Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs that
are 21–30 nucleotides in length and that interfere with their
target mRNAs, and there are approximately 2,000 of these sequences
in the human genome (1). Each
miRNA negatively regulates protein translation through the
degradation of mRNA cleaved by a miRNA-associated RNA-induced
silencing complex (2). It is now
apparent that an individual miRNA can modulate more than 200 miRNAs
(3,4), and greater than 2,000 miRNAs have
been identified at present (5).
Previous studies have demonstrated that miRNAs can
serve an important role in autoimmune diseases (AID) (6), including systemic lupus erythematosus
(7) and rheumatoid arthritis
(8). Several miRNAs have been
associated with the maturation of various immune cells and with the
regulation of their functions (9–11),
suggesting that certain miRNAs may affect the development and
etiology of AID. In addition, although it has been demonstrated
that miRNAs are involved in numerous types of liver disease, the
association between miRNA profiles and autoimmune liver diseases,
including the clinical relevance of miRNA in treating these
diseases, remains unclear.
Primary biliary cirrhosis (PBC) is a slow,
progressive, chronic cholestatic disease that is characterized by
the destruction of the intrahepatic bile ducts and fibrosis, and
with portal inflammation, it may develop into liver cirrhosis
(12). Several processes are
observed during the development of PBC: i) A specific immune
response including antimitochondrial antibodies (AMA), which are
directed towards the E2 component of the 2-oxo-dehydrogenase
pathway, particularly towards the pyruvate dehydrogenase-E2
complex; ii) abnormal innate immunity, in which the ligands of the
Toll-like receptors 3 and 4 stimulate type 1 T helper cell
responses, resulting in biliary cell destruction; and iii) a higher
frequency autoreactive T cell precursor in the liver (13,14).
During PBC treatment, it has been demonstrated that patients with
efficient biochemical responses to ursodeoxycholic acid (UDCA) have
a low risk of developing liver failure or cirrhosis in the long
term (15). In addition, the
normalization of alanine transaminase (ALT) with additional
bezafibrate treatment reduces the risk of occurrence of
liver-associated symptoms in patients with PBC with insufficient
responses to UDCA (16). At
present, no biomarker has been discovered for the prediction of
refractory PBC. Notably, alterations in hepatic miRNAs, including
miRNA-122a, miRNA-26a, miRNA-328 and miRNA-299-5p, have been
previously associated with PBC (17). However, the association between
these alterations and refractory PBC remains to be fully
elucidated.
The aim of the current study was to identify the
miRNA profiles associated with drug resistance to UDCA, bezafibrate
and prednisolone, in addition to the various clinical parameters in
patients with PBC.
Materials and methods
Patients
The current study involved 20 patients with PBC
treated at Kagawa University Hospital (Miki-Cho, Japan) between
2001 and 2013. The PBC diagnosis was established when two of the
following three criteria were met: i) Biochemical evidence of
cholestasis, primarily based on elevated alkaline phosphatase
levels (ALP), ii) the presence of AMA, and iii) histological
evidence of nonsuppurative destructive cholangitis and destruction
of the interlobular bile ducts (12).
Clinical presentation
The treatment-effective group was defined by
reductions in ALP (less than 600 IU/l) and γ-guanosine triphosphate
(γ-GTP) (less than 100 IU/l) within a year of being treated with
UDCA at a maximum dose of 900 mg/day. Bezafibrate administration
was decided upon within a year subsequent to initiation of the UDCA
medication, in accordance with the response to UDCA monotherapy
(n=8). The treatment-resistant group included the patients who did
not meet a condition of the treatment-effective group. Subsequent
to undergoing continuous treatment more than a year, serum samples
were collected from those patients. The baseline characteristics of
PBC at the time of miRNA sampling are presented in Table I. The present study was approved by
the ethics committee of Kagawa University Faculty of Medicine, and
informed consent was obtained from all patients.
 | Table ICharacteristics of the study
groups. |
Table I
Characteristics of the study
groups.
| Characteristic | Treatment-effective
group (n=15) | Treatment-resistant
group (n=5) |
|---|
| Gender (F/M) | 15/0 | 4/1 |
| Age (years) | 59.3±9.0 | 40±14.6 |
| TP (mg/dl) | 7.79±0.62 | 7.82±0.37 |
| Alb (mg/dl) | 4.01±0.35 | 3.9±0.44 |
| T-bil (mg/dl) | 0.61±0.24 | 0.88±0.48 |
| D-bil (mg/dl) | 0.21±0.15 | 0.32±0.11 |
| AST (IU/l) | 75±66.8 | 87.6±50.8 |
| ALT (IU/l) | 94.9±131.7 | 101.6±52.7 |
| ALP (IU/l) | 602.8±282.2 | 1080.4±504.4 |
| LDH (IU/l) | 246.1±56.5 | 246.1±65.7 |
| γ-GTP (IU/l) | 215.5±140.3 | 399±277.1 |
| IgA (mg/dl) | 272.4±124.6 | 341.2±140.5 |
| IgG (mg/dl) | 1908.3±598.2 | 1955.4±810.8 |
| IgM (mg/dl) | 381.9±258.4 | 404.4±126.1 |
| AMA
(positive/negative) | 9/6 | 3/2 |
| AMA-M2
(positive/negative) | 11/4 | 3/2 |
Analysis of the microRNA array
Total RNA was extracted from the samples derived
from the serum samples using a miRNeasy Mini kit (Qiagen GmbH,
Hilden, Germany), according to the manufacturer's instructions. RNA
samples typically exhibited A260/280 ratios
between 1.9 and 2.1, which were measured using an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
Subsequent to measuring the RNA with an RNA 6000
Nano kit (Agilent Technologies, Inc.), the samples were labeled
using a miRCURY Hy3/Hy5 Power Labeling kit (Takara Bio, Inc., Otsu,
Japan) and were hybridized onto a human miRNA Oligo chip (version
19.0; Toray Industries, Inc., Tokyo, Japan). Scanning was performed
with the 3D-Gene Scanner 3000 (Toray Industries, Inc.), and 3D-Gene
Extraction software (version 1.2; Toray Industries, Inc.) was used
to read the raw intensity of the image. To determine the
alterations in miRNA expression between the treatment-resistant and
treatment-effective groups, the raw data were analyzed with
GeneSpring GX software, version 10.0 (Agilent Technologies, Inc.).
Samples were first normalized relative to the 28S RNA, and then the
baseline was corrected to the median of all samples.
Replicate data were consolidated into two groups:
The treatment-resistant and treatment-effective groups.
Hierarchical clustering was performed with the farthest neighbor
method using the absolute Pearson's correlation coefficient as a
metric. The base-2 log-transformed intensities were median-centered
for each row (miRNA probe) and were color-coded, as presented on
the heat map. The P-value cutoff was set to 0.05. Only alterations
greater than 50% in a minimum of one of the time points for each
sample were considered to be significant. All analyzed data were
scaled by global normalization. The statistical significance of the
differentially expressed miRNAs was analyzed with Student's
t-test.
Statistical analysis
Statistical analyses were performed using the
computer-assisted StatFlex software, version 6.0 (Artec Co., Ltd.,
Osaka, Japan). A paired analysis between the groups was conducted
using Student's t-test and Pearson's correlation coefficient.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miRNA expression in the serum of patients
with PBC
miRNAs are present in human serum and are highly
stable due to the resistance to RNase digestion (18). An miRNA array was performed in the
current study using the serum of patients with PBC. The expression
patterns of 1,769 miRNAs were examined using extracted serum
miRNAs. As presented in Table II,
35 miRNAs were significantly upregulated in the treatment-resistant
group compared with the treatment-effective group. However, 23
miRNAs in the treatment-resistant group were significantly
downregulated compared with those in the treatment-effective group
(Table III). In addition,
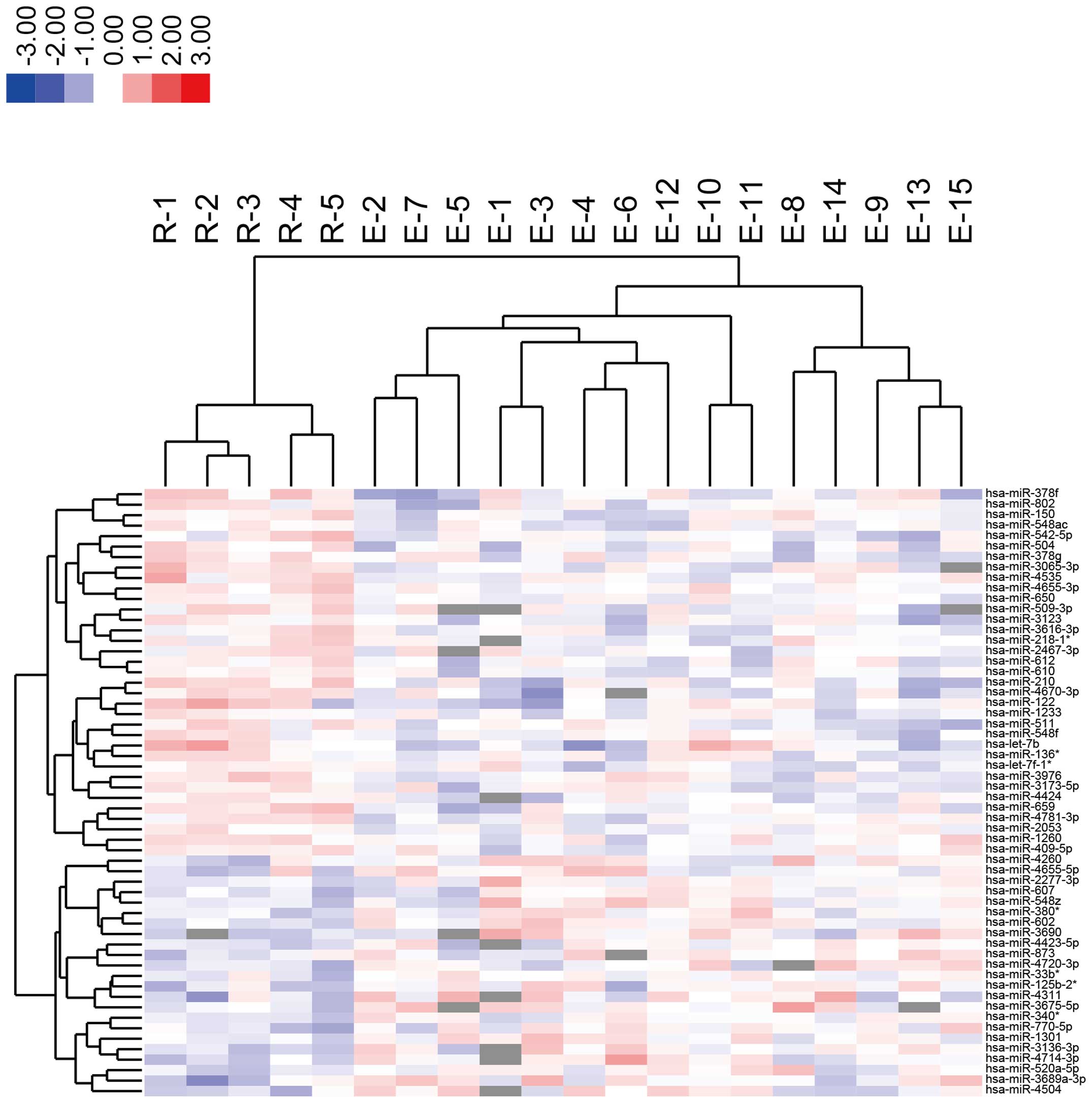
unsupervised hierarchical clustering analysis, using Pearson's
correlation, demonstrated that the treatment-effective group was
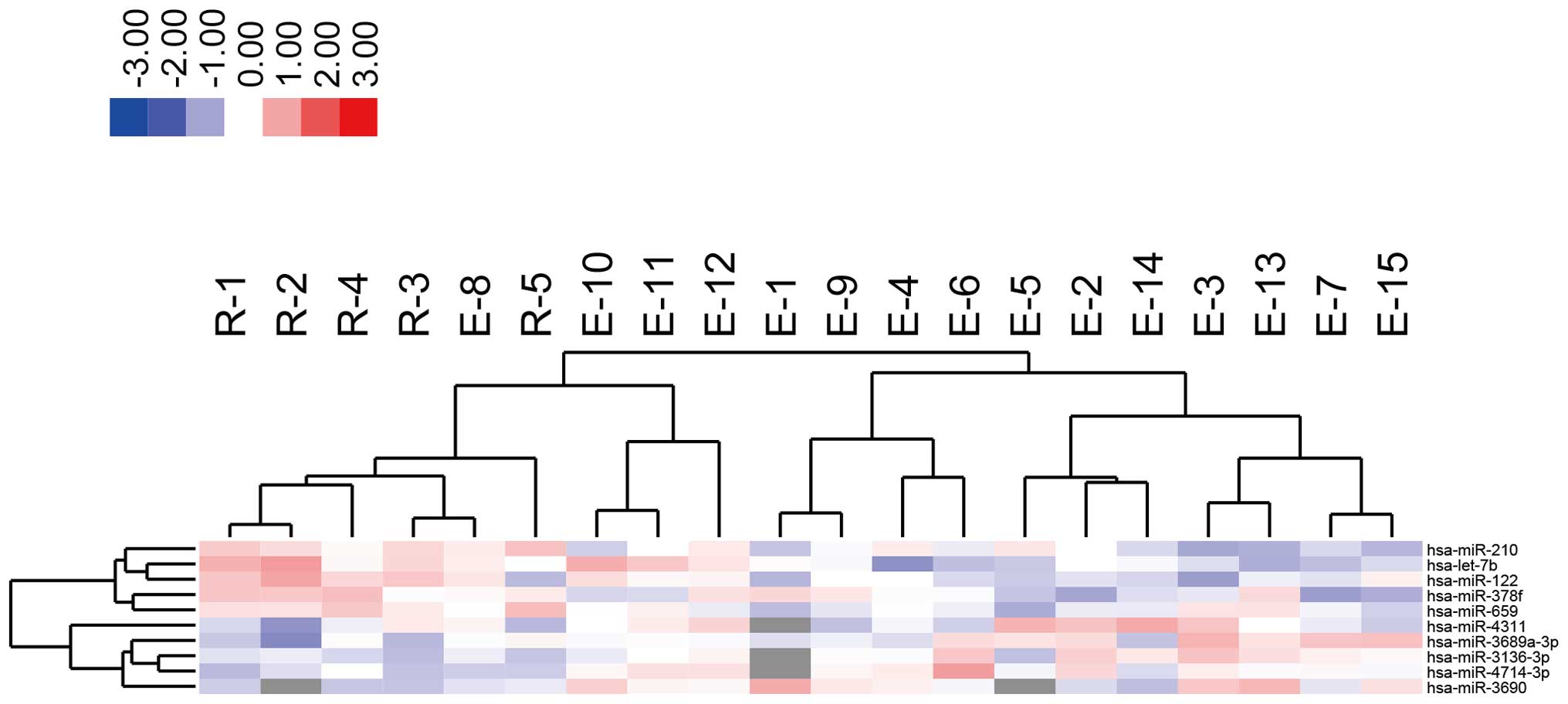
clustered separately from the treatment-resistant group (Fig. 1). Furthermore, highly altered
miRNAs (greater than 1.5-fold) were analyzed, and the two groups
were clustered separately using highly altered miRNAs (Fig. 2 and Tables I and II).
 | Table IIStatistical results of the miRNAs that
were upregulated in the serum of patients with primary biliary
cirrhosis. |
Table II
Statistical results of the miRNAs that
were upregulated in the serum of patients with primary biliary
cirrhosis.
| Upregulated
miRNA | P-value | Fold
(resistant/effective group) |
|---|
| hsa-let-7ba | 0.026804 | 1.567847 |
| hsa-let-7f-1* | 0.047298 | 1.215035 |
| hsa-miR-122a | 0.026881 | 1.561976 |
| hsa-miR-1233 | 0.038244 | 1.208806 |
| hsa-miR-1260 | 0.041029 | 1.227792 |
| hsa-miR-136* | 0.037889 | 1.249572 |
| hsa-miR-150 | 0.039072 | 1.253923 |
| hsa-miR-2053 | 0.039291 | 1.182613 |
| hsa-miR-210a | 0.003102 | 1.564477 |
| hsa-miR-218-1* | 0.045877 | 1.240002 |
|
hsa-miR-2467-3p | 0.035871 | 1.246314 |
|
hsa-miR-3065-3p | 0.003962 | 1.417027 |
| hsa-miR-3123 | 0.043136 | 1.298624 |
|
hsa-miR-3173-5p | 0.025762 | 1.276267 |
|
hsa-miR-3616-3p | 0.028351 | 1.273818 |
|
hsa-miR-378fa | 0.013278 | 1.527677 |
| hsa-miR-378g | 0.022622 | 1.330441 |
| hsa-miR-3976 | 0.014323 | 1.320455 |
| hsa-miR-409-5p | 0.030094 | 1.220102 |
| hsa-miR-4424 | 0.041228 | 1.236549 |
| hsa-miR-4535 | 0.007167 | 1.394004 |
|
hsa-miR-4655-3p | 0.005221 |
1.25409 |
|
hsa-miR-4670-3p | 0.007172 | 1.489381 |
|
hsa-miR-4781-3p | 0.012059 | 1.290433 |
| hsa-miR-504 | 0.026989 | 1.349747 |
| hsa-miR-509-3p | 0.037295 | 1.333371 |
| hsa-miR-511 | 0.01903 | 1.348944 |
| hsa-miR-542-5p | 0.03619 | 1.344654 |
| hsa-miR-548ac | 0.035305 | 1.230733 |
| hsa-miR-548f | 0.049632 | 1.239292 |
| hsa-miR-610 | 0.026332 | 1.228792 |
| hsa-miR-612 | 0.048399 | 1.251725 |
| hsa-miR-650 | 0.02361 | 1.182034 |
| hsa-miR-659a | 0.001558 | 1.500267 |
| hsa-miR-802 | 0.040635 | 1.305961 |
 | Table IIIStatistical results of the miRNAs
that were downregulated in the serum of the patients with primary
biliary cirrhosis. |
Table III
Statistical results of the miRNAs
that were downregulated in the serum of the patients with primary
biliary cirrhosis.
| Downregulated
miRNA | P-value | Fold
(resistant/effective group) |
|---|
|
hsa-miR-125b-2* | 0.014229 |
0.7001 |
| hsa-miR-1301 | 0.031384 |
0.77713 |
|
hsa-miR-2277-3p | 0.02819 | 0.772271 |
|
hsa-miR-3136-3pa | 0.003916 | 0.662795 |
| hsa-miR-33b* | 0.013788 | 0.815009 |
| hsa-miR-340* | 0.008022 |
0.78203 |
|
hsa-miR-3675-5p | 0.015949 | 0.685806 |
|
hsa-miR-3689a-3pa | 0.015287 | 0.659541 |
|
hsa-miR-3690a | 0.017489 | 0.614312 |
| hsa-miR-380* | 0.018128 | 0.755665 |
| hsa-miR-4260 | 0.046847 | 0.753042 |
|
hsa-miR-4311a | 0.044072 | 0.659274 |
|
hsa-miR-4423-5p | 0.035972 | 0.773386 |
| hsa-miR-4504 | 0.030226 | 0.696011 |
|
hsa-miR-4655-5p | 0.038655 | 0.752697 |
|
hsa-miR-4714-3pa | 0.007293 | 0.655707 |
|
hsa-miR-4720-3p | 0.027112 | 0.719039 |
|
hsa-miR-520a-5p | 0.012847 | 0.732509 |
| hsa-miR-548z | 0.046249 | 0.746099 |
| hsa-miR-602 | 0.003259 | 0.722861 |
| hsa-miR-607 | 0.043153 | 0.796838 |
| hsa-miR-770-5p | 0.012182 | 0.719448 |
| hsa-miR-873 | 0.040941 | 0.769026 |
miRNA expression and clinical
parameters
To examine the association between highly altered
miRNAs and clinical features, various parameters were analyzed. As
presented in Table IV, elevated
levels of direct bilirubin (D-bil), aspartate transaminase (AST),
and ALT were associated with miRNA-122 upregulation (Table IV). AST, ALT, and γ-GTP were also
related to miRNA-378f upregulation (Table IV). However, the reduction of
miRNA-4311 was related to the decreases of AST and ALT (Table V). miRNA-4714-3p was also
negatively correlated to total bilirubin and lactate dehydrogenase
(Table V). These results suggest
that these miRNAs may be new biomarkers for the development of
PBC.
 | Table IVAssociation between representative
upregulated miRNAs and clinical parameters in patients with primary
biliary cirrhosis. |
Table IV
Association between representative
upregulated miRNAs and clinical parameters in patients with primary
biliary cirrhosis.
| Parameter | let-7b | miR-122 | miR-210 | miR-378f | miR-659 |
|---|
| TP |
0.1038 |
0.4242 |
0.095 |
0.281 |
0.0973 |
| Alb |
0.1084 |
0.2757 | −0.0793 | −0.1152 | −0.1483 |
| T-Bil |
0.0636 |
0.1186 | −0.0581 | −0.1196 | −0.1074 |
| D-Bil |
0.3542 |
0.4811a |
0.1879 |
0.0569 | −0.0729 |
| AST |
0.3694 |
0.5539a |
0.2774 |
0.4814a |
0.3036 |
| ALT |
0.4883a |
0.7018a |
0.3105 |
0.5642a |
0.3424 |
| ALP |
0.3182 |
0.3793 |
0.2231 |
0.2928 |
0.0445 |
| LDH | −0.3468 | −0.2167 | −0.16 | −0.1538 | −0.3315 |
| γ-GTP |
0.2733 |
0.5706 |
0.296 |
0.5359a |
0.4495a |
| IgG | −0.1361 |
0.0219 | −0.0543 |
0.2627 |
0.1534 |
| IgM |
0.0957 |
0.1882 |
0.0436 | −0.0893 | −0.0768 |
 | Table VAssociation between representative
downregulated miRNAs and clinical parameters in patients with
primary biliary cirrhosis. |
Table V
Association between representative
downregulated miRNAs and clinical parameters in patients with
primary biliary cirrhosis.
| Parameter | miR-3136-3p | miR-3689a-3p | miR-3690 | miR-4311 | miR-4714-3p |
|---|
| TP | −0.1468 | −0.1996 | −0.2365 | −0.3657 | −0.1489 |
| Alb | 0.0189 | −0.1204 | −0.2857 | 0.2015 | −0.0633 |
| T-Bil | 0.3531 | 0.0764 | −0.2528 | 0.0557 | 0.4763a |
| D-Bil | 0.1435 | −0.2297 | −0.3576 | −0.0678 | 0.3744 |
| AST | −0.0728 | −0.3675 | −0.1569 | −0.5766a | 0.1138 |
| ALT | −0.1218 | −0.458a | −0.2569 | −0.559a | 0.0184 |
| ALP | 0.1701 | −0.1837 | −0.1857 | −0.3857 | 0.2868 |
| LDH | 0.3869 | 0.3149 | −0.0115 | −0.1767 | 0.5841a |
| γ-GTP | −0.3012 | −0.3801 | −0.3301 | −0.4519 | −0.0204 |
| IgG | −0.2189 | −0.0718 | 0.2479 | −0.2368 | −0.3262 |
| IgM | −0.0193 | 0.0584 | −0.2388 | −0.2487 | −0.0262 |
Discussion
It has been demonstrated that patients with PBC with
efficient biochemical responses to UDCA or bezafibrate have low
risks of developing liver failure or cirrhosis in the long term
(15,16). However, one third of patients with
PBC exhibited suboptimal responses to these treatment strategies
(19), and these patients
demonstrated poorer outcomes (20–22).
At present, no useful biomarker has been reported for the
development of PBC. In the present study, miRNA expression patterns
in the treatment-resistant group markedly differed compared with
those in the treatment-effective group. The expression levels of
highly altered miRNAs in the resistant group were also associated
with the clinical parameters of PBC. Notably, miRNA-122 was
significantly upregulated and was associated with D-bil, AST, and
ALT in Tables II and IV. Roderburg et al (23) demonstrated that elevated miRNA-122
serum levels are a potent and independent marker of liver injury
and are not seen in patients with liver cirrhosis without ongoing
liver damage. In addition, it has also been reported that miRNA-122
is upregulated in acute and chronic liver injury (24), myocardial infarction (25) and cancer (26). These results support those
indicating that miRNA-122 was enhanced in the serum of the
treatment-resistant group, due to the fact that clinical
parameters, including AST and ALT, were raised sufficiently to
indicate ongoing inflammation in the liver.
In addition, several miRNAs were identified to be
associated with cell cycle arrest, including let-7 and miR-210
among the upregulated miRNAs in the treatment-resistant group. It
has been previously demonstrated that let-7 and miR-210 targeted
cell cycle regulatory molecules (27,28).
Han et al (29) reported
that miR-194 suppresses cell proliferation through the inhibition
of insulin-like growth factor receptor 1. These reports suggest
that various miRNAs, including let-7, miR-210 and miR-194, may
inhibit hepatocyte regeneration by regulating cell cycle and cell
proliferation in the treatment control group.
Downregulated miRNAs, including miR-125b, miR-380
and miR-602, were detected in the treatment-resistant group.
Notably, it has been previously reported that miR-125b and miR-380
are negative regulators of p53 (30,31)
and miR-602 regulates the tumor-suppressive gene Ras-association
domain family 1, isoform A (32).
This suggests that downregulation of miR-125b, miR-380 and miR-602
may suppress cell proliferation and cell cycle acceleration, and
induce apoptosis in the hepatocytes of treatment-resistant
patients.
It has been previously reported that miRNAs are able
to regulate the efficacy of drugs interacting with the miRNA target
and protein (33). Several miRNAs,
including miR-125b, interact with the cytochrome P450, family 1,
member A1 gene and induce drug resistance (33). In the current study, miR-125b was
downregulated in the treatment-resistant group. The results of the
current study are in agreement with a previous study that indicated
that miRNAs are associated with drug resistance to UDCA or
bezafibrate in patients with PBC (33).
In conclusion, identification of miRNA profiles is
useful in characterizing the development of PBC, and representative
miRNAs, including miR-125b, let-7b and miR-520a-5p are suggested to
be potential biomarkers for refractory PBC.
Abbreviations:
|
UDCA
|
ursodeoxycholic acid
|
References
|
1
|
Morishita A and Masaki T: miRNA in
hepatocellular carcinoma. Hepatol Res. 45:128–141. 2015. View Article : Google Scholar
|
|
2
|
Suzuki H, Maruyama R, Yamamoto E and Kai
M: Epigenetic alteration and microRNA dysregulation in cancer.
Front Genet. 4:2582013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Griffiths-Jones S: miRBase: MicroRNA
sequences and annotation. Curr Protoc Bioinformatics. Chapter 12,
Unit 12. 19:1–10. 2010.
|
|
6
|
Iborra M, Bernuzzi F, Invernizzi P and
Danese S: MicroRNAs in autoimmunity and inflammatory bowel disease:
Crucial regulators in immune response. Autoimmun Rev. 11:305–314.
2012. View Article : Google Scholar
|
|
7
|
Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan
YH, Xu ZM and Yin YB: Microarray analysis of microRNA expression in
peripheral blood cells of systemic lupus erythematosus patients.
Lupus. 16:939–946. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pauley KM, Satoh M, Chan AL, Bubb MR,
Reeves WH and Chan EK: Upregulated miR-146a expression in
peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Res Ther. 10:R1012008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Neilson JR, Zheng GX, Burge CB and Sharp
PA: Dynamic regulation of miRNA expression in ordered stages of
cellular development. Genes Deve. 21:578–589. 2007. View Article : Google Scholar
|
|
10
|
Wu H, Neilson JR, Kumar P, Manocha M,
Shankar P, Sharp PA and Manjunath N: MiRNA profiling of naive,
effector and memory CD8 T cells. PloS One. 2:e10202007. View Article : Google Scholar
|
|
11
|
Zhou B, Wang S, Mayr C, Bartel DP and
Lodish HF: miR-150, a microRNA expressed in mature B and T cells,
blocks early B cell development when expressed prematurely. Proc
Natl Acad Sci USA. 104:7080–7085. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kaplan MM and Gershwin ME: Primary biliary
cirrhosis. N Engl J Med. 353:1261–1273. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shimoda S, Harada K, Niiro H, Shirabe K,
Taketomi A, Maehara Y, Tsuneyama K, Nakanuma Y, Leung P, Ansari AA,
et al: Interaction between Toll-like receptors and natural killer
cells in the destruction of bile ducts in primary biliary
cirrhosis. Hepatology. 53:1270–1281. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirschfield GM and Gershwin ME: The
immunobiology and pathophysiology of primary biliary cirrhosis.
Annu Rev Pathol. 8:303–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Corpechot C, Chazouillères O and Poupon R:
Early primary biliary cirrhosis: Biochemical response to treatment
and prediction of long-term outcome. J Hepatol. 55:1361–1367. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tanaka A, Hirohara J, Nakanuma Y,
Tsubouchi H and Takikawa H: Biochemical responses to bezafibrate
improve long-term outcome in asymptomatic patients with primary
biliary cirrhosis refractory to UDCA. J Gastroenterol. 50:675–682.
2015. View Article : Google Scholar
|
|
17
|
Padgett KA, Lan RY, Leung PC, Lleo A,
Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA and Gershwin ME:
Primary biliary cirrhosis is associated with altered hepatic
microRNA expression. J Autoimmun. 32:246–253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Poupon R: Primary biliary cirrhosis: A
2010 update. J Hepatol. 52:745–758. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Corpechot C, Abenavoli L, Rabahi N,
Chrétien Y, Andréani T, Johanet C, Chazouillères O and Poupon R:
Biochemical response to ursodeoxycholic acid and long-term
prognosis in primary biliary cirrhosis. Hepatology. 48:871–877.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kuiper EM, Hansen BE, de Vries RA, den
Ouden-Muller JW, van Ditzhuijsen TJ, Haagsma EB, Houben MH,
Witteman BJ, van Erpecum KJ and van Buuren HR; Dutch PBC Study
Group: Improved prognosis of patients with primary biliary
cirrhosis that have a biochemical response to ursodeoxycholic acid.
Gastroenterology. 136:1281–1287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pares A, Caballería L and Rodes J:
Excellent long-term survival in patients with primary biliary
cirrhosis and biochemical response to ursodeoxycholic Acid.
Gastroenterology. 130:715–720. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roderburg C, Benz F, Vargas Cardenas D,
Koch A, Janssen J, Vucur M, Gautheron J, Schneider AT, Koppe C,
Kreggenwinkel K, et al: Elevated miR-122 serum levels are an
independent marker of liver injury in inflammatory diseases. Liver
Int. 35:1172–1184. 2015. View Article : Google Scholar
|
|
24
|
Bihrer V, Friedrich-Rust M, Kronenberger
B, Forestier N, Haupenthal J, Shi Y, Peveling-Oberhag J, Radeke HH,
Sarrazin C, Herrmann E, et al: Serum miR-122 as a biomarker of
necroinflammation in patients with chronic hepatitis C virus
infection. Am J Gastroenterol. 106:1663–1669. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
D'Alessandra Y, Devanna P, Limana F,
Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC,
Spazzafumo L, De Simone M, et al: Circulating microRNAs are new and
sensitive biomarkers of myocardial infarction. Eur Heart J.
31:2765–2773. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Su Y, Xu F, Kong J, Yu H and Qian
B: Circulating microRNAs in relation to EGFR status and survival of
lung adenocarcinoma in female non-smokers. PloS One. 8:e814082013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mayr C, Hemann MT and Bartel DP:
Disrupting the pairing between let-7 and Hmga2 enhances oncogenic
transformation. Science. 315:1576–1579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He J, Wu J, Xu N, Xie W, Li M, Li J, Jiang
Y, Yang BB and Zhang Y: MiR-210 disturbs mitotic progression
through regulating a group of mitosis-related genes. Nucleic Acids
Res. 41:498–508. 2013. View Article : Google Scholar :
|
|
29
|
Han K, Zhao T, Chen X, Bian N, Yang T, Ma
Q, Cai C, Fan Q, Zhou Y and Ma B: microRNA-194 suppresses
osteosarcoma cell proliferation and metastasis in vitro and in vivo
by targeting CDH2 and IGF1R. Int J Oncol. 45:1437–1449.
2014.PubMed/NCBI
|
|
30
|
Swarbrick A, Woods SL, Shaw A,
Balakrishnan A, Phua Y, Nguyen A, Chanthery Y, Lim L, Ashton LJ,
Judson RL, et al: miR-380-5p represses p53 to control cellular
survival and is associated with poor outcome in MYCN-amplified
neuroblastoma. Nat Med. 16:1134–1140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B,
Korzh V, Lodish HF and Lim B: MicroRNA-125b is a novel negative
regulator of p53. Genes Dev. 23:862–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang L, Ma Z, Wang D, Zhao W, Chen L and
Wang G: MicroRNA-602 regulating tumor suppressive gene RASSF1A is
overexpressed in hepatitis B virus-infected liver and
hepatocellular carcinoma. Cancer Biol Ther. 9:803–808. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rukov JL, Wilentzik R, Jaffe I, Vinther J
and Shomron N: Pharmaco-miR: Linking microRNAs and drug effects.
Brief Bioinform. 15:648–659. 2014. View Article : Google Scholar :
|