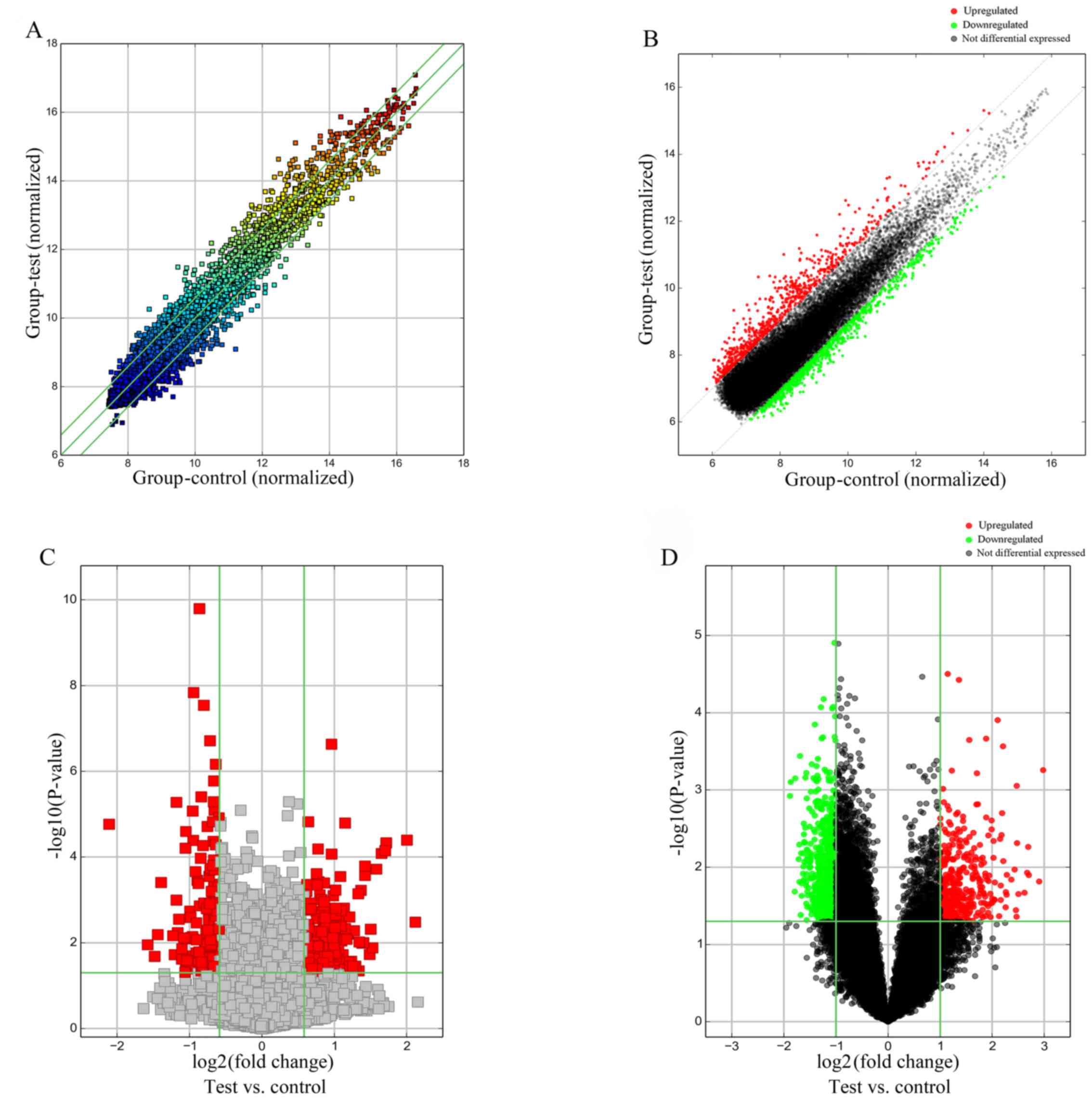
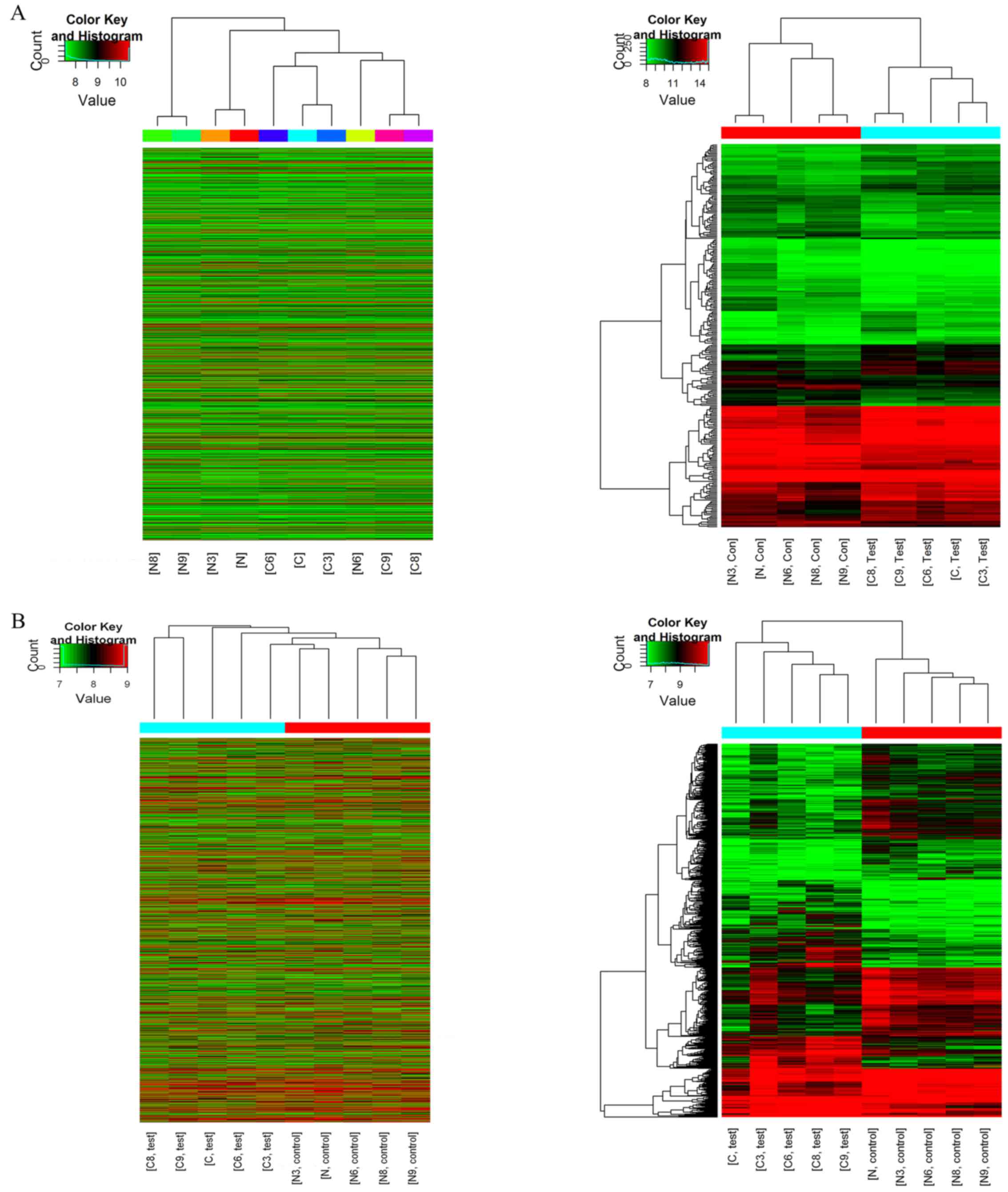
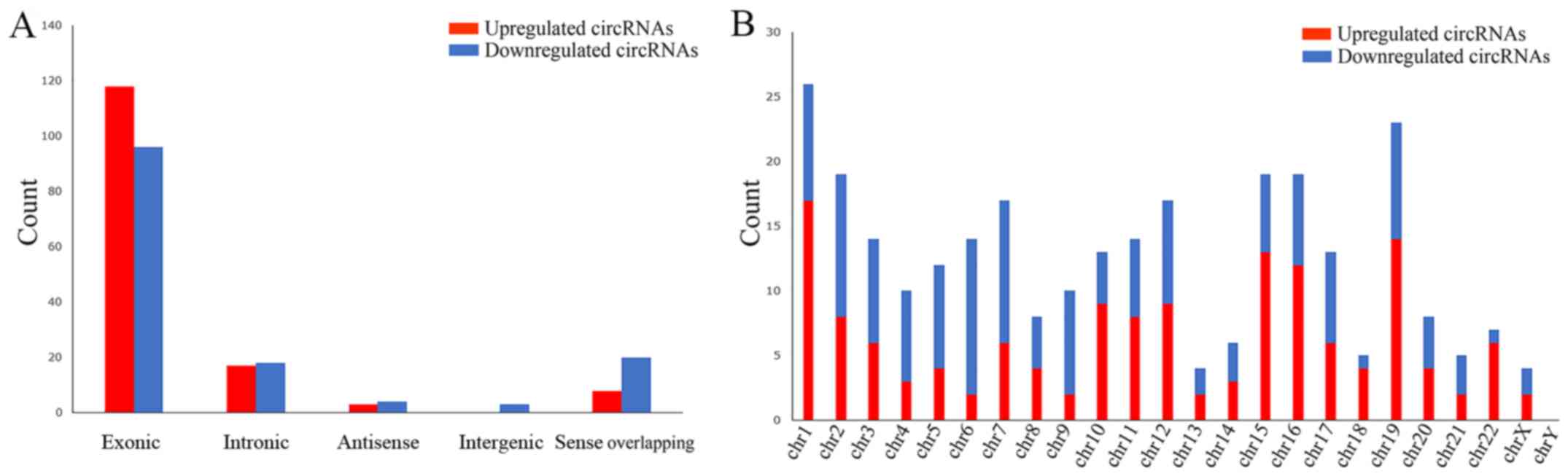
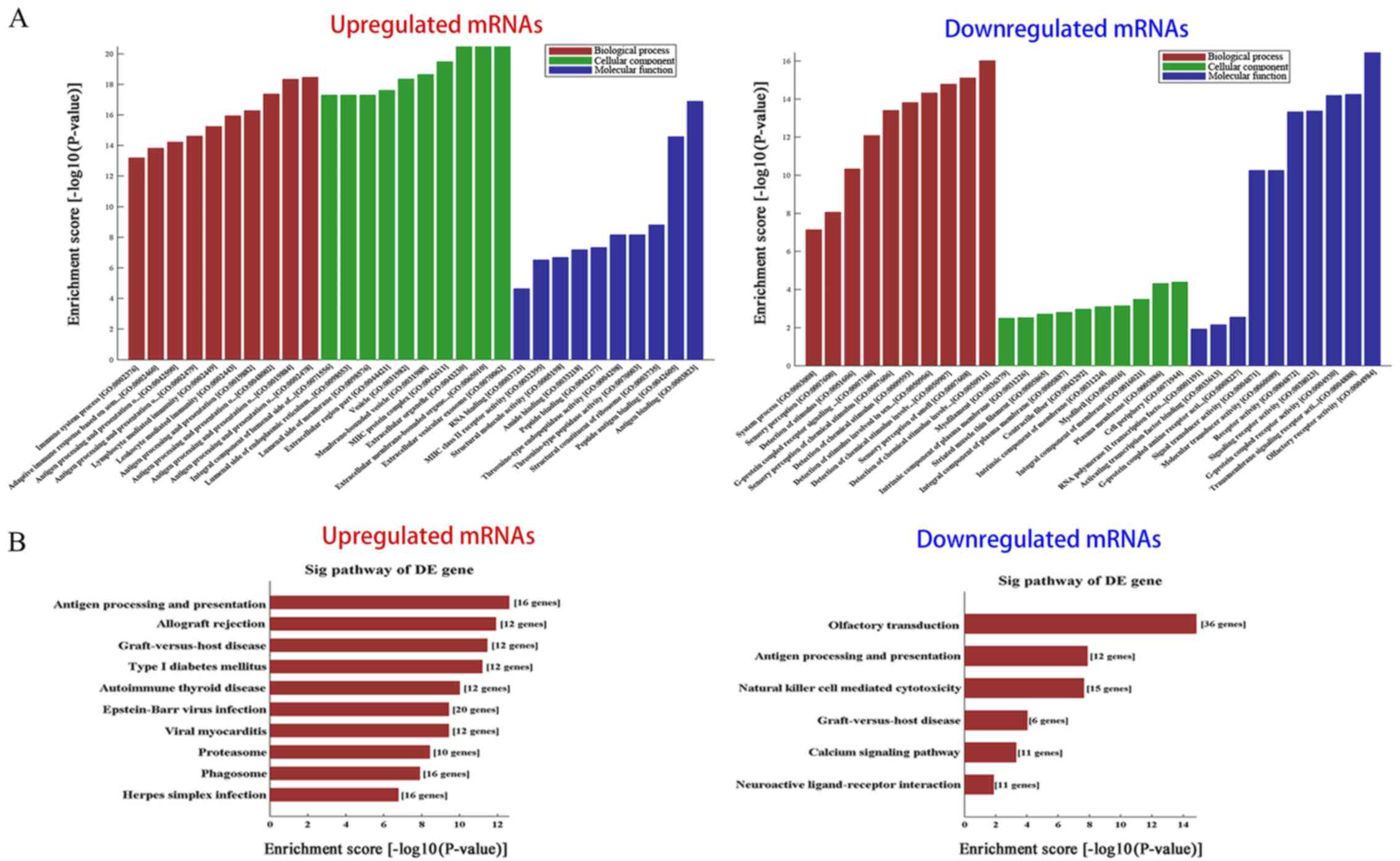
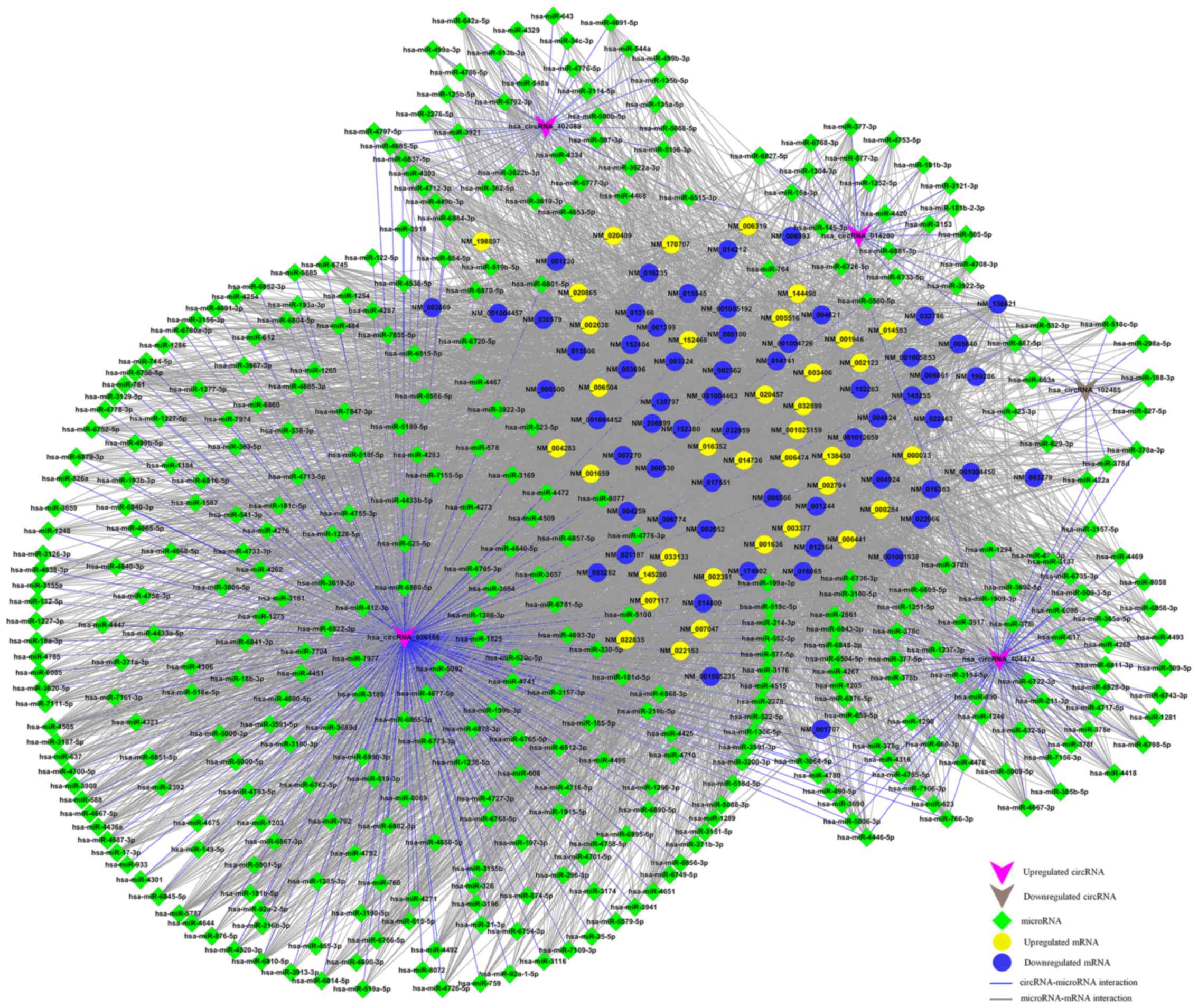
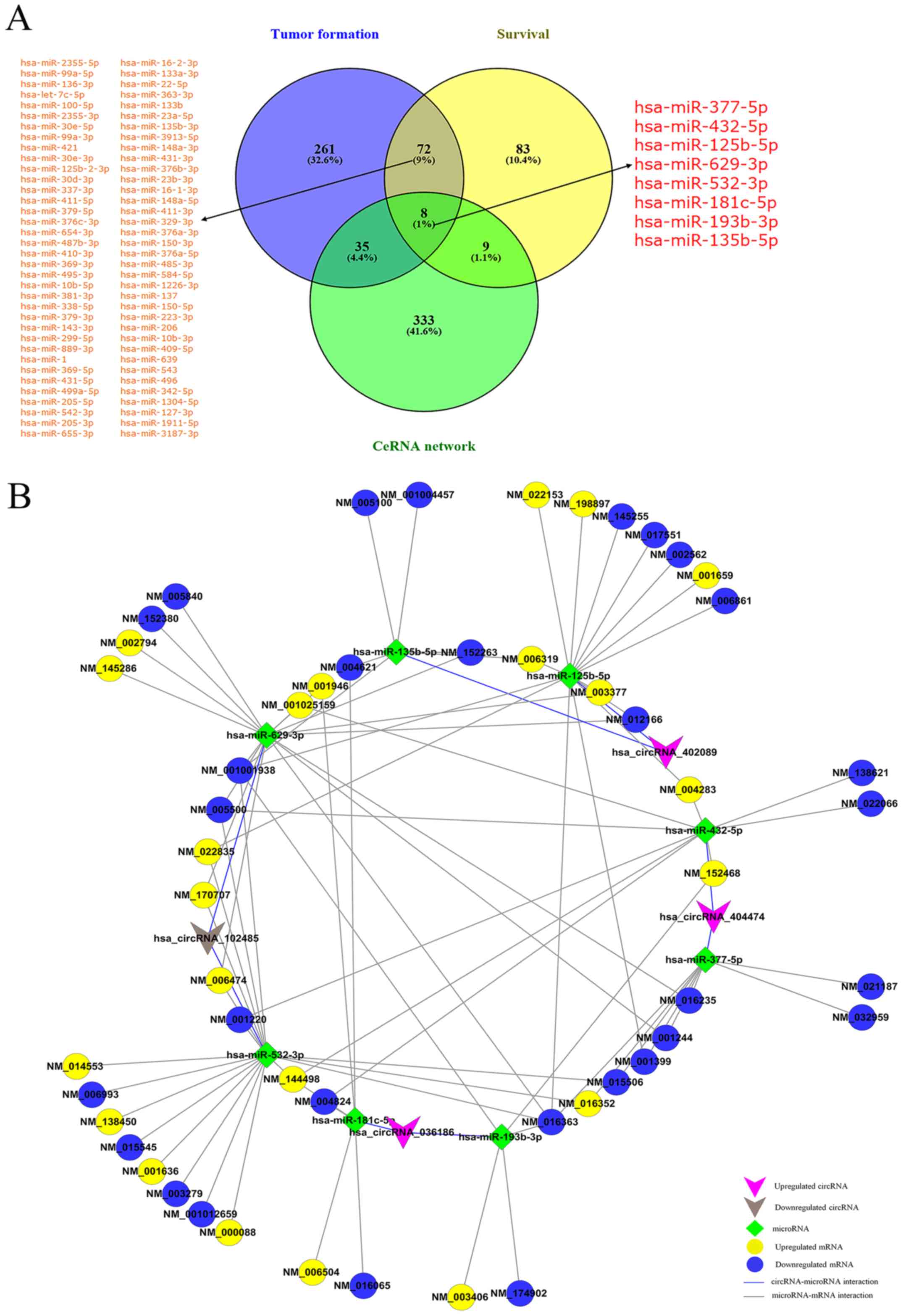
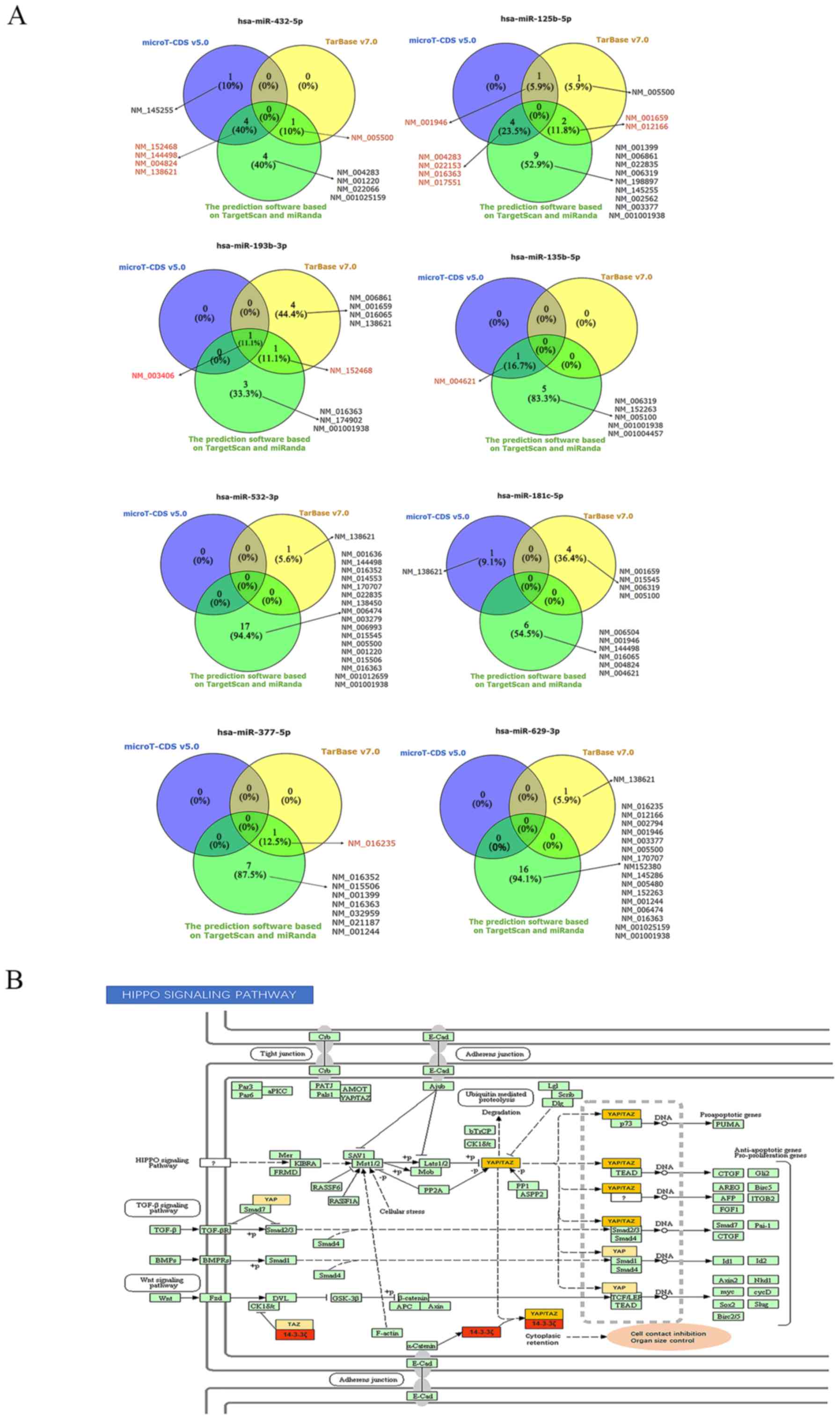
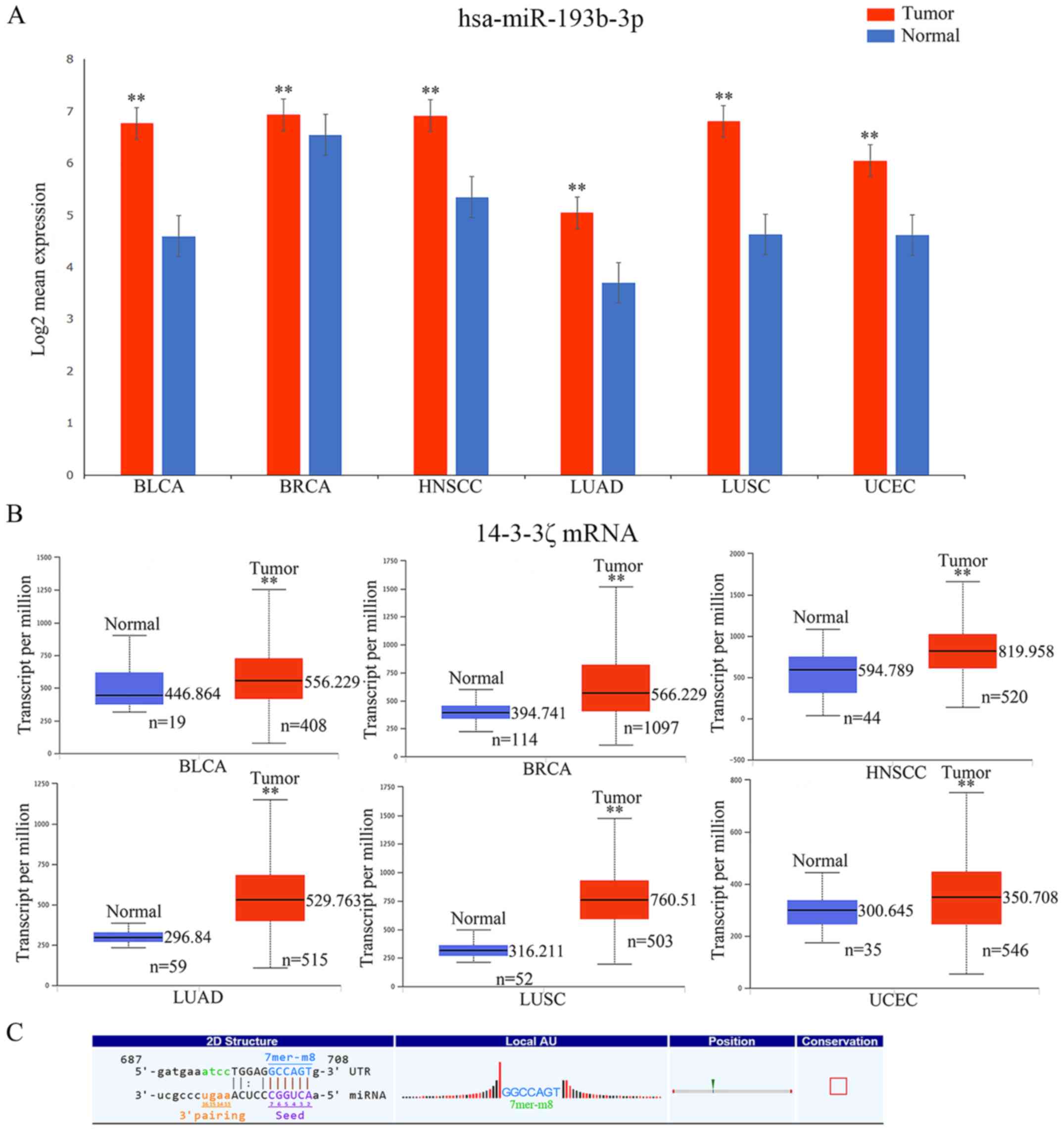
|
1
|
Marur S and Forastiere AA: Head and neck
squamous cell carcinoma: Update on epidemiology, diagnosis, and
treatment. Mayo Clin Proc. 91:386–396. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li H, Xie S, Liu M, Chen Z, Liu X, Wang L,
Li D and Zhou Y: The clinical significance of downregulation of
mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric
cancer tumorigenesis. Int J Oncol. 45:197–208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao J, Cai J, Huang D, Han Q, Yang Q, Li
T, Ding H and Wang Z: miR-335 represents an invasion suppressor
gene in ovarian cancer by targeting Bcl-w. Oncol Rep. 30:701–706.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
14
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen T, Han M, Wei G and Ni T: An
intriguing RNA species - perspectives of circularized RNA. Protein
Cell. 6:871–880. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vicens Q and Westhof E: Biogenesis of
circular RNAs. Cell. 159:13–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Westholm JO, Miura P, Olson S, Shenker S,
Joseph B, Sanfilippo P, Celniker SE, Graveley BR and Lai EC:
Genome-wide analysis of drosophila circular RNAs reveals their
structural and sequence properties and age-dependent neural
accumulation. Cell Reports. 9:1966–1980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suzuki H, Aoki Y, Kameyama T, Saito T,
Masuda S, Tanihata J, Nagata T, Mayeda A, Takeda S and Tsukahara T:
Endogenous multiple exon skipping and back-splicing at the DMD
mutation hotspot. Int J Mol Sci. 17:172016. View Article : Google Scholar
|
|
24
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar
|
|
30
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar
|
|
32
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Assenov Y, Ramírez F, Schelhorn SE,
Lengauer T and Albrecht M: Computing topological parameters of
biological networks. Bioinformatics. 24:282–284. 2008. View Article : Google Scholar
|
|
34
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wong NW, Chen Y, Chen S and Wang X:
OncomiR: An online resource for exploring pan-cancer microRNA
dysregulation. Bioinformatics. 34:713–715. 2018. View Article : Google Scholar
|
|
36
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vlachos IS, Zagganas K, Paraskevopoulou
MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and
Hatzigeorgiou AG: DIANA-miRPath v3.0: Deciphering microRNA function
with experimental support. Nucleic Acids Res. 43W1:W460–W466. 2015.
View Article : Google Scholar
|
|
38
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42D1:D92–D97. 2014. View Article : Google Scholar
|
|
39
|
Vlachos IS, Paraskevopoulou MD, Karagkouni
D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL,
Maniou S, Karathanou K, Kalfakakou D, et al: DIANA-TarBase v7.0:
indexing more than half a million experimentally supported
miRNA:mRNA interactions. Nucleic Acids Res. 43D:D153–D159. 2015.
View Article : Google Scholar
|
|
40
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C,
Dalamagas T and Hatzigeorgiou AG: DIANA-microT web server v5.0:
Service integration into miRNA functional analysis workflows.
Nucleic Acids Res. 41W:W169–W173. 2013. View Article : Google Scholar
|
|
41
|
Reczko M, Maragkakis M, Alexiou P, Grosse
I and Hatzigeorgiou AG: Functional microRNA targets in protein
coding sequences. Bioinformatics. 28:771–776. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
43
|
Alexander RP, Fang G, Rozowsky J, Snyder M
and Gerstein MB: Annotating non-coding regions of the genome. Nat
Rev Genet. 11:559–571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mei Y, Yang JP and Qian CN: For robust big
data analyses: A collection of 150 important pro-metastatic genes.
Chin J Cancer. 36:162017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qian CN, Mei Y and Zhang J: Cancer
metastasis: Issues and challenges. Chin J Cancer. 36:382017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Caiment F, Gaj S, Claessen S and Kleinjans
J: High-throughput data integration of RNA-miRNA-circRNA reveals
novel insights into mechanisms of benzo[a]pyrene-induced
carcinogenicity. Nucleic Acids Res. 43:2525–2534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J,
Xiang Y, Liu L, Zhong S, Han L, et al: Comprehensive
characterization of tissue-specific circular RNAs in the human and
mouse genomes. Brief Bioinform. 18:984–992. 2017.
|
|
51
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Granados-Riveron JT and Aquino-Jarquin G:
The complexity of the translation ability of circRNAs. Biochim
Biophys Acta. 1859:1245–1251. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang Y and Wang Z: Efficient backsplicing
produces translatable circular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar :
|
|
55
|
Sannigrahi MK, Sharma R, Panda NK and
Khullar M: Role of non-coding RNAs in head and neck squamous cell
carcinoma: A narrative review. Oral Dis. Sep 21–2017.Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Koshizuka K, Hanazawa T, Kikkawa N, Katada
K, Okato A, Arai T, Idichi T, Osako Y, Okamoto Y and Seki N:
Antitumor miR-150-5p and miR-150-3p inhibit cancer cell
aggressiveness by targeting SPOCK1 in head and neck squamous cell
carcinoma. Auris Nasus Larynx. 45:854–865. 2018. View Article : Google Scholar
|
|
57
|
Liu F, Zhao X, Qian Y, Zhang J, Zhang Y
and Yin R: miR-206 inhibits head and neck squamous cell carcinoma
cell progression by targeting HDAC6 via PTEN/AKT/mTOR pathway.
Biomed Pharmacother. 96:229–237. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Miao L, Wang L, Zhu L, Du J, Zhu X, Niu Y,
Wang R, Hu Z, Chen N, Shen H, et al: Association of microRNA
polymorphisms with the risk of head and neck squamous cell
carcinoma in a Chinese population: A case-control study. Chin J
Cancer. 35:772016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: circRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rauhala HE, Jalava SE, Isotalo J, Bracken
H, Lehmusvaara S, Tammela TL, Oja H and Visakorpi T: miR-193b is an
epigenetically regulated putative tumor suppressor in prostate
cancer. Int J Cancer. 127:1363–1372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Leivonen SK, Rokka A, Ostling P, Kohonen
P, Corthals GL, Kallioniemi O and Perälä M: Identification of
miR-193b targets in breast cancer cells and systems biological
analysis of their functional impact. Mol Cell Proteomics.
10:M110.0053222011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mitra AK, Chiang CY, Tiwari P, Tomar S,
Watters KM, Peter ME and Lengyel E: Microenvironment-induced
downregulation of miR-193b drives ovarian cancer metastasis.
Oncogene. 34:5923–5932. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhao GY, Ding JY, Lu CL, Lin ZW and Guo J:
The overexpression of 143-3ζ and Hsp27 promotes non-small cell lung
cancer progression. Cancer. 120:652–663. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rehman SK, Li SH, Wyszomierski SL, Wang Q,
Li P, Sahin O, Xiao Y, Zhang S, Xiong Y, Yang J, et al: 14-3-3ζ
orchestrates mammary tumor onset and progression via
miR-221-mediated cell proliferation. Cancer Res. 74:363–373. 2014.
View Article : Google Scholar
|
|
65
|
Li J, Kong F, Wu K, Song K, He J and Sun
W: miR-193b directly targets STMN1 and uPA genes and suppresses
tumor growth and metastasis in pancreatic cancer. Mol Med Rep.
10:2613–2620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mets E, Van der Meulen J, Van Peer G,
Boice M, Mestdagh P, Van de Walle I, Lammens T, Goossens S, De
Moerloose B, Benoit Y, et al: MicroRNA-193b-3p acts as a tumor
suppressor by targeting the MYB oncogene in T-cell acute
lymphoblastic leukemia. Leukemia. 29:798–806. 2015. View Article : Google Scholar
|
|
67
|
Zhang J, Qin J and Su Y: miR-193b-3p
possesses anti-tumor activity in ovarian carcinoma cells by
targeting p21-activated kinase 3. Biomed Pharmacother.
96:1275–1282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Matta A, Bahadur S, Duggal R, Gupta SD and
Ralhan R: Over-expression of 14-3-3zeta is an early event in oral
cancer. BMC Cancer. 7:1692007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Nishimura Y, Komatsu S, Ichikawa D, Nagata
H, Hirajima S, Takeshita H, Kawaguchi T, Arita T, Konishi H,
Kashimoto K, et al: Overexpression of YWHAZ relates to tumor cell
proliferation and malignant outcome of gastric carcinoma. Br J
Cancer. 108:1324–1331. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Murata T, Takayama K, Urano T, Fujimura T,
Ashikari D, Obinata D, Horie-Inoue K, Takahashi S, Ouchi Y, Homma
Y, et al: 14-3-3zeta, a novel androgen-responsive gene, is
upregulated in prostate cancer and promotes prostate cancer cell
proliferation and survival. Clin Cancer Res. 18:5617–5627. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lin M, Morrison CD, Jones S, Mohamed N,
Bacher J and Plass C: Copy number gain and oncogenic activity of
YWHAZ/14-3-3zeta in head and neck squamous cell carcinoma. Int J
Cancer. 125:603–611. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yao CB, Zhou X, Chen CS and Lei QY: The
regulatory mechanisms and functional roles of the Hippo signaling
pathway in breast cancer. Yi Chuan. 39:617–629. 2017.PubMed/NCBI
|
|
73
|
Patel SH, Camargo FD and Yimlamai D: Hippo
signaling in the liver regulates organ size, cell fate, and
carcinogenesis. Gastroenterology. 152:533–545. 2017. View Article : Google Scholar :
|
|
74
|
Ji XY, Zhong G and Zhao B: Molecular
mechanisms of the mammalian Hippo signaling pathway. Yi Chuan.
39:546–567. 2017.PubMed/NCBI
|
|
75
|
Pan D: The hippo signaling pathway in
development and cancer. Dev Cell. 19:491–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhang B, Gong A, Shi H, Bie Q, Liang Z, Wu
P, Mao F, Qian H and Xu W: Identification of a novel YAP-14-3-3ζ
negative feedback loop in gastric cancer. Oncotarget.
8:71894–71910. 2017.PubMed/NCBI
|