Introduction
Esophageal cancer, including esophageal squamous
cell carcinoma (ESCC) and esophageal adenocarcinoma, is one of the
most common cancer types in the world. It has the eighth highest
incidence of all cancer types and the sixth highest
cancer-associated mortality rate worldwide, based on GLOBOCAN 2012
(1). The incidence of esophageal
cancer varies widely among different regions. For example, it
ranges from 3 per 100,000 in low incidence regions to over 100 per
100,000 in high-incidence areas (2). ESCC is the most common histological
type of esophageal carcinoma in less economically developed
countries, accounting for more than 95% of esophageal cancer cases
(3). ESCC is typically diagnosed
at an advanced stage because of the lack of early symptoms, usually
resulting in poor prognosis, with an approximate 5-year overall
survival rate of 20% (4). Thus,
clarification of the pathogenic mechanisms and new methods for
prevention are urgently needed.
Factors such as an underdeveloped economy, low
nutrient intake and limited health resources are very common in
high-risk areas of ESCC. Epidemiological studies have demonstrated
that dietary zinc deficiency (ZD) is associated with the
pathogenesis of ESCC (5). Abnet
et al (6) indicated that
zinc concentration measured by X-ray fluorescence in biopsy samples
is negatively correlated with the subsequent risk of developing
ESCC. Zinc is an essential trace element and a critical component
of many enzymes (7). Zinc content
is low in most foods, except for red meat and seafood, and people
from high incidence areas of esophageal cancer whose primary diet
does not include red meat and seafood are likely to be
zinc-deficient. Zinc intake in high-incidence areas of esophageal
cancer has been reported at only 72 and 62% of the recommended
daily allowance in spring and autumn, respectively, and this may be
one of the factors contributing to the high risk of esophageal
cancer (8). Dietary ZD causes low
serum zinc levels. Our previous study demonstrated that serum zinc
levels in patients with ESCC were lower compared with those in
healthy people and that serum zinc levels in people from areas with
a high incidence of esophageal cancer were lower compared with
those in people from low-incidence areas (9).
Dietary ZD has been demonstrated to promote ESCC
development by inducing a distinct inflammatory signature.
Epidemiological and clinical studies have indicated that chronic
inflammation promotes the occurrence of cancer, including ESCC
(10,11). Taccioli et al (12) reported that ZD amplifies the
overexpression of pro-inflammatory mediators, including
cyclooxygenase-2 (COX-2), S100a8 and S100a9 in the esophagus with
accompanying esophageal epithelial hyperplasia in rats. This
inflammatory signature is already activated in the early dysplastic
stage. Taccioli et al (13)
also demonstrated that ZD amplifies the inflammatory response to
provide a microenvironment conducive to ESCC development.
Additionally, supplementation with zinc reversed the inflammatory
signature and prevented cancer formation. Thus, the molecular
mechanism of ZD-induced inflammation has important clinical
implications as a critical factor in ESCC development. The aim of
the current study was to investigate the mechanism of ZD-induced
inflammation and how it promotes esophageal cancer development.
Materials and methods
Ethical approval
The current study was approved by the Institutional
Human Ethics Committee of Hebei Medical University Fourth Hospital
(Shijiazhuang, China) and the Laboratory Animal Ethics Committee of
Hebei Medical University Fourth Hospital. Written informed consent
was obtained from all patients prior to their participation.
Animals and experimental design
A total of 120 6-week-old C57BL/6 mice (18±2 g; 1:1
male/female ratio) were obtained from Beijing Vital River
Laboratory Animal Technology Co., Ltd. (Beijing, China). The ZD and
zinc-sufficient (ZS) diets were identical with the exception of
zinc content, which was 1.6 and 60.4 mg/kg (Beijing HFK Bioscience
Co. Ltd. Beijing, China), respectively. The mice were randomly
divided into two dietary groups (ZD and ZS, n=60 mice/group). All
mice were treated with 4-Nitroquinoline 1-oxide (4NQO;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in drinking water
(100 µg/ml) from 6 weeks of age to establish an esophageal
cancer model. The animals were weighed weekly and monitored daily.
From each group, 6 mice were sacrificed at 16, 20, 24 and 32 weeks
after chemical carcinogen treatment, and at 28 weeks, 36 mice were
sacrificed in each group for tumor evaluation and gene expression
profiling.
Human ESCC tissue samples
ESCC tissue and serum samples were obtained from 52
patients from Cixian, which is a region in Hebei province with a
high incidence of histologically confirmed ESCC (2). Of the 52 patients, 38 were male (73%)
and 14 were female (27%). The median age was 58 years old (range,
40-76 years old). The inclusion criterion was that the patient must
have received a pathological diagnosis of primary ESCC. All
patients were surgically treated at The Fourth Hospital of Hebei
Medical University (also known as the Tumor Hospital of Hebei
Province, which is a large and comprehensive level three, grade A
hospital) from January 2011 to December 2012. All specimens were
obtained within 30 min of surgery from each patient and immediately
stored in liquid nitrogen; serum samples were collected prior to
surgery. All patients were pathologically confirmed to have
early-stage ESCC at the Hebei Medical University Fourth
Hospital.
Immunohistochemical (IHC) assay
Human and mouse esophageal tissues were fixed in
formalin and embedded in wax blocks. Paraffin sections (4 µm
thick) were deparaffinized and rehydrated, followed by treatment
with 0.02 M EDTA buffer (pH 9.0; Gene Tech, Shanghai, China). Then,
the sections were immersed in 3% H2O2 to
quench endogenous peroxidase activity and blocked with 5% normal
goat serum (Zhongshanjingqiao Biotechnical Co., Ltd., Beijing,
China), followed by incubation with a monoclonal anti-COX-2
antibody (#12282, 1:250; Cell Signaling Technology, Inc., Danvers,
MA, USA) overnight at 4°C. The antibody was diluted in PBS buffer
containing 5% normal goat serum. The negative control for each
slide was incubated with 5% normal goat serum without the
anti-COX-2 antibody. The sections were then incubated with
horseradish peroxidase-conjugated anti-rabbit IgG (cat. no.
SP-9001, Zhongshanjingqiao Biotechnical Co., Ltd., Beijing, China)
for 45 min at 37°C and visualized with diaminobenzidine
tetrahydrochloride. The stained slides were examined under a light
microscope and scored by three pathologists who were blinded to the
clinical diagnosis. An index of COX-2 labeling was implemented so
that samples were scored according to the percentage and intensity
of staining among tumor cells (14).
Cell culture
The human esophageal cancer cell lines KYSE170 and
Eca109, human esophageal epithelial cells NE1 and human embryonic
kidney cells 293T were donated by the MD Anderson Cancer Center,
University of Texas (Houston, TX, USA). KYSE170 and Eca109 cells
were cultured in RPMI-1640 medium (Hyclone; GE Healthcare, Logan,
UT, USA) with 10% heat-inactivated fetal bovine serum (FBS;
PAN-Biotech, Adenbach, Germany) at 37°C in a 5% CO2
humidified incubator. NE1 and 293T cells were cultured in
Dulbecco’s modified Eagle’s medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with sodium pyruvate and 10%
FBS. The membrane-permeable zinc chelator
N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN;
Sigma-Aldrich; Merck KGaA) was prepared in ethanol to reduce zinc
levels. Esophageal cancer cells were cultured with a gradient of
different TPEN concentrations (from 0.001 to 0.011 µmol/ml
in steps of 0.001 µmol/ml) to study the effect of ZD on the
expression level of COX-2. When the TPEN concentration increased to
0.011 µmol/ml, the expression of COX-2 began to increase. ZD
cells were cultured with 0.011 µmol/ml TPEN in subsequent
experiments.
Cell-Counting-Kit 8 (CCK-8) assays
Esophageal cancer cells were seeded in 96-well
plates (5×103 cells/well). Proliferation was analyzed at
12, 24, 48 and 72 h with a simple test in which 10 µl of
CCK-8 (Solarbio Science and Technology Co., Ltd., Beijing, China)
reagent was added to each well. The plates were then incubated at
37°C and 5% CO2 for 2-4 h, and the absorbance was
recorded at 450 nm.
Transwell assays
For the Transwell invasion assay, 2.5×104
cells were plated in the top chamber with a Matrigel-coated
membrane (24-well insert; pore size, 8 µm; BD Biosciences,
Franklin Lakes, NJ, USA). Cells were plated in RPMI-1640 medium
modified without FBS, and medium supplemented with 10% FBS was used
as a chemoattractant in the lower chamber. The cells were incubated
for 24 h, and cells that had not invaded through the pores were
gently removed with a cotton swab. Cells on the lower surface of
the membrane were fixed and stained with 0.5% Giemsa crystal violet
solution (Solarbio Science and Technology Co., Ltd.) at room
temperature for 15 min and counted under light microscopy.
Flow cytometry assays
Cells were harvested, washed twice with FBS and then
fixed in a 70% ethanol solution for 24 h at 4°C. Then, the fixed
cells were washed once with PBS and resuspended as a single-cell
suspension. The suspension was incubated with 500 µl
propidium iodide (50 µg/ml, Solarbio Science and Technology
Co., Ltd.) for 30 min and evaluated using a flow cytometer.
Cell transfection
KYSE170 and Eca109 cells were plated ~16 h prior to
transfection. Hsa-miR-128 mimics were synthesized by Shanghai
GeneChem Co., Ltd. (Shanghai, China), and the sequence of the
mature miR-128 was 5′-UCA CAG UGA ACC GGU CUC UUU U-3′. Hsa-miR-128
mimics (2 µg/ml) were transfected into KYSE170 and Eca109
cells with Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Hsa-miR-scramble was transfected into KYSE170
and Eca109 cells in the same way as a control, and the sequence of
the scrambled miR was 5′-TGG ATC CAA GGT CGG GCA GGA AGA G-3′. The
transfected cells were incubated for 4-6 h, and normal medium was
added. The cells were harvested for further analysis after 48
h.
Luciferase reporter assay
Cox2-associated miRNAs were queried via TargetScan
(http://www.targetscan.org/mamm_31/)
and The Cancer Genome Atlas (TCGA) database (https://cancerge-nome.nih.gov/). The predicted
3′-untranslated region (3′UTR) of COX-2, binding to miR-128, was
cloned from the genomic DNA of esophageal cancer cells. The
detailed sequence was as follows: GGT TGA ATG TTT GTC CTT AGG ATA
GGC CTA TGT GCT AGC CCA CAA AGA ATA TTG TCT CAT TAG CCT GAA TGT GCC
ATA AGA CTG ACC TTT TAA AAT GTT TTG AGG GAT CTG TGG ATG CTT CGT TAA
TTT GTT CAG CCA CAA TTT ATT GAG AAA ATA TTC TGT GTC AAG CAC TGT GGG
TTT TAA TAT TTT TAA ATC AAA CGC TGA TTA CAG ATA ATA GTA TTT ATA TAA
ATA ATT GAA AAA AAT TTT CTT TTG GGA AGA GGG AGA AAA TGA AAT AAA TAT
CAT TAA AGA TAA CTC AGG AGA ATC TTC TTT ACA ATT TTA CGT TTA GAA TGT
T. The sequence was inserted into the pmirGLO control luciferase
reporter vector (Shanghai GeneChem Co., Ltd.). The miR-128
mimics/blank vector and the wild-type/mutant COX-2 3′UTR were
transfected into 293T cells and ECA109 cells with Lipofectamine
2000 (Shanghai GeneChem Co., Ltd.). Luciferase reporter assays were
performed using the Dual-Glo luciferase assay system 24 h after
transfection (GeneChem Co., Ltd., Shanghai, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples, serum
samples and esophageal cancer cells using the miRVana™ PARIS™ kit
(Ambion; Thermo Fisher Scientific, Inc.), according to the
manufacturer′s protocol. Total RNA for analysis of COX-2, CDK4,
CDK6, cyclin D1 and p53 genes was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA reverse
transcription was performed with a RevertAid First Strand cDNA
Synthesis kit (cat. no. K1622, Thermo Fisher Scientific, Inc.), for
mRNA and an miRNAFirst Strand cDNA Synthesis kit (cat. no.
B532453-0020, Sangon Biotech Co., Ltd., Shanghai, China) for miRNA.
miRNA levels were analyzed using RT-qPCR with the miRcute Plus
miRNA qPCR Detection kit (cat. no. FP411, Tiangen Biotech Co.,
Ltd., Beijing, China), and mRNA levels were analyzed with
GoTaq® qPCR and RT-qPCR Systems (cat. no. A6001; Promega
Corporation, Madison, WI, USA), SYBR Green and appropriate primers.
Briefly, 10 ng of total RNA was used as the template for 15
µl reverse transcription reactions. Probes were designed for
specific mature miRNAs. For each miRNA, reactions were performed in
triplicate using the 7500 RT-PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.), and RNU66 (Applied Biosystems; Thermo
Fisher Scientific, Inc.; cat. no. 4427975) was used as the
normalization control (9). Primer
sequences are presented in Table
I.
 | Table IPrimer sequences for quantitative
polymerase chain reaction. |
Table I
Primer sequences for quantitative
polymerase chain reaction.
| Name | Forward | Reverse |
|---|
| miR-16 |
CAGCCTAGCAGCACGTAAAT |
GAGGTATTCGCACCAGAGGA |
| miR-128 |
AACAAATATTAACACCTTCATACAACA |
TGGTGTCGTGGAGTCG |
| miR-144 |
GCGCGCTACAGTATAGATGATG |
GCTGTCAACGATACGCTACG |
| miR-146a |
GCAGGGTCCGAGGTATTCG |
CGCGTGAGAACTGAATTCCAT |
| RNU66 |
GTGCTCGCTTCGGCAGCACATATAC |
AAAAATATGGAACGCTCACGAATTTG |
| COX-2 |
GCTTTATGCTGAAGCCCTATGA |
TCCAACTCTGCAGACATTTCC |
| CDK4 |
GTTCGTGAGGTGGCTTTACT |
ATGTCCTTAGGTCCTGGTCT |
| CDK6 |
TCTTCCGTGTGAGTTGTTTG |
TTGTGTGGCTCTATGTGTGC |
| Cyclin D1 |
ATAATAAAGGGGTAATGGGG |
GCGTTGTAGGAGAAAGGAAT |
| P53 |
GGGTATCAAAGAAGGGCACT |
ATTCAGCTTGGTTTACGGGC |
| Rb |
CAAGGGTCATTATGGGTTAG |
TTAGGTGTAGGGGAGGGGAG |
DNA methylation analysis
5-aza-dCyd (Solarbio Science and Technology Co.,
Ltd.) treatment inhibits methylation of DNA. Genomic DNA from
esophageal cancer tissue samples and cell lines was purified using
DNAzol (Qiagen, Inc., Valencia, CA, USA). Sodium bisulfite
conversion was conducted using a Qiagen Epitect Bisulfite kit
(Qiagen, Inc.) according to the manufacturer’s protocol.
Methylation status of miR-128 was determined by
methylation-specific PCR (MSP). PCR products were visualized by
electrophoresis on a 2% agarose gel. The sequences of primers were
as follows: Methylation-specific forward primer, 5′-TAG TAA AGC GAG
AAT TTC GC-3′ and reverse primer, 5′-CTA ACC GCC GAA AAT AAA C-3′;
non-methylation-specific forward primer, 5′-GTA GTA AAG TGA GAA TTT
TGT-3′ and reverse primer, 5′-ACT AAC CAC CAA AAA TAA AC-3′.
DNA methyltransferase (DNMT)
activity
To estimate the activity of DNMTs, cells were
collected and treated with a zinc concentration gradient (from
0.001 to 0.011 µmol/ml in steps of 0.001 µmol/ml),
then the cell pellet was obtained (6,000 × g, 4°C, 5 min) and lysed
by sonication (12,000 Hz, 4°C, 5 min). The cells were then
centrifuged at 13,200 × g for 30 min at 4°C. The supernatant was
collected, and the protein concentrations in cell lysates were
determined with a Bradford reagent (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) using bovine serum albumin as the standard. The
activity of DNMTs was determined using the EpiSeeker DNMT Activity
Quantification Assay kit (Abcam, Cambridge, UK). In the assay, 5
µg of cell extract was incubated with a universal DNMT
substrate coated onto microplate wells. Methylated DNA was probed
with detection antibodies provided with the kit. The amount of
methylated DNA was colorimetrically quantified using ELISA to
indicate enzyme activity (cat. no. CGE601Hu01, Wuhan USCN Business
Co., Ltd., Wuhan, China).
Western blot analysis
Cells were lysed with ice-cold lysis buffer (Keygen
Biotech Co., Ltd., Nanjing, China) for 30 min on ice. Cell lysates
were then collected after centrifugation at 13,200 × g for 5 min at
4°C. Extracted protein concentration was determined by the BCA
method. Lysate proteins (60 µg) were loaded onto gels and
separated by 12% SDS-PAGE, then transferred overnight at 4°C onto
polyvinylidene difluoride membranes. Membranes were blocked with
0.05 g/ml non-fat milk blocking solution for 1 h at room
temperature. The membranes were incubated with antibodies
[anti-COX-2 (cat. no. 12282), anti-cyclin D1 (cat. no. 3300) and
anti-Rb (cat. no. 9309), 1:1,500, Cell Signaling Technology, Inc.;
anti-DNMT1 (cat. no. ab13537), anti-DNMT3A (cat. no. ab2850) and
anti-DNMT3B (cat. no. ab2851), 1:1,500, Abcam; anti-GAPDH (cat. no.
G9545), 1:2,500, Sigma-Aldrich, Merck KGaA) at 4°C overnight,
washed three times with TBST with 0.1% Tween, and incubated with a
horseradish peroxidase-linked secondary antibody (goat anti-rabbit,
cat. no. 925-68071, 1:2,000, LI-COR Biosciences, Lincoln, NE, USA)
for 1 h at room temperature. The membranes were washed three times
with TBST with 0.1% Tween. The Odyssey imaging system (LI-COR
Biosciences) was used to detect gray values.
Statistical analysis
All statistical analyses were performed using SPSS
13.0 software (SPSS Inc., Chicago, IL, USA). Quantitative results
are presented as the mean ± standard deviation. Comparisons of
means between two groups was conducted using Student’s t-test,
while comparisons among more than two groups were conducted using
one-way analysis of variance with Student-Newman-Keuls tests for
pairwise comparisons. Pearson’s correlation coefficient analysis
was used to evaluate the correlation between miR-128 expression in
tissue with COX-2 IHC scores or miR-128 in serum. P<0.05 was
considered to indicate a statistically significant difference.
Results
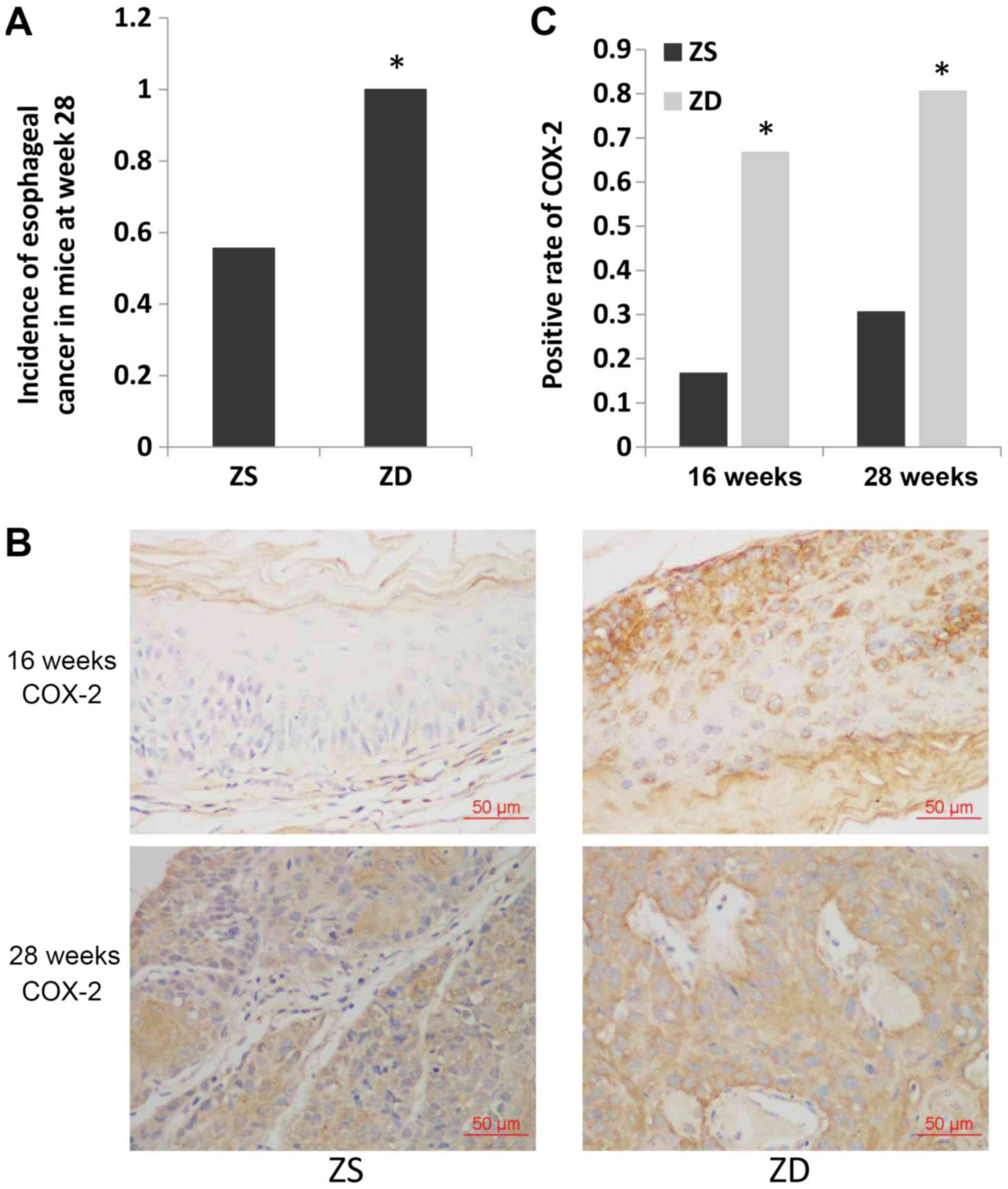
ZD diet increases the incidence of
esophageal cancer and increases the expression of COX-2 in
mice
Mice in the ZD and ZS groups were administered 4NQO
to induce esophageal tumorigenesis. Histological examination
revealed that esophageal cancer was first detected at 16 weeks in
mice in the ZD group (1/6) and at 20 weeks in mice in the ZS group
(1/6; data not shown). The cut-off date was set as week 28 after
4NQO treatment. The incidence of esophageal cancer was
significantly higher in mice in the ZD group compared with mice in
the ZS group at week 28 (36/36 vs. 20/36; P<0.05; Fig. 1A).
To investigate the temporal and spatial localization
of the key inflammation marker COX-2 in the ZD esophagus during
cancer development, IHC was performed. Both the precancerous
esophagus (16 weeks) and ESCC tissues (28 weeks) exhibited higher
expression of COX-2 with the ZD diet compared with the ZS diet in
mice (COX-2-positive rate: 4/6 vs. 1/6, P<0.05 and 29/36 vs.
11/36, P<0.05, respectively). These results demonstrated that
the expression of COX-2 increased due to ZD prior to the occurrence
of esophageal cancer.
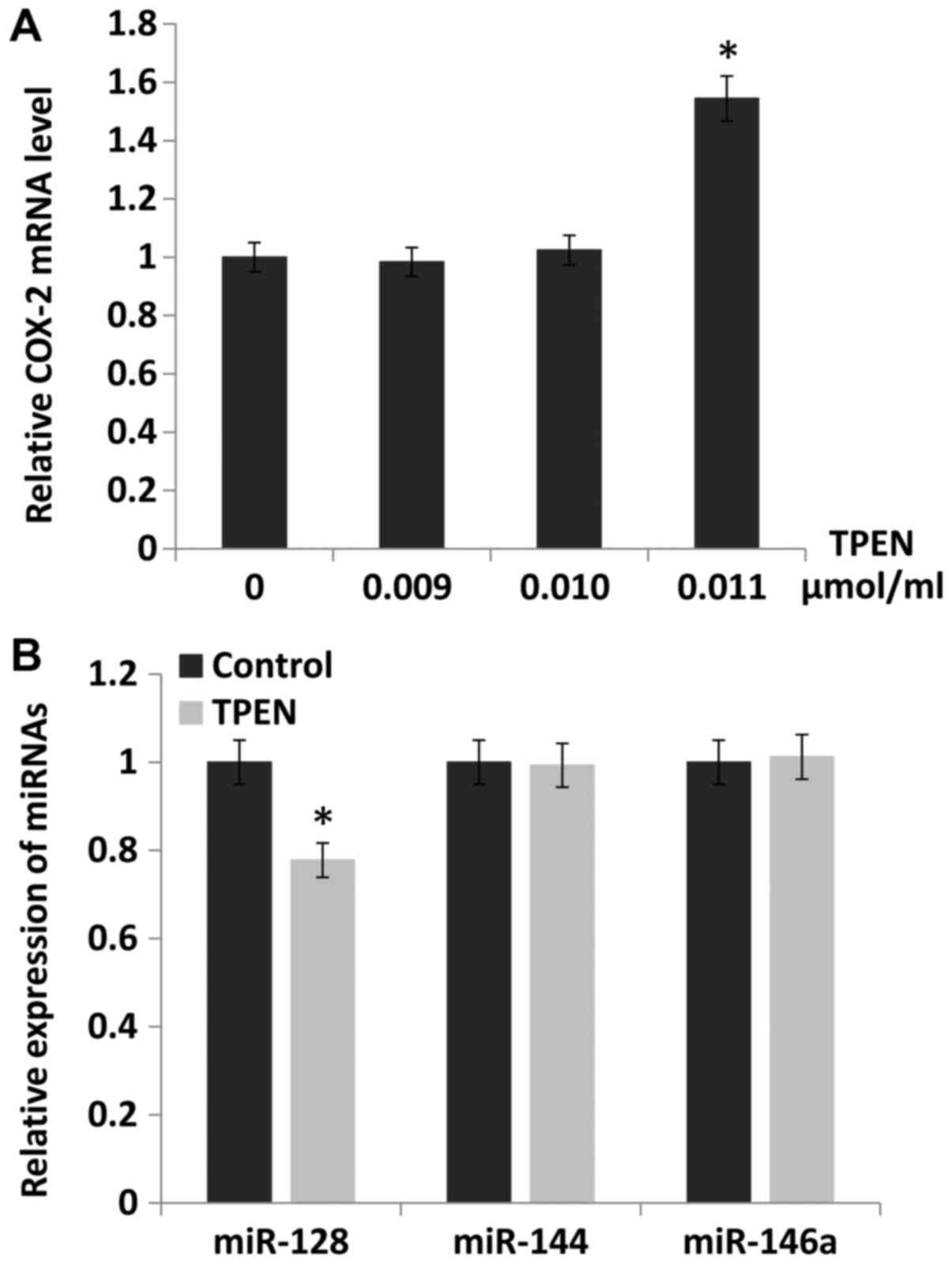
Downregulation of miR-128 by ZD increases
the expression of COX-2
To study the underlying mechanism by which COX-2
expression was increased by ZD, the public databases TargetScan and
TCGA were searched. It was identified that the expression of COX-2
could be directly regulated by miR-128, miR-144 and miR-146a. The
expression of COX-2 was evaluated in esophageal cancer cells
cultured with different zinc levels regulated by TPEN
concentration. The results indicated that the expression of COX-2
was increased by 54% when the concentration of TPEN was 0.011
µmol/ml (P<0.05; Fig.
2A). Furthermore, the expression of miR-128, miR-144 and
miR-146a were evaluated following treatment with 0.011
µmol/ml TPEN. It was identified that the expression of
miR-128 (P<0.05), but not miR-144 or miR-146a, was
downregulated, and the miR-128 expression level decreased by 22%
compared with the control (Fig.
2B).
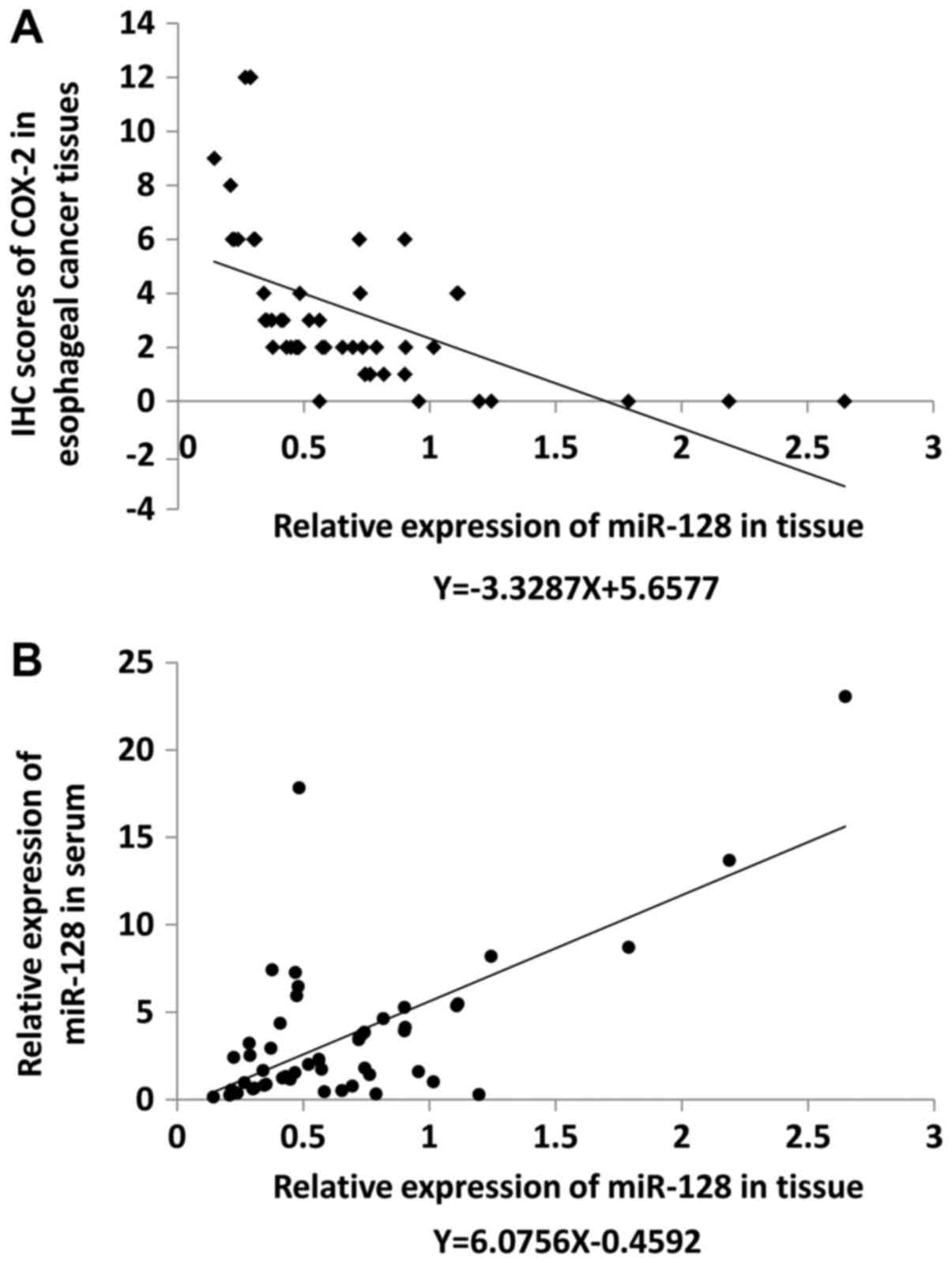
Expression of miR-128 in esophageal
cancer tissues was significantly lower compared with paracarcinoma
tissues and inversely related to the expression of COX-2
miR-128 expression levels were examined by qRT-PCR
and COX-2 expression level was evaluated by IHC in 52 pairs of
early esophageal cancer and paracarcinoma tissues. Compared with
the corresponding levels in paracarcinoma tissues, miR-128
expression level was upregulated in cancer tissues of 27% of
patients, and downregulated in cancer tissues of 73% of patients.
COX-2 expression was positive in 44% of adjacent tissues and 87% of
cancer tissues. The relative expression level of miR-128 was
negatively correlated with COX-2 expression as indicated by
regression analysis (R2=0.2749, F=18.954, P<0.05;
Fig. 3A). Relative miR-128
expression level in the serum was also detected using miR-16 as a
standard and was observed to be positively correlated with miR-128
expression levels in tissues (R2=0.4441, F=39.941,
P<0.05; Fig. 3B).
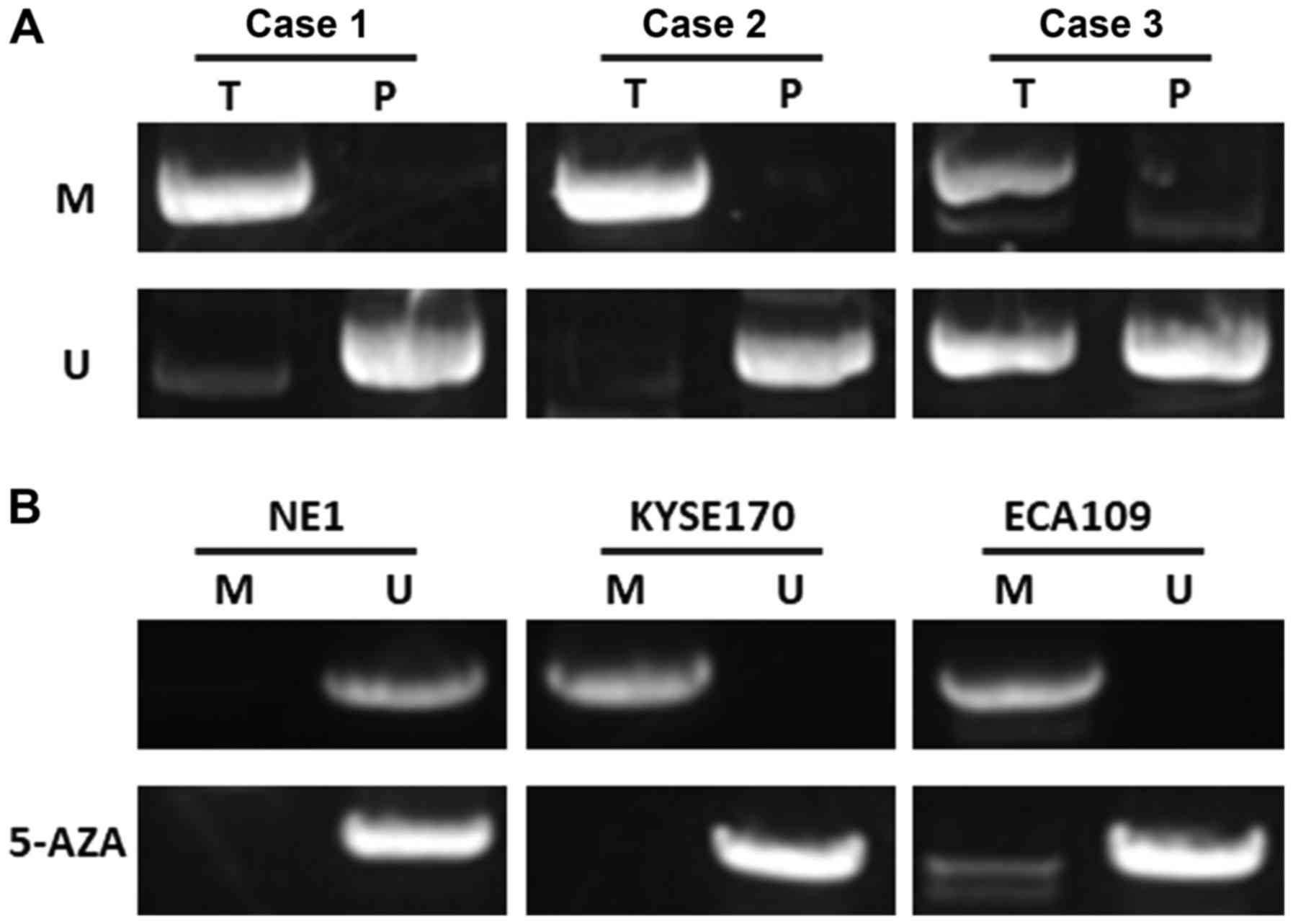
miR-128 is epigenetically silenced in
esophageal cancer
As a possible mechanism of suppression of miR-128,
the methylation status of CpG islands on promoter regions was
evaluated with MSP. It was identified that the CpG islands of
miR-128 were extensively methylated in esophageal cancer tumor
samples and cell lines. The methylation level in esophageal cancer
tissues was higher than that in paracarcinoma tissues (38/52 vs.
9/52; P<0.05; Fig. 4A). To
further determine whether the expression of miR-128 was silenced by
DNA methylation, miR-128 expression was determined in esophageal
cancer cell lines with or without 5-aza-dCyd treatment. The results
demonstrated that the miR-128 CpG islands were demethylated after
treatment with 5-aza-dCyd, indicating that miR-128 was
epigenetically silenced in esophageal cancer (Fig. 4B).
ZD decreases miR-128 expression levels by
increasing the methylation of miR-128 via enhancing DNMT
activity
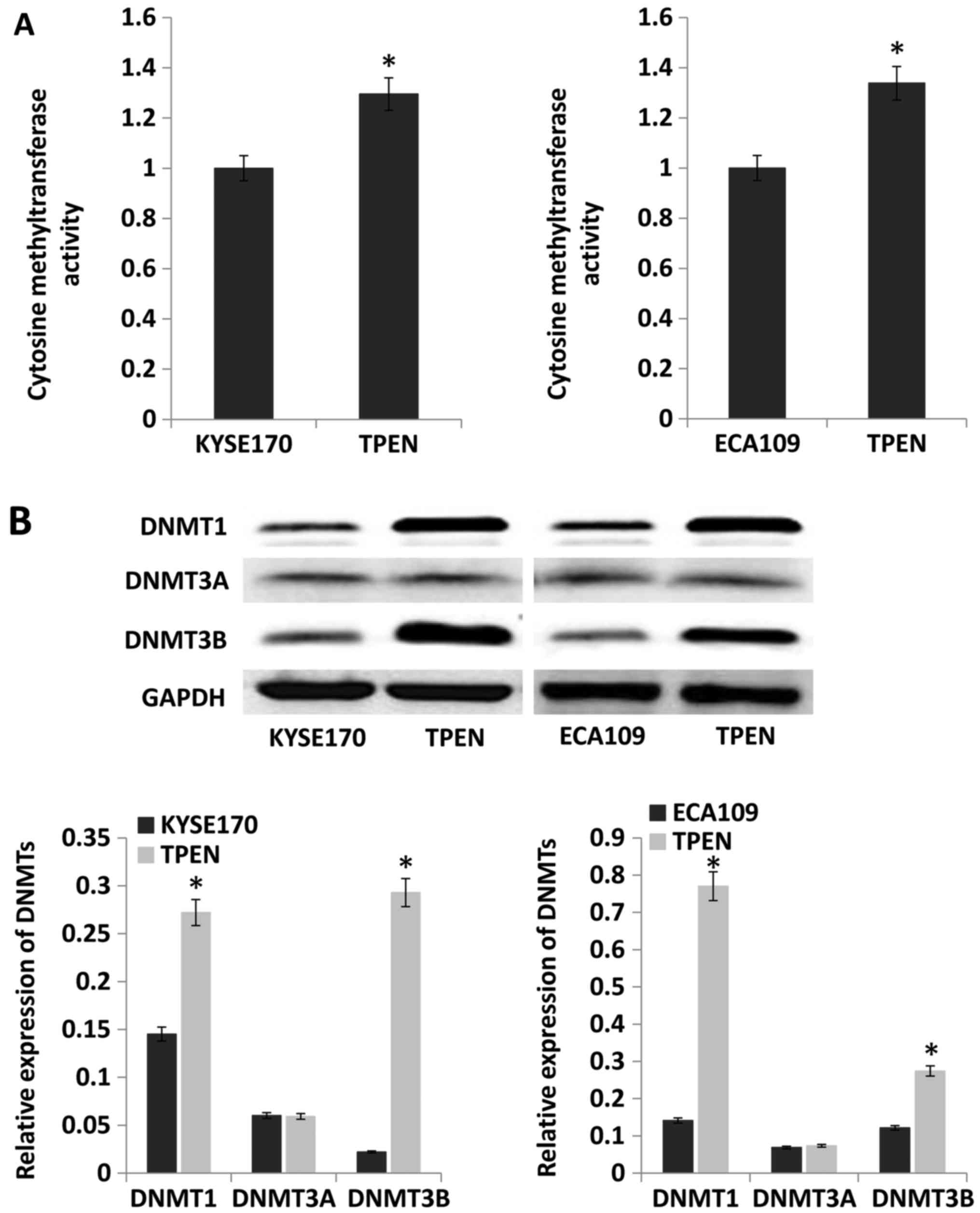
To examine the effect of ZD on DNMT activity, the
activity and expression level of DNMTs was evaluated. The results
indicated that compared with that in control cells, DNMT activity
was significantly enhanced by ~30 and 34% in KYSE170 and Eca109
cells cultured with TPEN, respectively (P<0.05; Fig. 5A). Protein expression levels of
DNMT1, DNMT3A and DNMT3B were evaluated by western blotting. The
expression of DNMT1 and DNMT3B was increased by TPEN, but there was
no difference in DNMT3A between cells cultured with and without
TPEN (Fig. 5B).
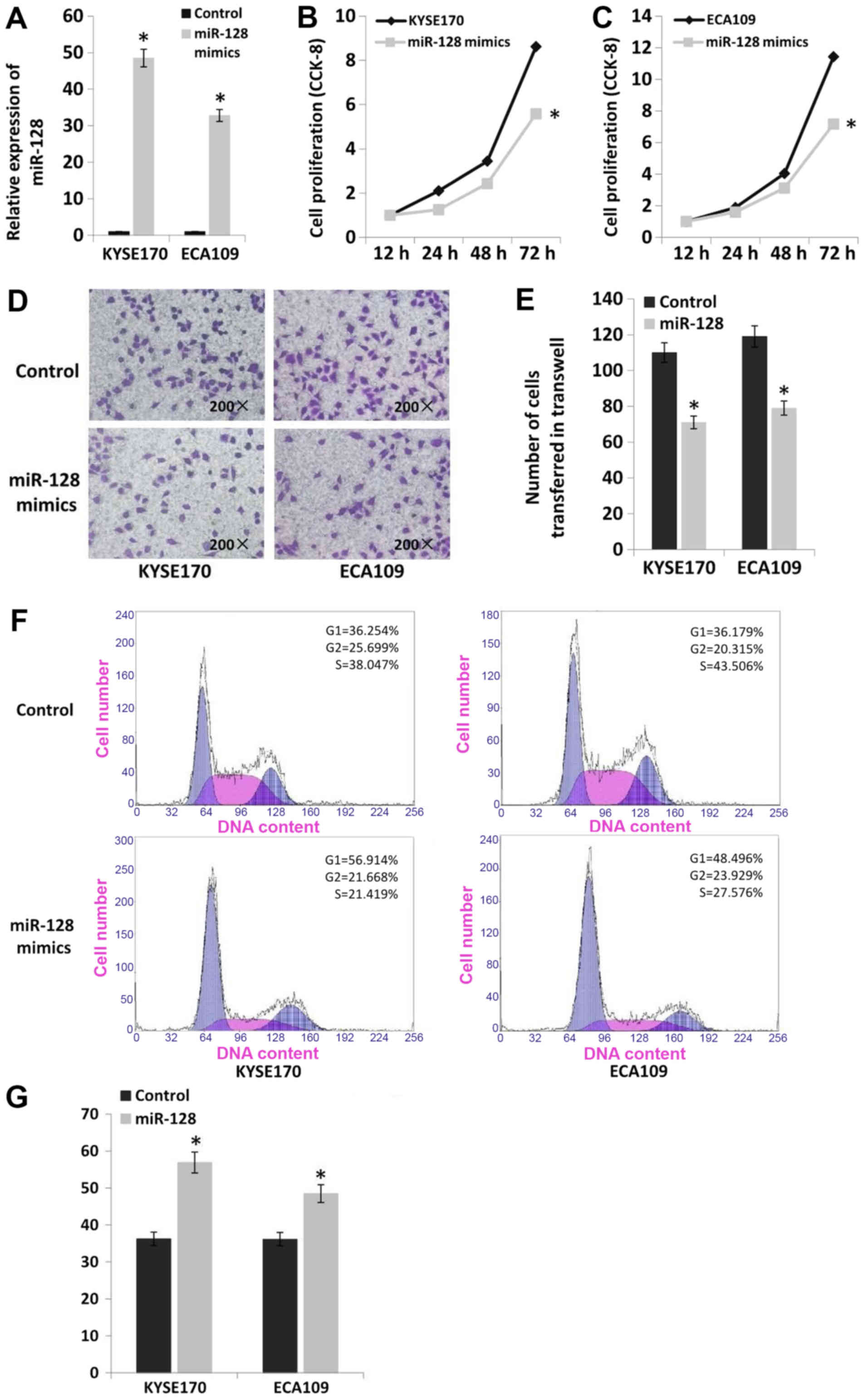
Upregulation of miR-128 inhibits the
proliferation and invasion of esophageal cancer cells
miR-128 mimics were transfected into the esophageal
cancer cell lines KYSE170 and Eca109 to upregulate the expression
of miR-128 (Fig. 6A). The
proliferation of cell lines with miR-128 upregulation was
significantly decreased compared with cells transfected with
scrambled controls according to CCK-8 assay results (P<0.05).
Proliferation was decreased by 35% in KYSE170 cells and 37% in
Eca109 cells (Fig. 6B and C).
Furthermore, upregulation of miR-128 significantly decreased the
invasion of esophageal cancer cells based on Transwell assays
(P<0.05). Invasion was decreased by 35% in KYSE170 and 34% in
Eca109 cells (Fig. 6D and E). The
results of flow cytometry analysis indicated that there was no
significant difference in apoptosis rate following miR-128
upregulation in the two cell lines (apoptosis rate <2%).
However, upregulation of miR-128 altered cell cycle distribution in
esophageal cancer cells. The number of cells in G1 phase was
increased by 20% in KYSE170 and 12% in Eca109 cells (Fig. 6F and G).
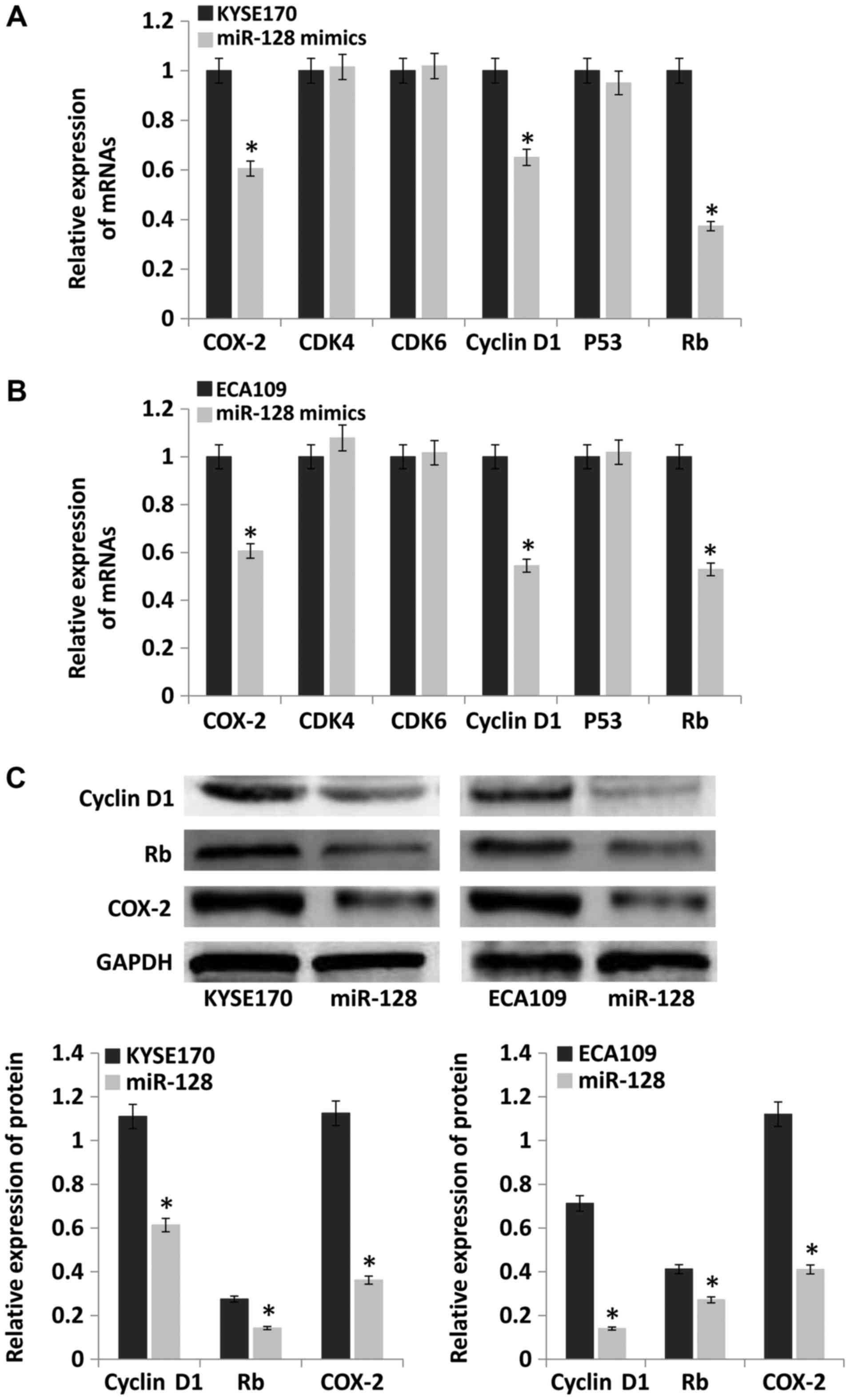
Upregulation of miR-128 inhibits the
expression of COX-2, cyclin D1 and retinoblastoma protein (Rb)
Cell cycle dysregulation is the main contributor to
tumorigenesis (15). The
expression of key cell cycle-related genes, cyclin-dependent kinase
(CDK)4, CDK6, cyclin D1, P53, Rb and COX-2, was evaluated by
RT-qPCR and western blotting. The results demonstrated that mRNA
levels of COX-2, cyclin D1 and Rb were significantly lower in the
miR-128 mimics group compared with the control group (P<0.05).
However, the mRNA levels of CDK4, CDK6 and P53 were not
significantly different between the groups (Fig. 7A and B). The protein levels of
cyclin D1, Rb and COX-2 were also detected, and the results were
consistent with mRNA levels (Fig.
7C).
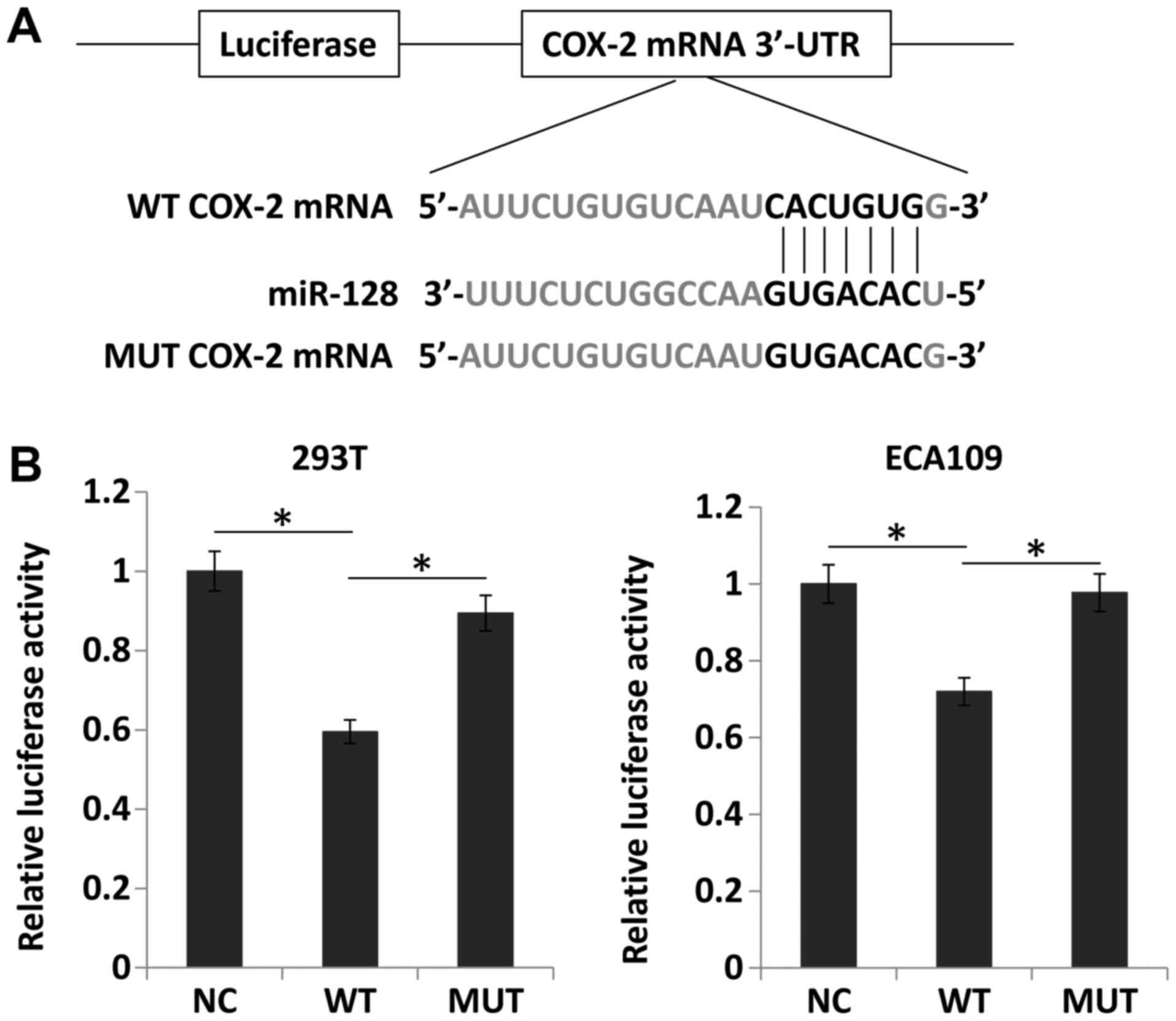
Upregulation of miR-128 directly inhibits
the expression of COX-2
The databases TargetScan and miRanda were searched
and it was identified that the seed sequence of miR-128 matched the
3′UTR of COX-2 mRNA (Fig. 8A).
Thus, miR-128 and luciferase reporter plasmids carrying the 3′UTR
of COX-2 containing the binding site of miR-128 were transfected
into 293T and ECA109 cells. Compared with the negative control,
miR-128 overexpression significantly downregulated luciferase
activity (P<0.05). However, miR-128 mimics did not alter the
luciferase activity of the mutant construct without miR-128 binding
sites (Fig. 8B). These results
suggested that miR-128 directly suppressed COX-2 expression by
targeting the 3′UTR of COX-2 mRNA.
Discussion
The majority of esophageal cancer cases worldwide
occur in the ‘Asian esophageal cancer belt’, which extends east
from northern Iran to China (16).
A well-known area for high incidence of esophageal cancer in China
is the Taihang Mountains, which are situated in Hebei and Henan
provinces. Cixian in Hebei province and its neighbor Linzhou in
Henan province are the areas with the highest incidence of
esophageal cancer in China and the world. The incidence of
esophageal cancer in Cixian is 176.9 per 100,000 among men and
108.8 per 100,000 among women (world age-standardized incidence
rate), which is much higher than the worldwide incidence (9.0 per
100,000 among men and 3.1 per 100,000 among women) (2). All of the patients in the current
study were from Cixian. Studies in areas with a high incidence of
esophageal cancer have implicated dietary ZD in the pathogenesis of
ESCC (5,9). Our previous study indicated that
serum zinc levels in ESCC patients were lower compared with those
in healthy people. Serum zinc levels in people from various
high-incidence areas of ESCC were also significantly lower compared
with those in people from low-incidence areas (9). To study the role of ZD in esophageal
cancer, mice were administered a ZD diet with 4NQO to establish an
esophageal cancer model. The results indicated that ZD could
promote the development of esophageal cancer. Certain inflammatory
factors were also evaluated and it was identified that ZD could
increase the expression of COX-2 prior to cancer initiation. These
results suggested that ZD could facilitate esophageal cancer
development by inducing certain inflammatory factors. These results
are consistent with the conclusions of Taccioli et al
(13).
The expression of inflammatory factors is regulated
by various mechanisms, including the posttranscriptional regulation
of gene expression by miRNAs. miRNAs are a family of short,
noncoding RNAs that play an important role in coordinating complex
programs of gene expression (17).
miRNAs alter many biological processes, including cellular
proliferation, apoptosis, immune response and signaling events
(18). The inflamed ZD esophagus
has a distinct miRNA signature that resembles human ESCC (19). Alder et al (20) demonstrated that chronic ZD induces
an inflammatory gene signature that fuels ESCC development and
induces a pro-tumorigenic miRNA signature. An inflamed ZD esophagus
exhibits dysregulation of specific miRNAs resembling the miRNA
signature of human ESCC, and zinc supplementation can prevent ESCC
by correcting aberrant miRNA expression. Fong et al
(21) also reported that miRNA
dysregulation and ESCC progression depend on the extent of dietary
ZD. Thus, ZD may promote the progression of esophageal cancer
through regulation of the expression of inflammatory cytokines by
changing the expression of miRNAs.
COX-2 is often induced during inflammation as a
lipid mediator of inflammation (22). To study the mechanisms leading to
the upregulated levels of COX-2 by ZD, in the current study the
expression levels of miRNAs that directly regulate COX-2 were
evaluated (23-25). The results indicated that miR-128
expression was decreased under ZD conditions, suggesting that the
expression of COX-2 may be regulated by miR-128 under ZD
conditions. Furthermore, the expression of miR-128 and COX-2 was
evaluated in clinical specimens and it was identified that the
expression of miR-128 in esophageal cancer tissues was negatively
associated with COX-2 expression. This result was consistent with
the results of the current in vitro experiments.
Furthermore, the effect of ZD on the expression of miR-128 was
evaluated. The results indicated that the expression of miR-128 was
downregulated by methylation and that ZD increased the methylation
of miR-128 by enhancing DNMT activity. DNA methylation can result
in altered miRNA expression. Numerous tumor suppressor genes have
already been identified to be methylated and transcriptionally
silenced in cancers (26). miR-128
has been reported to be downregulated by DNA methylation in
multiple cancers. For example, Yu et al (27) identified that miR-128 was
downregulated by DNA methylation in gastric cancer and induced
epithelial to mesenchymal transition through the P13K/AKT pathway.
Takahashi et al (28)
observed that miR-128 was silenced by DNA methylation in colorectal
cancer and played an important role via the NEK2 pathway. In the
current study, the role of miR-128 was evaluated in esophageal
cancer in vitro and it was demonstrated that upregulation of
miR-128 inhibited the proliferation and metastasis of esophageal
cancer and increased the proportion of cells in interphase during
cell division. Because cell cycle dysregulation is the main
contributor to the occurrence of esophageal cancer, the expression
of key cell cycle-associated genes was evaluated. The results
demonstrated that the upregulation of miR-128 inhibited the
expression of cyclin D1 and Rb. Cyclin D1 is a critical protein for
the regulation of the G1 phase of the cell cycle, and Rb promotes
the transition of cells from G1 to S phase (29). Therefore, these data indicate that
a decrease in miR-128 expression under ZD conditions promotes the
occurrence of esophageal cancer by regulating cyclin D1 and Rb
expression. COX-2 is a direct target of miR-128, and the expression
levels of cyclin D1 and Rb are altered by COX-2 expression
(23,30,31).
Thus, it is indicated that methylation-associated silencing of
miR-128 promotes the development of esophageal cancer by
upregulating the expression of cyclin D1 and Rb via targeting COX-2
in ZD areas with a high incidence of esophageal cancer.
The current study demonstrated the role of miR-128
and COX-2 in the early development of esophageal cancer and
therefore their value as biomarkers for the early diagnosis of
esophageal cancer could be considered. Although the expression
levels of miR-128 and COX-2 in esophageal cancer tissues may
reflect the progression of esophageal cancer, esophageal tissue
specimens cannot be obtained during screening or diagnosis of early
esophageal cancer. A good biomarker should be stable and easily
obtainable. The expression of miRNAs in serum is stable and
consistent with the expression of miRNAs in cancer tissues. For
example, Yoshida et al (32) identified that miR-25 was highly
expressed in the serum of patients with osteosarcoma and was also
observed in patient tissues. Komatsu et al (33) reported that miRNAs are stably
detectable in the serum and that circulating miRNAs may be tumor
markers for ESCC. Further research has indicated that circulating
miR-21 could be a useful biomarker for predicting chemoresistance
(34). These results suggest that
cancer may be diagnosable by detecting the levels of miRNAs in the
blood. In the current study, the expression level of miR-128 in the
serum of esophageal cancer patients was examined and was positively
associated with miR-128 expression levels in tissues. Therefore,
miR-128 expression levels in the serum may have potential use as a
biomarker for the diagnosis of early esophageal cancer.
Funding
This study was supported by grants from the National
Natural Scientific Foundation of China (grant no. 81272682) and the
Hebei Province Health Department (grant no. 20170745).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
JJ performed cell experiments. TG performed animal
experiments. YG analyzed the clinical specimens. JL performed flow
cytometry. FQ performed immunohistochemistry analysis. YH provided
theoretical guidance and supervised the study. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Institutional
Human Ethics Committee of Hebei Medical University Fourth Hospital
(Shijiazhuang, China) and the Laboratory Animal Ethics Committee of
Hebei Medical University Fourth Hospital. Written informed consent
was obtained from all patients prior to their participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Ferlay JSI, Ervik M, Dikshit R, Eser S,
Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: GLOBOCAN 2012
v10, Cancer Incidence and Mortality Worldwide: IARC CancerBase 11
Lyon, France International Agency for Research on Cancer. 2013,
http://globocan.iarc.fr.
Accessed December 12, 2013.
|
|
2
|
He J and Chen W: Chinese cancer registry
annual report. Press Mil Med Sci Beijing. 2012:160–161. 2012.
|
|
3
|
Anvari K, Sima HR, Seilanian Toussi M,
Anvari A, Shahidsales S, Memar B, Aledavoud SA, Forghani MN,
Abdollahi A and Ghaffarzadegan K: EGFR expression in patients with
esophageal squamous cell carcinoma and its association with
pathologic response to preoperative chemora-diotherapy: A study in
Northeastern Iran. Arch Iran Med. 20:240–245. 2017.PubMed/NCBI
|
|
4
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003-2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar
|
|
5
|
Hashemian M, Poustchi H, Abnet CC,
Boffetta P, Dawsey SM, Brennan PJ, Pharoah P, Etemadi A, Kamangar
F, Sharafkhah M, et al: Dietary intake of minerals and risk of
esophageal squamous cell carcinoma: Results from the Golestan
Cohort Study. Am J Clin Nutr. 102:102–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abnet CC, Lai B, Qiao YL, Vogt S, Luo XM,
Taylor PR, Dong ZW, Mark SD and Dawsey SM: Zinc concentration in
esophageal biopsy specimens measured by X-ray fluorescence and
esophageal cancer risk. J Natl Cancer Inst. 97:301–306. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gaither LA and Eide DJ: Eukaryotic zinc
transporters and their regulation. Biometals. 14:251–270. 2001.
View Article : Google Scholar
|
|
8
|
Zou XN, Taylor PR, Mark SD, Chao A, Wang
W, Dawsey SM, Wu YP, Qiao YL and Zheng SF: Seasonal variation of
food consumption and selected nutrient intake in Linxian, a high
risk area for esophageal cancer in China. Int J Vitam Nutr Res.
72:375–382. 2002. View Article : Google Scholar
|
|
9
|
He Y, Jin J, Wang L, Hu Y, Liang D, Yang
H, Liu Y and Shan B: Evaluation of miR-21 and miR-375 as prognostic
biomarkers in oesophageal cancer in high-risk areas in China. Clin
Exp Metastasis. 34:73–84. 2017. View Article : Google Scholar :
|
|
10
|
Balkwill F and Mantovani A: Inflammation
and cancer: back to Virchow? Lancet. 357:539–545. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Taccioli C, Wan S-G, Liu C-G, Alder H,
Volinia S, Farber JL, Croce CM and Fong LY: Zinc replenishment
reverses overex-pression of the proinflammatory mediator S100A8 and
esophageal preneoplasia in the rat. Gastroenterology. 136:953–966.
2009. View Article : Google Scholar
|
|
13
|
Taccioli, Chen H, Jiang Y, Liu XP, Huang
K, Smalley KJ, Farber JL, Croce CM and Fong LY: Dietary zinc
deficiency fuels esophageal cancer development by inducing a
distinct inflammatory signature. Oncogene. 31:4550–4558. 2012.
View Article : Google Scholar :
|
|
14
|
Jin J, Li Z, Liu J, Wu Y, Gao X and He Y:
Knockdown of zinc transporter ZIP5 (SLC39A5) expression
significantly inhibits human esophageal cancer progression. Oncol
Rep. 34:1431–1439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao S, Jiang Y, Zhao J, Li H, Yin X, Wang
Y, Xie Y, Chen X, Lu J, Dong Z, et al: Quercetin-3-methyl ether
inhibits esophageal carcinogenesis by targeting the AKT/mTOR/p70S6K
and MAPK pathways. Mol Carcinog. 57:1540–1552. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cortés González R and Villaseñor Caloca R:
Esophageal cancer. Rev Gastroenterol Mex. 62:149–159. 1997.In
Spanish.
|
|
17
|
Ambros V: MicroRNA pathways in flies and
worms: Growth, death, fat, stress, and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fong LY, Taccioli C, Jing R, Smalley KJ,
Alder H, Jiang Y, Fadda P, Farber JL and Croce CM: MicroRNA
dysregulation and esophageal cancer development depend on the
extent of zinc dietary deficiency. Oncotarget. 7:10723–10738. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alder H, Taccioli C, Chen H, Jiang Y,
Smalley KJ, Fadda P, Ozer HG, Huebner K, Farber JL, Croce CM, et
al: Dysregulation of miR-31 and miR-21 induced by zinc deficiency
promotes esophageal cancer. Carcinogenesis. 33:1736–1744. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fong LY, Taccioli C, Jing R, Smalley KJ,
Alder H, Jiang Y, Fadda P, Farber JL and Croce CM: MicroRNA
dysregulation and esophageal cancer development depend on the
extent of zinc dietary deficiency. Oncotarget. 7:10723–1038. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Grosser T, Yu Y and Fitzgerald GA: Emotion
recollected in tranquility: Lessons learned from the COX-2 saga.
Annu Rev Med. 61:17–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin Y and Wu Z: MicroRNA-128 inhibits
proliferation and invasion of glioma cells by targeting COX-2.
Gene. 658:63–69. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yao Q, Gu A, Wang Z and Xue Y:
MicroRNA-144 functions as a tumor suppressor in gastric cancer by
targeting cyclooxy-genase-2. Exp Ther Med. 15:3088–3095.
2018.PubMed/NCBI
|
|
25
|
Cornett AL and Lutz CS: Regulation of
COX-2 expression by miR-146a in lung cancer cells. RNA.
20:1419–1430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Espinosa-Parrilla Y, Muñoz X, Bonet C,
Garcia N, Venceslá A, Yiannakouris N, Naccarati A, Sieri S, Panico
S, Huerta JM, et al: Genetic association of gastric cancer with
miRNA clusters including the cancer-related genes MIR29, MIR25,
MIR93 and MIR106: Results from the EPIC-EURGAST study. Int J
Cancer. 135:2065–2076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu WW, Jiang H, Zhang CT and Peng Y: The
SNAIL/miR-128 axis regulated growth, invasion, metastasis, and
epithelial-to-mesen-chymal transition of gastric cancer.
Oncotarget. 8:39280–39295. 2017.PubMed/NCBI
|
|
28
|
Takahashi Y, Iwaya T, Sawada G, Kurashige
J, Matsumura T, Uchi R, Ueo H, Takano Y, Eguchi H, Sudo T, et al:
Up-regulation of NEK2 by microRNA-128 methylation is associated
with poor prognosis in colorectal cancer. Ann Surg Oncol.
21:205–212. 2014. View Article : Google Scholar
|
|
29
|
Feng Z, Xia Y, Gao T, Xu F 1, Lei Q 1,
Peng C 2, Yang Y 3, Xue Q 1, Hu X 1, Wang Q, et al: The
antipsychotic agent trifluoperazine hydrochloride suppresses
triple-negative breast cancer tumor growth and brain metastasis by
inducing G0/G1 arrest and apoptosis. Cell Death Dis. 9:10062018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhuang ZH, Tsao SW, Deng W, Wang JD, Xia
HH, He H, Feng HC, Wang LD, Gu Q, Lam SK, et al: Early upregulation
of cyclooxygenase-2 in human papillomavirus type 16 and
telom-erase-induced immortalization of human esophageal epithelial
cells. J Gastroenterol Hepatol. 23:1613–1620. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu B, Wen JK, Li BH, Fang XM, Wang JJ,
Zhang YP, Shi CJ, Zhang DQ and Han M: Celecoxib and
acetylbritannilactone interact synergistically to suppress breast
cancer cell growth via COX-2-dependent and -independent mechanisms.
Cell Death Dis. 28:e1852011. View Article : Google Scholar
|
|
32
|
Yoshida A, Fujiwara T, Uotani K, Morita T,
Kiyono M, Yokoo S, Hasei J, Nakata E, Kunisada T and Ozaki T:
Clinical and functional significance of intracellular and
extracellular microRNA-25-3p in osteosarcoma. Acta Med Okayama.
72:165–174. 2018.PubMed/NCBI
|
|
33
|
Komatsu S, Ichikawa D, Takeshita H,
Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H,
Shiozaki A, et al: Circulating microRNAs in plasma of patients with
oesophageal squamous cell carcinoma. Br J Cancer. 105:104–111.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Komatsu S, Ichikawa D, Kawaguchi T,
Miyamae M, Okajima W, Ohashi T, Imamura T, Kiuchi J, Konishi H,
Shiozaki A, et al: Circulating miR-21 as an independent predictive
biomarker for chemoresistance in esophageal squamous cell
carcinoma. Am J Cancer Res. 6:1511–1523. 2016.PubMed/NCBI
|