Introduction
Liver cirrhosis is one of the most common illnesses
compromising human health and no completely effective clinical
treatment exists. The mechanisms underlying the development of
hepatic fibrosis (HF) and effective treatments for this condition
remain to be elucidated. Receptor for advanced glycation end
products (RAGE), a member of the immunoglobulin superfamily
(1), is found on the surface of
various cell types, including mononuclear macrophages, neurons,
renal mesangial cells and vascular endothelial cells. This protein
contributes to various diseases, including type 2 diabetes,
Alzheimer’s disease, chronic kidney disease and atherosclerosis
(2–5). AGEs are implicated in the
pathogenesis of fibrosis in a number of tissue types (6,7) and
evidence indicates that AGEs and RAGE contribute to the
pathogenesis of chronic liver disease (6). RAGE can be significantly expressed on
several types of liver cell, including hepatic stellate cells
(HSCs) (8–10), which are the major effectors during
hepatic fibrogenesis (7). The
expression of RAGE is also increased in animal models exhibiting
chronic liver disease (11,12).
Therefore, RAGE may be important in the development of HF and
inhibiting RAGE may be a method to prevent or reverse HF.
Specific small interfering RNA (siRNA) targeting
RAGE, inhibits the expression levels of RAGE, α-smooth muscle actin
and collagen type I in T6 HSCs, indicating that RAGE may be
important for the activation of HSCs and the expression of collagen
(11). The present study aimed to
investigate the effect of RAGE-specific siRNAs on the development
of HF in primary rat HSCs.
Materials and methods
Materials
Three Healthy male Sprague-Dawley rats, aged 15
months and weighing between 400 and 500 g, were purchased from the
Nanjing Medical University Laboratory Animal Center (Nanjing,
China). The study was approved by the Animal Research Ethics
Committee of Zhongda Hospital, Southeast University (Nanjing,
China). The rats were fed a normal diet and had free access to food
and water; in addition, the rats were maintained at a temperature
of 22°C under a 12-h light/dark cycle. Type IV collagenase, pronase
E and Nycodenz were obtained from Sigma-Aldrich (St. Louis, MO,
USA) and DNase I was obtained from Gibco Life Technologies
(Carlsbad, CA, USA). The pAKD.CMV.bGlobin.egreen fluorescent
protein (GFP).H1.short hairpin (sh)RNA vector was purchased from
GenScript USA, Inc. (Piscataway, NJ, USA).
Isolation and culture of primary rat
HSCs
HSCs were obtained from rats as previously described
(13). Primary rat HSCs were
isolated by density gradient centrifugation at 400 × g for 5 min
and cultured with Dulbecco’s modified Eagle’s medium (DMEM;
Sigma-Aldrich) supplemented with 10% fetal bovine serum in
vitro. The cell viability was determined using trypan blue
staining (Sigma-Aldrich) and the expression of desmin in cells was
detected by immunohistochemistry. The primary HSCs were cultured
for 5 days and divided at random into five groups as follows: Cells
transfected with pAKD-GR125, pAKD-GR126, pAKD-GR127, pAKD-GR128 or
pAKD-GR129.
Preparation of specific siRNAs targeting
RAGE
Rat RAGE mRNA (GenBank accession no. NM-053336.1)
was used as the target sequence. siRNA Target Finder design
software (version 2.0; Ambion, Austin, TX, USA) was used to model
the secondary structures of the rat RAGE mRNA and five pairs of 19
nt siRNA sequences were designed in accordance with the target
sequences and their complementary sequences (the specificity of the
sequences were confirmed using BLAST). These sequences were then
converted into short RNA oligonucleotide sequences, which form
hairpin structures. BglII and KpnI restriction sites
and a 9 bp hairpin structure were added to the two ends of the
sequence. The final oligonucleotides were named GR125, GR126,
GR127, GR128 and GR129 (Table
1).
 | Table ISequences of specific small
interfering RNAs targeting receptor for advanced glycation end
products. |
Table I
Sequences of specific small
interfering RNAs targeting receptor for advanced glycation end
products.
|
Oligonucleotide | Sequence |
|---|
| GR125 |
| Sense |
5′-GATCCCCGCCAACCCAGAAGCTAGAATTCAAGAGATTCTAGCTTCTGGGTTGGCTTTTTTGTAC-3′ |
| Antisense |
5′-AAAAAAGCCAACCCAGAAGCTAGAATCTCTTGAATTCTAGCTTCTGGGTTGGCGGG-3′ |
| GR126 |
| Sense |
5′-GATCCCCGTGAATCCTGCCTCTGAACTTCAAGAGAGTTCAGAGGCAGGATTCACTTTTTTGTAC-3′ |
| Antisense |
5′-AAAAAAGTGAATCCTGCCTCTGAACTCTCTTGAAGTTCAGAGGCAGGATTCACGGG-3′ |
| GR127 |
| Sense |
5′-GATCCCCGCCTCTGAACTCACAGCCATTCAAGAGATGGCTGTGAGTTCAGAGGCTTTTTTGTAC-3′ |
| Antisense |
5′-AAAAAAGCCTCTGAACTCACAGCCATCTCTTGAATGGCTGTGAGTTCAGAGGCGGG-3′ |
| GR128 |
| Sense |
5′-GATCCCCGAAGGTGGAACAGTCGCTCTTCAAGAGAGAGCGACTGTTCCACCTTCTTTTTTGTAC-3′ |
| Antisense |
5′-AAAAAAGAAGGTGGAACAGTCGCTCTCTCTTGAAGAGCGACTGTTCCACCTTCGGG-3′ |
| GR129 |
| Sense |
5′-GATCCCCGCGAAAACGACAACCCAGATTCAAGAGATCTGGGTTGTCGTTTTCGCTTTTTTGTAC-3′ |
| Antisense |
5′-AAAAAAGCGAAAACGACAACCCAGATCTCTTGAATCTGGGTTGTCGTTTTCGCGGG-3′ |
| NC |
| Sense |
5′-GATCCCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTGATC-3′ |
| Antisense |
5′-AAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAGGG-3′ |
The BglII and KpnI restriction sites
and the hairpin structure sequence were also added to the two ends
of another pair of non-specific siRNAs (not homologous to rat RAGE
mRNA, as confirmed by BLAST). This construct was termed negative
control (NC; Table 1).
Construction of specific siRNA expression
vectors
The pAKD.CMV.bGlobin.eGFP.H1.shRNA vector was
linearized by restriction enzyme digestion using BglII and
KpnI and then ligated to the annealed double-stranded DNA
fragments GR125, GR126, GR127, GR128 and GR129, forming the
RAGE-specific siRNA expression vectors pAKD-GR125, pAKD-GR126,
pAKD-GR127, pAKD-GR128 and pAKD-GR129, respectively. The
non-specific siRNA expression vector, pAKD-NC, was constructed in
the same way and was used as a control.
Cell transfection
The pAKD-GR125, pAKD-GR126, pAKD-GR127, pAKD-GR128
and pAKD-GR129 RAGE siRNA expression vectors, were transfected into
primary rat HSCs individually at multiplicity of infections (MOIs)
of 20, 100, 200 and 1,000. Untreated and nonspecific
siRNA-transfected primary rat HSCs were used as controls. The
medium was replaced with serum-free DMEM prior to transfection. The
total RNA (0.5 μg) was extracted and the mRNA expression of
RAGE was determined by reverse transcription quantitative
polymerase chain reaction (RT-qPCR) following incubation for 48 h
at 37°C (11). RT reagent kit was
purchased from Takara Bio, Inc. (Dalian, China) and the primers
used were as follows: GR-129 sense,
5′-GATCCCCGTGAATCCTGCCTCTGAACTTCAAGAGAGTTCAGAGGCAGGATTCACTTTTTTGTAC-3′
and antisense,
5′-AAAAAAGTGAATCCTGCCTCTGAACTCTCTTGAAGTTCAGAGGCAGGATTCAC GGG-3′.
ABI-9700 PCR apparatus (Applied Biosystems, Waltham, MA, USA) was
used and the cycling parameters were as follows: 94°C for 3 min,
followed by 40 cycles of 94°C for 1 min, 55°C for 1 min and 72°C
for 1 min. β-actin served as the endogenous control, ΔΔCT method
for relative quantification was used to calculate differences in
the expression level of each target gene (14).
The specific siRNA expression vector pAKD-GR126 with
the maximum ability to inhibit the expression of RAGE was selected
and transfected into primary rat HSCs (0.2×106) cultured
in DMEM for 5 days at 37°C. Untreated and nonspecific
siRNA-transfected primary rat HSCs served as controls. The cells
were collected following incubation for 24, 48 and 72 h, and the
total RNA was extracted. The efficiency of RAGE gene silencing was
assessed by RT-qPCR, as described below.
Cell treatment
Primary rat HSCs (0.2×106) cultured for 5
days at 37°C were randomly divided into three groups (n=3/group):
Group A, blank group, treated with AGE-bovine serum albumen (BSA,
200 mg/l); the pAKD-GR126 group, treated with AGE-BSA (200 mg/l)
and pAKD-GR126 (MOI=1,000) and the pAKD-NC group, treated with
AGE-BSA (200 mg/l) and pAKD-NC (MOI=1,000). Each group had three
repeats. The medium was replaced with serum-free DMEM prior to
transfection.
Total RNA extraction and RT-qPCR
assay
The total RNA was extracted from the primary rat
HSCs using TRIzol reagent (Sigma-Aldrich) according to the
manufacturer’s instructions. The purity and concentration of the
RNA were determined prior to the RNA being reverse transcribed, as
previously described (11). The
cycling parameters were as follows: 94°C for 3 min and 40 cycles of
94°C for 1 min, 55°C for 1 min and 72°C for 1 min. β-actin served
as an internal control. The 2−ΔΔCT method for relative
quantification was used to calculate the differences in the
expression level of each target gene (11,14).
Western blot analysis
The cellular proteins were extracted using modified
radioimmunoprecipitation buffer (Sigma-Aldrich) containing 50 mM
Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl
sulphate (SDS), 0.5% deoxycholate, 1 mM EDTA, 2 mg/l leupeptin and
1 mM phenylmethylsulfonyl fluoride. The protein concentration was
determined using a bicinchoninic acid assay. The proteins (50
μg) were separated by electrophoresis on a 10% gradient
SDS-polyacrylamide gel (Sigma-Aldrich) and transferred onto
polyvinylidene fluoride membranes (Sigma-Aldrich). Following
protein transfer, the membranes were blocked with 5% non-fat milk
(Sigma-Aldrich) in Tris-buffered saline containing 0.1% Tween-20
(Sigma-Aldrich) for 1 h at room temperature and then incubated
overnight at 4°C with the following primary antibodies: Rabbit
anti-rat polyclonal RAGE (ab3611), rabbit anti-rat polyclonal
interleukin (IL)-6 (ab6672), mouse anti-rat monoclonal tumor
necrosis factor (TNF)-α (ab92324), mouse anti-rat monoclonal
transforming growth factor (TGF)-β1 (ab64715), rabbit anti-rat
polyclonal connective tissue growth factor (CTGF; ab6992), mouse
anti-rat monoclonal laminin (LN; ab8983), rabbit anti-rat
polyclonal hyaluronic acid (HA; ab20084), mouse anti-rat polyclonal
N-terminal procollagen III propeptide (PIIINP; ab169636) and mouse
anti-rat monoclonal β-actin (ab8226), which were all purchased from
Abcam (Cambridge, MA, USA). The membranes were subsequently washed
with TBST and then exposed to a secondary horseradish
peroxidase-labelled antibody [ab6721, goat polyclonal antibodies
against rabbit IgG - H&L (HRP) and ab193651, rabbit polyclonal
antibodies against mouse IgG - H&L (HRP)] in the blocking
solution for 1 h at room temperature. The band intensities were
measured using an enhanced chemiluminescent reagent
(Western-Lightening Plus; PerkinElmer Life Sciences, Waltham, MA,
USA) and the protein signals were normalized against β-actin.
Statistical analysis
Statistical analyses were performed using SPSS 13.0
statistical software (SPSS, Inc., Chicago, IL, USA). The data were
analyzed using one-way analysis of variance and
Student-Newman-Keuls multiple comparison test. P<0.05 was
considered to indicate a statistically significant difference. The
data are expressed as the mean ± standard deviation.
Results
Inhibitory effect of RAGE-specific siRNAs
on the expression of RAGE in primary rat HSCs
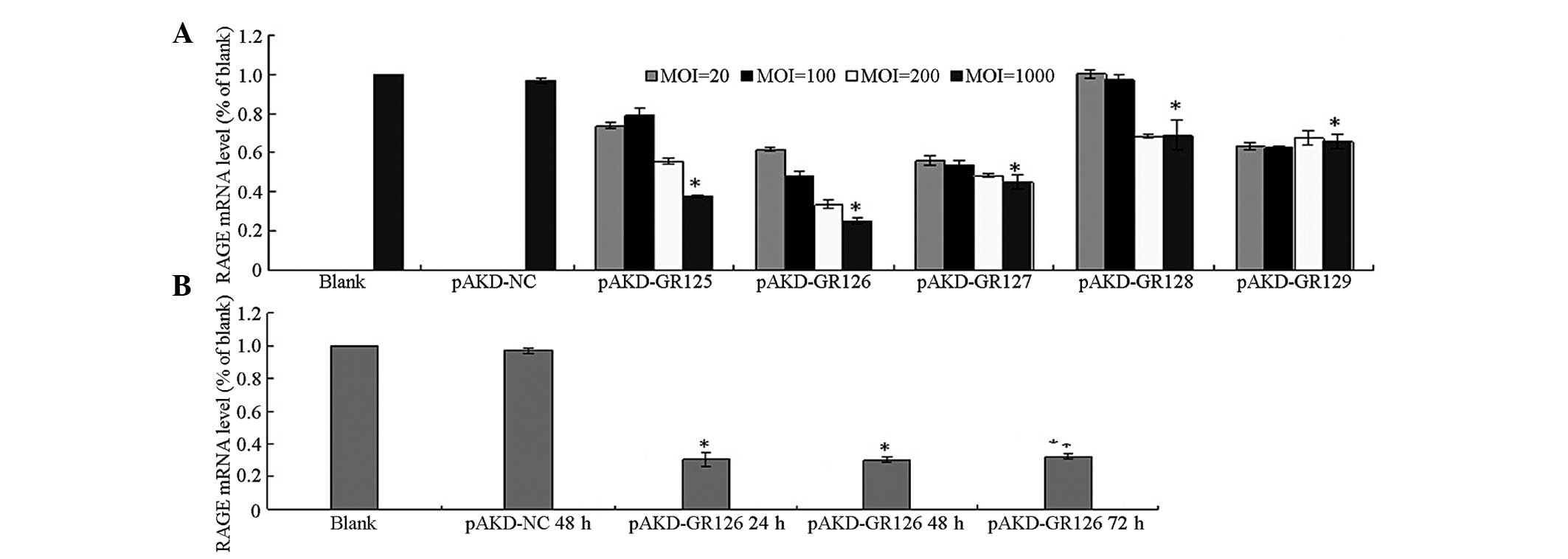
The mRNA expression of RAGE was significantly
downregulated in the primary rat HSCs treated with pAKD-GR125,
pAKD-GR126, pAKD-GR127, pAKD-GR128 and pAKD-GR129 (all P<0.05)
compared with the untreated primary rat HSCs and the HSCs treated
with pAKD-NC. The mRNA expression of RAGE decreased in a
dose-dependent manner in the MOI range of between 20 and 1,000. The
most marked decrease was observed in the HSCs treated with
pAKD-GR126 at an MOI of 1,000 (Fig.
1A). The mRNA expression of RAGE was downregulated in the
primary rat HSCs treated with pAKD-GR126 at 24, 48 and 72 h (all
P<0.05) compared with the untreated cells. However, inhibition
in the mRNA expression of RAGE exhibited no differences between 24
and 72 h (P>0.05; Fig. 1B).
These results indicated that the RAGE siRNA expressed by pAKD-GR126
effectively inhibited the expression of RAGE.
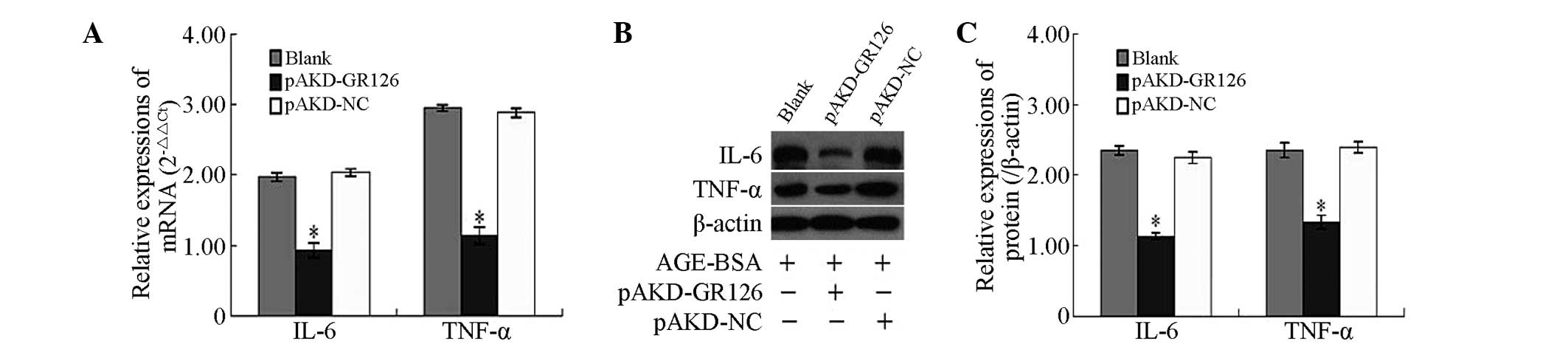
Inhibitory effect of pAKD-GR126 on the
mRNA and protein expression levels of the IL-6 and TNF-α
pro-inflammatory cytokines in primary HSCs
The mRNA and protein expression levels of the IL-6
and TNF-α pro-inflammatory cytokines, were significantly
downregulated in the primary rat HSCs treated with pAKD-GR126
(P<0.05) compared with the untreated primary rat HSCs and those
treated with the pAKD-NC (Fig.
2).
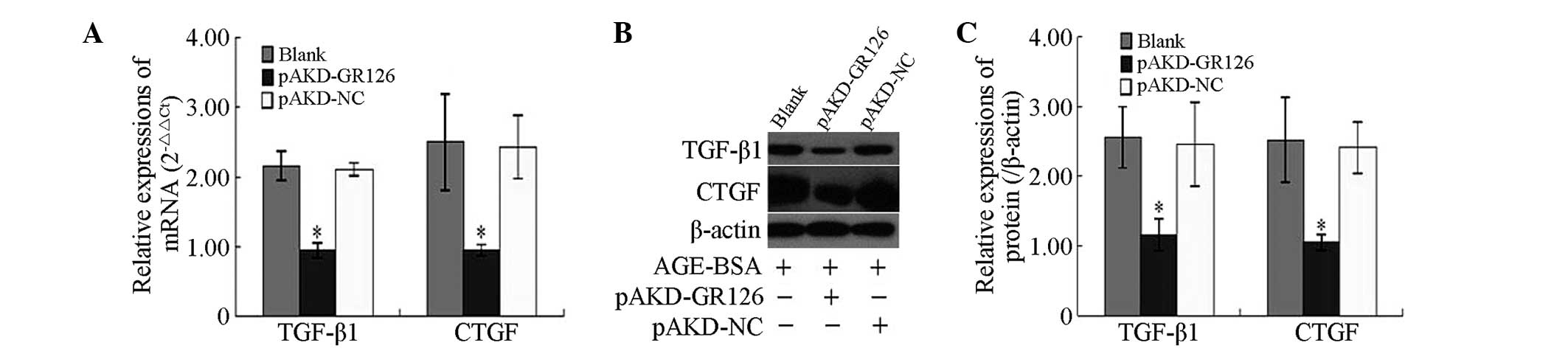
Inhibitory effect of pAKD-GR126 on the
mRNA and protein expression levels of the TGF-β1 and CTGF
profibrogenic cytokines in primary HSCs
The mRNA and protein expression levels of the TGF-β1
and CTGF profibrogenic cytokines were significantly downregulated
in the primary rat HSCs treated with pAKD-GR126 (P<0.05),
compared with the untreated primary rat HSCs and those treated with
the pAKD-NC (Fig. 3).
Inhibitory effect of pAKD-GR126 on the
mRNA and protein expression levels of the LN, HA and PIIINP
fibrosis markers in primary HSCs
The mRNA and protein expression levels of the LN, HA
and PIIINP fibrosis markers were significantly downregulated in the
primary rat HSCs treated with pAKD-GR126 (P<0.05), compared with
the untreated primary rat HSCs and those treated with the pAKD-NC
(Fig. 4).
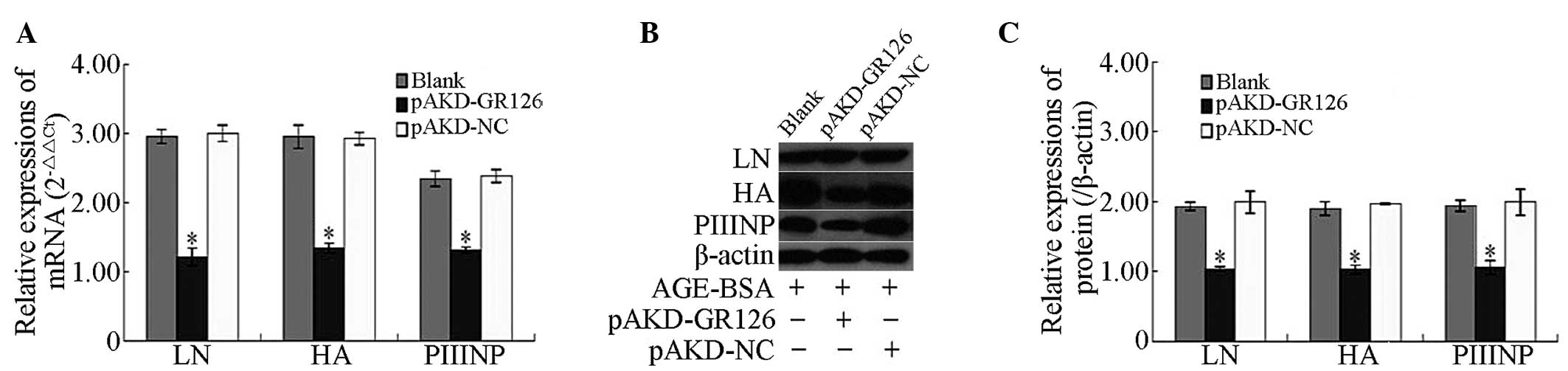 | Figure 4Inhibitory effect of pAKD-GR126 on
the mRNA and protein expression levels of the LN, HA and PIIINP
fibrosis markers in primary HSCs. (A) mRNA expression levels of LN,
HA and PIIINP were determined by reverse transcription quantitative
polymerase chain reaction. (B) Protein expression levels of LN, HA
and PIIINP were determined by western blot analysis. (C) Protein
expression levels of LN, HA and PIIINP are expressed as ratios
relative to the protein expression of β-actin and were determined
by densitometric scanning (*P<0.05, vs. blank and
pAKD-NC). LN, laminin; HA, hyaluronic acid; PIIINP, procollagen III
propeptide; NC, control; BSA, bovine serum albumen; HSC, hepatic
stem cell; Blank, untransfected cells; Ct, cycle threshold. |
Discussion
The present study demonstrated for the first time,
to the best of our knowledge, that specific siRNAs inhibited the
effect of RAGE on the development of HF in primary rat HSCs.
Increasing investigations are being performed on the biological
effects of AGEs and their receptors (15–18)
and the role of the AGE-RAGE axis as a cofactor in the development
of liver fibrosis (15–18). Honsawek et al revealed that
serum RAGE is associated with the severity of biliary atresia and,
therefore, may serve as an indicator reflecting the severity and
development of HF in individuals with postoperative biliary atresia
(19). Goodwin et al
demonstrated that AGEs were damaging to the liver and augmented HF
in an animal model exhibiting chronic liver disease, and these
effects of which were associated with the activation of
myofibroblasts (20). The
upregulation of RAGE, induced by AGE administration, may be
important in mediating these effects. Additionally, the serum AGE
levels are significantly increased in patients with liver cirrhosis
(21–23). RAGE is exclusively expressed in
HSCs and myofibroblasts in the rat liver and its expression is
upregu-lated during HSC activation and transdifferentiation into
myofibroblasts (24), during
which, the expression of TGF-β1 is significantly increased
(8). These previous findings
suggest that RAGE may be a major receptor involved in the
activation and transdifferentiation of HSCs into myofibroblasts and
that the expression of RAGE in the liver is important in liver
fibrosis.
Several methods to inhibit RAGE in liver cells have
been developed. Lin et al revealed that curcumin can
suppress the gene expression of RAGE by increasing the activity of
PPARγ and attenuating oxidative stress, leading to the elimination
of the effects of AGEs on the activation of HSCs (25). Our previous study demonstrated that
a specific siRNA targeting RAGE inhibited HF in a rat model
(5). The present study constructed
expression vectors for specific siRNAs targeting RAGE and
transfected these into primary rat HSCs. The results revealed that
the mRNA expression of RAGE was downregulated in the primary rat
HSCs treated with pAKD-GR126 compared with the untreated primary
rat HSCs and those treated with pAKD-NC. These results indicated
that the RAGE-specific siRNA expressed by pAKD-GR126 effectively
inhibited the gene expression of RAGE.
Furthermore, the activation of HSCs is important in
HF (26–30) and several cytokines are important
in the activation of HSCs (31–34).
Pro-inflammatory cytokines, including IL-6 and TNF-α, promote the
activity and proliferation of HSCs (35,36).
TGF-β1 and its downstream target, CTGF, are potent activators of
HSCs and important profibrogenic cytokines (37–40).
These cytokines promote the activation and transdifferentiation of
HSCs into myofibroblasts (41) and
promote the synthesis and secretion of extracellular matrix (ECM)
components (42,43). The ECM and its degradation
products, including LN, HA and PIIINP, are useful fibrosis markers
and the expression of these products are closely associated with
the degree of HF (18). The
present study indicated that the mRNA and protein expression levels
of RAGE, IL-6, TNF-α, TGF-β1, CTGF, LN, HA and PIIINP were
down-regulated in primary rat HSCs treated with pAKD-GR126 compared
with the untreated primary rat HSCs and those treated with pAKD-NC.
These results demonstrated that the RAGE-specific siRNA expressed
by pAKD-GR126 effectively inhibited gene expression of RAGE and
also inhibited the expression levels of IL-6, TNF-α, TGF-β1, CTGF,
LN, HA and PIIINP. Although the effect of RAGE on the development
of HF remains to be fully elucidated, these findings suggested that
RAGE may be a novel target for treating liver fibrosis and that
RAGE-specific siRNA may be an effective candidate for the
prevention of liver fibrogenesis.
In conclusion, the present study revealed for the
first time, to the best of our knowledge, that a RAGE-specific
siRNA can inhibit the effect of RAGE on the development of HF in
primary rat HSCs. The results demonstrated that the mRNA and
protein expression levels of pro-inflammatory cytokines,
profibrogenic cytokines and fibrosis markers were significantly
downregulated in cells treated with RAGE-specific siRNA, indicating
that RAGE may be a novel target for treatment of liver fibrosis,
and that RAGE-specific siRNA may be an effective candidate to
prevent liver fibrogenesis. However, there were limitations to the
present study, including the number of control groups and the lack
of repeats, therefore, the results obtained are not sufficient to
provide a firm conclusion.
Acknowledgments
The authors would like to thank Dr Changqing Yang of
the Department of Gastroenterology, Tongji Hospital of Tongji
University School of Medicine (Tongji, China) for their technical
assistance. This study was supported by the Natural Science
Foundation of Jiangsu Province (no. BK2009284).
References
|
1
|
Bierhaus A, Humpert PM, Morcos M, et al:
Nawroth, Understanding RAGE, the receptor for advanced glycation
end products. J Mol Med (Berl). 83:876–886. 2005. View Article : Google Scholar
|
|
2
|
Hegab Z, Gibbons S, Neyses L and Mamas MA:
Role of advanced glycation end products in cardiovascular disease.
World J Cardiol. 4:90–102. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yan SD, Bierhaus A, Nawroth PP and Stern
DM: RAGE and Alzheimer’s disease: a progression factor for
amyloid-beta-induced cellular perturbation? J Alzheimers Dis.
16:833–843. 2009.
|
|
4
|
Oleniuc M, Secara I, Onofriescu M, Hogas
S, et al: Consequences of advanced glycation end products
accumulation in chronic kidney disease and clinical usefulness of
their assessment using a non-invasive technique-skin
autofluorescence. Maedica (Buchar). 6:298–307. 2011.
|
|
5
|
Barlovic DP, Soro-Paavonen A and
Jandeleit-Dahm KA: RAGE biology, atherosclerosis and diabetes. Clin
Sci (Lond). 211:43–55. 2011. View Article : Google Scholar
|
|
6
|
Schwenger V, Morath C, Salava A, et al:
Damage to the peritoneal membrane by glucose degradation products
is mediated by the receptor for advanced glycation end-products. J
Am Soc Nephrol. 17:199–207. 2006. View Article : Google Scholar
|
|
7
|
Vlassara H, Striker LJ, Teichberg S, et
al: Advanced glycation end products induce glomerular sclerosis and
albuminuria in normal rats. Proc Natl Acad Sci USA. 91:11704–11708.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fehrenbach H, Weiskirchen R, Kasper M and
Gressner AM: Up-regulated expression of the receptor for advanced
glycation end products in cultured rat hepatic stellate cells
during transdifferentiation to myofibroblasts. Hepatology.
34:943–952. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guimarães EL, Empsen C, Geerts A and van
Grunsven LA: Advanced glycation end products induce production of
reactive oxygen species via the activation of NADPH oxidase in
murine hepatic stellate cells. J Hepatol. 52:389–397. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iwamoto K, Kanno K, Hyogo H, et al:
Advanced glycation end products enhance the proliferation and
activation of hepatic stellate cells. J Gastroenterol. 43:298–304.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia JR, Liu NF and Zhu NX: Specific siRNA
targeting the receptor for advanced glycation end products inhibits
experimental hepatic fibrosis in rats. Int J Mol Sci. 9:638–661.
2008. View Article : Google Scholar
|
|
12
|
Lohwasser C, Neureiter D, Popov Y, Bauer M
and Schuppan D: Role of the receptor for advanced glycation end
products in hepatic fibrosis. World J Gastroenterol. 15:5789–5798.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Friedman SL and Roll FJ: Isolation and
culture of hepatic lipocytes, Kuper cells, and sinusoidal
endothelial cells by density gradient centreifugation with
Stractan. Anal Biochem. 161:207–218. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PRC and
the 2 (− Delta Delta C(T) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Xie X, Zhao R and Shen GX: Impact of
cyanidin-3-glucoside on glycated LDL-induced NADPH oxidase
activation, mitochondrial dysfunction and cell viability in
cultured vascular endothelial cells. Int J Mol Sci. 13:15867–15880.
2012. View Article : Google Scholar
|
|
16
|
Sangle GV, Zhao R, Mizuno TM and Shen GX:
Involvement of RAGE, NADPH oxidase, and Ras/Raf-1 pathway in
glycated LDL-induced expression of heat shock factor-1 and
plasminogen activator inhibitor-1 in vascular endothelial cells.
Endocrinology. 151:4455–4466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Willems S, Verleden SE, Vanaudenaerde BM,
et al: Multiplex protein profiling of bronchoalveolar lavage in
idiopathic pulmonary fibrosis and hypersensitivity pneumonitis. Ann
Thorac Med. 8:38–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng A, Dong Y, Zhu F, et al: AGE-LDL
activates Toll like receptor 4 pathway and promotes inflammatory
cytokines production in renal tubular epithelial cells. Int J Biol
Sci. 9:94–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Honsawek S, Vejchapipat P, Payungporn S,
et al: Soluble receptor for advanced glycation end products and
liver stiffness in postoperative biliary atresia. Clin Biochem.
46:214–218. 2013. View Article : Google Scholar
|
|
20
|
Goodwin M, Herath C, Jia Z, et al:
Advanced glycation end products augment experimental hepatic
fibrosis. J Gastroenterol Hepatol. 28:369–376. 2013. View Article : Google Scholar
|
|
21
|
Sebeková K, Kupcová V, Schinzel R and
Heidland A: Markedly elevated levels of plasma advanced glycation
end products in patients with liver cirrhosis-amelioration by liver
transplantation. J Hepatol. 36:66–71. 2002. View Article : Google Scholar
|
|
22
|
Ahmed N, Lüthen R, Häussinger D, et al:
Increased protein glycation in cirrhosis and therapeutic strategies
to prevent it. Ann N Y Acad Sci. 1043:718–724. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yagmur E, Tacke F, Weiss C, et al:
Elevation of Nepsilon-(carboxymethyl) lysine-modified advanced
glycation end products in chronic liver disease is an indicator of
liver cirrhosis. Clin Biochem. 39:39–45. 2006. View Article : Google Scholar
|
|
24
|
Oldfield MD, Bach LA, Forbes JM, et al:
Advanced glycation end products cause epithelial-myofibroblast
transdifferentiation via the receptor for advanced glycation end
products (RAGE). J Clin Invest. 108:1853–1863. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin J, Tang Y, Kang Q, Feng Y and Chen A:
Curcumin inhibits gene expression of receptor for advanced
glycation end-products (RAGE) in hepatic stellate cells in vitro by
elevating PPARγ activity and attenuating oxidative stress. Br J
Pharmacol. 166:2212–2227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reynaert H, Thompson MG, Thomas T and
Geerts A: Hepatic stellate cells: role in microcirculation and
pathophysiology of portal hypertension. Gut. 50:571–581. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi GF and Li Q: Effects of oxymatrine on
experimental hepatic fibrosis and its mechanism in vivo. World J
Gastroenterol. 11:268–271. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reeves HL and Friedman SL: Activation of
hepatic stellate ceils-a key issue in liver fibrosis. Front Biosci.
7:d808–d826. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Campbell JS, Hughes SD, Gilbertson DG, et
al: Pletelet-derived growth factor C induces liver fibrosis,
steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci USA.
102:3389–3394. 2005. View Article : Google Scholar
|
|
30
|
Sakaida I, Hironaka K, Kimura T, et al:
Herbal medicine sho-saiko-to(TJ-9) increases expression matrix
metalloproteinases (MMPs) with reduced expression of tissue
inhibitor of metalloproteinases (TIMPs) in rat stellate cell. Life
Sci. 74:2251–2263. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Iredale JP: Hepatic stellate cell behavior
during resolution of liver injury. Semin Liver Dis. 21:427–436.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Benyon RC and Arthur MJ: Extracellular
matrix degradation and the role of hepatic stellate cells. Semin
Liver Dis. 21:373–384. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Borkham-Kamphorst E, Herrmann J, Stoll D,
et al: Dominent-negative solute PDGF-beta receptor inhibits hepatic
stellate cell activation and attenuates liver fibrosis. Lab Invest.
84:766–777. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Novosyadlyy R, Tron K, Dudas J, Ramadori G
and Scharf JG: Expression and regulation of the insulin-like growth
factor axis components in rat liver myofibroblasts. J Cell Physiol.
199:388–398. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Simeonova PP, Gallucci RM, Hulderman T, et
al: The role of tumor necrosis factor-alpha in liver toxicity,
inflammation, and fibrosis induced by carbon tetrachloride. Toxicol
Appl Pharmacol. 177:112–120. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
da Silva FM, Guimarães EL, Grivicich I, et
al: Hepatic stellate cell activation in vitro: cell cycle arrest at
G2/M and modification of cell motility. J Cell Biochem. 90:387–396.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang G and Brigstock DR: Integrin
expression and function in the response of primary culture hepatic
stellate cells to connective tissue growth factor (CCN2). J Cell
Mol Med. 15:1087–1095. 2011. View Article : Google Scholar
|
|
38
|
Galicia-Moreno M, Rodríguez-Rivera A,
Reyes-Gordillo K, et al: Trolox down-regulates transforming growth
factor-beta and prevents experimental cirrhosis. Basic Clin
Pharmacol Toxicol. 103:476–481. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen A: Acetaldehyde stimulates the
activation of latent transforming growth factor-betal and induces
expression of the type II receptor of the cytokine in rat cultured
hepatic stellate cells. Biochem J. 368:683–693. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sedlaczek N, Jia JD, Bauer M, et al:
Proliferating bile duct epithelial cells are a major source of
connective tissue growth factor in rat bilinry fibrosis. Am J
Pathol. 158:1239–1244. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Faouzi S, Le Bail B, Neaud V, et al:
Myofibroblasts are responsible for collagen synthesis in the stroma
of human hepatocellular carcinoma: an in vivo and in vitro study. J
Hepatol. 30:275–284. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Friedman SL: Molecular regulation of
hepatic fibrosis, an integrated cellular response to tissue injury.
J Biol Chem. 275:2247–2250. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pinzani M and Marra F: Cytokine receptors
and signaling in hepatic stellate cells. Semin Liver Dis.
21:397–416. 2001. View Article : Google Scholar : PubMed/NCBI
|