Introduction
Fragile X syndrome (FXS) is the most common
inherited developmental disorder. The disorder results from defects
in the fragile X mental retardation 1 (FMR1) gene, and lack
of FMR1 expression caused by hyper-expansion and methylation
of CGG repeats in the first exon of this gene (1–3). The
FMR protein family has three highly homologous members. The fragile
X mental retardation protein (FMRP) is coded by the FMR1
gene. Two other members are the fragile X related protein 1 (FXR1P)
(4) and FXR2P (5), which are coded by the fragile X
related 1 (FXR1) gene and FXR2, respectively. These
proteins possess two KH domains and a RGG box, which suggests that
they are involved in intracellular RNA transport and translation
regulation (6–10). They also contain a nuclear
localization signal and a nuclear export signal, which suggests
that they can shuttle between the nucleus and the cytoplasm
(11).
FMRP exhibits similar sequence identity (>60%) to
FXR1P/FXR2P, and also contains two KH domains and RGG box (12). The absence of FMRP is the cause of
FXS, however, it is not established whether FXR1P and FXR2P are
associated with FXS pathology or phenotype. Additionally, it is not
established whether FXR1P and FXR2P may compensate for the absence
of FMRP. Furthermore, no significant neuropathological
abnormalities were demonstrated in fetal brain tissue from FXS with
FXR1P and FXR2P are normally expressed (13). FMRP, FXR1P and FXR2P can interact
with themselves and each other (5,14,15),
however, their distributions demonstrated individual expression
patterns in certain mouse and human tissues (16,17),
which indicates that each protein also may function autonomously
(18).
FXR1P is expressed throughout the body, particularly
in muscle and heart tissue. There are seven FXR1P isoforms in mouse
(19,20), including three muscle-specific
isoforms (20) and one
cardiac-specific isoform (21).
FXR1P normal expression is essential for postnatal viability. A
previous study demonstrated that inactivation of FXR1P in mice
caused the death of neonates shortly after birth, which was likely
due to abnormal development of the myocardiac or respiratory muscle
(20). In Xenopus, FXR1P
regulates the development of somites, eyes and cranial cartilage,
however, reduction of FXR1P disrupted the rotation and segmentation
of somitic myotomal cells and hindered normal myogenesis (22). In a previous study in zebrafish,
inactivation of FXR1P caused abnormalities in striated muscle and
severe cardiomyopathy, and led to heart failure in embryos
(23). In humans, altered
expression of muscle-specific isoforms of FXR1P has been previously
implicated in facioscapulohumeral muscular dystrophy (24). Additionally, a previous study
demonstrated that inactivation of FXR1P enhanced the expression
level of tumor necrosis factor-α in mouse macrophages (25). Collectively, these previous studies
demonstrated that FXR1P has an important regulatory function in the
development of normal and cardiac muscle (26,27).
Materials and methods
Strains and cell lines
The Escherichia coli (E. coli)
strains, DH5α and TOP10 (China Center for Type Culture Collection,
Wuhan, China), were cultured in LB medium (1% tryptone, 0.5% yeast
extract, 1% NaCl, pH 7.0; Oxoid; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) at 37°C. The Saccharomyces cerevisiae
strains, Y187 and AH109, were cultured at 30°C in yeast extract
peptone dextrose (YPD) medium (2% tryptone, 1% yeast extract, and
2% glucose; Oxoid; Thermo Fisher Scientific, Inc.) or SC medium
(0.67% yeast nitrogen base without amino acids, 2% glucose)
supplemented with an amino acid mixture without the indicated amino
acids for selection. HeLa and HEK293T cells (Academy of Military
Medical Sciences, Beijing, China) were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal calf serum (Zhejiang Tianhang
Biotechnology Co., Ltd., Hangzhou, China) in a humidified 5%
CO2 incubator at 37°C. SH-SY5Y cells were cultured in
DMEM:nutrient mixture F12 supplemented with 10% fetal calf serum in
a humidified 5% CO2 incubator at 37°C.
Plasmids
For two-hybrid screening, the full open reading
frame of the human FXR1 (GenBank NM_005087.3) gene was
subcloned into the pGBKT7 vector (28) (Clontech Laboratories, Inc.,
Mountainview, CA, USA) to construct pGBKT7-FXR1 (Table I), which was used as the bait
plasmid. In the co-immunoprecipitation assay, the full open reading
frames of the human FXR1 gene and human
CMP-N-acetyl-neuraminic acid synthetase (CMAS) gene (GenBank
NM_018686.4) were subcloned into the pCMV-HA vector (28) (Clontech Laboratories, Inc.) and
pCMV-Myc vector (28) (Clontech
Laboratories, Inc.) to construct the recombinant vectors,
pCMV-HA-FXR1 and pCMV-Myc-CMAS. In the colocalization
assay, the full open reading frame of the human FXR1 gene
and human CMAS gene were subcloned into the pEGFP-N1 vector
(28) (Clontech Laboratories,
Inc.) and pDsRed-Monomer-N1 vector (28) (Clontech Laboratories, Inc.),
respectively, to construct pEGFP-N1-FXR1 and
pDsRed-Monomer-N1-CMAS (Table
I). Additionally, the full open reading frame of the human
FXR1 gene was inserted into the pcDNA3.1(−) vector (29) (Clontech Laboratories, Inc.) to
generate the recombinant vector, pcDNA3.1(−)-FXR1 (Table I), which was transfected into
SH-SY5Y cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and biological effect of the interaction between
FXR1P and CMAS was detected.
 | Table IPlasmids used in the current
study. |
Table I
Plasmids used in the current
study.
| Plasmid name | Vector | Insert(s) | Insertion site | Primers
(5′-3′) |
|---|
| pGBKT7-FXR1 | pGBKT7 | FXR1 | EcoR I |
ctagaattcatggcggacgtgacggtgctagaattcttatgaaacaccattc |
| pCMV-HA-FXR1 | pCMV-HA | FXR1 | EcoR I,
XhoI |
tatgaattcggatggcggagctgacggtggaggcgctcgagttatgaaacaccattcaggac |
| pCMV-Myc-CMAS | pCMV-Myc | BTF | EcoR I,
XhoI |
gtagaattccgatggactcggtggagcgtctcgagctatttttggcatgaat |
| pEGFP-N1-FXR1 | pEGFP-N1 | FXR1 | Xho I,
EcoR I |
tatctcgagctatggcggagctgacggtggaggcggaattcttatgaaacaccattcaggac |
|
pDsRed-Monomer-N1-CMAS | pDsRed-Monomer | BTF | Xho I,
EcoR I |
tatctcgagctatggactcggtggagggcgaattcctatttttggcatgaat |
|
pcDNA3.1(-)-FXR1 | pcDNA3.1(-) | FXR1 | EcoR I,
XhoI |
tatctcgagctatggcggagctgacggtggaggcggaattctatgaaacaccattcaggac |
Yeast two-hybrid screening
All procedures were performed out according to the
yeast two-hybrid system manufacturer's instructions (Clontech
Laboratories, Inc.). The yeast strain AH109 (Clontech Laboratories,
Inc.) was first transformed with the pGBKT7-FXR1 recombinant
plasmid, which was used as the bait to screen the library. The
Human Fetal Brain Matchmaker cDNA library (Clontech Laboratories,
Inc.) cloned into the vector pGADT7 was transformed into the yeast
strain Y187, which was used as prey for two-hybrid screening
(30). The yeast strain
AH109-pGBKT7-FXR1 was mated with Y187 pre-transformed with
the cDNA library and then cultured for 20 h in YPD adenine medium
supplemented with kanamycin to produce diploid zygotes. The
products were plated on leucine, tryptophan and histidine-free SD
medium (SD/LTH) and incubated for 3 days at 30°C. After 3 days,
colonies on individual SD/LTH plates were counted and the total
number of transformants was calculated. To verify putative
interactions between bait protein and prey proteins from the cDNA
libraries, the colonies present on SD/LTH medium were collected and
assayed for β-galactosidase (β-gal) activity (31). The β-gal activity assay was
conducted using a colony-lift filter assay according to the
manufacturer's instructions (Clontech Laboratories, Inc.).
Subsequently, the library from the positive colonies (positive
growth in SD/LTH media and positive β-gal activity) were extracted
and digested with Hind III using a Qiagen Plasmid Midi and
Maxi kit (Qiagen GmbH, Hilden, Germany) according to the
manufacturer's instructions. Plasmid fragment sizes were analyzed
by 1% agarose gel electrophoresis to segregate colonies. Plasmid
DNA purified from the positive yeast colonies was used to transform
E. coli TOP10. Finally, the identity of the positive clones
was determined by sequencing and Basic Local Alignment Search Tool
searches.
Validation of the interaction between
CMAS and FXR1 using the yeast two-hybrid system
The purified bait plasmid pGBKT7-FXR1 was
transformed into the AH109 strain and was then cultured on
tryptophan-free SD medium (SD/T) plates for toxicity and autonomous
activation assays (32). The
recombinant vector pGADT7-CMAS was transformed into the
yeast strain Y187 and then plated on SD medium lacking leucine
(SD/L) (33) (Oxoid; Thermo Fisher
Scientific, Inc.). Subsequently, AH109-pGBKT7-FXR1 was mated
to Y187-pGADT7-CMAS. The transformants were cultured in
yeast peptone dextrose adenine medium for 20 h and then plated on
the SD/LTH. The interaction between CMAS and FXR1P was confirmed
following colony formation on the plates and β-gal assays.
Co-immunoprecipitation assay
HEK293T cells were maintained in DMEM medium
supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin
and 100 mg/ml streptomycin. At 70% confluency, HEK293T cells were
seeded into 6-well plates with basal serum and antibiotic-free
DMEM. According to manufacturer's protocols, the plasmids
pCMV-HA-FXR1 and pCMV-Myc-CMAS were co-transfected
into HEK293 cells with Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), and pCMV-HA-FXR1 and
pCMV-Myc-CMAS were transfected as controls. After 5 h, the
cells were cultured with DMEM containing 10% FBS, 100 U/ml
penicillin and 100 mg/ml streptomycin. After 48 h, the transfected
cells were collected, and then lysed with RIPA lysis buffer
containing protease inhibitor. The cell lysates were harvested and
mixed with 2 µg anti-hemagglutinin (HA; CW0092; Beijing ComWin
Biotech Co., Ltd., Beijing, China) or anti-v-myc avian
myelocytomatosis viral oncogene homolog (Myc; CW0088; Beijing
ComWin Biotech Co., Ltd.) antibody and placed on a shaker for
overnight at 4°C. Subsequently, protein A/G agarose beads were
added and then rocked at 4°C for 4 h. The pulled-down proteins were
separated by 12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and then transferred to polyvinylidene
difluoride membranes. The membranes were blocked with dried skim
milk for 2 h at room temperature and the blot was probed with Myc
monoclonal antibody (1:1,000) overnight at 4°C. The membranes were
then incubated for 2 h at 4°C with horseradish
peroxidase-conjugated goat anti-mouse antibody (1:1,000; CW0102;
Beijing ComWin Biotech Co., Ltd.). Bands were visualized using an
ECL Western Blotting Detection System (Shanghai Majorbio Bio-Pharm
Technology Co., Ltd., Shanghai, China) and imaged (34).
Subcellular colocalization of FXR1P and
CMAS
HEK293T and HeLa cells were maintained in DMEM
medium supplemented with 10% FBS, 100 U/ml penicillin, and 100
mg/ml streptomycin. At 70% confluency, HEK293T and HeLa cells were
plated in laser scanning confocal petri dish with basal serum- and
antibiotic-free DMEM. The plasmids pEGFP-N1-FXR1 and
pDsRed-Monomer-N1-CMAS were single-transfected and
co-transfected into HEK293T cells and HeLa cells using
Lipofectamine 2000 (35). After 5
h, the medium was then replaced with complete DMEM containing 10%
FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin. After 48 h,
the transfected cells were collected and then washed twice with
phosphate-buffered saline. Subsequently, 4% paraformaldehyde was
used to fix the transfected cells and the nuclei were stained with
4′,6-diamidino-2-phenylindole (Beyotime Institute of Biotechnology,
Nantong, China) for 10 min. A laser confocal microscope (Carl Zeiss
AG, Oberkochen, Germany) was used to observe the localization and
colocalization of CMAS and FXR1P in the cells.
Monosialotetrahexosylganglioside 1 (GM1)
enzyme-linked immunosorbent assay (ELISA)
HEK293T and SH-SY5Y cells were maintained in basal
DMEM and DMEM/F12, respectively, lacking FBS and antibiotics.
HEK293T and SH-SY5Y cells were divided into normal cell, empty
vector and FXR1 overexpression group cells. The empty vector
group and FXR1 overexpression group cells were transfected
with the empty plasmid pcDNA3.1(−) and recombinant vector pcDNA3.1
(−)-FXR1, respectively, using Lipofectamine 2000 (36). Subsequently, cells from each group
were collected and lysed. Lysates were centrifuged to remove cell
debris (400 × g, 5 min, 4°C), and ELISA was performed by a human
anti-GM1 ELISA kit (YM-L0587; Shanghai YuanMu Biological Technology
Co., Ltd., Shanghai, China) according to the manufacturer's
protocol. Measurements in all groups were repeated six times.
Statistical analysis
Data are presented as the mean ± standard deviation.
The association among three groups was assessed by analysis of
variance using SPSS software, version 19.0 (IBM SPSS, Armonk, NY,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
FXR1P-interacting protein screening using
the yeast two-hybrid system
The bait plasmid pGBKT7-FXR1 was constructed
with complete human FXR1 and the prey plasmids pGADT7-cDNA
were constructed with the complete Human Fetal Brain Matchmaker
cDNA library. The results of sequencing analysis demonstrated that
the inserts were in-frame and the restriction sites were correct.
Subsequently, AH109-pGBKT7-FXR1 with Y187-pGADT7-cDNA
transformants were mated on SD/LTH medium and β-gal assays were
performed. In total, 10 positive colonies were formed on the SD/LTH
medium and exhibited positive results in the β-gal assay. Following
sequencing and analysis, a colony that produced a 434-amino acid
protein with high identity with human CMAS (99% identity) was
identified and used in the subsequent experiments.
Validation of the interaction between
FXR1P and CMAS using the yeast two-hybrid system
To validate the interaction between the bait protein
and prey protein from the cDNA library, Y187-pGADT7-cDNA colonies
and AH109-pGBKT7-FXR1 were mated again and cultured on
SD/LTH medium plates. Prey plasmid was recovered as described
previously from colonies that continued to grow after 5 days on
SD/LTH medium. Subsequently, the β-gal assay demonstrated a
positive result. Additionally, no β-gal activity was observed in
the yeast strain transformed with only the bait plasmid
(pGBKT7-FXR1).
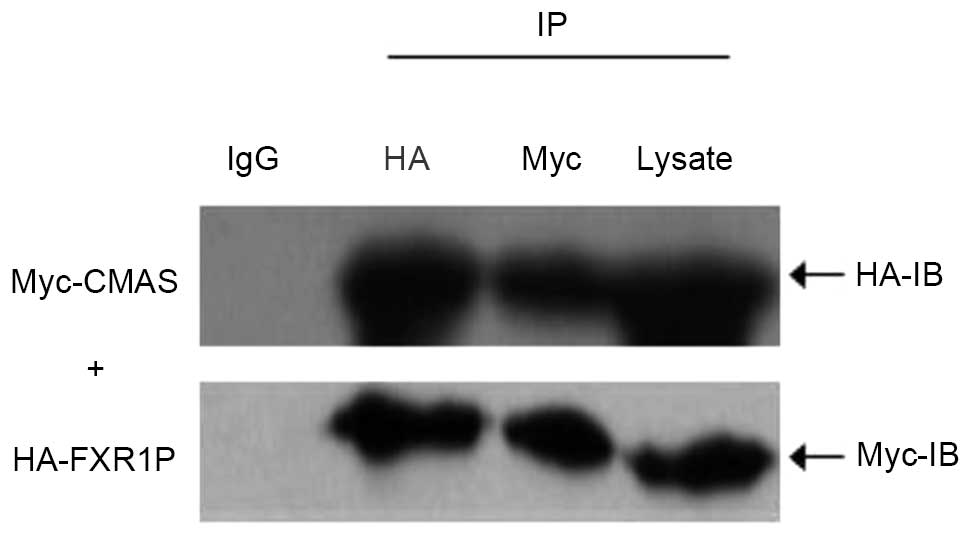
Directing verification of interaction
between FXR1P and CMAS by co-immunoprecipitation
To verify the direct interaction between FXR1P and
CMAS, two recombinant vectors, pCMV-HA-FXR1 and
pCMV-Myc-CMAS, were constructed and the results of
sequencing analysis suggested that the inserts were in-frame and
the restriction sites were correct. Subsequently,
pCMV-Myc-CMAS was co-transfected with pCMV-HA-FXR1
into HEK293T cells, and cell extracts were immunoprecipitated using
the polyclonal anti-HA antibody or anti-Myc antibody, and protein
A/G agarose beads. Following precipitation with anti-HA antibody,
CMAS-Myc was detected using anti-Myc antibody and FXR1P-HA using
anti-HA antibody. Furthermore, following precipitation with the
anti-Myc antibody, FXR1P-HA was detected by anti-HA antibody
(Fig. 1) and CMAS-Myc was detected
by anti-Myc antibody (Fig. 1).
Additionally, a negative control was performed using IgG (Fig. 1). The results indicated that the
demonstrated interaction between FXR1P and CMAS is not because of
nonspecific binding with IgG.
FXR1P colocalizes with CMAS in HEK293T
and HeLa cells
To measure the intracellular interaction between
FXR1P and CMAS, two recombinant eukaryotic expression vectors,
pEGFP-N1-FXR1 and pDsRed-N1-CMAS, were constructed.
pEGFP-N1-FXR1 contained a gene segment of FXR1 and
pDsRed-N1-CMAS contained a gene segment of CMAS. All
recombinant constructs were sequence-verified. HEK293T and HeLa
cells were transfected with the constructs, and the expression and
subcellular distribution of these proteins were analyzed by laser
confocal microscopy.
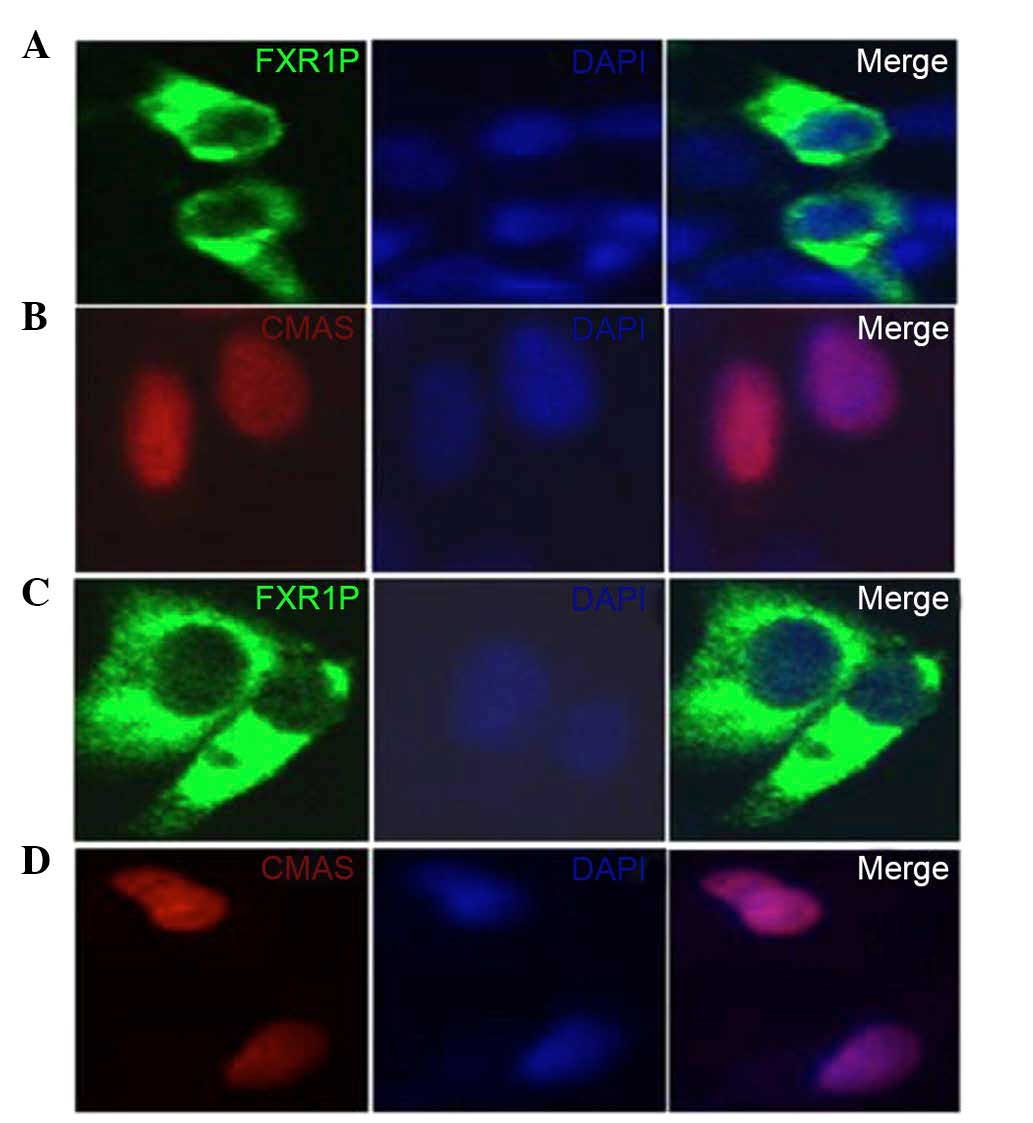
HEK293T and HeLa cells transfected with
pEGFP-N1-FXR1 exhibited strong cytoplasmic fluorescence,
whereas transfection with pDsRed-N1-CMAS produced
fluorescence in the nucleus (Fig.
2). Thus, the fusions to the N-terminus of EGFP or DsRed did
not affect the fluorescence properties of the native protein,
allowing the fusion protein to be correctly localized in
vivo.
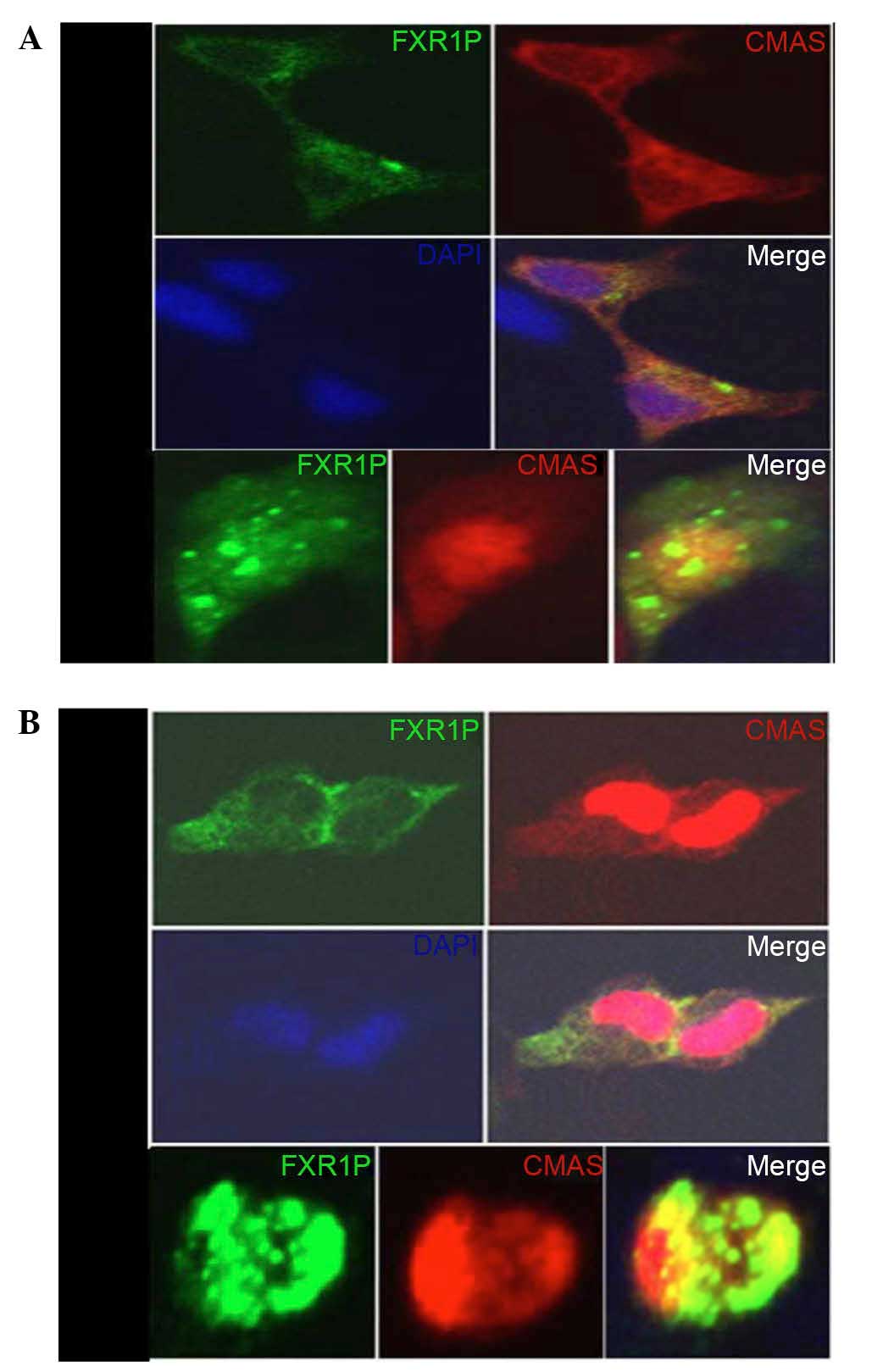
To evaluate the colocalization of FXR1P and CMAS,
pEGFP-N1-FXR1 and pDsRed-N1-CMAS plasmids were
transiently cotransfected into HEK293T and HeLa cells (Fig. 3). Transfection with
pEGFP-N1-FXR1 produced green fluorescence, predominantly in
the cytoplasm, whereas pDsRed-N1-CMAS transfection
predominantly produced red fluorescence in the nucleus and less in
the cytoplasm. The two polypeptides (pEGFP-N1-FXR1 and
pDsRed-N1-CMAS) maintained intracellular localization, with
yellow fluorescence demonstrating FXR1P and CMAS colocalization in
the cytoplasm and the edge of the nucleus (Fig. 3). Typically, these genes are
expressed in different parts of the cell, with no fluorescence
overlap, but co-overexpression leads to the formation of oligomers
in the cytoplasmic and nucleus, producing yellow fluorescence.
Overexpression of FXR1 increases the
concentration of GM1 in SH-SY5Y cells
The current study demonstrated that FXR1P interacts
with CMAS. CMAS catalyzes the synthesis of CMP-sialic acid, which
is an activated form of sialic and the raw material of ganglioside
synthesis. Accordingly, CMAS activity has been demonstrated to be
indirectly associated with amount of the sialic acid converted to
GM1 in cells. To estimate the effect of FXR1P overexpression on
CMAS activity, recombinant vector pcDNA (−) 3.1-FXR1 was
constructed and then the concentration of GM1 was measured in
SH-SY5Y and HEK293T cells transfected with pcDNA3.1
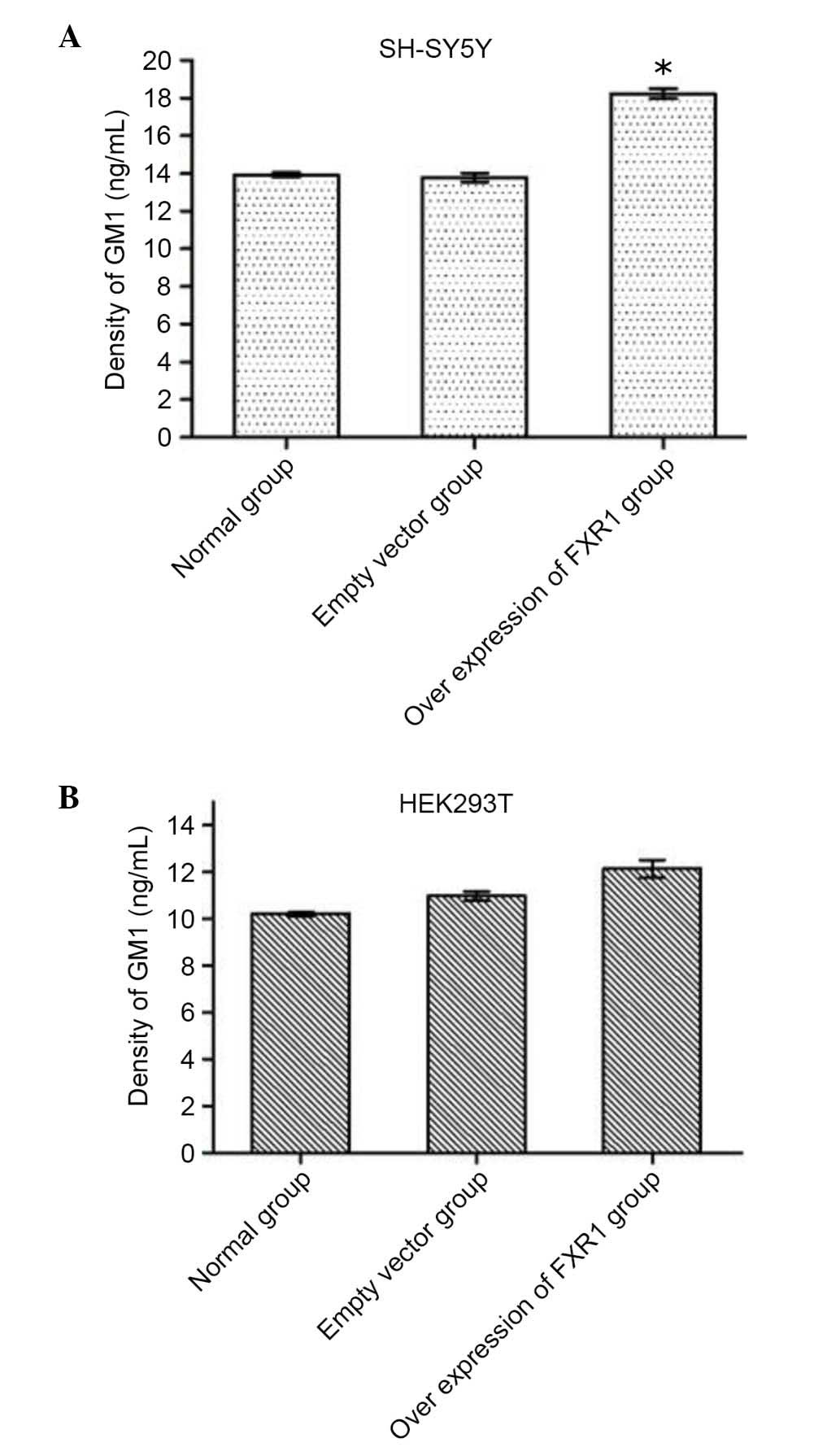
(−)-FXR1. ELISAs demonstrated that the concentration of GM1
was significantly increased in SH-SY5Y cells transfected with
recombinant vector compared with untransfected cells (P=0.016;
Table II; Fig. 4A), whereas there was no significant
difference between the empty vector-transfected SH-SY5Y cells
compared with untransfected cells (Table II; Fig. 4A). However, there was no
significant difference in GM1 concentration among the three groups
of HEK293 cells (Fig. 4B). These
results demonstrated that overexpression of FXR1 increases
the concentration of GM1 in SH-SY5Y cells by promoting CMAS
activity, but not in HEK293T cells (Table II; Fig. 4B).
 | Table IIGM1 concentration in two cell lines
under conditions of different expression level of FXR1. |
Table II
GM1 concentration in two cell lines
under conditions of different expression level of FXR1.
| Normal group | Empty vector
group | Overexpression of
FXR1 group |
|---|
| SH-SY5Y | 13.938±0.126 | 13.786±0.231 |
18.245±0.241a |
| HEK293T | 10.213±0.084 | 1 0.975±0.187 | 12.138±0.382 |
Discussion
FMRP and FXR1P members of the FMR protein family. It
has been previously demonstrated that FMRP is highly expressed in
neurons (37) and involved in the
development of dendritic spines, and the plasticity and structural
remodeling of synapses (38,39).
Deficiency of FMRP can cause mental retardation and impaired memory
and learning. It was previously demonstrated that FXR1P and FMRP
are co-expressed in certain tissues and regulated other proteins at
the translational level (40). A
previous study demonstrated that when FXR1 is expressed
normally in the brain tissue of patients with FXS, there were no
significant neuropathological abnormalities, which indicated that
FXR1P may rescue the function of FMRP (41). Currently, it remains unclear
whether the mechanism of FXS is associated with FXR1P, but a
variety of protein and factors are likely to be involved in this
process. Thus, the study of protein and factors that interact with
FXR1P will aid the clarification of the function of FXR1P in the
pathogenesis of FXS.
Various methods, including co-immunoprecipitation,
the yeast two-hybrid system, surface plasmon resonance and
pull-down technology, are used to investigate interactions between
proteins. The current study screened proteins using the yeast
two-hybrid system, and demonstrated that CMAS interacted with
FXR1P. However, the two-hybrid system tends to identify a high
proportion of false positives (protein hits that are unlikely to
associate in vivo) (42).
Thus, the results were validated using immunoprecipitation and
cellular colocalization analysis in subsequent experiments.
Co-immunoprecipitation is a powerful and simple
method to detect the interaction between proteins. Its principle is
that certain binding proteins are preserved intact when cells are
lysed under non-denaturing conditions. Based on the specificity
between the antibody and the antigen, a protein of interest
(protein X) can be immunoprecipitated with a targeted antibody,
then protein Y, which binds with protein X, can be also
precipitated. In the present study, the interaction between FXR1P
and CMAS was investigated using this method. SDS-PAGE and western
blotting demonstrated that CMAS was also precipitated when FXR1P
was pulled-down by a specific antibody from the total proteins of
cells co-transfected with pCMV-Myc-CMAS and
pCMV-HA-FXR1 (Fig. 1). This
demonstrated an interaction between FXR1P and CMAS. However,
co-immunoprecipitation does no fully to explain the specific
interaction between two proteins in vivo, because the
interaction may not be direct and may be an indirect interaction
through other proteins (43).
Thus, the result of co-immunoprecipitation was validated by
colocalization analysis of CMAS and FXR1P cells. The areas where
the proteins interact in cells were demonstrated by laser confocal
microscopy.
For intracellular colocalization experiments,
HEK-293T cells and HeLa cells were transfected with an
EGFP-FXR1 fusion vector and DsRed-CMAS fusion vector.
This demonstrated that FXR1P and CMAS were respectively located in
the cytoplasm and nuclei. However, when the cells were
co-transfected with both the fluorescent protein fusion vectors,
the merge signal was observed in the cytoplasm and nucleus. The
interaction between the two proteins was confirmed by
co-immunoprecipitation in vitro, and the overlapping
fluorescence signal indicated there was an interaction between
FXR1P and CMAS in live mammalian cells.
CMAS is an enzyme that can catalyzes the synthesis
of sialic acids, which are a family of nine-carbon sugars on cell
surface glycoproteins and glycolipids important for determining the
structure and function of various animal tissues, including
cell-cell communications and immune responses (44). Sialylation occurs in two stages;
activation of sialic acid by the enzyme CMAS and transfer to the
target molecule to generate various GMs (45,46).
The current study demonstrated that overexpression of FXR1
increased the concentration of GM1 in SH-SY5Y cells, which may be
caused by increased CMAS activity. However, this effect was limited
to SH-SY5Y cells, and the GM1 concentration in HEK293T cells was
not changed by FXR1 overexpression. It is well established
that gangliosides promote the occurrence, growth and
differentiation of nerve in the nervous system, however, HEK293T
are human embryonic kidney cells. Thus, it is speculated that the
regulation of GM1 level is tissue-specific.
Previous investigation has demonstrated that the
level of sialic acid is very high in the brain and important for
various cellular activities (47),
including cell adhesion and migration, neurite growth, neuron
differentiation and synapse formation. Additionally, it has been
previously demonstrated that high levels of sialic acid increased
the concentration of gangliosides and promoted the development of
cognitive ability (48). For some
patients with neurological diseases, including Alzheimer's disease
and schizophrenia, the levels of GM1 is decreased in the blood and
the brain (49,50).
In conclusion, FXR1P may enhance the activation of
sialic acid via interaction with CMAS, and increase the GM1 levels
to affect the development of the nervous system. The results of the
present study indicate that FXR1P interacts with CMAS in
vivo, and that FXR1P and CMAS can co-localize in the cytoplasm
around the nucleus. Additionally, FXR1P is able to enhance CMAS
activity to increase the concentration of GM1 in SH-SY5Y cells.
Taken wit the results of a previous study (51), the present study suggests that
FXR1P serves an important role in the development of the brain and
nervous system.
Acknowledgments
This research was supported by the National Natural
Science Foundation of China (grant no. 81200881) and the Hunan
Provincial Natural Science Foundation of China (grant no.
12JJ6073).
References
|
1
|
Vincent A, Heitz D, Petit C, Kretz C,
Oberlé I and Mandel JL: Abnormal pattern detected in fragile-X
patients by pulsed-field gel electrophoresis. Nature. 349:624–626.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Verkerk AJ, Pieretti M, Sutcliffe JS, Fu
YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang
FP, et al: Identification of a gene (FMR-1) containing a CGG repeat
coincident with a breakpoint cluster region exhibiting length
variation in fragile X syndrome. Cell. 65:905–914. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kremer EJ, Pritchard M, Lynch M, Yu S,
Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR and
Richards RI: Mapping of DNA instability at the fragile X to a
trinucleotide repeat sequence p(CCG)n. Science. 252:1711–1714.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siomi MC, Siomi H, Sauer WH, Srinivasan S,
Nussbaum RL and Dreyfuss G: FXR1, an autosomal homolog of the
fragile X mental retardation gene. EMBO J. 14:2401–2408.
1995.PubMed/NCBI
|
|
5
|
Zhang Y, O'Connor JP, Siomi MC, Srinivasan
S, Dutra A, Nussbaum RL and Dreyfuss G: The fragile X mental
retardation syndrome protein interacts with novel homologs FXR1 and
FXR2. EMBO J. 14:5358–5366. 1995.PubMed/NCBI
|
|
6
|
Ashley CT Jr, Wilkinson KD, Reines D and
Warren ST: FMR1 protein: Conserved RNP family domains and selective
RNA binding. Science. 262:563–566. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siomi H, Siomi MC, Nussbaum RL and
Dreyfuss G: The protein product of the fragile X gene, FMR1, has
characteristics of an RNA-binding protein. Cell. 74:291–298. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brown V, Small K, Lakkis L, Feng Y, Gunter
C, Wilkinson KD and Warren ST: Purified recombinant Fmrp exhibits
selective RNA binding as anintrinsic property of the fragile X
mental retardation protein. J Biol Chem. 273:15521–15527. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Darnell JC, Jensen KB, Jin P, Brown V,
Warren ST and Darnell RB: Fragile X mental retardation protein
targets G quartet mRNAs important for neuronal function. Cell.
107:489–499. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schaeffer C, Bardoni B, Mandel JL,
Ehresmann B, Ehresmann C and Moine H: The fragile X mental
retardation protein binds specifically to its mRNA via a purine
quartet motif. EMBO J. 20:4803–4813. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eberhart DE, Malter HE, Feng Y and Warren
ST: The fragile X mental retardation protein is a ribonucleoprotein
containing both nuclear localization and nuclear export signals.
Hum Mol Genet. 5:1083–1091. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Eberhart DE and Warren ST: The molecular
basis of fragile X syndrome. Cold Spring Harb Symp Quant Biol.
61:679–687. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Winograd C and Ceman S: Fragile X family
members have important and non-overlapping functions. Biomol
Concepts. 2:343–352. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Siomi MC, Zhang Y, Siomi H and Dreyfuss G:
Specific sequences in the fragile X syndrome protein FMR1 and the
FXR proteins mediate their binding to 60S ribosomal subunits and
the interactions among them. Mol Cell Biol. 16:3825–3832. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ceman S, Brown V and Warren ST: Isolation
of an FMRP-associated messenger ribonucleoprotein particle and
identification of nucleolin and the fragile X-related proteins as
components of the complex. Mol Cell Biol. 19:7925–7932. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tamanini F, Willemsen R, van Unen L,
Bontekoe C, Galjaard H, Oostra BA and Hoogeveen AT: Differential
expression of FMR1, FXR1 and FXR2 proteins in human brain and
testis. Hum Mol Genet. 6:1315–1322. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bakker CE, de Diego Otero Y, Bontekoe C,
Raghoe P, Luteijn T, Hoogeveen AT, Oostra BA and Willemsen R:
Immunocytochemical and biochemical characterization of FMRP, FXR1P,
and FXR2P in the mouse. Exp Cell Res. 258:162–170. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tamanini F, Van Unen L, Bakker C, Sacchi
N, Galjaard H, Oostra BA and Hoogeveen AT: Oligomerization
properties of fragile-X mental-retardation protein (FMRP) and the
fragile-X-related proteins FXR1P and FXR2P. Biochem J. 343:517–523.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kirkpatrick LL, Mcllwain KA and Nelson DL:
Alternative splicing in the murine and human FXR1 genes. Genomics.
59:193–202. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mientjes EJ, Willemsen R, Kirkpatrick LL,
Nieuwen-huizen IM, Hoogeveen-Westerveld M, Verweij M, Reis S,
Bardoni B, Hoogeveen AT, Oostra BA and Nelson DL: Fxr1 knockout
mice show a striated muscle phenotype: Implications for Fxr1p
function in vivo. Hum Mol Genet. 13:1291–1302. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khandjian EW, Bardoni B, Corbin F, Sittler
A, Giroux S, Heitz D, Tremblay S, Pinset C, Montarras D, Rousseau F
and Mandel J: Novel isoforms of the fragile X related protein FXR1P
are expressed during myogenesis. Hum Mol Genet. 7:2121–2128. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huot ME, Bisson N, Davidovic L, Mazroui R,
Labelle Y, Moss T and Khandjian EW: The RNA-binding protein fragile
X-related 1 regulates somite formation in Xenopus laevis. Mol Biol
Cell. 16:4350–4361. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Van't Padje S, Chaudhry B, Severijnen LA,
van der Linde HC, Mientjes EJ, Oostra BA and Willemsen R: Reduction
in fragile X related 1 protein causes cardiomyopathy and muscular
dystrophy in zebrafish. J Exp Biol. 212:2564–2570. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Davidovic L, Sacconi S, Bechara EG,
Delplace S, Allegra M, Desnuelle C and Bardoni B: Alteration of
expression of muscle specific isoforms of the fragile X related
protein 1 (FXR1P) in facioscapulohumeral muscular dystrophy
patients. J Med Genet. 45:679–685. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khera TK, Dick AD and Nicholson LB:
Fragile X-related protein FXR1 controls post-transcriptional
suppression of lipopolysaccharide-induced tumour necrosis
factor-alpha production by transforming growth factor-beta1. FEBS
J. 277:2754–2765. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zarnescu DC and Gregorio CC: Fragile
hearts: New insights into translational control in cardiac muscle.
Trends Cardiovasc Med. 23:275–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blech-Hermoni Y and Ladd AN: RNA binding
proteins in the regulation of heart development. Int J Biochem Cell
Biol. 45:2467–2478. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Wang C, Li B, Qin L, Su J, Yang M
and He S: Bcl-2-associated transcription factor 1 interacts with
fragile X-related protein 1. Acta Biochim Biophys Sin (Shanghai).
46:119–127. 2014. View Article : Google Scholar
|
|
29
|
Huang X, Qiu YL, Tan JY, Qiao YF and Li M:
Effect of UBE2C overexpression on the proliferation of 293Tcell
line. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 27:634–636. 2011.In
Chinese. PubMed/NCBI
|
|
30
|
Chen Y, Sittler A, Yu M, Bardoni B and Wu
G: Screening of proteins interact with FMR1 by yeast two-hybrid
system. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 20:173–178. 1998.In
Chinese.
|
|
31
|
Mockli N and Auerbach D: Quantitative
beta-galactosidase assay suitable for high-throughput applications
in the yeast two-hybrid system. Biotechniques. 36:872–876.
2004.PubMed/NCBI
|
|
32
|
Zou J, Mi L, Yu XF and Dong J: Interaction
of 14–3-3σ with KCMF1 suppresses the proliferation and colony
formation of human colon cancer stem cells. World J Gastroenterol.
19:3770–3780. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zheng Y, Zhang LP, Jia XQ and Wang HY:
Construction of a yeast two-hybrid cDNA library from the human
testis. Zhonghua Nan Ke Xue. 18:310–313. 2012.In Chinese.
PubMed/NCBI
|
|
34
|
Ma Y, He S, Yang Y, Chen Q, Xiao W, Li B,
Jiao S and Fu X: Ferritin, heavy polypeptide 1 interacts with
fragile X-related protein 1. Neural Regen Res. 6:790–796. 2011.
|
|
35
|
Duan Z, Chen J, He L, Xu H, Li Q, Hu S and
Liu X: Matrix protein of Newcastle disease virus interacts with
avian nucleophosmin B23.1 in HEK293T cells. Wei Sheng Wu Xue Bao.
53:730–736. 2013.In Chinese. PubMed/NCBI
|
|
36
|
Yuan SF, Shi CH, Yan W, Yao Q, Li NL, Wang
T, Wang L and Zhang YQ: Construction and expression of eukaryotic
coexpression plasmid containing human MUC1 gene and GM-CSF gene. Xi
Bao Yu Fen Zi Mian Yi Xue Za Zhi. 23:18–20. 242007.In Chinese.
|
|
37
|
Abitbol M, Menini C, Delezoide AL, Rhyner
T, Vekemans M and Mallet J: Nucleus basalis magnocellularis and
hippo-campus are the major sites of FMR-1 expression in the human
fetal brain. Nat Genet. 4:147–153. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dictenberg JB, Swanger SA, Antar LN,
Singer RH and Bassell GJ: A direct role for FMRP in
activity-dependent dendritic mRNA transport links filopodial-spine
morphogenesis to fragile X syndrome. Dev Cell. 14:926–939. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bassell GJ and Warren ST: Fragile X
syndrome: Loss of local mRNA regulation alters synaptic development
and function. Neuron. 60:201–214. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cook D, Sanchez-Carbente Mdel R, Lachance
C, Radzioch D, Tremblay S, Khandjian EW, DesGroseillers L and Murai
KK: Fragile X related protein 1 clusters with ribosomes and
messenger RNAs at a subset of dendritic spines in the mouse
hippocampus. PLoS One. 6:e261202011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Agulhon C, Blanchet P, Kobetz A, Marchant
D, Faucon N, Sarda P, Moraine C, Sittler A, Biancalana V, Malafosse
A and Abitbol M: Expression of FMR1, FXR1, and FXR2 genes in human
prenatal tissues. J Neuropathol Exp Neurol. 58:867–880. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo D, Rajamäki ML and Valkonen J:
Protein-protein interactions: The yeast two-hybrid system. Methods
Mol Biol. 451:421–439. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Monti M, Orrù S, Pagnozzi D and Pucci P:
Interaction proteomic. Biosci Rep. 25:45–56. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Horsfall LE, Nelson A and Berry A:
Identification and characterization of important residues in the
catalytic mechanism of CMP-Neu5Ac synthetase from Neisseria
meningitides. FEBS J. 277:2779–2790. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qasba PK, Ramakrishnan B and Boeggeman E:
Substrate-induced conformational changes in glycosyltransferases.
Trends Biochem Sci. 30:53–62. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Samuels NM, Gibson BW and Miller SM:
Investigation of the kinetic mechanism of cytidine 5′-monophosphate
N-acetylneuraminic acid synthetase from Haemophilus ducreyi with
new insights on rate-limiting steps from product inhibition
analysis. Biochemistry. 38:6195–6203. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sánchez-Felipe L, Villar E and
Muñoz-Barroso I: α2-3- and α2-6-N-linked sialic acids allow
efficient interaction of Newcastle Disease Virus with target cells.
Glycoconj J. 29:539–549. 2012. View Article : Google Scholar
|
|
48
|
Gurnida DA, Rowan AM, Idjradinata P,
Muchtadi D and Sekarwana N: Association of complex lipids
containing gangli-osides with cognitive development of 6-month-old
infants. Early Hum Dev. 88:595–601. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hirano-Sakamaki W, Sugiyama E, Hayasaka T,
Ravid R, Setou M and Taki T: Alzheimer's disease is associated with
disordered localization of ganglioside GM1 molecular species in the
human dentate gyrus. FEBS Lett. 589:3611–3616. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sommer BR, Cohen BM, Satlin A, Cole JO,
Jandorf L and Dorsey F: Changes in tardive dyskinesia symptoms in
elderly patients treated with ganglioside GM1 or placebo. J Geriatr
Psychiatry Neurol. 7:234–237. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ma Y, Qin L, Don X, Li B, Wang C, Xu C,
Wang S and He S: Biological effect of the interaction between
mental retardation related protein (FXR1P) and CMAS. Prog Biochem
Biophys. 40:1124–1131. 2013.
|