Introduction
Comparative genomics has identified numerous genetic
regions in Mycobacterium tuberculosis and M. bovis
that are deleted in M. bovis Bacillus Calmette-Guérin (BCG),
such as region of difference 1 (RD1) and RD2 (1). RD1 was lost during the original
derivation of BCG between 1908 and 1921 (2). Proteins encoded in these regions have
the potential to form the basis of novel specific T-cell-based
blood tests that do not cross-react with BCG. Among these antigens,
early secretory antigenic target 6 (ESAT-6; ESXA, Rv3875),
ESAT-6-like protein esxB (CFP10; ESXB, Rv3874), Pro-Pro-Glu 68
(PPE68; Rv3873) and Pro-Glu 35 (PE35; Rv3872) are immunodominant
(3–5). The former two antigens (ESAT-6 and
CFP10) have been investigated in detail in humans and are known to
be predominant virulence factors (6,7) and
in addition are good candidates for the diagnosis of tuberculosis
(TB) (8). The latter two, PE35 and
PPE68, are members of the PE/PPE family and exhibit immunodominance
(9). The PE/PPE proteins are
secreted or associated with the mycobacterial cell envelope, and
mediate interactions at the host-pathogen interface (10–12).
PE35 and PPE68 have been demonstrated to be associated with
cellular immune responses to mycobacterial infections (9).
Numerous previous studies have demonstrated high
sequence variation of PE/PPE genes in M. tuberculosis
strains (13,14), an observation that suggests
involvement in antigenic variation. To improve the understanding of
the genetic diversity of PE35 and PPE68 belonging to the PE-PPE
genes in the RD1 region, and to explore the effect of immune
recognition on the sequence variation of these two genes, the
current study selected 161 clinical M. tuberculosis isolates
in China, amplified the PE35 and PPE68 genes and compared the
sequences. The effect of the polymorphisms in PE35 and PPE68 were
investigated at the protein level to identify whether alterations
in the function of these proteins occurs as this may potentially
affect strain virulence. In addition, the variation in the human
T-cell epitopes of PE35 and PPE68 were investigated to explore
whether the two antigens are involved in diversifying selection to
evade host immunity.
Materials and methods
Ethics statement
The study obtained approval from the Ethics
Committee of National Institute for Communicable Disease Control
and Prevention, Chinese Center for Disease Control and Prevention
(Beijing, China). The patients with TB included in the present
study were provided with a subject information sheet and written
informed consent was obtained.
Strains and DNA preparation
A total of 161 strains were selected from 2,346
M. tuberculosis complex (MTBC) strains isolated in Beijing
Municipality and 12 provinces and autonomous regions in China
(Table I), which were genotyped by
spoligotyping as described previously (15–18).
Strains belonging to all major and rare spoligotypes in China were
included. Considering the predominance of the Beijing family
strains in China, approximately half of the Beijing family strains
(82 strains) and half non-Beijing family strains (79 strains) were
selected. The 82 Beijing family strains were randomly selected from
the 1,738 Beijing strains among the total 2,346 strains. The
remaining 79 strains were selected from 608 non-Beijing family
isolates. Furthermore, it was attempted to include strains
representing different spoligotypes that were isolated from
different locations. Table I
presents the numbers of strains used in the present study that were
obtained from different provinces in China. The spoligotype
patterns of the 161 strains are presented in Table II.
 | Table INumber of strains in different
locations in China. |
Table I
Number of strains in different
locations in China.
| Location | Number of
isolates |
|---|
| Anhui | 11 |
| Shanxi | 16 |
| Beijing | 11 |
| Fujian | 24 |
| Gansu | 12 |
| Guangxi Zhuang
Autonomous Region | 29 |
| Sichuan | 1 |
| Henan | 12 |
| Hunan | 7 |
| Xizang (Tibet)
Autonomous Region, | 4 |
| Xinjiang Uygur
Autonomous Region | 11 |
| Jilin | 12 |
| Zhejiang | 11 |
 | Table IINumber of strains of each spoligotype
pattern. |
Table II
Number of strains of each spoligotype
pattern.
| Spoligotype | Number of
strains |
|---|
| Beijing | 82 |
| T | 12 |
| U | 27 |
| MANU | 10 |
| Haarlem | 4 |
| EAI | 2 |
| LAM | 2 |
| S | 1 |
| CAS | 3 |
| New | 18 |
A total of 2,346 M. tuberculosis isolates
were randomly collected between 2005 and 2007 from 2,346 patients
at 13 different provincial tuberculosis hospitals across China
(16). Subsequently, 161 strains
were selected from those 2,346 isolates. Sputum specimens were
collected from the TB patients and used to inoculate
Löwenstein-Jensen slants. The strains were cultured using a
standard Löwenstein-Jensen medium (Baso Diagnostics, Inc., Zhuhai,
China) method (15), heat
inactivated and then used directly in polymerase chain reactions
(PCRs).
Primers
The nucleotide sequences of the primers used in the
present study were designed with DNASTAR software (version 7.0;
DNASTAR, Inc., Madison, WI, USA) according to the M.
tuberculosis H37Rv genomic sequence and were as follows: PE35,
forward 5′-GTAATCGAGTTCGGGCAATG-3′ and reverse
5′-AGGCTTCTCCCAGAGAGTT-3′; PPE68, forward
5′-GACATTGGCACGCAAGTGAG-3′ and reverse
5′-TAGCGGCATCGGTCTTCATC-3′.
PCR
The PCRs were performed in a total volume of 20
µl. The PCR mix contained 10 µl PCR buffer (Tiangen
Biotech (Beijing) Co., Ltd., Beijing, China), 100 nM primer, 200
µM each of the four dNTPs (Tiangen (Beijing) Co., Ltd.) and
0.5 units DNA Taq Polymerase (Takara Bio, Inc., Otsu, Japan). An
initial denaturation of 5 min at 94°C was followed by 35 cycles of
denaturation at 94°C for 45 sec, annealing at 62°C for 45 sec and
extension at 72°C for 1 min, followed by a final extension at 72°C
for 10 min in a Bio-Rad Thermal Cycler (Bio-Rad Laboratories, Inc.,
Hercules, CA).
Negative controls (reagents only, no DNA) were
included each time the PCR was performed. The positive control was
500 pg DNA from the M. tuberculosis reference strain H37Rv.
The presence and size of each PCR product was determined by
electrophoresis on 2% agarose gel in Tris/boric
acid/ethyl-enediaminetetraacetic acid buffer (Tiangen Biotech
(Beijing) Co., Ltd.) followed by staining with ethidium bromide
(SBS Genetech Co., Ltd., Beijing, China).
The PCRs were conducted a minimum of two times to
validate the reproducibility. The variants were confirmed by the
sequencing of the new PCR products.
Sequence and data analysis
The sequences of the PCR products were determined
using an ABI 3730xl DNA Analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
The sequences were first aligned using ClustalW
software (19) with the PE35 and
PPE68 gene sequences from the M. tuberculosis H37Rv genome
to determine the PE35 and PPE68 region. Sequence comparisons and
translations were conducted using Bioedit software, version 7.1.3.0
(20). The Immune Epitopes
Database (IEBD) (http://www.iedb.org/) was used and 1
human T-cell epitope in PE35 and 62 in PPE68 were found (21). In addition, SPSS software, version
14.0 (SPSS, Inc., Chicago, IL, USA) was used to conduct
χ2 analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
Mutation and deletion in gene
sequences
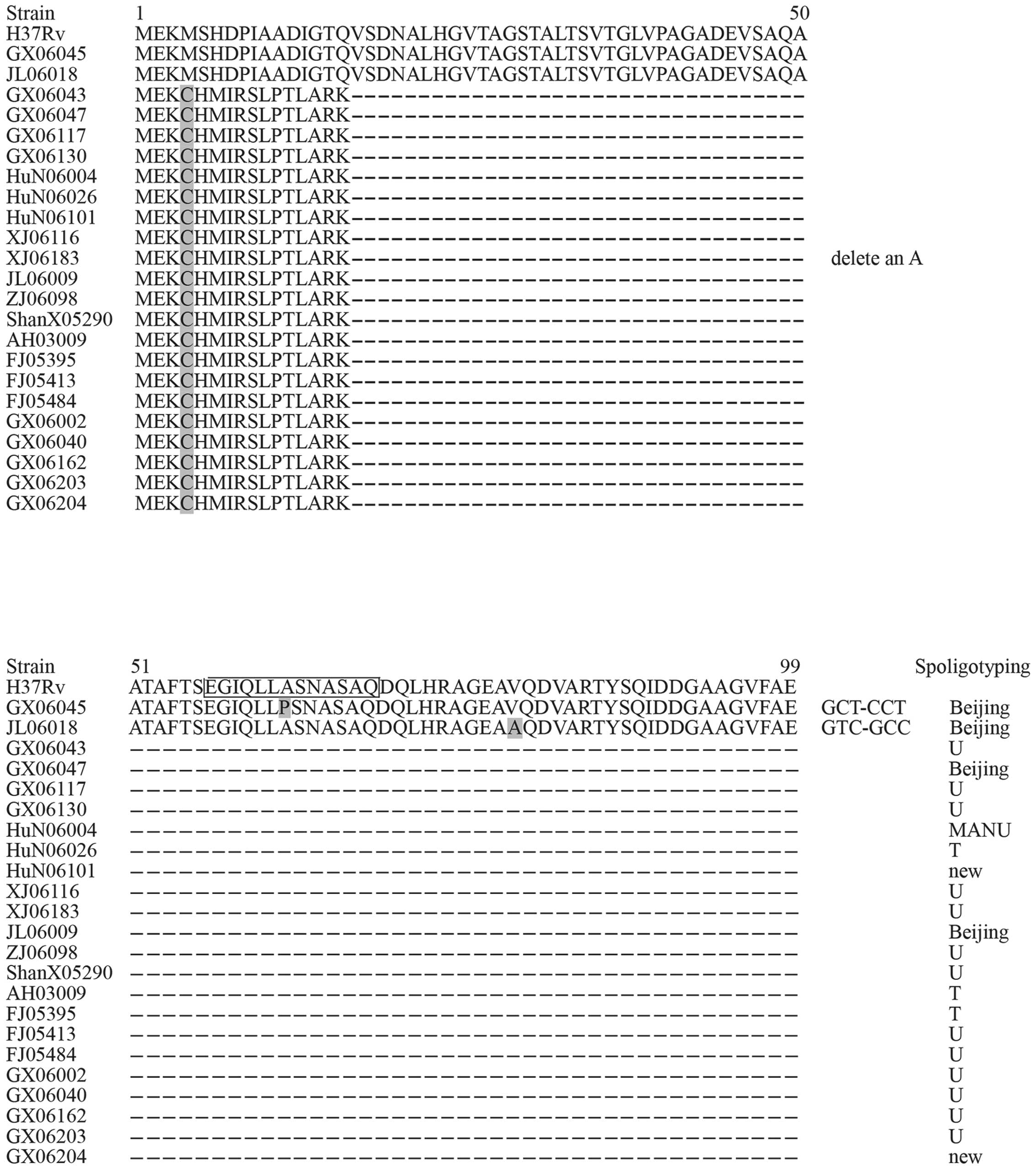
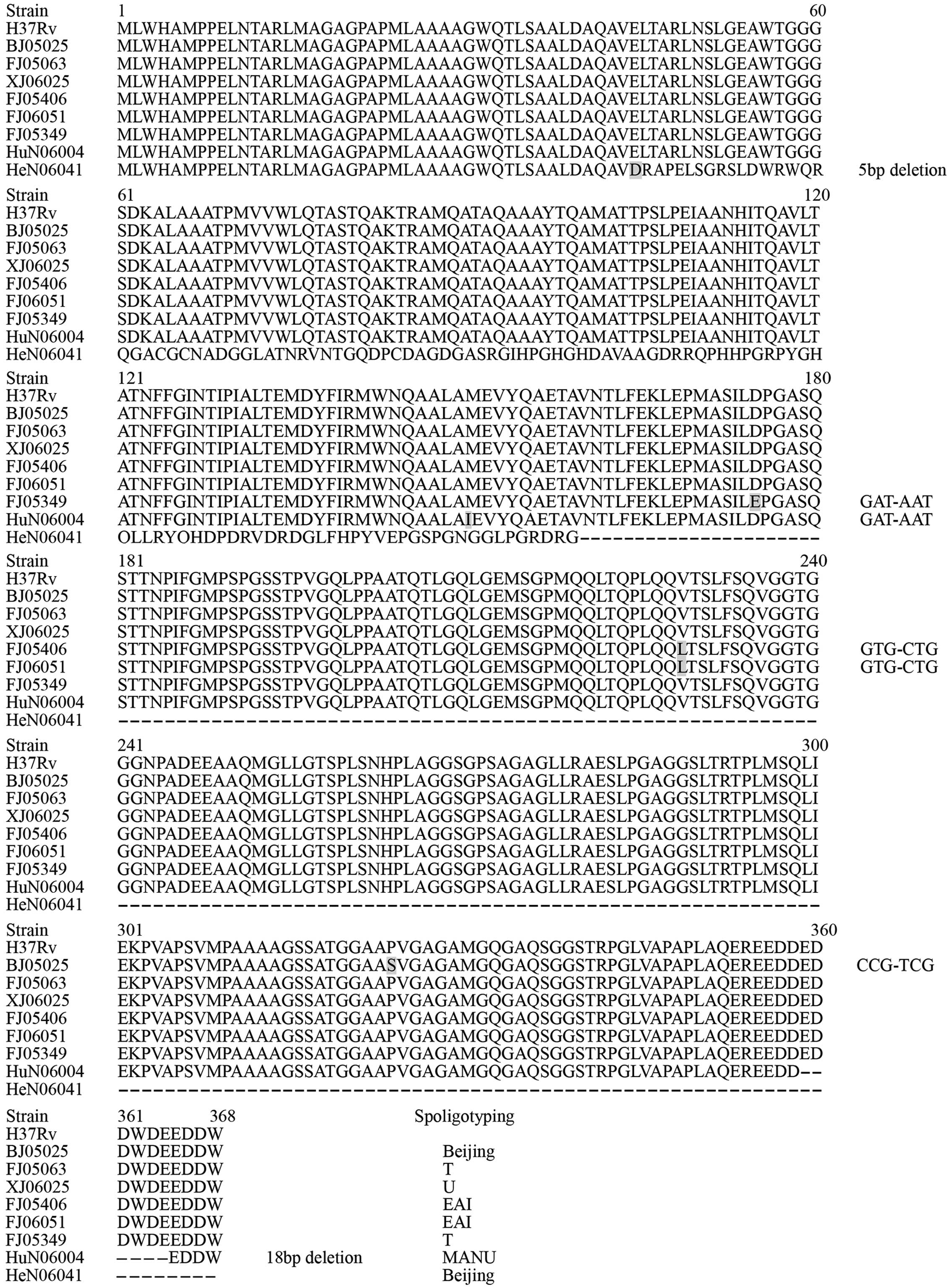
The genes encoding PE35 and PPE68 were amplified and
the sequences compared. All 161 strains yielded PCR products of
these two antigens. Among the 161 M. tuberculosis strains,
23 isolates exhibited polymorphisms in the gene sequence of PE35
(Fig. 1) and 8 strains exhibited
polymorphisms in PPE68. For PE35, there were 21 strains containing
an A deletion and the remaining 2 strains harbored two different
nonsynonymous mutations. For PPE68, two isolates had two different
deletions and six strains showed five nonsynonymous mutations
(Fig. 2).
Changes at the protein level
Figs. 1 and
2 present the amino acid (AA)
alterations and their positions in the PE35 and PPE68 antigens. All
the alterations resulted in an AA change. A total of 21 strains
with an A deletion in PE35 resulted in a frameshift, and therefore
the premature termination of the protein, preventing it's
production and thereby impacting upon protein function. HeN06041
contained a 5 base pair (bp) deletion located at the fifth AA of
PPE68, which additionally resulted in premature termination, and
may impact upon protein function due to the deletion abolishing the
production of the protein. HuN06004 contained an 18 bp deletion,
which resulted in a six AA deletion in PPE68.
Spoligotyping of variant strains
For PE35, 23 variant strains were identified,
including 4 Beijing strains, 13 U family strains, 3 T strains, 1
MANU strain and 2 new spoligotype strains. The two strains with
nonsynonymous mutations were members of the Beijing family. For
PPE68, 8 variant strains including 2 Beijing strains, 2 T strains,
2 EAI strains, 1 U strain and 1 MANU strain were identified. The
two EAI strains, FJ06051 and FJ05406, exhibited the same mutation
in 229(V-L) in the AA sequence of PPE68, which may represent a
unique mutation in EAI strains. HuN06004 exhibited polymorphisms in
PE35 and PPE68.
The prevalence of strains containing a PE35 mutation
in the non-Beijing family is significantly greater compared with
the Beijing family (Table III,
P<0.01). The prevalence of strains with the PPE68 mutation in
the non-Beijing family is greater compared with the Beijing family,
however this was identified to be a significant difference.
 | Table IIIComparison of the Mycobacterium
tuberculosis strains containing mutations in the Beijing family
and the non-Beijing family. |
Table III
Comparison of the Mycobacterium
tuberculosis strains containing mutations in the Beijing family
and the non-Beijing family.
| Strain | Beijing family | Non-Beijing
family |
|---|
| Strains with PE35
mutation | 4 | 19a |
| Strains with PPE68
mutation | 2 | 6b |
| All | 82 | 79 |
Alterations in T-cell epitopes
There is 1 human T-cell epitope in PE35 and 62 in
PPE68 according to the IEBD (http://www.iedb.org/) (21). Table
IV presents the alterations in the T-cell epitopes of the two
antigens. All mutations observed in PE35, except for that in
JL06018, affected the T-cell epitopes. For PPE68, there were no
nonepitope regions in the gene, as the 62 T-cell epitopes covered
the whole gene sequence. This additionally indicates the importance
of the PPE68 antigen for the development of T-cell immune responses
following infection. Among all of the strains, 58/62 T-cell
epitopes in PPE68 (93.5%), exhibited AA alterations resulting from
nucleotide alterations (Table
IV).
 | Table IVAmino acid alterations of human
T-cell epitopes in the antigens, PE35 and PPE68a. |
Table IV
Amino acid alterations of human
T-cell epitopes in the antigens, PE35 and PPE68a.
| Epitope ID | Epitope peptide
sequence | Rv locus | Gene | Amino acid
alteration |
|---|
| 144881 |
EGIQLLASNASAQ | Rv3872 | PE35 | GCT(A)-CCT(P);
Frameshift |
| 183 |
AAGSSATGGAAPVGAGAMGQGAQSG | Rv3873 | PPE68 | CCG(P)-TCG(S);
Frameshift |
| 191 |
AAGWQTLSAALDAQAVELTARLNSL | Rv3873 | PPE68 | Frameshift(5 bp
deletion) |
| 265 |
AALAMEVYQAETAVNTLF | Rv3873 | PPE68 | ATG(M)-ATA(I);
Frameshift |
| 2434 |
ALAMEVYQAETAVNTLFEKLEPMAS | Rv3873 | PPE68 | ATG(M)-ATA(I);
Frameshift |
| 2922 |
ALTEMDYFIRMWNQAALAMEVYQAE | Rv3873 | PPE68 | ATG(M)-ATA(I);
Frameshift |
| 3098 |
AMGQGAQSGGSTRPGLVA | Rv3873 | PPE68 | Frameshift |
| 4186 |
ARLMAGAGPAPMLAAAAG | Rv3873 | PPE68 | No change |
| 4187 |
ARLMAGAGPAPMLAAAAGWQTLSAA | Rv3873 | PPE68 | No change |
| 4727 |
ASQSTTNPIFGMPSPGSSTPVGQLP | Rv3873 | PPE68 | Frameshift |
| 4969 |
ATGGAAPVGAGAMGQGAQ | Rv3873 | PPE68 | CCG(P)-TCG(S);
Frameshift |
| 5063 | ATNFFGINTIPIAL | Rv3873 | PPE68 | Frameshift |
| 11486 |
EEAAQMGLLGTSPLSNHP | Rv3873 | PPE68 | Frameshift |
| 14339 |
ETAVNTLFEKLEPMASIL | Rv3873 | PPE68 | Frameshift |
| 15812 | FFGINTIPIA | Rv3873 | PPE68 | Frameshift |
| 16010 |
FGMPSPGSSTPVGQLPPA | Rv3873 | PPE68 | Frameshift |
| 18685 |
GAMGQGAQSGGSTRPGLVAPAPLAQ | Rv3873 | PPE68 | Frameshift |
| 18776 |
GASQSTTNPIFGMPSPGS | Rv3873 | PPE68 | Frameshift |
| 19860 |
GGGSDKALAAATPMVVWLQTASTQA | Rv3873 | PPE68 | Frameshift |
| 20016 |
GGSGPSAGAGLLRAESLP | Rv3873 | PPE68 | Frameshift |
| 20048 |
GGTGGGNPADEEAAQMGL | Rv3873 | PPE68 | Frameshift |
| 20997 |
GLLGTSPLSNHPLAGGSGPSAGAGL | Rv3873 | PPE68 | Frameshift |
| 21179 |
GLVAPAPLAQEREEDDEDDWDEEDD | Rv3873 | PPE68 | 18 bp deletion;
Frameshift |
| 21707 |
GPMQQLTQPLQQVTSLFS | Rv3873 | PPE68 | GTG(V)-CTG(L);
Frameshift |
| 22351 |
GSGPSAGAGLLRAESLPGAGGSLTR | Rv3873 | PPE68 | Frameshift |
| 22531 |
GSSTPVGQLPPAATQTLGQLGEMSG | Rv3873 | PPE68 | Frameshift |
| 22657 |
GTGGGNPADEEAAQMGLLGTSPLSN | Rv3873 | PPE68 | Frameshift |
| 29846 |
KALAAATPMVVWLQTAST | Rv3873 | PPE68 | Frameshift |
| 35251 |
LDPGASQSTTNPIFG | Rv3873 | PPE68 | GAT(D)-AAT(N);
Frameshift |
| 35652 |
LEPMASILDPGASQSTTN | Rv3873 | PPE68 | GAT(D)-AAT(N);
Frameshift |
| 35819 |
LFEKLEPMASILDPGASQSTTNPIF | Rv3873 | PPE68 | GAT(D)-AAT(N);
Frameshift |
| 37727 |
LLRAESLPGAGGSLTRTP | Rv3873 | PPE68 | Frameshift |
| 38448 |
LPEIAANHITQAVLTATN | Rv3873 | PPE68 | Frameshift |
| 38492 |
LPGAGGSLTRTPLMSQLIEKPVAPS | Rv3873 | PPE68 | Frameshift |
| 39817 |
LTATNFFGINTIPIA | Rv3873 | PPE68 | Frameshift |
| 41291 |
MDYFIRMWNQAALAMEVY | Rv3873 | PPE68 | ATG(M)-ATA(I);
Frameshift |
| 42095 |
MLWHAMPPELNTARLMAG | Rv3873 | PPE68 | No change |
| 46130 |
NTIPIALTEMDYFIRMWN | Rv3873 | PPE68 | Frameshift |
| 48567 |
PMLAAAAGWQTLSAALDA | Rv3873 | PPE68 | No change |
| 49875 |
PVGQLPPAATQTLGQLGE | Rv3873 | PPE68 | Frameshift |
| 50366 |
QATAQAAAYTQAMATTPSLPEIAAN | Rv3873 | PPE68 | Frameshift |
| 51367 |
QLIEKPVAPSVMPAAAAGSSATGGA | Rv3873 | PPE68 | Frameshift |
| 52167 |
QQVTSLFSQVGGTGGGNP | Rv3873 | PPE68 | GTG(V)-CTG(L);
Frameshift |
| 52556 |
QTLGQLGEMSGPMQQLTQ | Rv3873 | PPE68 | Frameshift |
| 60492 |
SQLIEKPVAPSVMPAAAA | Rv3873 | PPE68 | Frameshift |
| 62250 |
SVMPAAAAGSSATGGAAP | Rv3873 | PPE68 | CCG(P)-TCG(S);
Frameshift |
| 64822 |
TLGQLGEMSGPMQQLTQPLQQVTSL | Rv3873 | PPE68 | Frameshift |
| 65054 |
TLSAALDAQAVELTARLN | Rv3873 | PPE68 | Frameshift (5 bp
deletion) |
| 65767 |
TPSLPEIAANHITQAVLTATNFFGI | Rv3873 | PPE68 | Frameshift |
| 65912 |
TQPLQQVTSLFSQVGGTGGGNPADE | Rv3873 | PPE68 | GTG(V)-CTG(L);
Frameshift |
| 66074 |
TRPGLVAPAPLAQEREED | Rv3873 | PPE68 | Frameshift |
| 66364 |
TSPLSNHPLAGGSGPSAG | Rv3873 | PPE68 | Frameshift |
| 68284 |
VELTARLNSLGEAWTGGG | Rv3873 | PPE68 | Frameshift(5 bp
deletion) |
| 68285 |
VELTARLNSLGEAWTGGGSDKALAA | Rv3873 | PPE68 | Frameshift (5 bp
deletion) |
| 69128 |
VITMLWHAMPPELNTARLMAGAGPA | Rv3873 | PPE68 | Frameshift |
| 69795 |
VLTATNFFGINTIPIALT | Rv3873 | PPE68 | Frameshift |
| 69796 |
VLTATNFFGINTIPIALTEMDYFIR | Rv3873 | PPE68 | Frameshift |
| 71944 |
VWLQTASTQAKTRAMQAT | Rv3873 | PPE68 | Frameshift |
| 71945 |
VWLQTASTQAKTRAMQATAQAAAYT | Rv3873 | PPE68 | Frameshift |
| 73833 |
YFIRMWNQAALAMEV | Rv3873 | PPE68 | ATG(M)-ATA(I);
Frameshift |
| 121011 |
TPMVVWLQTASTQAKTR | Rv3873 | PPE68 | Frameshift |
| 144907 |
IALTEMDYFIRMWNQAALAMEVY | Rv3873 | PPE68 | ATG(M)-ATA(I);
Frameshift |
| 144964 | TNFFGINTIPIALT | Rv3873 | PPE68 | Frameshift |
The 5 bp deletion in HeN06041 resulted in a
frameshift in the PPE68 protein code, leading to alterations in the
corresponding T-cell epitopes including IEDB_ID 191, 65054, 68284,
68285 and further downstream epitopes. The 18 bp deletion in
HuN06004 resulted in a 6 AA deletion in IEDB_ID21179.
Discussion
In the present study, 161 clinical M.
tuberculosis strains in China were selected which originated
from a large geographical area and exhibited different
spoligotyping patterns. This strategy was selected so that the data
provided would be representative of the genetic diversity that may
be present within China, at least to some extent.
In previous studies, genetic approaches coupled with
biochemical analyses have indicated that proteins encoded by the
RD1 locus are part of a secretion system required for ESAT-6 and
CFP-10 export (22–26), hereafter referred to as the ESAT-6
system-1 (ESX-1). PE35 (Rv3872) and PPE68 (Rv3873) are encoded by
RD1 and exhibited immunodominance (9). PE35 is an important antigen that
stimulates human peripheral blood mononuclear cells in protective
Th1 cell assays, demonstrating antigen-induced proliferation and
γ-interferon secretion (4). PPE68
is predominantly associated with the cell wall (27) and forms complexes with the RD1
locus proteins Rv3866, Rv3868, CFP-10 and ESAT-6 (28,29).
A recent study (9) indicated that
PE35 and PPE68 may serve a major role in RD1-associated
pathogenesis, and may contribute to the establishment and
maintenance of M. tuberculosis infection. Among the 161
strains investigated in the present study, 14.3% of strains with an
A deletion in PE35 resulted in premature termination leading to a
16 AA peptide as opposed to the full length protein of 99 AA. This
deletion would result in the prevention of protein production, and
consequently lead to the complete loss of PE35 function. In
addition, the 5 bp deletion in HeN06041 of PPE68 resulted in
premature termination, and therefore may exert an effect on protein
function via the abolition of protein production. Strains carrying
mutations that lead to alterations in the functions of PE35 and
PPE68 may be significantly compromised with regards to their
virulence. Therefore, polymorphisms in PE35 and PPE68 may result in
alterations in the functions of these proteins, which may
potentially affect strain virulence. Furthermore, as PE35 has been
demonstrated to be essential for ESXA/B secretion and RD1-mediated
virulence (30), the null mutant
of Rv3872 may influence the release of the ESX-1 antigens, CFP-10
and ESAT-6. PPE68 is a gating protein that regulates the release of
ESX-1 antigens (30) therefore,
abolition of PPE68 protein may additionally affect the release of
CFP-10 and ESTA-6. To investigate this further, virulence
comparison of mutant strains and wild strains of PE35 and PPE68
should be conducted.
PE/PPE genes are known to vary and to encode cell
surface-exposed proteins, which has led to the hypothesis that they
may be involved in antigenic variation (31) and have been suggested to be a
'molecular mantra' to aid in the escape of host immunity. Comas
et al (32) reported that
the human T-cell epitopes of M. tuberculosis are
evolutionarily hyperconserved and suggested that M.
tuberculosis lacks antigenic variation and immune evasion
ability, however, the study excluded PE/PPE genes. In the current
study, there were 63 human T-cell epitopes identified in PE35 and
PPE68 according to the IEDB (21).
Among the strains, 59/63 T-cell epitopes (93.7%), exhibited AA
alterations resulting from nucleotide alterations. The large number
of amino acid alterations in these T-cell epitopes may reflect
ongoing immune evasion. The data from the current study supports
the view that certain PE/PPE genes exhibiting high sequence
variation may be involved in antigenic variation induced immune
evasion.
The prevalence of strains with PE35 mutations in the
non-Beijing family was significantly greater compared with the
Beijing family, indicating that the Beijing family strains are less
changeable in the T-cell epitopes of PE35 than the non-Beijing
family strains. This is supported by a previous study, which
demonstrated that Beijing strains from different geographic areas
exhibited a high degree of genetic conservation compared with the
other M. tuberculosis strains (33). There is evidence that T-cell
responses may contribute directly to human-to-human transmission of
MTBC (34). The current study
indicated that the Beijing strains were more likely to be
recognized by host T-cells in PE35 than the non-Beijing strains,
which may render them easier to transmit than the non-Beijing
strains. Furthermore, analysis of sequences of PE35 and PPE68 in
isolates of these various lineages that have been isolated from
non-Chinese populations could provide interesting information and
insight.
In conclusion, it has been previously reported that
PE35 had potential as a serodiagnostic candidate for M.
tuberculosis (35). The
results of the current study indicate that PE35 harbors a
comparatively high number of AA alterations, suggesting that strain
diversity should be considered during the further development of
novel serodiagnostic candidates that contain PE35.
Acknowledgments
The current study was supported by the projects of
the Natural Science Foundation of China (grant no. 81401647) and
the Chinese National Key Program of Mega Infectious Disease (grant
nos. 2013ZX10003006 and 2013ZX10003002-001).
The authors would like to thank the staff at the
institutes in the Beijing municipality, and the 13 provinces and
autonomous regions in China for their contribution to the study,
and in particular the help of Professor Lishui Zhang (Fuzhou
Pulmonary Hospital of Fujian; Fujian, China), Professor Yunhong Tan
(Hunan Chest Hospital; Hunan, China), Mr. Xiujun Yang (Jilin
Institute for Tuberculosis Control; Jilin, China), Mrs. Chongxiang
Tong (Gansu Institute for Tuberculosis Control; Gansu, China), Mrs.
Feiying Liu (Guangxi Center for Disease Control and Prevention;
Guangxi, China), Mr. Yingcheng Qi (Xinjiang Center for Disease
Control and Prevention; Xinjiang, China), Professor Qing Wang
(Anhui Chest Hospital; Anhui, China), Professor Xiaohui Cao
(Haidian District Center for Disease Control and Prevention;
Beijing, China), Professor Ping Zhao (Chaoyang District Center for
Disease Control and Prevention; Beijing, China), Mr. Haitao Li
(Henan Provincial Chest Hospital; Henan, China), Mrs. Jun Yang
(Sichuan Center for Disease Control and Prevention; Sichuan,
China), Mr. Xuanmin Zhang (Xi'an Chest Hospital; Xi'an, China),
Professor Li Shi (Xizang Center for Disease Control and Prevention;
Xizang, China) and Professor Xiaomeng Wang (Zhejiang Center for
Disease Control and Prevention; Zhejiang, China).
References
|
1
|
Liu XQ, Dosanjh D, Varia H, Ewer K, Cockle
P, Pasvol G and Lalvani A: Evaluation of T-cell responses to novel
RD1- and RD2-encoded Mycobacterium tuberculosis gene products for
specific detection of human tuberculosis infection. Infect Immun.
72:2574–2581. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mahairas GG, Sabo PJ, Hickey MJ, Singh DC
and Stover CK: Molecular analysis of genetic differences between
Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol.
178:1274–1282. 1996.PubMed/NCBI
|
|
3
|
Hanif SN, El-Shammy AM, Al-Attiyah R and
Mustafa AS: Whole blood assays to identify Th1 cell antigens and
peptides encoded by Mycobacterium tuberculosis-specific RD1 genes.
Med Princ Pract. 17:244–249. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mustafa AS, Al-Attiyah R, Hanif SN and
Shaban FA: Efficient testing of large pools of Mycobacterium
tuberculosis RD1 peptides and identification of major antigens and
immunodominant peptides recognized by human Th1 cells. Clin Vaccine
Immunol. 15:916–924. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanif SN, Al-Attiyah R and Mustafa AS:
Species-specific antigenic Mycobacterium tuberculosis proteins
tested by delayed-type hypersensitivity response. Int J Tuberc Lung
Dis. 14:489–494. 2010.PubMed/NCBI
|
|
6
|
Pym AS, Brodin P, Brosch R, Huerre M and
Cole ST: Loss of RD1 contributed to the attenuation of the live
tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium
microti. Mol Microbiol. 46:709–717. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lewis KN, Liao R, Guinn KM, Hickey MJ,
Smith S, Behr MA and Sherman DR: Deletion of RD1 from Mycobacterium
tuberculosis mimics bacille Calmette-Guérin attenuation. J Infect
Dis. 187:117–123. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Malaghini M, Thomaz-Soccol V, Probst CM,
Krieger MA, Preti H, Kritski A and Soccol CR: Recombinant antigen
production for assays of intradermoreaction for diagnosis and
surveillance of tuberculosis. J Biotechnol. 156:56–58. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tiwari B, Soory A and Raghunand TR: An
immunomodulatory role for the Mycobacterium tuberculosis region of
difference 1 locus proteins PE35 (Rv3872) and PPE68 (Rv3873). FEBS
J. 281:1556–1570. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mukhopadhyay S and Balaji KN: The PE and
PPE proteins of Mycobacterium tuberculosis. Tuberculosis (Edinb).
91:441–447. 2011. View Article : Google Scholar
|
|
11
|
Sampson SL: Mycobacterial PE/PPE proteins
at the host-pathogen interface. Clin Dev Immunol. 2011:4972032011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akhter Y, Ehebauer MT, Mukhopadhyay S and
Hasnain SE: The PE/PPE multigene family codes for virulence factors
and is a possible source of mycobacterial antigenic variation:
Perhaps more? Biochimie. 94:110–116. 2012. View Article : Google Scholar
|
|
13
|
McEvoy CR, Cloete R, Müller B, Schürch AC,
van Helden PD, Gagneux S, Warren RM and Gey van Pittius NC:
Comparative analysis of Mycobacterium tuberculosis pe and ppe genes
reveals high sequence variation and an apparent absence of
selective constraints. PLoS One. 7:e305932012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Copin R, Coscollá M, Seiffert SN,
Bothamley G, Sutherland J, Mbayo G, Gagneux S and Ernst JD:
Sequence diversity in the pe_pgrs genes of Mycobacterium
tuberculosis is independent of human T-cell recognition. MBio.
5:e00960–e13. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boehme CC, Nabeta P, Hillemann D, Nicol
MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee
R, et al: Rapid molecular detection of tuberculosis and rifampin
resistance. N Engl J Med. 363:1005–1015. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong H, Liu Z, Lv B, Zhang Y, Liu J, Zhao
X, Liu J and Wan K: Spoligotypes of Mycobacterium tuberculosis from
different provinces of China. J Clin Microbiol. 48:4102–4106. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kamerbeek J, Schouls L, Kolk A, van
Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H,
Shaw R, Goyal M and van Embden J: Simultaneous detection and strain
differentiation of Mycobacterium tuberculosis for diagnosis and
epidemiology. J Clin Microbiol. 35:907–914. 1997.PubMed/NCBI
|
|
18
|
Brudey K, Driscoll JR, Rigouts L,
Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J,
Baumanis V, et al: Mycobacterium tuberculosis complex genetic
diversity: mining the fourth international spoligotyping database
(SpolDB4) for classification, population genetics and epidemiology.
BMC Microbiol. 6:232006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Larkin MA, Blackshields G, Brown NP,
Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm
A, Lopez R, et al: Clustal W and Clustal X version 2.0.
Bioinformatics. 23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hall TA: BioEdit: A user-friendly
biological sequence alignment editor and analysis program for
Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98. 1999.
|
|
21
|
Ernst JD, Lewinsohn DM, Behar S, Blythe M,
Schlesinger LS, Kornfeld H and Sette A: Meeting Report: NIH
workshop on the tuberculosis immune epitope database. Tuberculosis
(Edinb). 88:366–370. 2008. View Article : Google Scholar
|
|
22
|
Hsu T, Hingley-Wilson SM, Chen B, Chen M,
Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, et
al: The primary mechanism of attenuation of bacillus
Calmette-Guerin is a loss of secreted lytic function required for
invasion of lung interstitial tissue. Proc Natl Acad Sci USA.
100:12420–12425. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pym AS, Brodin P, Majlessi L, Brosch R,
Demangel C, Williams A, Griffiths KE, Marchal G, Leclerc C and Cole
ST: Recombinant BCG exporting ESAT-6 confers enhanced protection
against tuberculosis. Nat Med. 9:533–539. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stanley SA, Raghavan S, Hwang WW and Cox
JS: Acute infection and macrophage subversion by Mycobacterium
tuberculosis require a specialized secretion system. Proc Natl Acad
Sci USA. 100:13001–13006. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brodin P, Rosenkrands I, Andersen P, Cole
ST and Brosch R: ESAT-6 proteins: Protective antigens and virulence
factors? Trends Microbiol. 12:500–508. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guinn KM, Hickey MJ, Mathur SK, Zakel KL,
Grotzke JE, Lewinsohn DM, Smith S and Sherman DR: Individual
RD1-region genes are required for export of ESAT-6/CFP-10 and for
virulence of Mycobacterium tuberculosis. Mol Microbiol. 51:359–370.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Okkels LM, Brock I, Follmann F, Agger EM,
Arend SM, Ottenhoff TH, Oftung F, Rosenkrands I and Andersen P: PPE
protein (Rv3873) from DNA segment RD1 of Mycobacterium
tuberculosis: Strong recognition of both specific T-cell epitopes
and epitopes conserved within the PPE family. Infect Immun.
71:6116–6123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okkels LM and Andersen P: Protein-protein
interactions of proteins from the ESAT-6 family of. Mycobacterium
tuberculosis J Bacteriol. 186:2487–2491. 2004.
|
|
29
|
Teutschbein J, Schumann G, Möllmann U,
Grabley S, Cole ST and Munder T: A protein linkage map of the
ESAT-6 secretion system 1 (ESX-1) of Mycobacterium tuberculosis.
Microbiol Res. 164:253–259. 2009. View Article : Google Scholar
|
|
30
|
Brodin P, Majlessi L, Marsollier L, de
Jonge MI, Bottai D, Demangel C, Hinds J, Neyrolles O, Butcher PD,
Leclerc C, et al: Dissection of ESAT-6 system 1 of Mycobacterium
tuberculosis and impact on immunogenicity and virulence. Infect
Immun. 74:88–98. 2006. View Article : Google Scholar :
|
|
31
|
Vordermeier HM, Hewinson RG, Wilkinson RJ,
Wilkinson KA, Gideon HP, Young DB and Sampson SL: Conserved immune
recognition hierarchy of mycobacterial PE/PPE proteins during
infection in natural hosts. PLoS One. 7:e408902012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Comas I, Chakravartti J, Small PM, Galagan
J, Niemann S, Kremer K, Ernst JD and Gagneux S: Human T-cell
epitopes of Mycobacterium tuberculosis are evolutionarily
hyperconserved. Nat Genet. 42:498–503. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Parwati I, van Crevel R and van Soolingen
D: Possible underlying mechanisms for successful emergence of the
Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect
Dis. 10:103–111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kwan CK and Ernst JD: HIV and
tuberculosis: A deadly human syndemic. Clin Microbiol Rev.
24:351–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mukherjee P, Dutta M, Datta P, Dasgupta A,
Pradhan R, Pradhan M, Kundu M, Basu J and Chakrabarti P: The
RD1-encoded antigen Rv3872 of Mycobacterium tuberculosis as a
potential candidate for serodiagnosis of tuberculosis. Clin
Microbiol Infect. 13:146–152. 2007. View Article : Google Scholar : PubMed/NCBI
|