Introduction
Major depressive disorder (MDD) is a complex
heterogeneous mood disorder, and an estimated one in four women and
one in six men are likely to experience depression during their
lifetime (1). Previous findings
have established a close association between inflammation and
depression (2). As compared with
euthymic individuals, patients with MDD exhibit an increased
inflammatory response (3), and
patients with increased inflammatory cytokine levels suffer from
increased rates of depression (4).
Furthermore, in rodent models, the systemic administration of the
pro-inflammatory endotoxin lipopolysaccharide (LPS) triggers
depressive-like behavioral alterations (5) and promotes interleukin (IL)-1β, IL-6,
and tumor necrosis factor α (TNF-α) inflammatory cytokine
production in the brain (6).
LPS-induced depressive-like behavior may be observed even after the
acute inflammation reaction characteristic of the disease in
normalized LPS-treated mice (6).
Therefore, increases in inflammatory cytokine levels may be
associated with the development of depression (7). However, the mechanisms underlying
this phenomenon remain to be elucidated.
A previous study reported that the prefrontal cortex
(PFC) has an important role in the pathogenesis of MDD (8). Patients with MDD exhibit gray matter
density abnormalities in the right dorsolateral prefrontal cortex
(9). Our previous metabolic study
demonstrated the presence of amino acid metabolic dysfunction in
the PFC of a rodent model of depression (10). Although PFC dysfunction has been
associated with the pathophysiology of depression, the association
between the PFC and depressive behavior under inflammatory
conditions requires further investigation. Therefore, the present
study used pro-inflammatory bacterial endotoxin LPS to induce acute
an inflammatory reaction and depressive-like behavior in mice to
comparatively analyze the PFC proteomes by two-dimensional
electrophoresis (2-DE) and matrix-assisted laser desorption
ionization-time of flight-tandem mass spectrometry
(MALDI-TOF-MS/MS). Western blotting was then used to investigate
the expression levels of creatine kinase B-type (CKB) and
dihydropyrimidinase-related protein 3 (DPYSL3), proteins which are
involved in the pathophysiology of MDD.
Materials and methods
Animals
Male CD-1 mice (n=48; weight, 35–40 g; age, 10–14
weeks) were obtained from the animal facility at the Chongqing
Medical University (Chongqing, China). The animals were housed
under standard laboratory conditions (12 h light/dark cycle;
temperature, 23±1°C; humidity, 45–55%), in a single cage with a
shelter, and food and water was available ad libitum. The
present study was approved by the Ethics Committee of the Chongqing
Medical University and all experiments were conducted in accordance
with the National Institutes of Health (Bethesda, MD, USA)
Guidelines for Animal Research (Guide for the Care and Use of
Laboratory Animals).
Drug administration
The mice were randomly divided into three groups: A
control group (Con; n=16) treated with 1 mg/kg sterile saline, an
LPS-induced acute inflammatory reaction group (AIR; n=16) treated
with 0.83 mg/kg LPS, and an LPS-induced depressive-like behavior
group (Dep; n=16) treated with 0.83 mg/kg LPS. For the AIR and Dep
mice, LPS (L-3129, serotype 055:B5; Sigma-Aldrich, St. Louis, MO,
USA) was dissolved in sterile saline and intraperitoneally injected
at a dose of 0.83 mg/kg to induce acute inflammatory reaction
(11) and depressive-like behavior
(12).
Open-field test (OFT)
Prior to experimentation, the mice were placed in
the testing room for 30 min to allow adaptation. The animals were
then individually placed in the center of the open field area
(44.5×44.5×45 cm) and after 30 sec adaptation, a 6 min period of
free exploration was recorded using a Sony DCR-SR45E digital camera
(Sony, Tokyo, Japan). The total distance was recorded during the
last 5 min. Following each trial, the apparatus was wiped and
cleaned with 80% alcohol. The videos of the test were analyzed
using a Smart video-tracking system (PanLab Harvard Apparatus,
Holliston, MA, USA).
Forced swimming test (FST)
An FST was conducted as previously described
(13) with minor modifications.
Briefly, the mice were individually placed into transparent glass
cylinders (diameter, 15 cm; height, 30 cm) containing 15 cm of
water at 23±1°C. Each mouse was required to swim for 6 min, and the
total duration of immobility was recorded during the last 5 min of
testing. Immobility was defined as the absence of active,
escape-oriented behaviors, such as swimming, jumping, rearing,
sniffing, or diving (14). The
water was replaced with fresh water between each test. Following
completion of each test, the mouse was gently removed from the
cylinder, dried, and returned to its cage.
Tail suspension test (TST)
The TST is one of the most widely-used models for
assessing antidepressant-like activity in mice. The test was
conducted as previously described (15) with minor modifications. Briefly,
each mouse was suspended by its tail using adhesive tape placed 2
cm from the tip of the tail in an acoustically and
visually-isolated suspension box (22×21×33 cm) for 6 min, and the
duration of immobility was recorded during the last 5 min of
testing. The mice were regarded as immobile only when they hung
passively or stayed completely motionless.
Protein sample preparation
Our previous studies established a highly effective
protein sample preparation protocol (16,17).
Briefly, 48 mice (n=16) were decapitated under 100% diethyl ether
[Chongqing Chuandong Chemical (Group) Co., Ltd., Chongqing, China]
anesthesia. PFC tissue samples were rapidly dissected from the
brain and subsequently frozen in liquid nitrogen. The tissue
samples of each experimental group were stored at −80°C until
use.
The PFC tissue samples from each experimental group
(n=10) were removed from the liquid nitrogen and suspended in 2 ml
acetone solution supplemented with 0.2% (w/v) dithiothreitol (DTT)
and 10% (w/v) trichloroacetic acid. Following tissue homogenization
(Eppendorf, Hamburg, Germany), the cell suspension was incubated at
−20°C overnight prior to centrifugation at 35,000 × g for 30 min at
4°C. The supernatant was decanted, and the cell pellet was
suspended in 2 ml pre-cooled acetone solution supplemented with
0.2% (w/v) DTT, and incubated at −20°C for 1 h, prior to being
centrifuged at 35,000 × g for 30 min at 4°C. The supernatant was
then decanted, and the cell pellet was dried in a fuming cupboard.
Each sample was dissolved in 2 ml 2-D lysis buffer (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) containing 7 M urea, 2 M
thiourea, 4% CHAPS, 40 mM Tris, 65 mM DTT, 0.3 mg/ml EDTA, 35
µg/ml phenylmethylsulfonyl fluoride, 0.7 µg/ml
pepstatin, 0.5 µg/ml leupeptin, and 0.5% v/v CA, and
centrifuged at 40,000 × g for 60 min at 15°C. The protein
concentration was calculated using a Bradford Protein Assay kit
(Bio-Rad Laboratories, Inc.). Bovine serum albumin (Sangon Biotech
Co., Ltd., Shanghai, China) was used as a standard control.
2-DE
2-DE was conducted as previously described (18). Isoelectric focusing (IEF) was
performed using 17 cm ReadyStrip™ IPG Strips (pH 3–10; Bio-Rad
Laboratories, Inc.), and 350 µl rehydration buffer
containing 0.1 mg protein were loaded onto the strips. Each strip
was rehydrated for 12 h (at 30 V) using 350 µl rehydration
solution containing 7 M urea, 2 M thiourea, 4% CHAPS, 50 mM DTT,
0.2% Bio-Lyte, and 0.001% bromophenol blue (Bio-Rad Laboratories,
Inc.) (19). IEF was performed on
a Protean IEF cell (Bio-Rad Laboratories, Inc.) as follows: 50 V
for 12 h, 250 V for 30 min, 1,000 V for 1 h, 1,000–10,000 V over a
5 h step-up period, and 10,000 V for 6 h. Following IEF, the IPG
strips were equilibrated in the reduction buffer containing 0.375 M
Tris-HCl (pH 8.8), 6 M urea, 20% glycerol, and 2% SDS. The first
equilibration was performed using equilibration buffer supplemented
with 2% DTT, and then the second equilibration was performed using
equilibration buffer supplemented with 2.5% indole-3-acetic acid.
The second dimension was performed by running the strips on 1-mm
thick 10% SDS-PAGE using a Protean® IIG xi Multi-Cell
(Bio-Rad Laboratories, Inc.). Six gels (three gels per group) were
simultaneously run at 12.5 mA/gel for 30 min and then 25 mA/gel for
5.0–5.5 h at 20°C. The protein spots were visualized by silver
staining as previously described by Yan et al (20).
Gel image analysis
Following visualization, images were scanned using
an Epson 10000XL scanner (Epson Co., Ltd. Beijing, China) at an
optical resolution of 300 dpi. Image analysis and spot detection
were conducted using PDQuest 8.0.1 (Bio-Rad Laboratories, Inc.)
with Gaussian spot modeling. For quantitative comparison of the
spots across the gels, replicate images of the gels were created.
To correct for the variability in silver staining, the individual
spot volumes were normalized by dividing the optical density (OD)
of each spot value by the sum the total OD of all spots in the
respective gels. This method controlled for differences in sample
loading and color intensities among the gels. Automated and manual
spot matching were also performed. Integrated intensities
demonstrating a ≥1.5-fold change were applied to determine the
statistical differences in protein expression between the two
groups (17).
MALDI-TOF/TOF
Protein samples were separated by 2-DE and
visualized with silver staining. In-gel protein digestion was
performed as previously described by Zhou et al (21) with minor modifications. The protein
spots of interest were excised from the gels and destained.
Following reduction and alkylation, the gel sections were digested
overnight with Sequencing Grade Modified Trypsin (Promega
Corporation, Madison, WI, USA). The digested peptides were
extracted using 100 µl 60% CAN (Merck Millipore, Darmstadt,
Germany) supplemented with 0.1% TFA (Merck Millipore) and
concentrated in a SpeedVac® vacuum concentrator (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The peptides were
re-dissolved in a matrix solution and spotted on a MALDI target
plate (Applied Biosystems Life Technologies, Foster City, CA, USA).
The peptides were then analyzed using the 4800 Plus MALDI TOF/TOF
Analyzer (Applied Biosystems Life Technologies) in default
mode.
Western blotting
To investigate the differential protein expression
in the PFC, two proteins, creatine kinase B-type (CKB) and
dihydropyrimidinase-related protein 3 (DPYSL3) were selected for
western blotting. β-tubulin was used as a loading control
(1:1,000). The PFC samples from each group (n=6) were homogenized
using a standard lysis buffer containing 50 mM Tris (pH 7.4), 150
mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, sodium
orthovanadate, sodium fluoride, EDTA, and leupeptin (Beyotime
Institute of Biotechnology, Haimen, China), prior to protein
extraction. A bicinchoninic acid assay was used to analyze the
protein concentrations. A total of 30 µg protein samples
were separated by 10% SDS-PAGE and transferred onto polyvinylidene
fluoride membranes. The membranes were blocked using 5% (w/v)
skimmed milk for 1 h at room temperature, and then incubated
overnight at 4°C with anti-CKB rabbit monoclonal antibody (1:3,000;
cat. no. ab92452; Abcam, Cambridge, UK) and anti-DPYSL3 rabbit
monoclonal antibody (1:1,000; cat. no. ab126787; Abcam). The
membranes (Merck Millipore) were washed in Tris-buffered saline
with 0.05% Tween 20 containing 150 mM NaCl, 0.05% Tween 20, and 10
mM Tris-HCl (pH 7.5), and incubated with their respective
horseradish peroxidase-conjugated secondary antibodies (1:10,000;
Bio-Rad Laboratories, Inc., Hercules, CA, USA) for 2 h at room
temperature. The membranes were visualized using enhanced
chemiluminescence and exposed to autoradiography films. Band
intensity was determined using Quantity One software (version
4.6.2; Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analyses were conducted using SPSS 17.0
(SPSS, Inc., Chicago, IL, USA). The results from the western
blotting were analyzed using a one-way analysis of variance
followed by a Bonferroni's test. Body weight and behavioral test
results were compared using the Student's t-tests. P<0.05 was
considered to indicate a statistically significant difference.
Results
LPS decrease body weight and
mobility
LPS-induced changes in body weight and mobility were
measured 6 and 24 h post-injection by assessing changes in body
weight loss and OFT (22). The
weights of the Dep mice were significantly reduced, as compared
with their weights prior to injection (Fig. 1A), and the AIR mice exhibited a
significant decrease in the total distance travelled in the OFT
test, as compared with the control group (Fig. 1B).
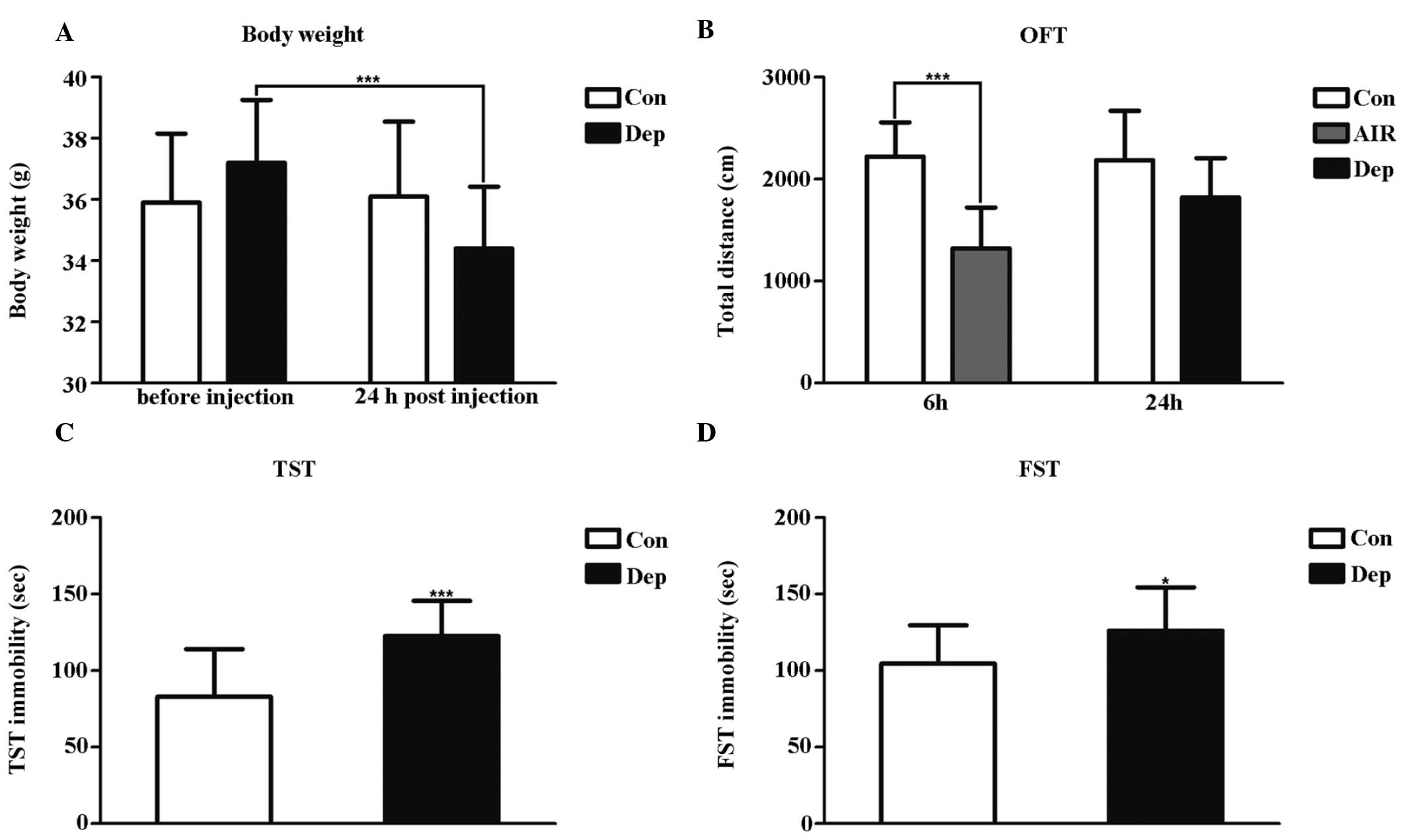 | Figure 1Quality assessment of an LPS-induced
mouse model 6 h and 24 h post-LPS peritoneal injection. (A) Body
weight change, (B) total distance travelled in the OFT, (C)
immobility time in the TST, and (D) immobility time in the FST.
Data are presented as the mean ± standard deviation (n=16 subjects
per group). *P<0.05, vs. the Con group, and
***P<0.001. LPS, lipopolysaccharide; OFT, open-field
test; TST, tail suspension test; FST, forced swimming test; AIR,
LPS-induced acute inflammatory reaction group; Dep, LPS-induced
depressive-like behavior group; Con, control group. |
The Dep mice exhibited a significant increase in
immobility in the TST and FST conducted 24 h post-LPS injection
(Fig. 1C and D). This was the time
point when typical acute sickness behavior was no longer apparent
(22). These results suggest that
acute activation of the peripheral innate immune system by LPS
induces depressive-like symptoms in mice that are not biased by
acute sickness behaviors.
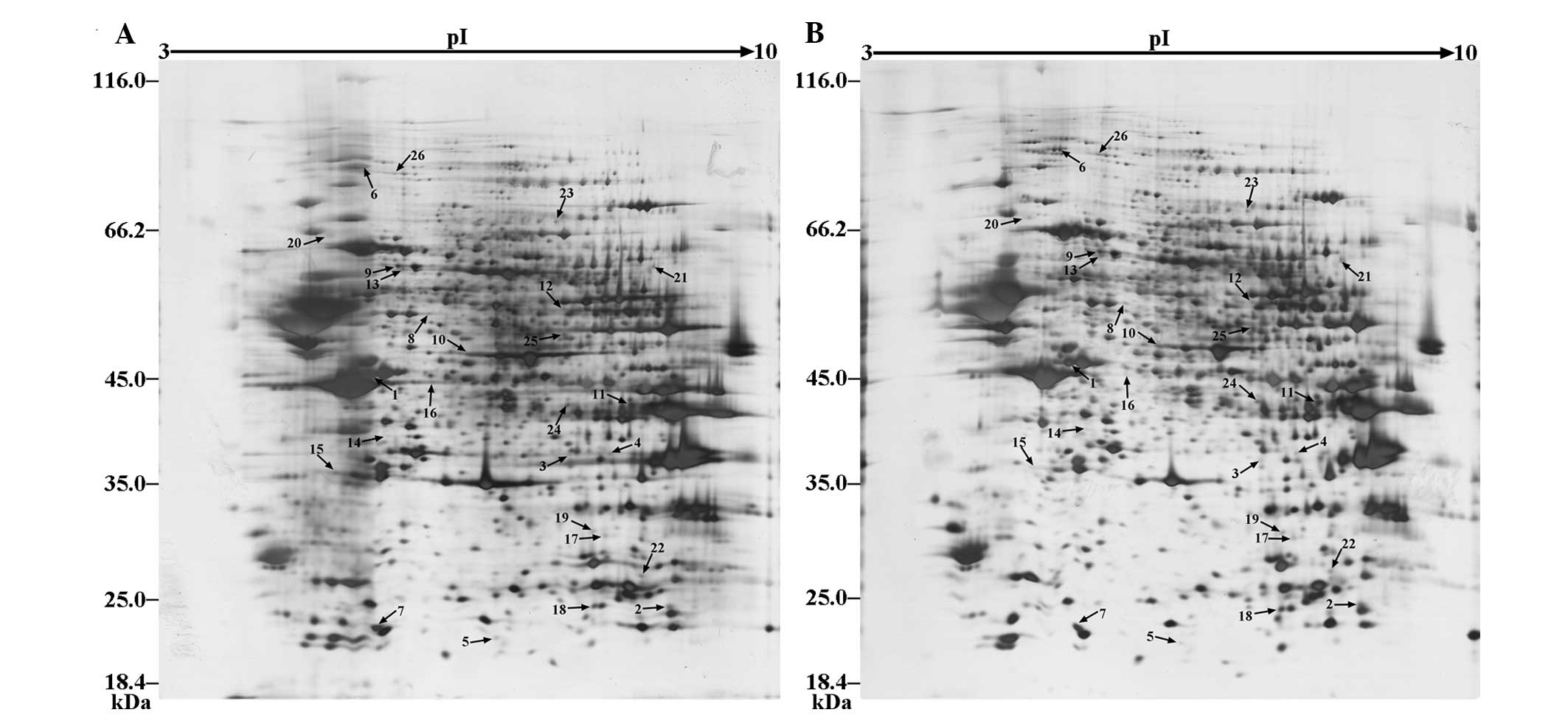
Differentially expressed protein identification by
2-DE and MALDI-TOF-MS/MS were conducted to examine whether PFC
protein expression was altered by LPS. Wide-range pH 3–10 strips
were used to investigate the differential protein expression in the
AIR and Dep mice, as compared with the Con mice. A total of ~1,780
protein spots were visualized by silver staining (Fig. 2). To identify the differential
proteins in the 2-DE gels, a 1.5-fold change was used as a
threshold (Table I). The
functional classification of differentially expressed proteins was
energy metabolism, neurogenesis, cytoskeleton, signal transducer,
redox homeostasis, nuclein metabolism and molecular chaperones
(Table I).
 | Table IDifferentially expressed proteins in
the prefrontal cortex, as determined by 2-DE and
MALDI-TOF-MS/MS. |
Table I
Differentially expressed proteins in
the prefrontal cortex, as determined by 2-DE and
MALDI-TOF-MS/MS.
| Spot no. | Accession no. | Gene | Protein score
(CI%) | MW (Da) | PI | Fold-change
|
|---|
| AIR/Con | Dep/Con |
|---|
| Energy
metabolism | | | | | | | |
| 1 | Q04447 | Ckb | 100 | 42971.4 | 5.4 | 2.19 | 8.24 |
| 2 | P19157 | Gstp1 | 100 | 23765.2 | 7.68 | 2.42 | 2.83 |
| 3 | P45376 | Akr1b1 | 100 | 36037.6 | 6.71 | 2.46 | 2.81 |
| 4 | Q9CZ42 | Carkd | 100 | 35651.4 | 8.4 | 2.06 | 1.73 |
| 5 | Q64520 | Guk1 | 100 | 22018.3 | 6.12 | 2.52 | 1.58 |
| 6 | Q02053 | Uba1 | 100 | 118931.5 | 5.43 | 0.56 | 1.98 |
| 7 | Q9R0Y5 | Ak1 | 100 | 21640.2 | 5.67 | 0.65 | 0.44 |
| 8 | P62814 |
Atp6v1b2 | 100 | 56874.9 | 5.57 | 0.64 | 0.47 |
| 9 | Q8BMF4 | Dlat | 100 | 68468.9 | 8.81 | 0.41 | 0.56 |
| 10 | P17182 | Eno1 | 100 | 47453.3 | 6.37 | 0.54 | 0.31 |
| 11 | P05201 | Got1 | 100 | 46488.6 | 6.68 | 0.36 | 0.34 |
| 12 | Q64332 | Syn2 | 100 | 52817.6 | 7.62 | 0.24 | 0.25 |
| Neurogenesis | | | | | | | |
| 13 | Q3TT92 | Dpysl3 | 100 | 62140.1 | 6.04 | 3.72 | 7 |
| Cytoskeleton | | | | | | | |
| 14 | P05213 | Tuba1b | 100 | 50787.8 | 4.94 | 0.47 | 0.44 |
| 15 | E9Q6X0 | Mapre2 | 100 | 32472.9 | 5.14 | 0.46 | 0.32 |
| 16 | P60710 | Actb | 100 | 42065.9 | 5.3 | 0.32 | 0.06 |
| Signal
transducer | | | | | | | |
| 17 | Q9DBS2 | Tprg1l | 100 | 30023.5 | 6.92 | 1.94 | 2.19 |
| Redox
homeostasis | | | | | | | |
| 18 | Q9D6Y7 | Msra | 100 | 23679.7 | 8.6 | 2.09 | 2.45 |
| 19 | O09131 | Gsto1 | 100 | 27708 | 6.92 | 1.86 | 2.92 |
| 20 | P08003 | Pdia4 | 100 | 65388.3 | 5.91 | 0.14 | 0.45 |
| Nuclein
metabolism | | | | | | | |
| 21 | P50544 | Acadvl | 100 | 71229.8 | 8.91 | 3.75 | 5.42 |
| 22 | Q80U88 |
mKIAA0038 | 100 | 25511.7 | 8.64 | 2.29 | 2.56 |
| 23 | Q8K1M6 | Dnm1l | 100 | 83119.7 | 6.61 | 2.31 | 3.97 |
| 24 | Q9Z1S5 | Sept3 | 100 | 38913 | 6.35 | 1.62 | 2.19 |
| 25 | Q8C1B7 | Sept11 | 100 | 49991.4 | 6.14 | 0.44 | 0.51 |
| Molecular
chaperones | | | | | | | |
| 26 | P48722 | Hspa4l | 100 | 95177.8 | 5.54 | 1.76 | 4.86 |
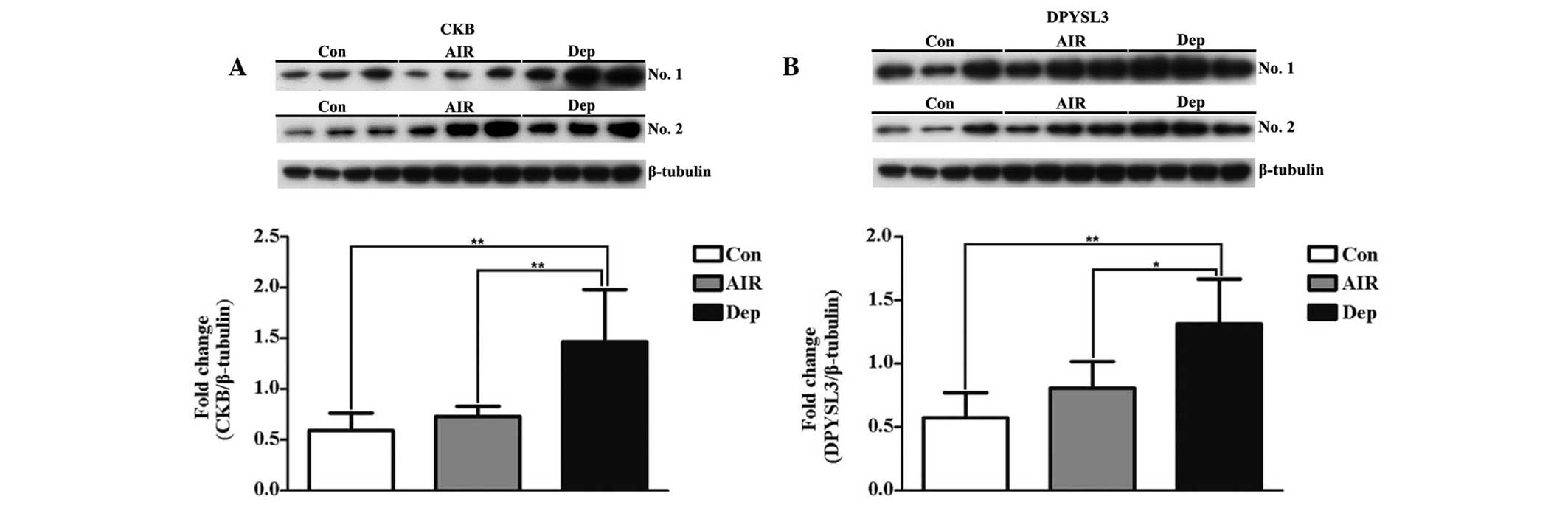
Western blot analysis
CKB and DPYSL3 were selected for analysis by western
blot analysis. The expression levels of CKB and DPYSL3 were
significantly increased in the Dep mice, as compared with the Con
and AIR mice (Fig. 3).
Discussion
Accumulating evidence suggests that inflammation may
have an important role in the pathophysiology of MDD. Meta-analysis
demonstrated that significantly higher blood expression levels of
pro-inflammatory cytokines, such as TNFα and IL-6, were present in
drug-free depressive patients, as compared with healthy controls
(23). Numerous conditions such as
cardiovascular disease (24), type
2 diabetes (25) and obesity
(26) are characterized by chronic
inflammation and high prevalence of depression. The bacterial
endotoxin LPS is widely used in preclinical research as a tool to
study neuroinflammation, and numerous studies have identified the
mechanisms by which LPS is able to induce inflammatory responses in
the brain (27). It is generally
accepted that neuroinflammation induces depressive-like behavior in
rats (28). In the present study,
the systemic administration of LPS resulted in acute inflammatory
reaction and depressive-like behavior in CD1 mice. These results
suggest that the experimental animals exhibited negative side
effects and depressive-like behaviors, such as weight loss, a
decrease in the total distance travelled in the OFT, and an
increase in immobility time both in the TST and FST. These results
were concordant with those of O'Connor et al (22). Based on these results, the
LPS-induced model may be established as a reliable animal model of
depression, and may be used to better understand the
pathophysiological mechanisms underlying depression (29,30).
The present study used a proteomic approach to
examine PFC protein expression in an LPS-induced mouse model of
depression. A total of 26 proteins were identified that may have
roles in the pathogenesis of MDD. Based on the MOTIF database
(http://www.genome.jp/tools/motif/)
and previous studies (16), a
biological function classification on the differentially expressed
proteins in the PFC was established. Energy metabolic-associated
proteins constitute the majority of the differentially expressed
proteins. In a previous study, we demonstrated that energy
metabolic signaling pathways were significantly altered in the PFC
of a chronic unpredictable mild stress rat model of depression, as
determined by KEGG analysis (17).
As determined by positron emission tomography of glucose
metabolism, energy metabolic dysfunction in the PFC has been
associated with depression (31).
CKB was one of the energy metabolic-associated proteins of the
present study, and is a cytosolic CK of the brain. There are two
cytosolic CKs [brain-type CK (CKB), and muscle-type CK (CKM)], and
two mitochondrial CKs [ubiquitous mitochondrial CK (uMtCK) and
muscle-specific sarcomeric mtCK] (32). CKs regulate ATP regeneration via
the transfer of high-energy phosphate from create phosphocreatine
to adenosine diphosphate (33,34).
The phosphocreatine/CK energy circuit is important for the
maintenance of normal energy homeostasis (35,36),
and has a number of integrated functions, such as temporary energy
buffering and energy transfer, as well as regulating metabolic
capacity (37). The results from
the western blot analysis indicated that the expression levels of
CKB were significantly elevated in the Dep mice, suggesting that
LPS-induced depressive-like behaviors in mice may be associated
with changes in the energy metabolism of the PFC.
In 2000, a previous study proposed for the first
time a neurogenic theory of depression, and demonstrated that
impaired adult hippocampal neurogenesis (AHN) triggers depression,
and restoration of AHN lead to the alleviation of depressive
symptoms (38). Previously, we
reported that alterations in hippocampal neurogenesis may have a
role in mediating the pathogenesis of depression (16). However, to the best of our
knowledge, few studies have investigated neurogenesis in the PFC.
The results of the present study demonstrated that DPYSL3, which is
a neurogenesis-associated protein, was one of the differentially
expressed proteins of the PFC. DPYSL3 is also known as collapsin
response mediator protein 4 (CRMP4), which belongs to the cytosolic
phosphoprotein family, and is involved in neurite and axonal
outgrowth (39–42), neuronal differentiation (39,43,44),
axonal guidance (45) and
regeneration (46). Previous
studies demonstrated that DPYSl3 interacts with cytoskeletal
proteins tubulin and actin, suggesting DPYSl3 is involved in cell
assembly and migration (40,47).
DPYSl3 is also involved in developmental neurogenesis,
neuroregeneration following nerve lesion, and growth cone collapse
during neuronal cell injury, by organizing filamentous actin into
tight bundles (43,46,48).
Immunocytochemistry revealed that CRMP-4 is transiently expressed
in post-mitotic neurons during rat brain development (49). Therefore, DPYSL3 was selected for
further investigation. The results of the present study demonstrate
that the expression levels of DPYSL3 were significantly elevated in
the Dep mice, suggesting that LPS-induced depressive-like behavior
in mice may be associated with changes in neurogenesis in the
PFC.
In conclusion, the present study has demonstrated
that LPS is able to induce depressive-like behavior independently
of changes in motor activity. Using a 2-DE-MALDI-TOF-MS/MS-based
proteomic approach, 26 differentially expressed proteins were
identified in the PFC of the Dep mice, as compared with the Con
mice. The expression levels of CKB and DPYSL3 were significantly
elevated in the Dep mice, as determined by western blotting. These
results suggest that energy metabolic dysfunction and neurogenesis
are significantly altered in the PFC of Dep mice. Investigation
into these processes and proteins in the PFC is crucial to obtain a
further understanding of the pathophysiological mechanisms
underlying MDD.
Acknowledgments
The authors would like to thank Dr N.D. Melgiri for
editing and proofreading the manuscript of the present study, as
well as Jack Cheng (Chongqing Medical University). The present
study was supported by a grant from the National Basic Research
Program of China (973 program; grant no. 2009CB918300).
References
|
1
|
Kessler RC, Birnbaum H, Bromet E, Hwang I,
Sampson N and Shahly V: Age differences in major depression:
Results from the National Comorbidity Survey Replication (NCS-R).
Psychol Med. 40:225–237. 2010. View Article : Google Scholar
|
|
2
|
Raison CL, Capuron L and Miller AH:
Cytokines sing the blues: Inflammation and the pathogenesis of
depression. Trends Immunol. 27:24–31. 2006. View Article : Google Scholar
|
|
3
|
Zorrilla EP, Luborsky L, McKay JR,
Rosenthal R, Houldin A, Tax A, McCorkle R, Seligman DA and Schmidt
K: The relationship of depression and stressors to immunological
assays: A meta-analytic review. Brain Behav Immun. 15:199–226.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yirmiya R, Pollak Y, Morag M, Reichenberg
A, Barak O, Avitsur R, Shavit Y, Ovadia H, Weidenfeld J, Morag A,
et al: Illness, cytokines, and depression. Ann NY Acad Sci.
917:478–487. 2000. View Article : Google Scholar
|
|
5
|
Dantzer R, O'Connor JC, Freund GG, Johnson
RW and Kelley KW: From inflammation to sickness and depression:
When the immune system subjugates the brain. Nat Rev Neurosci.
9:46–56. 2008. View
Article : Google Scholar
|
|
6
|
Frenois F, Moreau M, O'Connor J, Lawson M,
Micon C, Lestage J, Kelley KW, Dantzer R and Castanon N:
Lipopolysaccharide induces delayed FosB/DeltaFosB immunostaining
within the mouse extended amygdala, hippocampus and hypothalamus,
that parallel the expression of depressive-like behavior.
Psychoneuroendocrinology. 32:516–531. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hannestad J, DellaGioia N and Bloch M: The
effect of antidepressant medication treatment on serum levels of
inflammatory cytokines: A meta-analysis. Neuropsychopharmacology.
36:2452–2459. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Koenigs M and Grafman J: The functional
neuroanatomy of depression: Distinct roles for ventromedial and
dorsolateral prefrontal cortex. Behav Brain Res. 201:239–243. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye T, Peng J, Nie B, Gao J, Liu J, Li Y,
Wang G, Ma X, Li K and Shan B: Altered functional connectivity of
the dorsolateral prefrontal cortex in first-episode patients with
major depressive disorder. Eur J Radiol. 81:4035–4040. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen G, Yang D, Yang Y, Li J, Cheng K,
Tang G, Zhang R, Zhou J, Li W, Liu Z, Fan S and Xie P: Amino acid
metabolic dysfunction revealed in the prefrontal cortex of a rat
model of depression. Behav Brain Res. 278:286–292. 2015. View Article : Google Scholar
|
|
11
|
Mormède C, Palin K, Kelley KW, Castanon N
and Dantzer R: Conditioned taste aversion with lipopolysaccharide
and peptidoglycan does not activate cytokine gene expression in the
spleen and hypothalamus of mice. Brain Behav Immun. 18:186–200.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lestage J, Verrier D, Palin K and Dantzer
R: The enzyme indoleamine 2,3-dioxygenase is induced in the mouse
brain in response to peripheral administration of
lipopolysaccharide and superantigen. Brain Behav Immun. 16:596–601.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Porsolt RD, Bertin A and Jalfre M:
Behavioral despair in mice: A primary screening test for
antidepressants. Arch Int Pharmacodyn Ther. 229:327–336.
1977.PubMed/NCBI
|
|
14
|
Porsolt RD, Anton G, Blavet N and Jalfre
M: Behavioural despair in rats: A new model sensitive to
antidepressant treatments. Eur J Pharmacol. 47:379–391. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Steru L, Chermat R, Thierry B and Simon P:
The tail suspension test: A new method for screening
antidepressants in mice. Psychopharmacology (Berl). 85:367–370.
1985. View Article : Google Scholar
|
|
16
|
Mu J, Xie P, Yang ZS, Yang DL, Lv FJ, Luo
TY and Li Y: Neurogenesis and major depression: Implications from
proteomic analyses of hippocampal proteins in a rat depression
model. Neurosci Lett. 416:252–256. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Y, Yang D, Tang G, Zhou C, Cheng K,
Zhou J, Wu B, Peng Y, Liu C, Zhan Y, et al: Proteomics reveals
energy and glutathione metabolic dysregulation in the prefrontal
cortex of a rat model of depression. Neuroscience. 247:191–200.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu Y, Zhou J, Fang L, Liu H, Zhan Q, Luo
D, Zhou C, Chen J, Li Q and Xie P: Hippocampal synaptic
dysregulation of exo/endocytosis-associated proteins induced in a
chronic mild-stressed rat model. Neuroscience. 230:1–12. 2013.
View Article : Google Scholar
|
|
19
|
Hwang YY and Li MD: Proteins
differentially expressed in response to nicotine in five rat brain
regions: Identification using a 2-DE/MS-based proteomics approach.
Proteomics. 6:3138–3153. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yan JX, Wait R, Berkelman T, Harry RA,
Westbrook JA, Wheeler CH and Dunn MJ: A modified silver staining
protocol for visualization of proteins compatible with
matrix-assisted laser desorption/ionization and electrospray
ionization-mass spectrometry. Electrophoresis. 21:3666–3672. 2000.
View Article : Google Scholar
|
|
21
|
Zhou J, Xiong J, Li J, Huang S, Zhang H,
He Q, Lin Y, Chen P, Wang X and Liang S: Gel absorption-based
sample preparation for the analysis of membrane proteome by mass
spectrometry. Anal Biochem. 404:204–210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
O'Connor JC, Lawson MA, André C, Moreau M,
Lestage J, Castanon N, Kelley KW and Dantzer R:
Lipopolysaccharide-induced depressive-like behavior is mediated by
indoleamine 2,3-dioxygenase activation in mice. Mol Psychiatry.
14:511–522. 2009. View Article : Google Scholar :
|
|
23
|
Dowlati Y, Herrmann N, Swardfager W, Liu
H, Sham L, Reim EK and Lanctôt KL: A meta-analysis of cytokines in
major depression. Biol Psychiatry. 67:446–457. 2010. View Article : Google Scholar
|
|
24
|
Celano CM and Huffman JC: Depression and
cardiac disease: A review. Cardiology (in review).
|
|
25
|
Alexandraki K, Piperi C, Kalofoutis C,
Singh J, Alaveras A and Kalofoutis A: Inflammatory process in type
2 diabetes: The role of cytokines. Ann NY Acad Sci. 1084:89–117.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Capuron L, Poitou C, Machaux-Tholliez D,
Frochot V, Bouillot JL, Basdevant A, Layé S and Clément K:
Relationship between adiposity, emotional status and eating
behaviour in obese women: Role of inflammation. Psychol Med.
41:1517–1528. 2011. View Article : Google Scholar
|
|
27
|
Kellom M, Basselin M, Keleshian VL, Chen
M, Rapoport SI and Rao JS: Dose-dependent changes in
neuroinflammatory and arachidonic acid cascade markers with
synaptic marker loss in rat lipopolysaccharide infusion model of
neuroinflammation. BMC Neurosci. 13:502012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hurley LL and Tizabi Y: Neuroinflammation,
neurodegeneration, and depression. Neurotox Res. 23:131–144. 2013.
View Article : Google Scholar :
|
|
29
|
Ji WW, Wang SY, Ma ZQ, Li RP, Li SS, Xue
JS, Li W, Niu XX, Yan L, Zhang X, et al: Effects of perillaldehyde
on alternations in serum cytokines and depressive-like behavior in
mice after lipopolysaccharide administration. Pharmacol Biochem
Behav. 116:1–8. 2014. View Article : Google Scholar
|
|
30
|
Lawson MA, Parrott JM, McCusker RH,
Dantzer R, Kelley KW and O'Connor JC: Intracerebroventricular
administration of lipopolysaccharide induces
indoleamine-2,3-dioxygenase-dependent depression-like behaviors. J
Neuroinflammation. 10:872013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reininghaus EZ, Reininghaus B, Ille R,
Fitz W, Lassnig RM, Ebner C, Annamaria P, Hofmann P, Kapfhammer HP,
Reingard A, et al: Clinical effects of electroconvulsive therapy in
severe depression and concomitant changes in cerebral glucose
metabolism - an exploratory study. J Affect Disord. 146:290–294.
2013. View Article : Google Scholar
|
|
32
|
Wyss M and Kaddurah-Daouk R: Creatine and
creatinine metabolism. Physiol Rev. 80:1107–1213. 2000.PubMed/NCBI
|
|
33
|
Wallimann T, Walzthöny D, Wegmann G, Moser
H, Eppenberger HM and Barrantes FJ: Subcellular localization of
creatine kinase in Torpedo electrocytes: Association with
acetylcholine receptor-rich membranes. J Cell Biol. 100:1063–1072.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wallimann T: Bioenergetics. Dissecting the
role of creatine kinase. Curr Biol. 4:42–46. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Khuchua ZA, Qin W, Boero J, Cheng J, Payne
RM, Saks VA and Strauss AW: Octamer formation and coupling of
cardiac sarcomeric mitochondrial creatine kinase are mediated by
charged N-terminal residues. J Biol Chem. 273:22990–22996. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schlattner U and Wallimann T: Octamers of
mitochondrial creatine kinase isoenzymes differ in stability and
membrane binding. J Biol Chem. 275:17314–17320. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saks VA, Kuznetsov AV, Kupriyanov VV,
Miceli MV and Jacobus WE: Creatine kinase of rat heart
mitochondria. The demonstration of functional coupling to oxidative
phosphorylation in an inner membrane-matrix preparation. J Biol
Chem. 260:7757–7764. 1985.PubMed/NCBI
|
|
38
|
Jacobs BL, van Praag H and Gage FH: Adult
brain neurogenesis and psychiatry: A novel theory of depression.
Mol Psychiatry. 5:262–269. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Charrier E, Reibel S, Rogemond V, Aguera
M, Thomasset N and Honnorat J: Collapsin response mediator proteins
(CRMPs): Involvement in nervous system development and adult
neurodegenerative disorders. Mol Neurobiol. 28:51–64. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rosslenbroich V, Dai L, Baader SL, Noegel
AA, Gieselmann V and Kappler J: Collapsin response mediator
protein-4 regulates F-actin bundling. Exp Cell Res. 310:434–444.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schmidt EF and Strittmatter SM: The CRMP
family of proteins and their role in Sema3A signaling. Adv Exp Med
Biol. 600:1–11. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Alabed YZ, Pool M, Ong Tone S, Sutherland
C and Fournier AE: GSK3 beta regulates myelin-dependent axon
outgrowth inhibition through CRMP4. J Neurosci. 30:5635–5643. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Goshima Y, Nakamura F, Strittmatter P and
Strittmatter SM: Collapsin-induced growth cone collapse mediated by
an intracellular protein related to UNC-33. Nature. 376:509–514.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gaetano C, Matsuo T and Thiele CJ:
Identification and characterization of a retinoic acid-regulated
human homologue of the unc-33-like phosphoprotein gene (hUlip) from
neuroblastoma cells. J Biol Chem. 272:12195–12201. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Byk T, Dobransky T, Cifuentes-Diaz C and
Sobel A: Identification and molecular characterization of
Unc-33-like phosphoprotein (Ulip), a putative mammalian homolog of
the axonal guidance-associated unc-33 gene product. J Neurosci.
16:688–701. 1996.PubMed/NCBI
|
|
46
|
Minturn JE, Fryer HJ, Geschwind DH and
Hockfield S: TOAD-64, a gene expressed early in neuronal
differentiation in the rat, is related to unc-33, a C. elegans gene
involved in axon outgrowth. J Neurosci. 15:6757–6766.
1995.PubMed/NCBI
|
|
47
|
Franken S, Junghans U, Rosslenbroich V,
Baader SL, Hoffmann R, Gieselmann V, Viebahn C and Kappler J:
Collapsin response mediator proteins of neonatal rat brain interact
with chondroitin sulfate. J Biol Chem. 278:3241–3250. 2003.
View Article : Google Scholar
|
|
48
|
Yuasa-Kawada J, Suzuki R, Kano F, Ohkawara
T, Murata M and Noda M: Axonal morphogenesis controlled by
antagonistic roles of two CRMP subtypes in microtubule
organization. Eur J Neurosci. 17:2329–2343. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Minturn JE, Geschwind DH, Fryer HJ and
Hockfield S: Early postmitotic neurons transiently express TOAD-64,
a neural specific protein. J Comp Neurol. 355:369–379. 1995.
View Article : Google Scholar : PubMed/NCBI
|