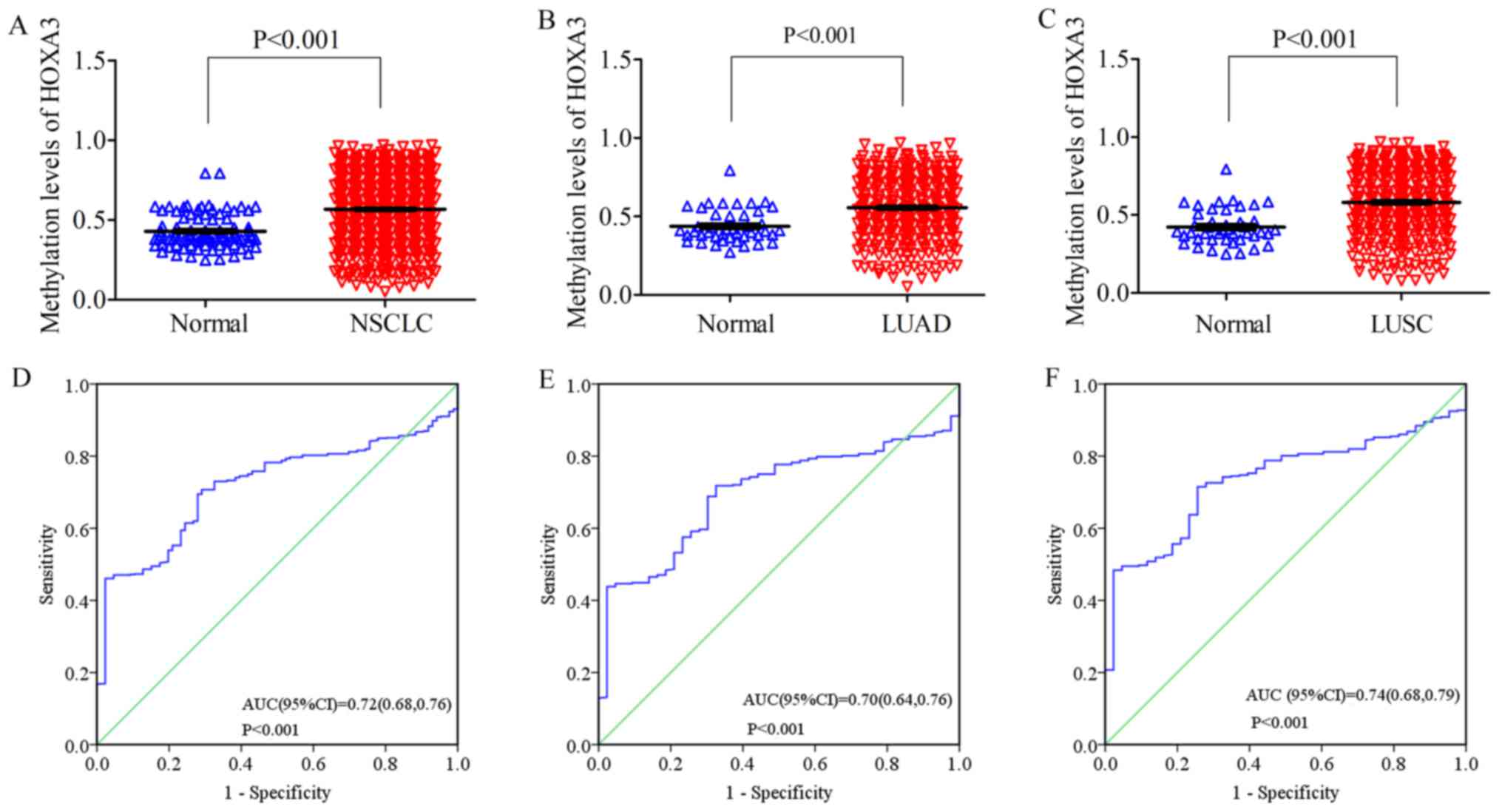
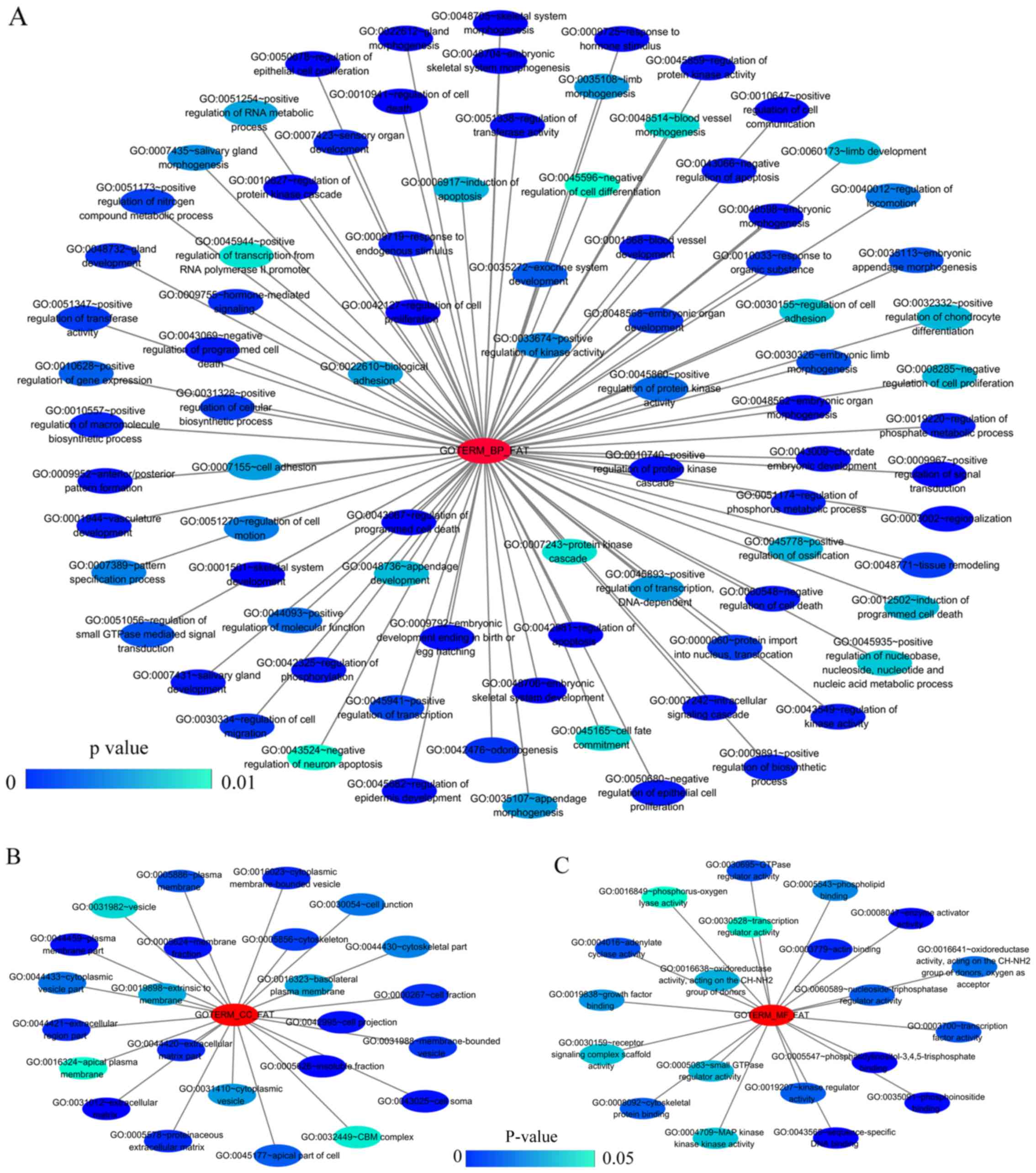
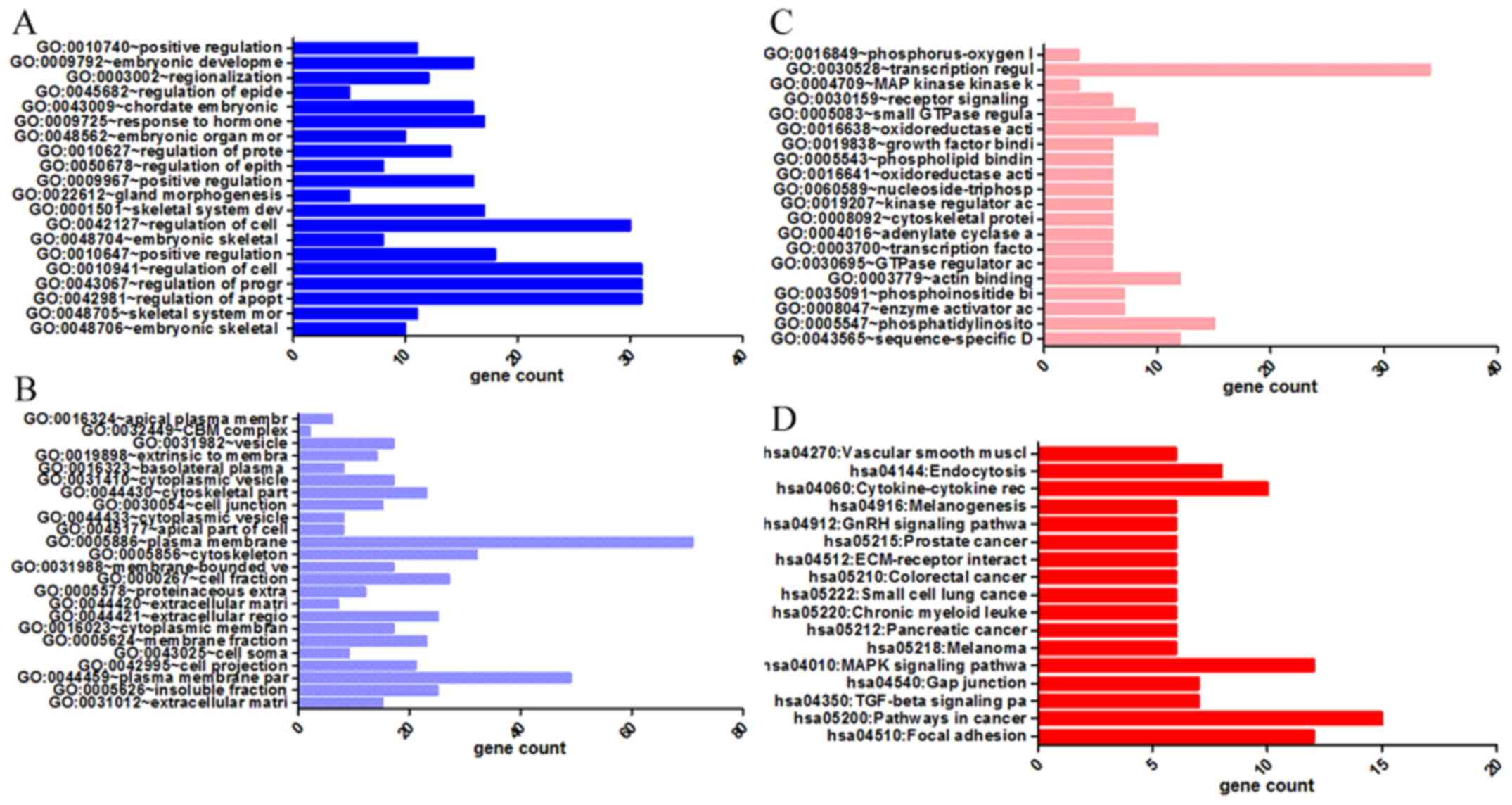
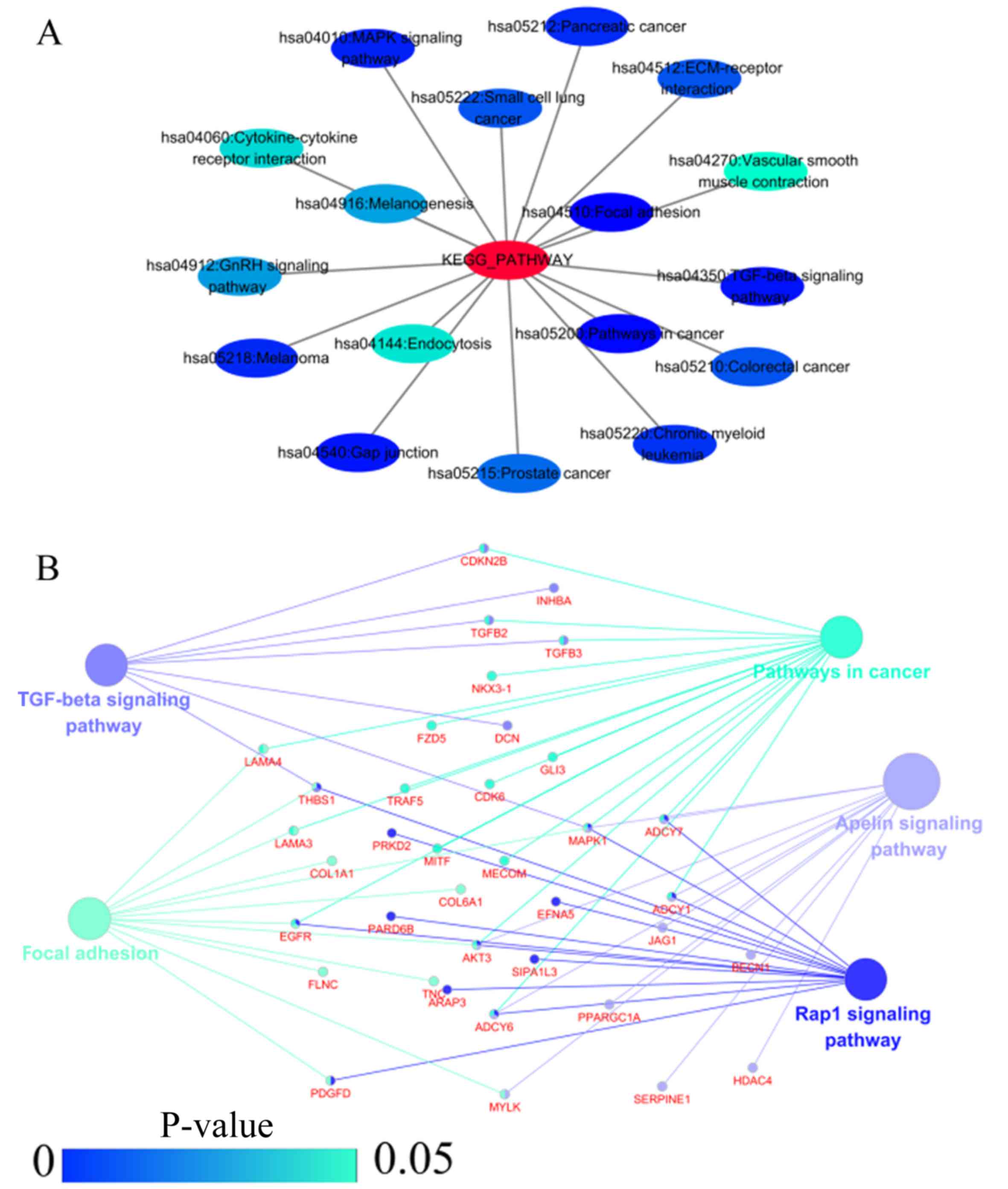
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer UK: Lung cancer mortality
statistics. 2014
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Govindan R, Page N, Morgensztern D, Read
W, Tierney R, Vlahiotis A, Spitznagel EL and Piccirillo J: Changing
epidemiology of small-cell lung cancer in the United States over
the last 30 years: Analysis of the surveillance, epidemiologic, and
end results database. J Clin Oncol. 24:4539–4544. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
O'Brien TD, Jia P, Caporaso NE, Landi MT
and Zhao Z: Weak sharing of genetic association signals in three
lung cancer subtypes: Evidence at the SNP, gene, regulation, and
pathway levels. Genome Med. 10:162018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abraham C, Garsa A, Badiyan SN, Drzymala
R, Yang D, DeWees T, Tsien C, Dowling JL, Rich KM, Chicoine MR, et
al: Internal dose escalation is associated with increased local
control for non-small cell lung cancer (NSCLC) brain metastases
treated with stereotactic radiosurgery (SRS). Adv Radiat Oncol.
3:146–153. 2017. View Article : Google Scholar
|
|
8
|
Fernandes P, Lareiro S, Vouga L, Guerra M
and Miranda J: Uniportal video-assisted thorascoscopic surgery -
The new paradigm in the surgical treatment of lung cancer. Rev Port
Cir Cardiotorac Vasc. 24:1272017.
|
|
9
|
Jean F, Tomasini P and Barlesi F:
Atezolizumab: Feasible second-line therapy for patients with
non-small cell lung cancer? A review of efficacy, safety and place
in therapy. Ther Adv Med Oncol. 9:769–779. 2017. View Article : Google Scholar
|
|
10
|
Jiang Q, Xie M, He M, Yan F, Zhang X and
Yu S: Anti-PD-1/PD-L1 antibodies versus docetaxel in patients with
previously treated non-small-cell lung cancer. Oncotarget.
9:7672–7683. 2017.
|
|
11
|
Byrne M, Martinez P and Morris V:
Evolution of a pentameral body plan was not linked to translocation
of anterior Hox genes: The echinoderm HOX cluster revisited. Evol
Dev. 18:137–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim HS, Kim BM, Lee BY, Souissi S, Park HG
and Lee JS: Identification of Hox genes and rearrangements within
the single homeobox (Hox) cluster (192.8 kb) of the cyclopoid
copepod (Paracyclopina nana). J Exp Zoolog B Mol Dev Evol.
326:105–109. 2016. View Article : Google Scholar
|
|
13
|
Schiemann SM, Martín-Durán JM, Børve A,
Vellutini BC, Passamaneck YJ and Hejnol A: Clustered brachiopod Hox
genes are not expressed collinearly and are associated with
lophotrochozoan novelties. Proc Natl Acad Sci USA. 114:E1913–E1922.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang X, Bu J, Liu X, Wang W, Mai W, Lv B,
Zou J, Mo X, Li X, Wang J, et al: miR-133b suppresses metastasis by
targeting HOXA9 in human colorectal cancer. Oncotarget.
8:63935–63948. 2017.PubMed/NCBI
|
|
15
|
Wang K, Jin J, Ma T and Zhai H: MiR-139-5p
inhibits the tumorigenesis and progression of oral squamous
carcinoma cells by targeting HOXA9. J Cell Mol Med. 21:3730–3740.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao
X, Li W, Zheng S, Ye H, Wang L, et al: LncRNA HOTTIP modulates
cancer stem cell properties in human pancreatic cancer by
regulating HOXA9. Cancer Lett. 410:68–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen S, Yu J, Lv X and Zhang L: HOXA9 is
critical in the proliferation, differentiation, and malignancy of
leukaemia cells both in vitro and in vivo. Cell Biochem Funct.
35:433–440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao
Z, Li Q and Zhang W: Up-regulation of long non-coding HOTTIP
functions as an oncogene by regulating HOXA13 in non-small cell
lung cancer. Am J Transl Res. 8:2022–2032. 2016.PubMed/NCBI
|
|
19
|
Daugaard I, Dominguez D, Kjeldsen TE,
Kristensen LS, Hager H, Wojdacz TK and Hansen LL: Identification
and validation of candidate epigenetic biomarkers in lung
adenocarcinoma. Sci Rep. 6:358072016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Chen WJ, Gan TQ, Zhang XL, Xie
ZC, Ye ZH, Deng Y, Wang ZF, Cai KT, Li SK, et al: Clinical
significance and effect of lncRNA HOXA11-AS in NSCLC: A study based
on bioinformatics, in vitro and in vivo verification. Sci Rep.
7:55672017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang RX, Chen WJ, He RQ, Zeng JH, Liang L,
Li SK, Ma J, Luo DZ and Chen G: Identification of a RNA-Seq based
prognostic signature with five lncRNAs for lung squamous cell
carcinoma. Oncotarget. 8:50761–50773. 2017.PubMed/NCBI
|
|
22
|
Li Z, Xie Y, Zhong T, Zhang X, Dang Y, Gan
T and Chen G: Expression and clinical contribution of MRGD mRNA in
non-small cell lung cancers. J BUON. 20:1101–1106. 2015.PubMed/NCBI
|
|
23
|
Muraoka T, Soh J, Toyooka S, Aoe K,
Fujimoto N, Hashida S, Maki Y, Tanaka N, Shien K, Furukawa M, et
al: The degree of microRNA-34b/c methylation in serum-circulating
DNA is associated with malignant pleural mesothelioma. Lung Cancer.
82:485–490. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh J, Batish VK and Grover S:
Simultaneous detection of Listeria monocytogenes and Salmonella
spp. in dairy products using real time PCR-melt curve analysis. J
Food Sci Technol. 49:234–239. 2012. View Article : Google Scholar
|
|
25
|
Xu D, Yang Z, Zhang D, Wu W, Guo Y, Chen
Q, Xu D and Cui W: Rapid detection of immunoglobulin heavy chain
gene rearrangement by PCR and melting curve analysis using combined
FR2 and FR3 primers. Diagn Pathol. 10:1402015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zaghloul H, El Morsi AA, Soweha HE,
Elsayed A, Seif S and El-Sharawy H: A simple real-time polymerase
chain reaction assay using SYBR Green for hepatitis C virus
genotyping. Arch Virol. 162:57–61. 2017. View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Tang R, Liang L, Luo D, Feng Z, Huang Q,
He R, Gan T, Yang L and Chen G: Downregulation of miR-30a is
associated with poor prognosis in lung cancer. Med Sci Monit.
21:2514–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren F, Ding H, Huang S, Wang H, Wu M, Luo
D, Dang Y, Yang L and Chen G: Expression and clinicopathological
significance of miR-193a-3p and its potential target astrocyte
elevated gene-1 in non-small lung cancer tissues. Cancer Cell Int.
15:802015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Liu G, Ding L, Jiang T, Shao S,
Gao Y and Lu Y: HOXA3 promotes tumor growth of human colon cancer
through activating EGFR/Ras/Raf/MEK/ERK signaling pathway. J Cell
Biochem. 119:2864–2874. 2018. View Article : Google Scholar
|
|
31
|
Shen ZH, Zhao KM and Du T: HOXA10 promotes
nasopharyngeal carcinoma cell proliferation and invasion via
inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci.
21:945–952. 2017.PubMed/NCBI
|
|
32
|
Kuasne H, Cólus IM, Busso AF,
Hernandez-Vargas H, Barros-Filho MC, Marchi FA, Scapulatempo-Neto
C, Faria EF, Lopes A, Guimarães GC, et al: Genome-wide methylation
and transcriptome analysis in penile carcinoma: Uncovering new
molecular markers. Clin Epigenetics. 7:462015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang X, Liu G, Ding L, Jiang T, Shao S,
Gao Y and Lu Y: HOXA3 promotes tumor growth of human colon cancer
through activating EGFR/Ras/Raf/MEK/ERK signaling pathway. J Cell
Biochem. 119:2864–2874. 2018. View Article : Google Scholar
|
|
34
|
Zhang YW, Zheng Y, Wang JZ, Lu XX, Wang Z,
Chen LB, Guan XX and Tong JD: Integrated analysis of DNA
methylation and mRNA expression profiling reveals candidate genes
associated with cisplatin resistance in non-small cell lung cancer.
Epigenetics. 9:896–909. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jin X, Liu X, Li X and Guan Y: Integrated
analysis of DNA methylation and mRNA expression profiles data to
identify key Genes in lung adenocarcinoma. Biomed Res Int.
2016:43694312016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tai YL, Chen LC and Shen TL: Emerging
roles of focal adhesion kinase in cancer. Biomed Res Int.
2015:6906902015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Min A, Zhu C, Wang J, Peng S, Shuai C, Gao
S, Tang Z and Su T: Focal adhesion kinase knockdown in
carcinoma-associated fibroblasts inhibits oral squamous cell
carcinoma metastasis via downregulating MCP-1/CCL2 expression. J
Biochem Mol Toxicol. 29:70–76. 2015. View Article : Google Scholar
|
|
38
|
Eke I and Cordes N: Focal adhesion
signaling and therapy resistance in cancer. Semin Cancer Biol.
31:65–75. 2015. View Article : Google Scholar
|
|
39
|
Zhou B, Wang GZ, Wen ZS, Zhou YC, Huang
YC, Chen Y and Zhou GB: Somatic mutations and splicing variants of
focal adhesion kinase in non-small cell lung cancer. J Natl Cancer
Inst. 110:195–204. 2017. View Article : Google Scholar
|
|
40
|
Tang KJ, Constanzo JD, Venkateswaran N,
Melegari M, Ilcheva M, Morales JC, Skoulidis F, Heymach JV,
Boothman DA and Scaglioni PP: Focal adhesion kinase regulates the
DNA damage response and its inhibition radiosensitizes mutant KRAS
lung cancer. Clin Cancer Res. 22:5851–5863. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang HJ, Tao J, Sheng L, Hu X, Rong RM,
Xu M and Zhu TY: Twist2 promotes kidney cancer cell proliferation
and invasion by regulating ITGA6 and CD44 expression in the
ECM-receptor interaction pathway. Onco Targets Ther. 9:1801–1812.
2016.PubMed/NCBI
|
|
42
|
Zhang H, Ye J, Weng X, Liu F, He L, Zhou D
and Liu Y: Comparative transcriptome analysis reveals that the
extracellular matrix receptor interaction contributes to the venous
metastases of hepatocellular carcinoma. Cancer Genet. 208:482–491.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lv M and Wang L: Comprehensive analysis of
genes, pathways, and TFs in nonsmoking Taiwan females with lung
cancer. Exp Lung Res. 41:74–83. 2015. View Article : Google Scholar
|
|
44
|
GE ZH, Cheng QS, LI WM, Feng Z, Wen MM,
Wang WC and Zhang ZP: Expression of TNC in NSCLC and its clinical
significance. Xiandai Shengwu Yixue Jinzhan. 31:6103–6106. 2014.In
Chinese.
|
|
45
|
Gocheva V, Naba A, Bhutkar A, Guardia T,
Miller KM, Li CM, Dayton TL, Sanchez-Rivera FJ, Kim-Kiselak C,
Jailkhani N, et al: Quantitative proteomics identify Tenascin-C as
a promoter of lung cancer progression and contributor to a
signature prognostic of patient survival. Proc Natl Acad Sci USA.
114:E5625–E5634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Edlund K, Lindskog C, Saito A, Berglund A,
Pontén F, Göransson-Kultima H, Isaksson A, Jirström K, Planck M,
Johansson L, et al: CD99 is a novel prognostic stromal marker in
non-small cell lung cancer. Int J Cancer. 131:2264–2273. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Oleksiewicz U, Liloglou T, Tasopoulou KM,
Daskoulidou N, Gosney JR, Field JK and Xinarianos G: COL1A1,
PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell
lung cancer. J Cancer Res Clin Oncol. 143:1133–1141. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Q, Liu S, Zhao X, Wang Y, Tian D and
Jiang W: MiR-372-3p promotes cell growth and metastasis by
targeting FGF9 in lung squamous cell carcinoma. Cancer Med.
6:1323–1330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Syring I, Bartels J, Holdenrieder S,
Kristiansen G, Müller SC and Ellinger J: Circulating serum miRNA
(miR-367-3p, miR-371a-3p, miR-372-3p and miR-373-3p) as biomarkers
in patients with testicular germ cell cancer. J Urol. 193:331–337.
2015. View Article : Google Scholar
|