80D were decreased in INO80D-knockdown
cells. The expression levels of NONHSAT098146 and
ANXA5 were decreased in NONHSAT098146- and
ANXA5-knockdown NSCLC cells. It was found that there were
independent promoters in CR933609 and INO80D,
however, NONHSAT098146 hybridization signals were
undetectable. Furthermore, it was found that INO80D was
downregulated by endogenous miRNA-5096 in A549 cells but not in
CR933609-overexpressing A549 cells. Therefore, the lncRNA
CR933609 may act as a decoy to protect INO80D from
downregulation by miRNA-5096 in NSCLC cells. Thus, a protocol was
established to identify novel lncRNAs in 3′-UTRs.
The interaction of RNA-binding proteins and miRNAs
with the 3′-UTRs of mRNAs is known to affect the expression of
eukaryotic genes by regulating mRNA translation, stability, and
subcellular localization (44,45).
The present study tested the hypothesis that, in addition to
miRNAs, lncRNAs are involved in posttranscriptional gene
regulation. A genome-wide computational pipeline identified 12
human lncRNA transcripts that showed high sequence matches with the
3′-UTRs of protein-coding mRNAs, suggesting that lncRNAs may be
miRNA decoys (Fig. 1B and C). The
protein-coding genes matching this set of 12 lncRNAs were involved
in regulation of translational initiation and receptor binding, and
their promoters contained similar transcription factor binding
sites. Two of these lncRNA/coding-gene pairs, CR933609 and
INO80D, and NONHSAT098146 and ANXA5 were
investigated. Notably, the transcription unit of the lncRNA
CR933609 was found to be embedded within the 3′-UTR of the
INO80D gene (Fig. 3A),
whereas CR933609 had a distinct promoter region (Fig. 6). Furthermore, the lncRNA
NONHSAT098146 was identified as an artificial gene (Figs. 7 and 8). The results indicated that certain
lncRNAs embedded in coding genes may not be transcripts, and a
distinct promoter region requires identification.
To examine the biological functions of
CR933609 and INO80D, RNA interference experiments
were performed using artificial siRNAs (targeting sequences common
to both transcripts) and shRNAs (specific for INO80D). The
lncRNA CR933609 was expressed at significantly lower levels
in three types of tumor tissue, compared with adjacent normal
tissues (Fig. 4A). In addition,
cell migration decreased in A549 clonal lines overexpressing
CR933609 (Fig. 9).
Therefore, CR933609 may act as a tumor suppressor gene in
A549 cells. Notably, the expression levels of INO80D were
downregulated in the CR933609- and INO80D-knockdown
cells, whereas the lncRNA CR933609 was downregulated in
CR933609-knockdown NSCLC cells (Fig. 5A). However, no significant changes
were observed in the expression level of INO80D following
CR933609 knockdown in CR933609-overexpressing cells
(Fig. 11A). Additionally,
endogenous miRNA-5096 downregulated CR933609 and
INO80D in A549 cells, but a different pattern was observed
in CR933609-overexpressing A549 cells (Fig. 13). Previous data have shown that
the expression level of miRNA-5096 was reduced, whereas the
expression level of miRNA-4694-3p was not altered significantly in
lung cancer (46). CR933609
acted as a decoy to prevent the downregulation of
INO80D.
Chromatin remodeling is the dynamic modification of
chromatin architecture to allow condensed genomic DNA access to the
regulatory transcription machinery, thereby controlling gene
expression (47). It also provides
fine-tuning at crucial cell growth and division steps to suppress
tumor development. Targeting of the chromatin remodeling pathways
is currently emerging as a major therapeutic strategy for the
treatment of several types of cancer. INO80D encodes subunit
D of the INO80 chromatin remodeling complex and is involved in
transcriptional regulation, DNA replication and DNA repair, and may
also be crucial in tumorigenesis (48). The data in the present study
suggested that the lncRNA CR933609 regulated the mRNA
stability of INO80D by competing for transcriptional
enhancers or repressors. Therefore, the lncRNA CR933609 is
likely to be involved in chromatin remodeling through the
regulation of INO80D. Furthermore, the present study found
that the lncRNA CR933609 may act as a tumor suppressor by
regulating NSCLC cell migration (Fig.
9), and may serve as a repressor decoy to prevent the
downregulation of INO80D (Fig.
11). Chromatin remodeling is one of the most important
mechanisms for DNA repair, particularly in DNA DSBs. DNA DSBs
result in cell cycle checkpoint arrest and apoptosis. This response
can suspend tumorigenesis. The appropriate regulation of the
mechanisms involved in DNA DSBs, including chromatin remodeling, is
critical (49). Therefore, the
lncRNA CR933609, which acts as a decoy to protect
INO80D, a chromatin remodeling regulator, is essential for
cell survival.
3′-UTRs can be independently expressed as
developmentally regulated lncRNAs, which further blurs the
distinction between coding and non-coding RNAs (50). This serves as a reminder that the
traditional concept of the gene is becoming increasingly outmoded
(51,52). However, compared with coding RNA
and miRNA, significant gaps in the understanding of lncRNA function
remain. To examine whether other lncRNAs, which may likewise be
embedded within protein-coding genes, also function as repressor
decoys, similar experiments on the remaining 11 lncRNA/coding-gene
pairs examined in in silico analyses can be performed. When
the NONCODE database updated to the second edition, NONHSAT143029
and NONHSAT055977 had been removed from the database (30). This indicated that the novel lncRNA
from the database required confirmation. The present study
established a protocol to identify novel lncRNAs in the 3′-UTR and
confirmed the existence of novel lncRNAs using promoter activity or
northern blotting.
As there are defects in all of the bioinformatics
analytical methods, a protocol was established in the present study
to verify the results of the analysis. It was also found that the
lncRNAs published in databases may not exist. It is necessary to
detect the existence of an lncRNA prior to investigating its
function. This may be the reason that certain lncRNAs were removed
from the database a number of years following publication.
In conclusion, the present first constructed a
bioinformatics pipeline to investigate genome-wide 3′-UTR lncRNAs
and then identified their clinical significance. The results
indicated that the lncRNA CR933609 may act as a repressor
decoy to protect INO80D from downregulation in NSCLC.
Abbreviations:
|
ANXA5
|
annexin A5
|
|
CMTM4
|
CKLF-like MARVEL trans-membrane
domain containing 4
|
|
DSB
|
double strand break
|
|
EIF3B
|
eukaryotic translation initiation
factor 3, subunit B
|
|
EIF4G2
|
eukaryotic translation initiation
factor 4γ, 2
|
|
INO80D
|
INO80 complex subunit D
|
|
NSCLC
|
non-small cell lung cancer
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TAP2
|
transporter 2, ATP-binding cassette,
subfamily B (MDR/TAP)
|
|
lncRNA
|
long non-coding RNA
|
Acknowledgments
Not applicable.
Funding
This study was supported by the Ministry of Science
and Technology of the Republic of China (grant nos.
NSC-99-2320-B-037-006-MY3, MOST-105-2632-E-468-002 and
MOST-106-2221-E-468-018), the National Health Research Institutes
(grant no. NHRI-EX103-10326BI) and the Asia University and China
Medical University Hospital (grant nos. DMR-107-206,
ASIA104-CMUH-22 and ASIA105-CMUH-15).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
CCC, TYL, YTL, YSC, YLC, YCC, CCL and PCL detected
the biological function of cells, performed the RT-qPCR assays,
performed the cell proliferation and luciferase reporter assays,
performed the statistical analysis, and assisted in drafting the
manuscript. WLC conceived the study, participated in the
bioinformatics analysis, and coordinated and drafted the
manuscript. KTY provided the tissue samples and the clinical data.
YSC, WLC, JGC and TCL conceived the study, participated in its
design and coordination, and assisted in drafting the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All participants provided their written informed
consent to participate in the present study. The institutional
review boards of CMUH and CCH approved the study (CMUH102-RECJ-015
and CCH-IRB-080322) and the consent procedure.
Consent for publication
All participants provided their written informed
consent to participate in the present study. The publication of
such data does not compromise anonymity.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ilott NE and Ponting CP: Predicting long
non-coding RNAs using RNA sequencing. Methods. 63:50–59. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hung T and Chang HY: Long noncoding RNA in
genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar
|
|
9
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen W, Böcker W, Brosius J and Tiedge H:
Expression of neural BC200 RNA in human tumours. J Pathol.
183:345–351. 1997. View Article : Google Scholar
|
|
11
|
Iacoangeli A, Lin Y, Morley EJ, Muslimov
IA, Bianchi R, Reilly J, Weedon J, Diallo R, Böcker W and Tiedge H:
BC200 RNA in invasive and preinvasive breast cancer.
Carcinogenesis. 25:2125–2133. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berteaux N, Lottin S, Adriaenssens E, Van
Coppenolle F, Leroy X, Coll J, Dugimont T and Curgy JJ: Hormonal
regulation of H19 gene expression in prostate epithelial cells. J
Endocrinol. 183:69–78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brannan CI, Dees EC, Ingram RS and
Tilghman SM: The product of the H19 gene may function as an RNA.
Mol Cell Biol. 10:28–36. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gabory A, Jammes H and Dandolo L: The H19
locus: Role of an imprinted non-coding RNA in growth and
development. BioEssays. 32:473–480. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hibi K, Nakamura H, Hirai A, Fujikake Y,
Kasai Y, Akiyama S, Ito K and Takagi H: Loss of H19 imprinting in
esophageal cancer. Cancer Res. 56:480–482. 1996.PubMed/NCBI
|
|
16
|
Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li
LY, Guan GH, Liu Q, Qian YH and Xie D: The lncRNA H19 mediates
breast cancer cell plasticity during EMT and MET plasticity by
differentially sponging miR-200b/c and let-7b. Sci Signal.
10:102017. View Article : Google Scholar
|
|
17
|
Guo F, Li Y, Liu Y, Wang J, Li Y and Li G:
Inhibition of metastasis-associated lung adenocarcinoma transcript
1 in CaSki human cervical cancer cells suppresses cell
proliferation and invasion. Acta Biochim Biophys Sin (Shanghai).
42:224–229. 2010. View Article : Google Scholar
|
|
18
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar
|
|
20
|
Krüger J and Rehmsmeier M: RNAhybrid:
microRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34(Web Server): W451–4. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zuo ZK, Gong Y, Chen XH, Ye F, Yin ZM,
Gong QN and Huang JS: TGFβ1-induced lncRNA UCA1 upregulation
promotes gastric cancer invasion and migration. DNA Cell Biol.
36:159–167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Zhang H, Li X and Chen W:
Differential expression profile analysis of lncRNA UCA1α regulated
mRNAs in bladder cancer. J Cell Biochem. 119:1841–1854. 2018.
View Article : Google Scholar
|
|
24
|
Nakagawa T, Endo H, Yokoyama M, Abe J,
Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T and Satoh K: Large
noncoding RNA HOTAIR enhances aggressive biological behavior and is
associated with short disease-free survival in human non-small cell
lung cancer. Biochem Biophys Res Commun. 436:319–324. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ülger Y, Dadaş E, Yalinbaş Kaya B, Sümbül
AT, Genç A and Bayram S: The analysis of lncRNA HOTAIR rs12826786
C>T polymorphism and gastric cancer susceptibility in a Turkish
population: Lack of any association in a hospital-based
case-control study. Ir J Med Sci. 186:859–865. 2017. View Article : Google Scholar
|
|
26
|
Wu X, Cao X and Chen F: LncRNA-HOTAIR
activates tumor cell proliferation and migration by suppressing
miR-326 in cervical cancer. Oncol Res. Aug 31–2017.Epub ahead of
print. View Article : Google Scholar
|
|
27
|
Han L, Kong R, Yin DD, Zhang EB, Xu TP, De
W and Shu YQ: Low expression of long noncoding RNA GAS6-AS1
predicts a poor prognosis in patients with NSCLC. Med Oncol.
30:6942013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chan WL, Yuo CY, Yang WK, Hung SY, Chang
YS, Chiu CC, Yeh KT, Huang HD and Chang JG: Transcribed pseudogene
ψPPM1K generates endogenous siRNA to suppress oncogenic cell growth
in hepatocellular carcinoma. Nucleic Acids Res. 41:3734–3747. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bu D, Yu K, Sun S, Xie C, Skogerbø G, Miao
R, Xiao H, Liao Q, Luo H, Zhao G, et al: NONCODE v3.0: Integrative
annotation of long noncoding RNAs. Nucleic Acids Res.
40D:D210–D215. 2012. View Article : Google Scholar
|
|
31
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39(Database): D152–D157. 2011. View Article : Google Scholar :
|
|
32
|
Karolchik D, Barber GP, Casper J, Clawson
H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L,
Haeussler M, et al: The UCSC Genome Browser database: 2014 update.
Nucleic Acids Res. 42D:D764–D770. 2014. View Article : Google Scholar
|
|
33
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al: miRTarBase: A
database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39(Suppl 1): D163–D169. 2011.
View Article : Google Scholar
|
|
34
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res.
42D:D78–D85. 2014. View Article : Google Scholar
|
|
35
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
36
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human microRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
40
|
Sherman BT, Huang da W, Tan Q, Sherman BT,
Huang W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane
HC, Lempicki RA, et al: DAVID Knowledgebase: A gene-centered
database integrating heterogeneous gene annotation resources to
facilitate high-throughput gene functional analysis. BMC
Bioinformatics. 8:4262007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jeggo PA and Downs JA: Roles of chromatin
remodellers in DNA double strand break repair. Exp Cell Res.
329:69–77. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Andrews NW, Corrotte M and Castro-Gomes T:
Above the fray: Surface remodeling by secreted lysosomal enzymes
leads to endocytosis-mediated plasma membrane repair. Semin Cell
Dev Biol. 45:10–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kuersten S and Goodwin EB: The power of
the 3′ UTR: Translational control and development. Nat Rev Genet.
4:626–637. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Seiler J, Breinig M, Caudron-Herger M,
Polycarpou-Schwarz M, Boutros M and Diederichs S: The lncRNA VELUCT
strongly regulates viability of lung cancer cells despite its
extremely low abundance. Nucleic Acids Res. 45:5458–5469. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xie L, Yang Z, Li G, Shen L, Xiang X, Liu
X, Xu D, Xu L, Chen Y, Tian Z, et al: Genome-wide identification of
bone metastasis- related microRNAs in lung adenocarcinoma by
high-throughput sequencing. PLoS One. 8:e612122013. View Article : Google Scholar
|
|
47
|
Chen T and Dent SY: Chromatin modifiers
and remodellers: Regulators of cellular differentiation. Nat Rev
Genet. 15:93–106. 2014. View Article : Google Scholar
|
|
48
|
Ho L and Crabtree GR: Chromatin
remodelling during development. Nature. 463:474–484. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Delia D and Mizutani S: The DNA damage
response pathway in normal hematopoiesis and malignancies. Int J
Hematol. 106:328–334. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dinger ME, Pang KC, Mercer TR and Mattick
JS: Differentiating protein-coding and noncoding RNA: Challenges
and ambiguities. PLOS Comput Biol. 4:e10001762008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dinger ME, Amaral PP, Mercer TR and
Mattick JS: Pervasive transcription of the eukaryotic genome:
Functional indices and conceptual implications. Brief Funct
Genomics Proteomics. 8:407–423. 2009. View Article : Google Scholar
|
|
52
|
Gerstein MB, Bruce C, Rozowsky JS, Zheng
D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S and Snyder
M: What is a gene, post-ENCODE? History and updated definition.
Genome Res. 17:669–681. 2007. View Article : Google Scholar : PubMed/NCBI
|