Introduction
Human mesenchymal stem cells (hMSCs) are
characteristically multipotent, and are therefore considered
potential cellular therapies for tissue regeneration (1,2). In
response to osteoblast-induction medium, hMSCs can differentiate
into an osteogenic lineage and exhibit a typical osteoblast
phenotype. Runt-related transcription factor 2 (RUNX2), which is an
important transcription factor that regulates osteogenic gene
expression, is activated during the differentiation of hMSCs into
osteoblasts and triggers the expression of various
osteoblast-associated markers, including collagen I, alkaline
phosphatase and osteocalcin (3,4);
therefore, has a key role in bone formation regulation. Previous
studies have reported that RUNX2 binds to the promoter of several
genes, which are predominantly expressed in osteoblasts (5,6);
however, RUNX2-regulated genes in hMSCs have yet to be fully
elucidated.
The passage of molecules into the nucleus is
facilitated by the activity of nuclear pore complexes (NPCs)
(7). The classical nuclear import
pathway involves two components, namely importin-β, and the adaptor
protein, importin-α. The binding of importin-β to importin-α
triggers a conformational change in importin-α, which increases its
affinity to cargo proteins that possess a nuclear localization
signal (NLS) (8). Importin-β is
the largest evolutionarily conserved family of nuclear transport
receptors. In addition to its role in the transport of
nucleocytoplasmic materials, importin-β is considered a global
regulatory factor of transport-associated cellular functions during
the cell cycle (9).
The importin 8 (IPO8) gene, which belongs to
the importin-β family and is located at chromosomal region
12p11.21, encodes a 1,037-amino acid protein (10). Previous studies have reported that
the IPO8 gene is one of the most commonly used reference
genes for clinicopathological analysis (11–13).
However, other studies have revealed that IPO8 interacts in
an RNA-dependent manner with Argonaute (AGO) proteins, which are
involved in microRNA (miRNA) processing and function (14). Furthermore, HeLa and HEK293 cells
with an IPO8 knockdown exhibited a redistribution of AGO2,
which was originally located in the nucleus, into the cytoplasm,
which in turn led to a moderate increase in target mRNAs (14). These findings indicate that
IPO8 may possess specific functions that are yet to be
elucidated; however, information on the control of IPO8 gene
expression is currently inadequate.
The present study conducted a RUNX2 chromatin
immunoprecipitation (ChIP)-on-chip analysis in hMSCs, which
revealed IPO8 as a target gene of RUNX2. Promoter deletion
and mutation assays demonstrated that RUNX2 is essential in
attaining the maximum level of IPO8 promoter activity in
Saos-2 human osteosarcoma cells. In addition, a ChIP assay
confirmed the specific binding of RUNX2 to the IPO8
promoter. The present study also confirmed the synchronization of
IPO8 and RUNX2 expression during the process of osteoblast
differentiation. The present study is the first, to the best of our
knowledge, to provide insight into understanding the
transcriptional regulation of IPO8 by RUNX2, in which
IPO8 may be involved in osteoblast differentiation.
Materials and methods
ChIP
hMSCs (American Type Culture Collection, Manassas,
VA, USA) were chemically cross-linked with 1% formaldehyde. The
fixed cells were washed twice with phosphate-buffered saline and
were lysed using a lysis buffer [0.1% sodium dodecyl sulfate (SDS),
0.5% Triton X-100, 20 mM Tris-HCl, pH 8.1] that contained 150 mM
NaCl and a protease inhibitor. The lysed cells were subsequently
subjected to sonication in ice water. The resulting sonicated
fragments were within the size range of 200-1,000 bp. Following
sonication, the samples were centrifuged at 13,000 × g for 10 min
at 4°C, and the supernatant was pre-absorbed by 50 µl
protein G beads and was incubated with magnetic beads conjugated to
RUNX2 antibodies [RUNX2 (C-19) cat. no. sc-8566; Santa Cruz
Biotechnology, Inc., Dallas TX, USA; or cat. no. OALA04014; Aviva
Systems Biology Corporation, San Diego, CA, USA] overnight at 4°C.
IgG (H-270) antibody (cat. no. sc-66931, Santa Cruz Biotechnology,
Inc.) served as a negative control. The magnetic beads were then
rinsed four times with lysis buffer, twice with LiCl buffer, and
three times with Tris-EDTA buffer. The bound immunocomplex was
eluted by adding 300 µl of fresh elution buffer [10 mM Tris;
1 mM EDTA, (pH 8.0)]. Subsequently, 20 µl 5 M NaCl was mixed
with the eluted product, which was incubated at 65°C overnight to
reverse the crosslinking. Immunoprecipitated genomic DNA was then
purified and dissolved in EB buffer for ChIP-on-chip analysis.
The immunoaffinity-enriched DNA was subjected to
amplification using a whole genome amplification kit (WGA kit;
Sigma-Aldrich, St. Louis, MO, USA). The amplified products were
submitted to purification using a QIAquick Polymerase Chain
Reaction (PCR) Purification kit (Qiagen GmbH, Hilden, Germany). To
assess ChIP enrichment, qPCR for known specific transcription
factor binding sites was performed
ChIP-on-chip assays
To label DNA, the NimbleGen Dual-Color DNA Labeling
kit (Roche NimbleGen, Inc., Madison, WI, USA) was used, according
to the manufacturer's protocol. DNA (~1 µg) from each sample
was then incubated for 10 min at 98°C, with ~1 OD Cy5-9mer primer
(IP sample) or Cy3-9mer primer (input sample; Roche Applied
Science, Penzberg, Germany). Subsequently, 100 pmol
deoxynucleo-side triphosphates were mixed with 100 U Klenow
fragment (New England Biolabs, Ipswich, MA, USA), which was
incubated at 37°C for 2 h. The labeling reaction was terminated
following the addition of a 0.1 volume of 0.5 M EDTA, which was
followed by DNA purification and precipitation using
isopropanol/ethanol [precipitation with isopropanol (12,000 × g for
15 min); washing with 1 ml 70% ethanol (12,000 × g for 5 min)]. The
microarrays were then hybridized to 4 µg Cy3/5-labeled DNA
dissolved in NimbleGen hybridization buffer/hybridization component
A at 42°C for 16–20 h in a hybridization chamber (Roche NimbleGen,
Inc.). Washing was performed using the NimbleGen Wash Buffer kit
(Roche NimbleGen, Inc.). To conduct array hybridization, the Human
ChIP-chip 3X720K RefSeq Promoter Array (Roche NimbleGen, Inc.) was
used.
Cell cultures
The Saos-2 human osteosarcoma cell line (American
Type Culture Collection) was cultured in McCoy's 5A (Gibco; Thermo
Fisher Scientific, Inc. Waltham, MA, USA) supplemented with 15%
(v/v) fetal bovine serum (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C in an atmosphere containing 5% CO2. Cells
were passaged according to standard cell culture techniques.
5′ rapid amplification of cDNA ends
(RACE)
A 5′ RACE system was used, according to the
manufacturer's protocol (Takara Biotechnology Co., Ltd., Dalian,
China). Briefly, 10 µg total RNA was extracted from Saos-2
cells using TRIzol Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and was treated with calf intestinal phosphatase, in order to
remove any free 5′-phosphate groups. Tobacco acid pyrophosphatase
was used to specifically detach the cap structure from the
full-length mRNA, which in turn isolates the 5′-monophosphate
segment. An RNA oligonucleotide adaptor was then attached to the
newly de-capped mRNA strand at the 5′ end using a T4 RNA ligase.
Using the ligated RNA as a template, IPO8 cDNA synthesis via
reverse transcription was conducted using Murine Moloney Virus
(M-MLV) reverse transcriptase and random primers (reaction volume,
20 µl per well). Briefly, 400 ng total RNA was reverse
transcribed using the PrimeScript™ RT reagent kit with gDNA Eraser
(Takara Biotechnology Co., Ltd.). The protocol was as follows: 25°C
for 5 min, 42°C for 1 h and 70°C for 5 min. The generated cDNA was
subsequently amplified via nested PCR using the LA Taq DNA
polymerase (Takara Biotechnology Co., Ltd.). The reaction
conditions were as follows: 94°C Initial denaturation (5 min); 94°C
denaturation for 30 sec and 56°C annealing for 30 sec, and 72°C
elongation for 40 sec, for 30 cycles. The gene-specific antisense
outer primer 5′-CTTCTTACACTTCCACCATA-3′ (+789 to +770) and inner
primer 5′-CACCAGTTGATACAGGCATA-3′ (+471 to +452) were based on the
IPO8 cDNA sequence. The PCR products were then analyzed by
agarose gel electrophoresis and were cloned into the PMD-18T vector
(Takara Biotechnology Co., Ltd.) prior to direct sequencing
(Invitrogen Biotechnology Co. Ltd., Shanghai, China), in order to
identify the transcription start site (TSS).
Plasmid constructs
PCR analyses were conducted using Pyrobest DNA
polymerase (Takara Biotechnology Co., Ltd.). Briefly, chromosome
DNA was isolated according to the manufacturer's instructions
(Tiangen Biotech, Co., Ltd., Beijing, China). A series of deletion
DNA fragments consisting of the 5′ flanking region of the
IPO8 gene were PCR amplified (reaction volume, 20 µl
per well; initial denaturation, 94°C for 5 min; denaturation, 94°C
for 30 sec; annealing, 56°C for 30 sec; elongation, 72°C for 40
sec, for 30 cycles) using genomic DNA extracted from Saos-2 cells.
The following primers were used: Upstream,
5′-TTCCACGCGTAATCAAAGGGTTAGGAATGT-3′ for P-3302/+134,
5′-TTCCACGCGTGTGGGTCAAGGCTAGAGTTA-3′ for P-2193/+134,
5′-TTCCACGCGTTGTGGCTCGCT TCT TCAGTG-3′ for P-1337/+134,
5′-TTCCACGCGTGTGTCAGTCCTCACCTAGGT-3′ for P-732/+134,
5′-TTCCACGCGTCGATGCCCATACAGTTCTCG-3′ for P-330/+134,
5′-TTCCACGCGTGACGGGAGGCGGTCATAGC-3′ for P-97/+134, and the
downstream primer was 5′-AAGGAGATCTCGACC CCTGGATTACCTCAC-3′ for
all. The PCR products were subjected to gel purification and were
then subcloned into the pGL3-basic firefly luciferase vector
(Promega Corporation, Madison, WI, USA). Site-directed mutagenesis,
which inactivates the RUNX2 binding site, was conducted, according
to the protocol of the MutanBEST kit (Takara Biotechnology Co.,
Ltd.). All constructs were confirmed by DNA sequencing.
Transient transfections and dual
luciferase reporter assays
Cells at a density of 5×104 cells per
well were seeded onto 24-well culture dishes and transiently
transfected using Fugene 6.0 (Promega Corporation), according to
the manufacturer's protocol. Each well received 1.0 µg
IPO8 reporter plasmids and 0.2 µg Renilla
luciferase construct (pRL-TK) as an internal control. All
transfection experiments were conducted in triplicate. A total of
24 h post-transfection, the cells were lysed and used for the dual
luciferase assays (Dual-Luciferase® Reporter assay
system; Promega Corporation).
RNA interference
Cells were transfected with RUNX2 short hairpin
(sh)RNA lentiviral particles (sc-37145-V; Santa Cruz Biotechnology,
Inc.) or control shRNA lentiviral particles (sc-108080; Santa Cruz
Biotechnology, Inc.), according to the manufacturer's protocol.
Briefly, 5×105 target cells were plated in a 6-well
plate alongside 2 ml complete optimal medium with siRNA
Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
and were incubated overnight at 37°C. The medium was then removed
from the wells and was replaced with 2 ml 5 µg/ml polybrene
media mixture and 2 µl shRNA lentiviral particles per well.
A total of 48 h post-infection, the shRNA-infected cells were
harvested and subjected to quantitative (q)PCR or western blot
analysis.
qPCR
Saos-2 cells treated with osteogenic differentiation
media (Invitrogen; Thermo Fisher Scientific, Inc.) were harvested
and total RNA was extracted, as described previously. RNA (2
µg) was used to generate cDNA using M-MLV reverse
transcriptase (Promega Corporation). qPCR analysis was conducted
using SYBR Green PCR Master Mix (Qiagen GmbH) and an ABI 7500
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The PCR reaction conditions were as follows: 94°C Initial
denaturation (10 min); 94°C denaturation for 15 sec and 60°C
annealing and elongation for 60 sec, for 40 cycles. The
quantitative data were determined using the 2−ΔΔCq
method (15). The primer pairs
(Sangon Biotech Co., Ltd., Shanghai, China) used in the present
study were as follows: IPO8, forward
5′-TGTTCAGCTCCTTCCTGATTC-3′, reverse 5′-CTTCTTACACTTCCACCATAC-3′;
RUNX2, forward 5′-GCCTTCAAGGTGGTAGCCC-3′, reverse
5′-AAGGTGAAACTCTTGCCTCGTC-3′. glyceraldehyde 3-phosphate
dehydrogenase (GAPDH), forward 5′-AGAAGGCTGGGGCTCATTTG-3′
and reverse 5′-AGGGGCCATCCACAGTCTTC-3′.
Western blot analysis
Cell lysis was conducted using 1X SDS sample buffer
(1% SDS). Proteins (25 µl) were separated by 10%
SDS-polyacrylamide gel electrophoresis, and were electroblotted
onto a nitrocellulose membrane. The membranes were then blocked
with 5% non-fat milk in tris-buffered saline for 2 h, and
hybridized to an anti-RUNX2 mouse antibody (cat. no. sc-390715 and
dilution, 1:500; Santa Cruz Biotechnology, Inc.), anti-IPO8 rabbit
antibody (cat. no. ab72109 and dilution, 1:1,000; Abcam, Cambridge,
UK), or anti-GAPDH mouse antibody (cat. no. sc-365062 and dilution,
1:1,000; Santa Cruz Biotechnology, Inc.) overnight at 4°C.
Following washing, the membranes were incubated with horseradish
peroxidase-conjugated rabbit anti-mouse IgG (dilution, 1:10,000;
cat. no. sc-358920; Santa Cruz Biotechnology, Inc.) or goat
anti-rabbit IgG (dilution, 1:10,000; cat. no. sc-2004; Santa Cruz
Biotechnology, Inc.) secondary antibodies for 1 h at room
temperature. The blots were developed using the Pierce enhanced
chemiluminescence system (Pierce Biotechnology, Inc., Rockford, IL,
USA), according to the manufacturer's protocol.
Statistical analysis
Data are presented as the mean ± standard error of
the mean. Comparisons among three or more experimental groups were
performed by one-way analysis of variance using SPSS 11.0 software
(SPSS, Inc., Chicago, IL, USA) and P<0.05 was considered to
indicate a statistically significant difference.
Results
RUNX2 targets the IPO8 gene in hMSCs
RUNX2 is considered a key transcription factor that
binds to the promoter region of various genes, which are
predominantly expressed in osteoblasts. In the present study, two
independent biological replicates of the ChIP-on-chip experiment
were performed on hMSCs using two types of anti-RUNX2 antibodies.
To assess ChIP enrichment, qPCR for known specific transcription
factor binding sites was performed on input DNA, ChIP DNA and mock
IP DNA from each sample. The promoter regions of osterix were
amplified from RUNX2-immunoprecipitated DNA, confirming chromatin
enrichment by ChIP. For array hybridization, the Roche NimbleGen
Human ChIP-chip 3X720K RefSeq Promoter Array was used, which is a
single array design that includes 22,542 gene promoter regions
(from −3,200 to +800 bp of the TSSs) covering ~720,000 50–75mer
probes that are spaced apart by ~100 bp, dependent on the
nucleotide sequence of the segment. Notably, RUNX2 binding was
observed for the human IPO8 gene in both types of RUNX2 ChIP
assay.
Analysis of promoter activity within the
5′ flanking region
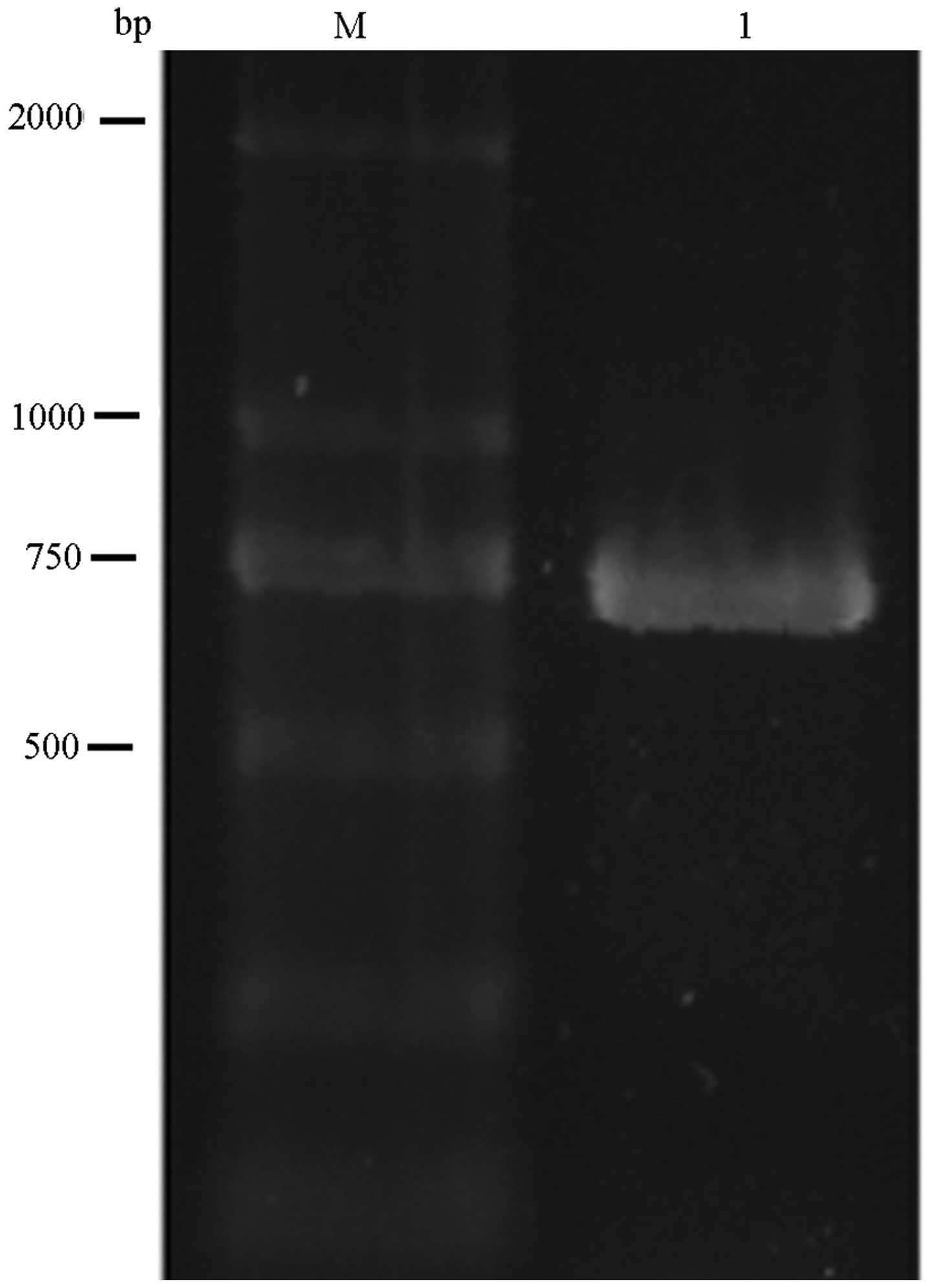
To characterize the IPO8 gene TSS, a 5′ RACE
system was used to amplify the 5′ terminus of IPO8 cDNA
generated from the mRNA of Saos-2 cells. Sequencing of the 700-bp
PCR product generated by 5′ RACE identified a single TSS (+1),
which was located −333 bp upstream of the initiation codon
(Fig. 1).
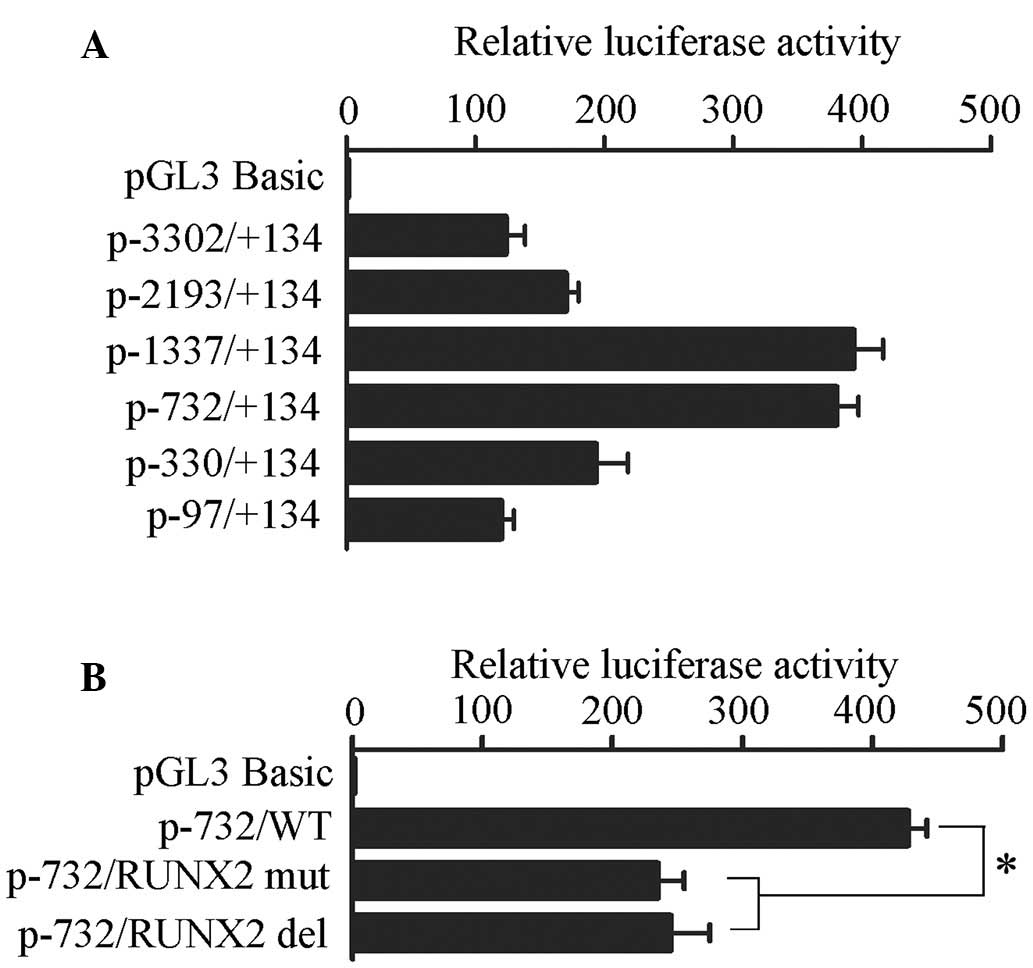
Based on the results of the TSS analysis, a 3,436-bp
region (−3,302 to +134 bp) that encompasses the TSS was amplified
from human genomic DNA and inserted into the promoter-deleted
pGL3-basic firefly luciferase vector. Post-transfection into Saos-2
cells, the P-3302 construct exhibited significant promoter activity
that was >100-fold greater than the activity of the control
plasmid pGL3-basic, thus suggesting that the −3,302 to +134 bp
region had strong basal promoter activity. To identify the regions
controlling IPO8 promoter activity, a set of 5′ truncated
constructs were inserted in the upstream region of the luciferase
reporter gene of the pGL3-basic vector. No significant luciferase
activity differences were detected between the P-2193 and P-3302
constructs, suggesting the absence of activation or repression
motifs between −2,193 and −3,302 bp. In addition, the constructs
P-1337 and P-732 exerted similar promoter activities, which were
the highest among all of the constructs in Saos-2 cells. Further
truncation from bp −732 to −330 resulted in significant decreases
in luciferase activity. These results indicated that the region
−732 to −330 bp possessed the critical element(s) necessary for
maximum activity of the IPO8 promoter (Fig. 2A).
TRANSFAC analysis (http://www.gene-regulation.com) demonstrated that the
region between −732 and −330 bp contained a TGTGGT sequence (−496
to −501 bp), which has been identified as a component of the
binding site for RUNX2 (16,17).
To evaluate the role of this DNA motif in IPO8 gene
transcription, site-directed mutation and deletion was introduced
into the RUNX2 binding site and promoter activity was determined.
As presented in Fig. 2B,
disruption of the RUNX2 binding site reduced luciferase gene
expression by 50% compared with the wild-type.
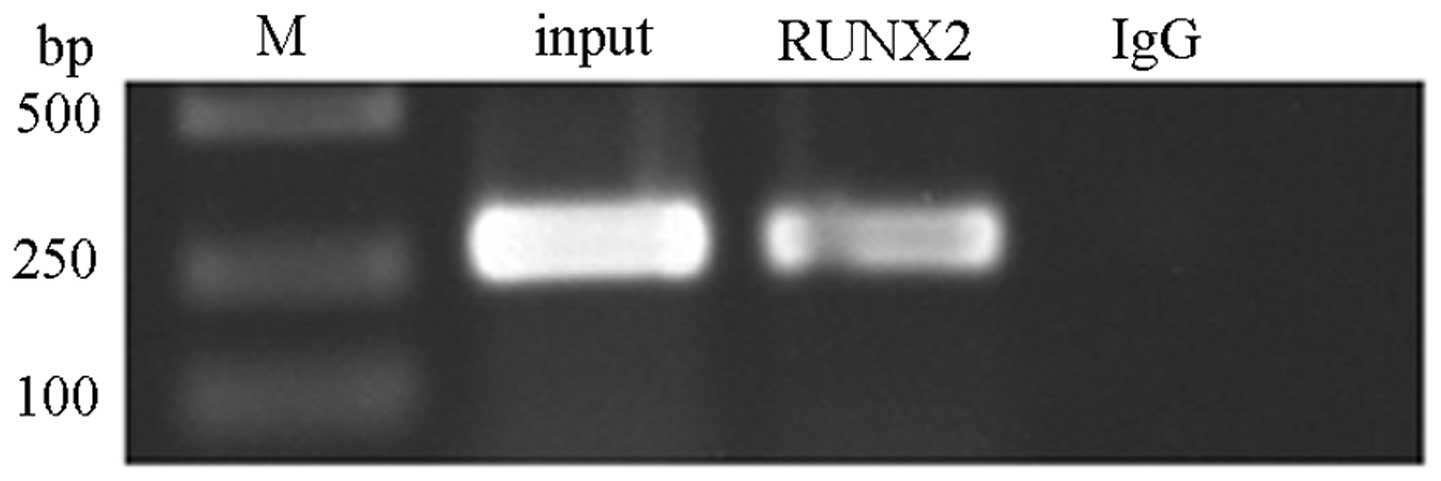
RUNX2 binds to the promoter region of the
IPO8 gene in Saos-2 cells
To determine whether cellular RUNX2 binds to the
IPO8 promoter region in Saos-2 cells, ChIP was performed
using RUNX2 (C-19) antibody, which specifically reacts to human
RUNX2. Immunoprecipitation of cross-linked chromatin with the
anti-RUNX2 antibody, followed by PCR amplification of the region
(the primers were as follows: 5′-TTCACTTCGCCCATCCCT-3′;
5′-CATTCATTCATTCGCTTTCA-3′), determined that the endogenous RUNX2
polypeptide was indeed bound to this particular segment (size, 239
bp) of the IPO8 promoter of Saos-2 cells (Fig. 3).
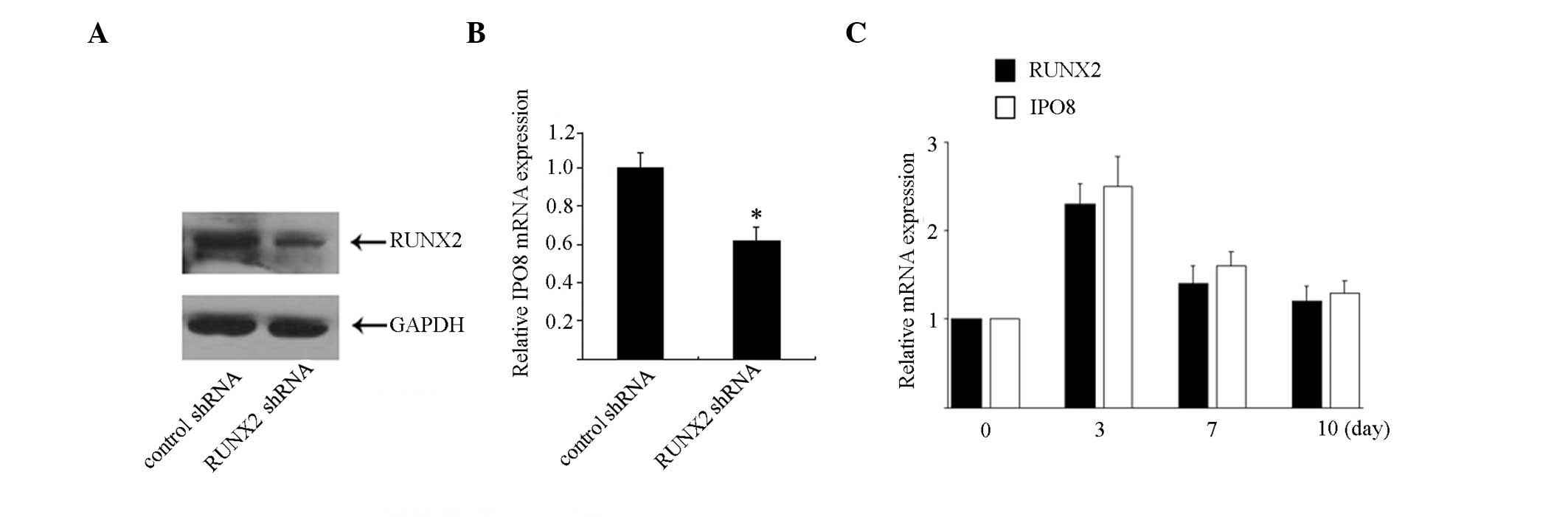
RUNX2 has a role in regulating IPO8
transcription
To examine the involvement of RUNX2 in the
transcriptional regulation of IPO8, RUNX2 expression was
suppressed using lentivirus-mediated shRNA. Saos-2 cells were
infected with shRNA that specifically targets RUNX2 or control
shRNA for 48 h, and were harvested for RUNX2 detection. As shown in
Fig. 4A, the protein expression
levels of RUNX2 were significantly reduced to <25% compared with
the control cells treated with a control shRNA. Subsequently, the
endogenous IPO8 mRNA expression levels were evaluated by
qPCR. As shown in Fig. 4B,
treatment with RUNX2-specific shRNA induced a significant reduction
in the IPO8 mRNA expression by ~50%.
Since IPO8 is the downstream target of RUNX2,
it was hypothesized that IPO8 may be involved in osteoblast
differentiation. Therefore, RUNX2 and IPO8 mRNA
expression levels were detected in Saos-2 cells following treatment
with osteogenic differentiation media. As shown in Fig. 4C, the mRNA expression levels of
IPO8 were significantly increased after 3 days of treatment
and subsequently decreased at day 7. RUNX2 expression was also
observed to be in synchronization with this trend.
Discussion
The present study generated the following key
findings: i) IPO8 is a novel target gene of RUNX2 in hMSCs;
ii) the IPO8 TSS was identified in Saos-2 cells and
characterization of its 5′ flanking regions was conducted; iii) the
RUNX2 binding site was required for maximum IPO8 promoter
activity; iv) RUNX2 regulated IPO8 transcription; and v) the
mRNA expression levels of IPO8 are synchronized with RUNX2
expression during osteoblast differentiation in human osteosarcoma
cells.
hMSCs are self-renewing multipotent cells that may
potentially be used as cellular therapies for tissue regeneration.
Under appropriate conditions, hMSCs have the ability to
differentiate into osteoblasts; however, the control of these
cellular pathways has yet to be fully established. RUNX2 is a
runt-related transcription factor, which is activated during
osteoblast differentiation, and triggers the expression of
osteoblast-specific markers, including collagen I, alkaline
phosphatase and osteocalcin, during the initial stages of
osteoblast maturation. Therefore, RUNX2 is considered to have an
essential role in the control of bone development (3,4).
RUNX2-deficient mice are incapable of developing an ossified
skeleton due to an insufficiency in the number of osteoblasts, thus
indicating the essential function of RUNX2 in determining the
osteoblastic cell fate of MSCs (18). The aim of the present study was to
examine the novel target genes regulated by RUNX2 in hMSCs.
ChIP-on-chip is a technique that facilitates the
large-scale determination of genomic regions interacting with
DNA-binding proteins under physiologically essential in vivo
conditions. As a member of the RUNX family, RUNX2 forms
heterodimers with core-binding factor subunit β and recognizes the
consensus sequences PyGPyGGTPy (16,17),
thus regulating gene transcription, including that of osterix
(19). Previously, genome-wide
occupancy analysis of RUNX2 at gene loci in Saos-2 human
osteosarcoma cells was performed, which revealed several novel
potential target genes, including Talin-1 and cyclic AMP-responsive
element-binding protein 3 (20).
The present study performed a genome-wide ChIP-on-chip analysis of
promoters bound by RUNX2 in hMSCs, the results of which identified
a 22,542 gene promoter region (from −3,200 to +800 bp of the TSSs),
which was completely encompassed by ~720,000 50–75 mer probes at
intervals of ~100 bp, dependent on the sequence of that particular
region. In addition, various novel potential target genes regulated
by RUNX2 were identified. Among these genes, IPO8 exhibited
a higher maximum score in two types of RUNX2 immunoprecipitation
assay, these findings have not been described in previous
studies.
IPO8 belongs to the importin β family. The
importin-α/β complex, in combination with the GTPase ras-related
nuclear protein (Ran), mediates nuclear import of proteins that
possess a classic NLS. IPO8 undergoes binding to the NPC.
Furthermore, along with RanGTP and Ran binding protein 1,
IPO8 prevents GTPase-activating protein induction of Ran
GTPase (21). Numerous proteins
have been reported to be transported by IPO8, including
Smads (22), signal recognition
particle protein 19 (21), and
glucocorticoid receptor (23);
however, the mechanisms leading to IPO8 transcriptional
activation remain elusive. To examine the mechanism underlying
transcriptional regulation, the present study cloned and analyzed
the 5′ flanking segment of the IPO8 gene. Through a series
of deletion studies, a crucial DNA fragment was identified at the
5′ flanking promoter region (−732/−330 bp) responsible for maximum
gene activation. Sequence analysis of the region between −732 and
−330 bp identified a consensus sequence (TGTGGT) for RUNX2-binding
sites (16,17). Since RUNX2 is a bifunctional
regulator that has the ability to activate or inhibit gene
transcription based on the promoter context as well as cellular
milieu, the RUNX2 binding site was mutated to investigate its
effects on IPO8 promoter activity. Mutation studies revealed
that this RUNX2 binding site is required for maximum IPO8
promoter activity. These data indicated that the RUNX2 binding site
imparts a positive regulatory role in IPO8 transcription.
Confirmation of RUNX2 binding in vivo by ChIP indicated that
RUNX2 binds to the critical promoter region of Saos-2 cells, thus
suggesting the physiological relevance of this DNA-protein
interaction in IPO8 gene regulation.
The present study also investigated the effects of
RUNX2 on IPO8 expression. shRNA-mediated knockdown of RUNX2
induced a marked reduction in the mRNA expression of IPO8
compared with that observed in the control cells, thus suggesting
that the RUNX2 transcription factor has a positive role in
regulating IPO8 promoter activity.
Since RUNX2 acts as an upstream regulator for
IPO8 transcription, it may be hypothesized that IPO8
functions are associated with the function of RUNX2. The most
significant function of RUNX2 is the control of osteoblast
differentiation during the process of bone development. The present
study demonstrated that IPO8 expression is synchronized with
RUNX2 expression following osteoblast differentiation in Saos-2
cells, which may be due to the fact that IPO8 transcription
is regulated by RUNX2. IPO8 has long been considered a
potential reference in gene expression investigations of human
glioma, subcutaneous adipose tissues, and differentiated primary
preadipocytes (12,13). However, the results of the present
study indicated that the degree of IPO8 expression may
involve more variability that largely contributes to osteogenic
differentiation following media stimulation. Until now, the
association between IPO8 and osteoblast differentiation has
not yet been verified; however, some reports on IPO8
function may provide indirect clues. As previously mentioned,
translocation of Smad protein into the nucleus is mediated by
IPO8. Smad proteins have previously been implicated as
downstream effectors of transforming growth factor-β/bone
morphogenetic protein signaling (24,25),
which is the key pathway in the transcriptional control of bone
formation. Furthermore, a previous study revealed that IPO8
is required for AGO2 binding to a large series of target mRNAs and
is necessary for miRNA-guided gene suppression (14). miRNAs are currently considered to
be key regulators of a wide range of physiological and pathological
processes, including cell proliferation, apoptosis, cancer, and
osteogenic differentiation. It is therefore likely that the
expression of IPO8 has an unknown important role in cellular
biology.
In conclusion, the findings of the present study
provided novel insights into the control of IPO8
transcription, and may enhance understanding regarding RUNX2
regulatory mechanisms in osteoblast differentiation, bone
development, and degenerative bone disease. These results may
assist in uncovering novel strategies of modulating IPO8
gene expression, particularly during the design of treatment
strategies for osteoporosis, bone repair, and other physiological
processes.
Acknowledgments
The present study was supported by grants from the
National Nature Science Foundation of China (grant no. 81260140 and
81360143) and the Key Projects Grant of Jiangxi Province Education
Office (grant no. GJJ12683).
References
|
1
|
Bruder SP, Jaiswal N and Haynesworth SE:
Growth kinetics, self-renewal, and the osteogenic potential of
purified human mesenchymal stem cells during extensive
subcultivation and following cryopreservation. J Cell Biochem.
64:278–294. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Engler AJ, Sen S, Sweeney HL and Discher
DE: Matrix elasticity directs stem cell lineage specification.
Cell. 126:677–689. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Geoffroy V, Kneissel M, Fournier B, Boyde
A and Matthias P: High bone resorption in adult aging transgenic
mice overexpressing cbfa1/runx2 in cells of the osteoblastic
lineage. Mol Cell Biol. 22:6222–6233. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shui C, Spelsberg TC, Riggs BL and Khosla
S: Changes in Runx2/Cbfa1 expression and activity during
osteoblastic differentiation of human bone marrow stromal cells. J
Bone Miner Res. 18:213–221. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ducy P, Zhang R, Geoffroy V, Ridall AL and
Karsenty G: Osf2/Cbfa1: A transcriptional activator of osteoblast
differentiation. Cell. 89:747–754. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Teplyuk NM, Haupt LM, Ling L, Dombrowski
C, Mun FK, Nathan SS, Lian JB, Stein JL, Stein GS, Cool SM and van
Wijnen AJ: The osteogenic transcription factor Runx2 regulates
components of the fibroblast growth factor/proteoglycan signaling
axis in osteoblasts. J Cell Biochem. 107:144–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stewart M: Molecular mechanism of the
nuclear protein import cycle. Nat Rev Mol Cell Biol. 8:195–208.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kutay U, Bischoff FR, Kostka S, Kraft R
and Görlich D: Export of importin alpha from the nucleus is
mediated by a specific nuclear transport factor. Cell.
90:1061–1071. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cook A, Bono F, Jinek M and Conti E:
Structural biology of nucleocytoplasmic transport. Annu Rev
Biochem. 76:647–671. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Görlich D, Dabrowski M, Bischoff FR, Kutay
U, Bork P, Hartmann E, Prehn S and Izaurralde E: A novel class of
RanGTP binding proteins. J Cell Biol. 138:65–80. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nguewa PA, Agorreta J, Blanco D, Lozano
MD, Gomez-Roman J, Sanchez BA, Valles I, Pajares MJ, Pio R,
Rodriguez MJ, et al: Identification of importin 8 (IPO8) as the
most accurate reference gene for the clinicopathological analysis
of lung specimens. BMC Mol Biol. 9:1032008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kreth S, Heyn J, Grau S, Kretzschmar HA,
Egensperger R and Kreth FW: Identification of valid endogenous
control genes for determining gene expression in human glioma.
Neuro Oncol. 12:570–579. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hurtado del Pozo C, Calvo RM,
Vesperinas-García G, Gómez-Ambrosi J, Frühbeck G, Corripio-Sánchez
R, Rubio MA and Obregon MJ: IPO8 and FBXL10: New reference genes
for gene expression studies in human adipose tissue. Obesity
(Silver Spring). 18:897–903. 2010. View Article : Google Scholar
|
|
14
|
Weinmann L, Höck J, Ivacevic T, Ohrt T,
Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H and Meister G:
Importin 8 is a gene silencing factor that targets argonaute
proteins to distinct mRNAs. Cell. 136:496–507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Komori T: Regulation of skeletal
development by the Runx family of transcription factors. J Cell
Biochem. 95:445–453. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Little GH, Noushmehr H, Baniwal SK, Berman
BP, Coetzee GA and Frenkel B: Genome-wide Runx2 occupancy in
prostate cancer cells suggests a role in regulating secretion.
Nucleic Acids Res. 40:3538–3547. 2012. View Article : Google Scholar :
|
|
18
|
Komori T, Yagi H, Nomura S, Yamaguchi A,
Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al:
Targeted disruption of Cbfa1 results in a complete lack of bone
formation owing to maturational arrest of osteoblasts. Cell.
89:755–764. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nishio Y, Dong Y, Paris M, O'Keefe RJ,
Schwarz EM and Drissi H: Runx2-mediated regulation of the zinc
finger Osterix/Sp7 gene. Gene. 372:62–70. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van der Deen M, Akech J, Lapointe D, Gupta
S, Young DW, Montecino MA, Galindo M, Lian JB, Stein JL, Stein GS
and van Wijnen AJ: Genomic promoter occupancy of runt-related
transcription factor RUNX2 in Osteosarcoma cells identifies genes
involved in cell adhesion and motility. J Biol Chem. 287:4503–4517.
2012. View Article : Google Scholar :
|
|
21
|
Dean KA, von Ahsen O, Görlich D and Fried
HM: Signal recognition particle protein 19 is imported into the
nucleus by importin 8 (RanBP8) and transportin. J Cell Sci.
114:3479–3485. 2001.PubMed/NCBI
|
|
22
|
Xu L, Yao X, Chen X, Lu P, Zhang B and Ip
YT: Msk is required for nuclear import of TGF-{beta}/BMP-activated
Smads. J Cell Biol. 178:981–994. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Freedman ND and Yamamoto KR: Importin 7
and importin alpha/importin beta are nuclear import receptors for
the gluco-corticoid receptor. Mol Biol Cell. 15:2276–2286. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Retting KN, Song B, Yoon BS and Lyons KM:
BMP canonical Smad signaling through Smad1 and Smad5 is required
for endochondral bone formation. Development. 136:1093–1104. 2009.
View Article : Google Scholar : PubMed/NCBI
|