Introduction
Due to their high rates of incidence and mortality,
cardiovascular diseases have become a primary health concern
worldwide, and increasing attention has focussed on the prevention
and treatment of cardiovascular diseases.
The release of Weibel-Palade bodies (WPBs) in
endothelial cells is associated with the occurrence and development
of several types of heart disease, including atherosclerosis and
acute coronary syndrome (1–3).
N-ethylamaleimide-sensitive factor (NSF) is a key protein in the
process of WPB membrane fusion with cell membranes, leading to the
release of WPBs (4,5). Small interfering RNA (siRNA) can be
used to repress the expression of specific target genes (6,7).
Reports on the use of siRNA to inhibit the expression of NSF, to
then downregulate WPB release, are limited. In the present study,
small hairpin RNA (shRNA) was designed, which exhibited
complimentary matching to the functional region of the NSF gene, to
examine the mechanism of WPB release, based on previous
investigations of NSF (8,9). The aim of the present study was to
establish whether NSF-shRNA may offer a valuable method for the
prevention and therapy of atherosclerosis and coronary heart
disease, and for the results to provide a valuable reference in
this.
Materials and methods
Design and synthesis of NSF-specific
shRNA
RNA-Seq and Hierarchical Indexing for Spliced
Alignment of Transcripts (HISAT) was used for analyzing the NSF
sequence. The sequence of the human NSF gene was selected from the
GeneBank database (http://www.insdc.org; gene accession no. NM_006178),
and a 21 nt interference oligonucleotide sequence
(5′-TAGGACTGGTTGTTGGAAACA-3′; https://rnaidesigner.thermofisher.com/rnaiexpress/)
was used to target the N′-terminal region of the NSF gene using
siRNA Target Designer-Version 1.51 software.
Two single strands encoding NSF-shRNA DNA were
synthesized by GenePharma Co., Ltd. (Suzhou, China). followed by
the termination sites of RNA polymerase III. The restriction enzyme
sites, MluI and HindIII (Thermo Fisher Scientific,
Inc., Waltham, MA, USA), were located at the ends of the
strands.
NSF-shRNA adenovirus expression vector
construction and identification
The pRNAT-H1.1/Adeno (SD1219; Thermo Fisher
Scientific Inc.) adenovirus vector was digested using MluI
and HindIII restriction enzymes. Following identification by
agarose gel electrophoresis and purification, the vector was
connected to the NSF-shRNA DNA template, which contained
MluI and HindIII enzyme loci. 1% Agarose gel (Gene
Company Ltd., Hong Kong, China) was prepared and used for
electrophoresis for analysis of NSF-shRNA products. The results
were recorded using a gel imaging system (Tanon 1600, Tanon Science
& Technology Co., Ltd., Shanghai, China). The purification of
NSF-shRNA was conducted using a DNA purification kit (Thermo Fisher
Scientific Inc.), according to the manufacturer's instructions.
Ligation of the products was performed by incubating 10 µl
of reaction solution containing 50 ng of the annealed products, 1.5
µl of 10X T4 buffer (Thermo Fisher Scientific Inc.) and 0.5
µl T4 DNA ligase (Thermo Fisher Scientific Inc.) at 16°C
overnight. The recombinant vector was transformed into 50 µl
DH5α-competent cells (American Type Culture Collection, Rockville,
MD, USA). These cells (1×106 cells/ml) were inoculated
into 400 µl lysogeny broth (Guangdong Huankai Microbial Sci
& Tech., Co., Ltd. Guangzhou, China) supplemented with
kanamycin in an incubator overnight (16–18 h) with agitation, at
37°C. Following incubation, the DNA plasmids were extracted using a
Plasmid DNA Mini-Preparation kit (Thermo Fisher Scientific Inc.),
according to the manufacturer's protocol, and identified using 1%
agarose gel electrophoresis. Following identification and
sequencing, the plasmid was stored at −80°C.
Viral packaging and preparation
Conventional resuscitation and subculture of HEK293
cells (American Type Culture Collection) were performed until 80%
of the cells were fused. The cells were prepared by seeding
2×106–2×107 cells/ml into 24-well plates,
with each well containing 0.5 ml cell suspension Dulbecco's
modified Eagle's medium (DMEM, Gibco, Thermo Fisher Scientific
Inc.) containing 10% fetal bovine serum (Gibco, Thermo Fisher
Scientific Inc.). Transfection of the cells with the vector was
performed using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.)
transfection reagent. Briefly, 80 µg DNA and 2 µl
Lipofectamine 2000 were added to 50 µl serum-free DMEM.
Following gentle mixing, the mixture was incubated at room
temperature for 20 min., following which a further 1,000 µl
DMEM was added to the DNA-Lipofectamine 2000 mixture. After 30 min,
the mixture was used for transfection of the HEK293 cells. At 10
days post-transfection, the virus was released from the cells using
a repeated freeze-thaw method, and stored at −80°C.
Viral titer 50% tissue culture infective
dose (TCID50) analysis
HEK293 cells (10 ml; 5×106) and 0.5 ml of
the primitive viral presentation solution were added to a culture
bottle at 37°C in 5% CO2 for 90 min, and 5% (9 ml) DMEM
was added. After 72 h, the cells were collected and centrifuged at
600 × g for 5 min, and the supernatant was discarded. Subsequently,
virus preservation solution (1 ml) was added and freeze-thawing was
repeated three times at −20 and 37°C. The supernatant was collected
by centrifugation at 1,000 × g for 5 min. Using the TCID50 method
for the measurement of viral interference, the pRNAT-H1.1 (SD1219)
adenovirus vector carrying a cGFP gene was transduced into the
HEK293 cells at concentrations of 1, 0.1, 0.01 and 0.001 µl
respectively. Infected cells carrying the GFP tag, were green under
the fluorescent microscope (XSP-63B, Shanghai Optical Instrument
Factory, Co., Ltd., Shanghai, China). The number of green cells was
calculated according to Reed-Muench or Karber methods. After 48 h,
the effects were observed using a fluorescent microscope. In
addition, QBC939 cells (1×106 cells/ml; American Type
Culture Collection) were transduced with the recombinant adenovirus
at 37°C and 5% CO2. Fluorescence polymerase chain
reaction (PCR) analysis was used to detect the viral interference
efficiency, and cells were frozen at −80°C following
assessment.
Transduction of human aortic endothelial
cells (HAECs)
HAECs (1×106 cells/ml; American Type
Culture Collection) were transduced using Lipofectamine 2000 at
37°C and 5% CO2. The HAECs were plated in 6-well plates
and allowed to grow to 70% confluence. Subsequently, 60 µl
viral preservation solution was added to each well, periodically,
at 0, 24, 48 and 72 h following administration of each sample. The
NSF-shRNA adenovirus-transduced group was considered the
experimental group, whereas the negative control group was
transduced with the SD1219 adenovirus vector, and the blank control
group was without interference.
Detection of target gene expression by
reverse transcription-quantitative PCR (RT-qPCR) analysis
Extraction of total RNA
Total RNA was isolated using TRIzol reagent
(Sigma-Aldrich, St. Louis, MO, USA). Each group of cells were mixed
with 200 µl reagent at 4°C, and centrifuged at 8,000 × g for
5 min. The supernatant was discarded, and 50 µl
H2O-dissolved RNA was added at 55–60°C for 5–10 min. The
RNA concentration was quantified by measuring the absorbance, and
total RNA was detected using electrophoresis.
Designation and synthesis of
primers
According to the NSF mRNA sequence, the PCR primers
were as follows: Upstream, TGGGCTGGGCTTTCTATTG; downstream,
TGCCATCTTGTCGGTGTCA; with a PCR amplification fragment length of
150 bp. β-actin was used as a reference and had the following
sequences: Upstream primer, TGACGTGGACATCCGCAAAG; downstream
primer, CTGGAAGGTGGACAGCGAGG; with a PCR amplification fragment
length of 205 bp. The primers were synthesized by Shanghai
Shinegene Molecular Biotech, Inc. (Shanghai, China).
Quantification of mRNA levels of
NSF
The entire process of RT-qPCR was monitored through
real time fluorescent signal accumulation. All reactions were
completed using the PCR System (StepOnePlusTM, Applied Biosystems,
Thermo Fisher Scientific, Inc.). The 50 µl reaction mixture
contained the following: 4 µl cDNA was, 1 µl primers,
4 µl dNTPs, 0.5 µl PCR enzyme, and 5 µl PCR
buffer (Thermo Fisher Scientific Inc.). The reverse transcription
step was conducted under the following conditions: 25°C for 10 min,
42°C for 60 min and 85°C for 5 min, followed by cooling at 4°C. By
detecting the signals of the experimental group, negative control
group and blank control group at 0, 24, 48 and 72 h, the relative
gene expression levels were calculated, using the following method:
ΔCq (relative expression) = Cq (gene)-Cq (internal reference)
(10). The procedures were
performed in accordance with the kit protocol. The reaction
conditions were as follows: 25°C for 10 min, 42°C for 60 min, 85°C
for 5 min and 4°C cooling. The fluorescence qPCR amplification
conditions were as follows: 35 cycles of 94°C for 4 min, 60°C for
30 sec and 72°C for 30 sec, with β-actin as an internal reference,
this was repeated three times and the average of the recorded Cq1,
Cq2, Cq3 values was calculated.
Western blot analysis
The cells were digested and the proteins were
extracted with radioimmunoprecipitation assay buffer (Beijing
Solarbio Science and Technology Co., Ltd., Beijing, China). The
quantity of the protein was confirmed by a bicinchoninic acid
protein assay kit (Beyo-time Institute of Biotechnology, Shanghai,
China). The cells were washed in PBS (3 ml; 4°C; 0.01 mol/l; pH
7.2–7.3) and homogenized in Triton-based lysis buffer (Wuhan Boster
Biological Technology, Ltd., Wuhan, China). Equal quantities of
protein (10 µl) were separated by sodium dodecyl sulfate
polyacrylamide gel electrophoresis and transferred onto a nylon
membrane (Merck KGaA, Darmstadt, Germany). Following blocking with
5% bovine serum albumin (Gen-View Scientific, Inc., Jacksonville,
FL, USA), the membranes were incubated with rabbit polyclonal
anti-NSF antibody (1:5,000, cat. no. 87155; Abcam, Cambridge, MA,
USA) and rabbit anti-glyceraldehyde 3-phosphate dehydrogenase
(GAPDH; 1:10,000, cat. no. 10494-1-AP, Proteintech Group Inc.,
Wuhan, China) primary antibodies overnight at 4°C. Membranes were
then incubated with goat anti-rabbit secondary antibody (1:3,000,
cat. no. 111-035-003, Jackson ImmunoResearch Laboratories, Inc.,
West Grove, PA, USA) was used as secondary antibody. The
membrane-bound antibodies were visualized by incubation with
horseradish peroxidase-conjugated secondary antibodies (1:1,000
dilution) at 25°C for 1 h. The membrane was visualized using an
enhanced chemiluminescence system (Merck KGaA) and X-ray film
(FUJIFILM (China) Investment Co., Ltd., Shanghai, China). The
expression levels were quantified by densitometry.
Immunofluorescence staining
Immunofluorescence analysis of the transfected cells
was performed 72 h following transfection. The cells were washed
with PBS and cultivated with 4 U/l thrombin (Beijing Solarbio
Science and Technology Co., Ltd.) for 1 h. The permeabilized cells
were then incubated (at 25°C) with a mouse anti-VWF antibody
(1:1,000) in PBS for 1 h at room temperature. Following washing
with PBS, the cells were incubated with a fluorescein
isothiocyanate-conjugated anti-mouse IgG antibody for 1 h at room
temperature. The cells were then observed using a Leica AF7000
fluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany)
following washing with PBS, with data expressed as the mean ±
standard deviation of three independent experiments.
Statistical analysis
Data are presented as the mean ± standard deviation.
A homogeneity test of variance was performed using SPSS 16.0
statistical software (SPSS, Inc., Chicago, IL, USA). If variances
were uneven, variable conversion was performed to reach
homogeneity. Student's t-test was used for inter-group comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of NSF-shRNA
sequences
The interfering oligonucleotide sequence targeting
the N-terminal region of the NSF gene was designed as
5′-TAGGACTGGTCGTTGGAAACA-3′ (21 nt). Following database analysis,
no homologous sequences with other molecules were identified. The
content of guanylate and cytosine was close to 50%. The positive
and antisense strands were connected with a looped oligonucleotide
(10 nt) to constitute shRNA. The sequences of NSF-shRNA had
MluI and HindIII enzyme loci at the 5′ and 3′
terminals.
Sequencing results of the NSF-shRNA
recombinant adenovirus vector
The products of the NSF-shRNA sequences were
connected to the adenovirus vector, pRNAT-H1.1/Adeno (SD1219), and
eukaryotic plasmid positive clones were established. Sequencing
data were confirmed by sequence alignment results. Electrophoresis
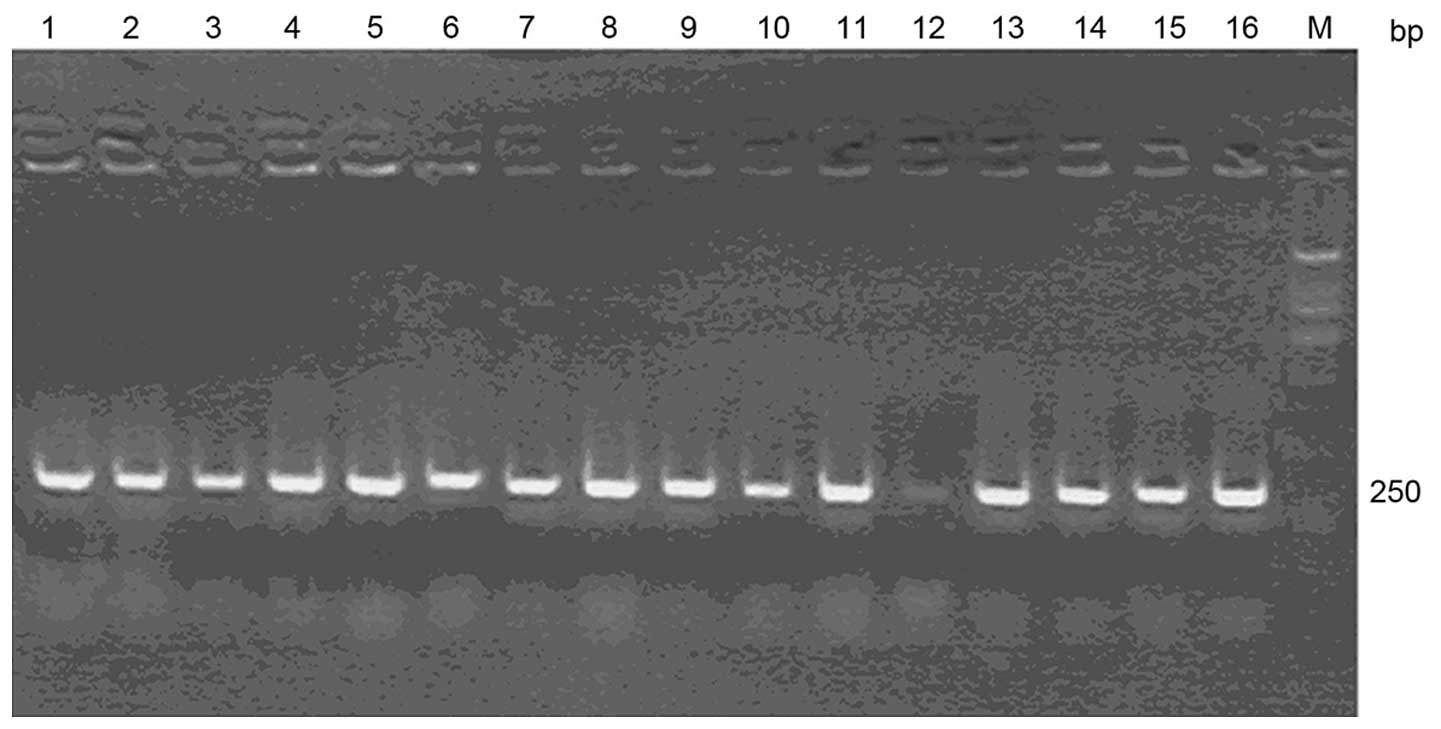
was used for identification of the PCR products, which showed that
the sizes of the positive clones in the 1–11 and 13–16 lanes were
~250 bp. No specific amplification bands of the negative clone were
identified (Fig. 1). The specific
primers were as follows: Sense 5′-TAATACGACTCACTATAGGG-3′ and
antisense 5′-CAAAACTACATAAGACCCCCAC-3′.
Identification of the recombinant
adenovirus
The numbers of fluorescent cells were counted under
an inverted microscope. The TCID50 value was determined using a
classical method and the viral titer of recombinant adenovirus was
determined to be 2×109 TU/ml.
Detection of NSF mRNA
The total RNA in the purified samples were detected
using an ultraviolet spectrophotometer, the optical density
(OD)260/OD280 value was ~1.8–2.0. The bands of 18S/28S were bright
and clear.
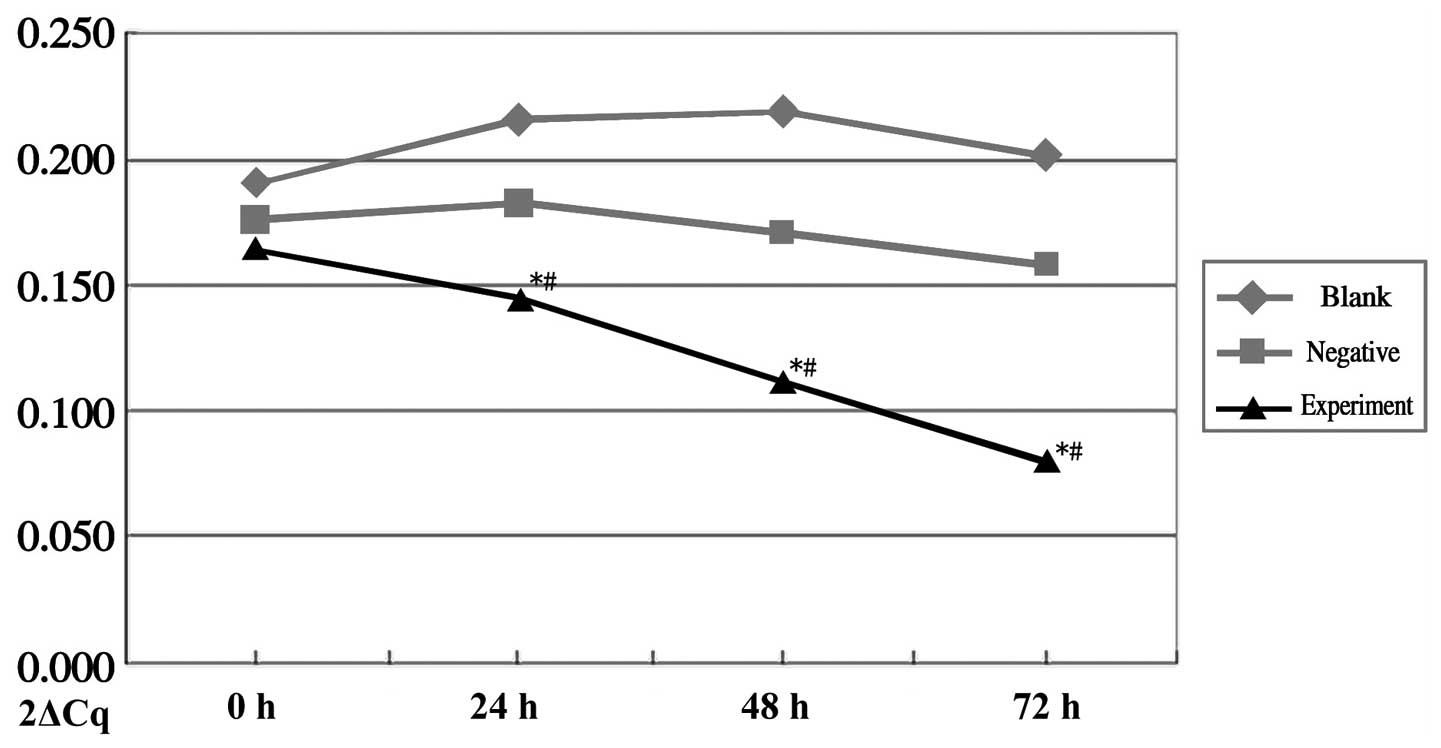
mRNA expression levels of NSF
The mRNA expression of NSF in the experimental group
was markedly reduced and showed statistical difference, compared
with the negative control group (P=0.035) and blank control group
(P=0.02). The mRNA levels of NSF did not differ significantly in
the negative or blank groups between the time points. In the
experimental group, the mRNA level of NSF decreased in a
time-dependent manner, with differences between the 24, 48 and 72 h
groups (P<0.05; Fig. 2).
Protein expression of NSF
The protein expression of NSF was significantly
decreased in the experimental group, compared with the negative
control group (P=0.004) and blank control group (P=0.031), and the
decreased protein expression level of NSF in the experimental group
occurred in a time-dependent manner. No changes were observed in
the protein levels of NSF and GAPDH in the blank and negative
control groups, and no statistical differences were observed
between the blank and negative control groups (P=0.249; Fig. 3).
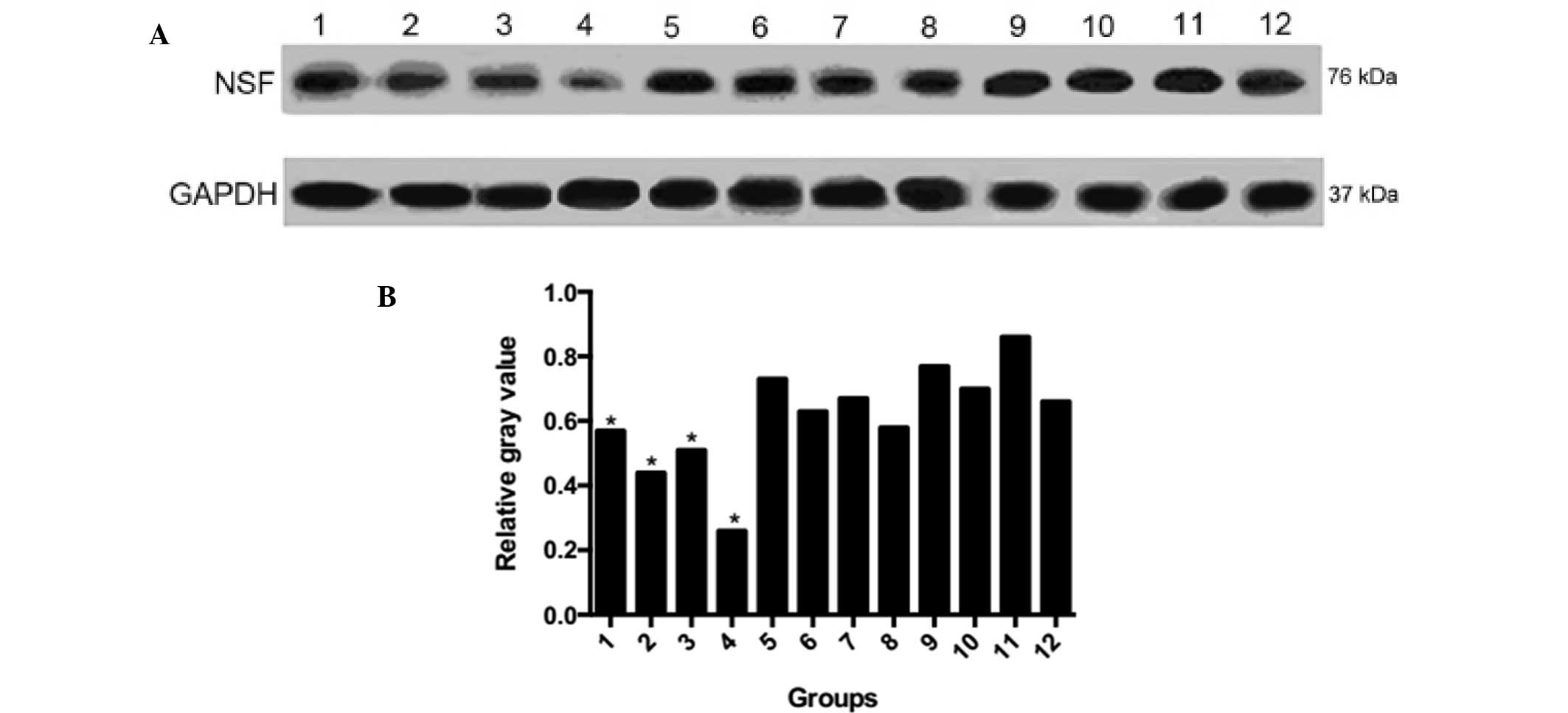 | Figure 3Protein expression levels of NSF. (A)
Western blot analysis of NSF protein. (B) Relative gray values.
Lane 1, experimental group 0 h; 2, experimental group 24 h; 3,
experimental group 48 h; 4, experimental group 72 h; 5, negative
control 0 h; 6, negative control 24 h; 7, negative control 48 h; 8,
negative control 72 h; 9, blank control 0 h; 10, blank control 24
h; 11, blank control 48 h; 12, blank control 72 h. GAPDH served as
the internal control. NSF, N-ethylmaleimide sensitive factor;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
*P<0.05 compared with the negative control. |
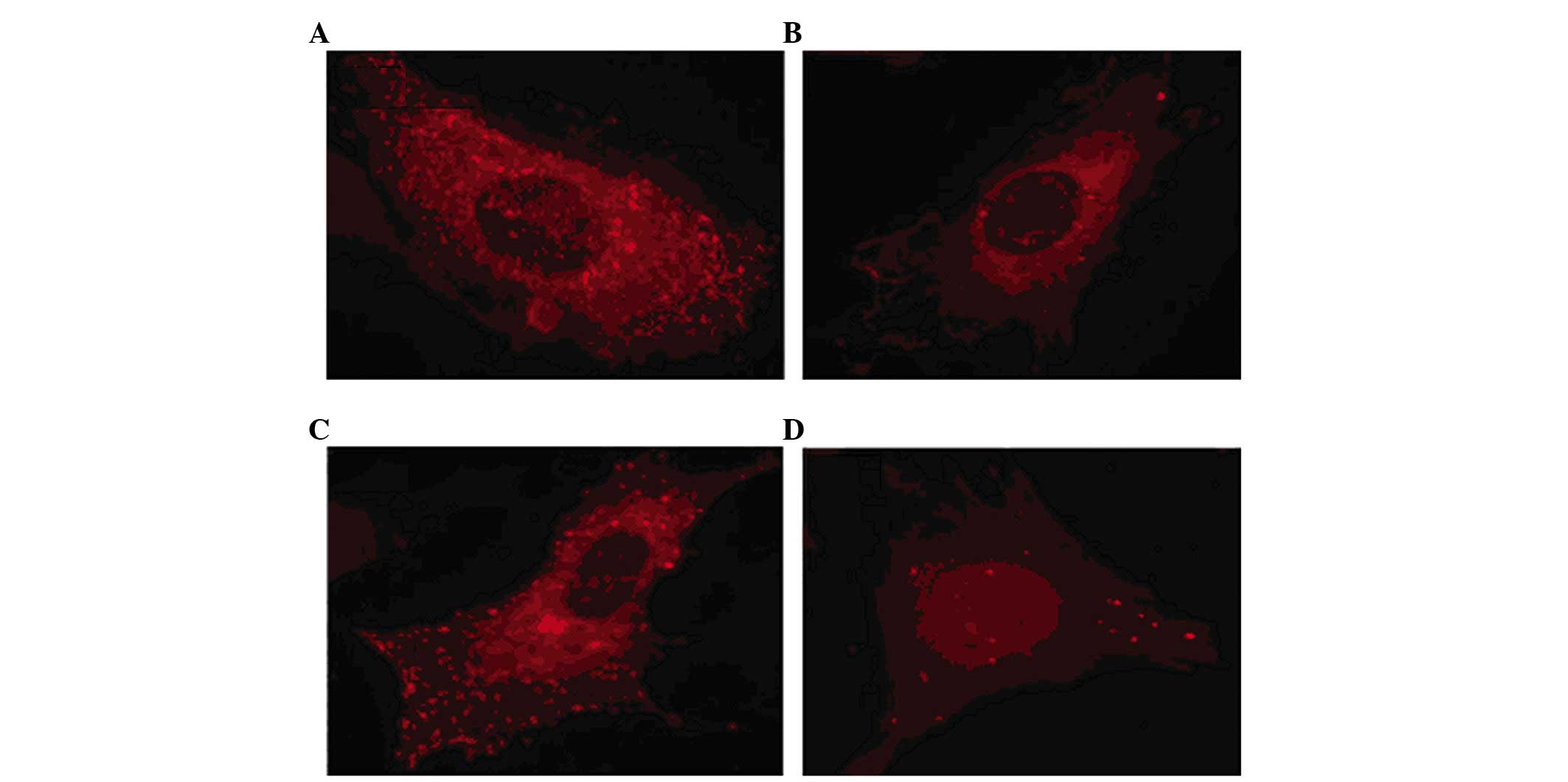
Fluorescence observation of WPB release
following infection with shRNA
The immunofluorescence staining showed that the
release of WPBs in the thrombin-induced endothelial cells was
markedly inhibited following transduction with the virus carrying
NSF-shRNA, but was not affected in the negative or blank control
groups (Fig. 4).
Discussion
Using siRNA for the interference and silencing of
genes has become an area of investigations in clinical treatment
(11,12). In the present study, following the
introduction of a recombinant adenovirus vector, the expression of
NSF was downregulated in HAECs, indicating that NSF-shRNA inhibited
WPB release.
Palade and de Duve first observed the release of WPB
particles from human endothelial cells using immunoelectron
microscopy. There are several bioactive substances present in WPBs,
including P-selectin, endothelin-1 (ET-1) and interleukin-8 (IL-8).
The elevation of P-selectin reflects endogenous platelet activation
(13). ET-1 is a vasoconstrictor
peptide, which can cause coronary vasospasm and may lead to
myocardial ischemia (14). IL-8,
as one of the leukocyte chemoattractants, stimulates the activation
of leukocyte integrins (15).
Therefore, the release of WPBs leads to platelet aggregation, which
may cause the adhesion of neutrophils to the vascular endothelium
and result in vasospasm. A series of inflammatory reactions occur
at the blood vessel wall, and this pathophysiological process is
closely associated with atherosclerosis and acute coronary syndrome
(16).
NSF was first identified as a cytosolic protein,
which is necessary for the in vitro reconstitution of Golgi
transportation, and has been subsequently shown to regulate
intracellular transport in several species (17–19).
Soluble NSF receptor (SNARE) proteins, which are localized to
vesicle and target membranes, assemble into stable ternary
complexes. NSF and the family of soluble NSF attachment proteins
(SNAPs), are critical in regulating vesicle trafficking by
hydrolyzing ATP and disassembling SNARE. NSF comprises three
domains: An N-terminal domain and two homologous ATP binding
domains. The N-terminal domain (residues 1–205) interacts with
members of the SNAP family, which in turn interact with SNARE
molecules (20).
As WPBs are critical in the regulation of
cardiovascular diseases, the present study aimed to identify
methods to inhibit NSF in order to regulate the release of WPBs. A
report by Lowenstein et al suggested that thioredoxin
increases exocytosis by denitrosylating NSF (5). Nitric oxid inhibits the platelet
secretion of granules by targeting NSF, leading to diverse effects
in inhibiting thrombosis and limiting vascular inflammation
(8). Three specific SNAREs
molecules, vesicle-associated membrane protein (VAMP)-1, VAMP-2 and
syntaxin-4, regulate cardiac myocyte exocytosis of atrial
natriuretic peptide, which may affect natriuresis and blood
pressure (21,22). Circulating levels of ET-1 are
increased by NSF in aged arteries (23). In our previous study, it was
confirmed that in addition to the increase in WPBs mediated by
thrombin, the binding activity of Rac-GTP and reactive oxygen
species (ROS) were upregulated. The expression mechanism of WPBs
was regarded as an Rac1-dependent ROS regulatory process (9). Therefore, the present study examined
how NSF siRNA inhibits the release of WPBs.
An important finding of the present study was that
the transcription of NSF mRNA in the experimental group decreased
significantly, compared with the blank and negative control groups,
with similar results for NSF protein. This result demonstrates that
RNA interference (RNAi) induced by NSF-shRNA repressed target gene
expression.
Following the transduction of HASCs with a
recombinant adenovirus, shRNAs are amplified under the control of a
recombinant adenovirus H1 promoter. shRNA is cleaved to yield
siRNA, which targets NSF. The NSF-siRNA duplex is unwound to form
the RNA-induced silencing complex, which recognizes target mRNA.
Target recognition is dependent on complementarity to the target
region. Finally, translational inhibition or, in certain cases,
mRNA degradation, contributes to downregulation in the mRNA level
of NSF. At the same time, the gene encoding the N-terminal
functional region of NSF is inactivated, SNARE compounds cannot
depolymerize, and WPB release is inhibited.
Previous investigations have demonstrated that
vector-based RNAi has time- and dose-dependent effects in mammalian
cells, and observed that decreases in mRNA or protein levels were
not marked at 12 h, but gradually became more evident between 24
and 48 h. The decrease reached a maximal degree between 48 and 60
h, and then became less marked prior to being restored (24). The reason for this may be due to
viral replication interference by cell division. The efficiency of
RNAi had a similar time-dependent effect in the present study. In
the experimental group, the mRNA and protein levels of NSF declined
gradually as the duration of co-culture increased, and the maximum
inhibition was observed 72 h following transfection. This may be
affected by several factors, including transfection efficiency,
co-culture duration, cell type and quantity of vector.
In conclusion, the present study demonstrated the
inhibition of WPB release in endothelial cells with siRNA, mediated
by a recombinant adenovirus. The biology of RNAi represents a
relatively novel area of research. However, further investigations
are required prior to the use of RNAi-based therapeutic strategies
in humans (25). The recombinant
adenovirus components may induce immunoreactions in the host. The
selection of an appropriate vector, helper virus or plasmid, and
the viral purification and biosecurity are major challenges,
although RNAi may offer an effective strategy in the treatment of
cardiovascular diseases.
Acknowledgments
The current study was supported by the Beijing
Natural Science Foundation (grant no. 7083108).
References
|
1
|
Valentijn KM, van Driel LF, Mourik MJ,
Hendriks GJ, Arends TJ, Koster AJ and Valentijn JA: Multigranular
exocytosis of Weibel-Palade bodies in vascular endothelial cells.
Blood. 116:1807–1816. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kiskin NI, Hellen N, Babich V, Hewlett L,
Knipe L, Hannah MJ and Carter T: Protein mobilities and P-selectin
storage in Weibel-Palade bodies. J Cell Sci. 123:2964–2975. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Berriman JA1, Li S, Hewlett LJ, Wasilewski
S, Kiskin FN, Carter T, Hannah MJ and Rosenthal PB: Structural
organization of Weibel-Palade bodies revealed by cryo-EM of
vitrified endothelial cells. Proc Natl Acad Sci USA.
106:17407–17412. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lowenstein CJ and Tsuda H:
N-ethylmaleimide-sensitive factor: A redox sensor in exocytosis.
Biol Chem. 387:1377–1383. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lowenstein CJ, Morrell CN and Yamakuchi M:
Regulation of Weibel-Palade body exocytosis. Trends in Cardiovasc
Med. 15:302–308. 2005. View Article : Google Scholar
|
|
6
|
Sioud M: Promises and challenges in
developing RNAi as a research tool and therapy. Methods in Mol
Biol. 703:173–187. 2011. View Article : Google Scholar
|
|
7
|
Shimizu H and Fujita T: New short
interfering RNA-based therapies for glomerulonephritis. Nat Rev
Nephrol. 7:407–415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qian Z, Gelzer-Bell R, Yang SX, Cao W,
Ohnishi T, Wasowska BA, Hruban RH, Rodriguez ER, Baldwin WM III and
Lowenstein CJ: Inducible nitric oxide synthase inhibition of
weibel-palade body release in cardiac transplant rejection.
Circulation. 104:2369–2375. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang SX, Yan J, Deshpande SS, Irani K and
Lowenstein CJ: Rac1 regulates the release of Weibel-Palade bodies
in human aortic endothelial cells. Chin Med J (Engl).
117:1143–1150. 2004.
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-timequantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
11
|
Ming X: Cellular delivery of siRNA and
antisense oligonucleotides via receptor-mediated endocytosis.
Expert Opin Drug Deliv. 8:435–449. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Samuel-Abraham S and Leonard JN: Staying
on message: Design principles for controlling nonspecific responses
to siRNA. FEBS J. 277:4828–4836. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amin HM, Ahmad S, Walenga JM, Hoppensteadt
DA, Leitz H and Fareed J: Soluble P-selectin in human plasma:
Effect of anticoagulant matrix and its levels in patients with
cardiovascular disorders. Clin Appl Thromb Hemost. 6:71–76. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Russell FD, Skepper JN and Davenport AP:
Evidence using immunoelectron microscopy for regulated and
constitutive pathways in the transport and release of endothelin. J
Cardiovasc Pharmacol. 31:424–430. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wolff B, Burns AR, Middleton J and Rot A:
Endothelial cell 'memory' of inflammatory stimulation: Human
venular endothelial cells store interleukin 8 in Weibel-Palade
bodies. J Exp Med. 188:1757–1762. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alberto P1, Francesca I, Chiara S and
Ranuccio N: Acute coronary syndromes: from the laboratory markers
to the coronary vessels. Biomark Insights. 1:123–130.
2007.PubMed/NCBI
|
|
17
|
Ito T, Yamakuchi M and Lowenstein CJ:
Thioredoxin increases exocytosis by denitrosylating
N-ethylmaleimide-sensitive factor. J Biol Chem. 1:11179–11184.
2011. View Article : Google Scholar
|
|
18
|
Lowenstein CJ: Nitric oxide regulation of
protein trafficking in the cardiovascular system. Cardiovasc Res.
75:240–246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ferlito M, Fulton WB, Zauher MA, Marbán E,
Steenbergen C and Lowenstein CJ: VAMP-1, VAMP-2 and syntaxin-4
regulate ANP release from cardiac myocytes. J Mol Cell Cardiol.
49:791–800. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Matsushita K, Morrell CN, Cambien B, Yang
SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O'Rourke B, et
al: Nitric oxide regulates exocytosis by S-nitrosylation of
N-ethylmaleimdide-sensitive factor. Cell. 115:139–150. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamakuchi M, Ferlito M, Morrell CN,
Matsushita K, Fletcher CA, Cao W and Lowenstein CJ: Exocytosis of
endothelial cells is regulated by N-ethylmaleimide-sensitive
factor. Methods Mol Biol. 440:203–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Calvert JW, Gundewar S, Yamakuchi M, Park
PC, Baldwin WM III, Lefer DJ and Lowenstein CJ: Inhibition of
N-ethylmaleimide-sensitive factor protects against myocardial
ischemia/reperfusion injury. Circ Res. 101:1247–1254. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Goel A, Su B, Flavahan S, Lowenstein CJ,
Berkowitz DE and Flavahan NA: Increased endothelial exocytosis and
generation of endothelin-1 contributes to constriction of aged
arteries. Circ Res. 107:242–251. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martinez J, Patkaniowska A, Urlaub H,
Lührmann R and Tuschl T: Single-stranded antisense siRNAs guide
target RNA cleavage in RNAi. Cell. 110:563–574. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng Y, Wang CC, Choy KW, Du Q, Chen J,
Wang Q, Li L, Chung TK and Tang T: Therapeutic potentials of gene
silencing by RNA interference: Principles, challenges and new
strategies. Gene. 538:217–227. 2014. View Article : Google Scholar : PubMed/NCBI
|